Abstract
Rearrangements of the MLL gene located at 11q23 are common chromosomal abnormalities associated with acute leukemia, especially infant and therapy-related leukemias. A variety of chimeric oncoproteins resulting from these rearrangements has been described; all of these include the N-terminal region of MLL implicated in protein-protein interactions and transcriptional repression. While the molecular basis for the oncogenic activity of MLL chimeric proteins is incompletely understood, it appears to be derived, at least in part, through activation of clustered homeobox (HOX) genes. Here, we survey MLL gene rearrangements that are associated with acute leukemia and discuss molecular pathways leading to these rearrangements.
Keywords: MLL, chromosomal translocation, infant leukemia, DNA topoisomerase II, non-homologous end-joining, homeobox
Introduction
Chromosomal rearrangements involving the MLL (MLL1, ALL1, TRX, HTRX) gene, including balanced and unbalanced translocations, inversions, insertions, and a partial tandem duplication, have been associated with a heterogeneous group of lymphoid, myeloid, and mixed lineage leukemias (1). The MLL gene, a homologue of the Drosophila trithorax, is located at chromosome 11q23 , consists of 36 exons, and encodes a protein of 3969 amino acid residues, with an estimated molecular weight of 430 kDa (1). Most MLL gene rearrangements map to an 8.3 kb breakpoint cluster region (bcr) (2), and result in production of a chimeric onco-protein which fuses the amino terminal portion of MLL with the carboxy terminal portion of a partner gene (Figure 1). MLL can be regarded as a highly “promiscuous” oncogene, since more than 70 different 11q23 chromosomal partners have been identified, and at least 50 of these have been cloned and characterized on a molecular level (http://atlasgeneticsoncology.org/Genes/MLL.html).
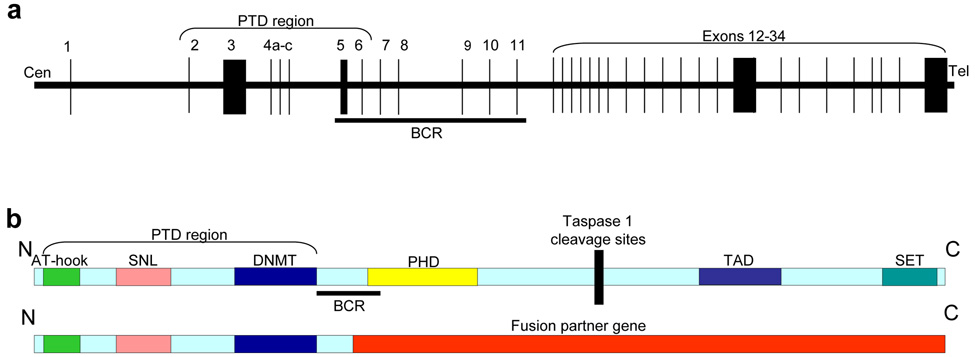
Figure 1. Structure of the MLL gene and protein.
(a) Schematic representation of MLL gene showing location of the PTD (partial tandem duplication, see text) and BCR (breakpoint cluster region, which encompasses almost all known MLL translocation breakpoints). Exon size and intronic distances are not to scale. (b) Location of MLL protein domains in relation to BCR, PTD, and fusion partners. AT-hook [DNA binding motif that binds adenosine-thymidine (AT) rich DNA], SNL (speckled nuclear localization sites), DNMT (DNA methyltransferase domain), PHD (plant homeodomains), TAD (transactivation domain), SET [SET (for Suppressor of variegation/Enhancer of zeste/Trithorax) domain]. Cleavage by Taspase (Threonine-aspartase) 1 divides MLL in N- and C-terminal fragments.
Clinical Findings
Although MLL rearrangements are associated with a wide spectrum of leukemias, and numerous translocation partners, there are several unique subsets of MLL leukemias that are defined by associated clinical and laboratory findings. These sub-groups include infant leukemias, therapy-related leukemia, MLL-amplified leukemia, and T-cell ALL.
Infant leukemia
Leukemia in infancy (<12 months of age), whether classified as ALL, AML, or mixed lineage, is often associated with MLL gene rearrangements (3). It seems likely that infant leukemia with and without MLL are different diseases with different clinical characteristics at presentation, different responses to therapy, and very different genetic profiles (4). When molecular diagnostic techniques are employed, MLL gene fusions can be identified in approximately 80% of all infant leukemia patients (4). Although greater than 90% of infant ALL patients achieve a complete remission, despite the use of aggressive intensification regimens and allogeneic stem cell transplantation, event free survival (EFS) for MLL-rearranged infant ALL patients remain in the 30–40 % range (4). Several lines of evidence have demonstrated that the MLL rearrangement in most infants with leukemia occurs in utero. First, MLL rearrangements have been identified in congenital infant leukemia, and even in aborted fetuses. Second, the concordance rate for infant leukemia between identical twins is very high, suggesting that the MLL rearrangement occurred initially in one twin in utero, and subsequently “metastasized” to the second twin (5). This hypothesis has been confirmed for numerous twin pairs that had identical immunoglobulin gene rearrangements (5). Finally, MLL gene fusions have been identified in archived neonatal blood spots (“Guthrie” cards) of infants who subsequently developed leukemia (6).
Therapy-related acute myeloid leukemia (t-AML)
In the 1990s, the use of the epipodophylotoxins became linked to the development of therapy-related acute myeloid leukemia (t-AML) (7). Remarkably, a very high proportion of patients who developed t-AML following therapy with topoisomerase II (topo II) poisons had translocations involving the MLL gene (7). This observation led to speculation that topo II poisons were directly involved in the generation of MLL rearrangements. The MLL partner genes involved in t-AML, including AF9, AF4, and ENL, are similar to those involved in de novo AML, except for the CBP gene, which is almost always associated with t-AML (8). Although less common than t-AML, therapy-related ALL and lymphoblastic lymphoma with MLL gene rearrangement have also been reported.
MLL amplification
Amplification of the MLL gene, either at 11q23, distant chromosomal regions (referred to as segmental jumping translocations), or double minute chromosomes were identified using FISH probes (9). In these cases, no rearrangement of the MLL gene has been reported, rather, MLL has been amplified as part of a large amplicon encompassing up to 10 Mb of genomic sequence. Of note, a wild-type MLL transcript is reported to be over-expressed in these cases, as is expression of several documented target genes for leukemogenic MLL fusion proteins, such as HOXA9 and MEIS1.
T-cell ALL
Although T-cell ALL with MLL gene rearrangements are uncommon, this is an interesting subgroup as this group of translocations involve relatively few partner genes, almost exclusively t(11; 19) that generate a MLL-ENL fusion. In addition, these patients tend to have a good prognosis, with close to 90% long term EFS in one published series (10).
Effects of MLL fusion genes
The MLL protein is widely expressed during development, and continues to be expressed in most adult tissues, including myeloid and lymphoid cells (1). MLL is required for normal development and body pattern formation, as deletion of MLL in mice leads to homeotic transformation (1 ). MLL is processed in the cytoplasm by the threonine- aspartase (TASPASE1) enzyme into a 320 kD amino-terminal fragment (MLL-N) and an 180 kD carboxytermial fragment (MLL-C), which remain non-covalently bound (1 ) (Figure 1). The MLL-N fragment is thought to bind DNA as part of a multi-subunit complex that includes components of the basal transcription machinery (1). Recent experiments have suggested that MLL-N binds to regulatory regions of clustered homeobox (HOX) genes, including HOXA9 and HOXC8. Upon binding DNA, MLL-N can mediate transcriptional repression of the target gene, likely dependent on recruitment of additional co-factors such as BMI1. However, in the presence of MLL-C, the MLL-N complex can lead to transcriptional activation (1).
Although no single theme connects all of the MLL translocation partners, several are known or putative transcription factors (AF9, ENL, CBP, and P300). Other partners, such as AF1P and GAS7, have no known transcription factor motifs, and have instead hydrophobic coiled-coil domains with the potential to form oligomers. Useful clues as to how MLL fusion proteins might be leukemogenic have been identified by gene expression profiling. Leukemic cells that express MLL gene fusions have a gene expression profile that distinguishes them from ALL and AML without MLL gene rearrangements; among the genes most differentially expressed are HOXA5 and HOXA9(1). Expression of MLL fusion proteins such as MLL-ENL or MLL-CBP in mouse bone marrow leads to overexpression of Hoxa7, Hoxa9, and Meis1, and it is thought that overexpression of these genes is important for leukemic transformation (11) . Hoxa9 is normally expressed in primitive hematopoietic cells, becomes downregulated as cells differentiate, and promotes hematopoietic stem cell self-renewal (12). Enforced expression of Hoxa9 in mouse bone marrow cells is leukemogenic, and leukemic transformation is accelerated by co-expression of Meis1(12). However, expression of Hoxa9 is not required for leukemic transformation by MLL fusion proteins, as expression of an MLL-GAS7 fusion in Hoxa9-deficient bone marrow remains leukemogenic (13). Given the redundancy of the clustered Hox genes, it is possible that upregulation of other Hox genes (such as Hoxa7 or Hoxa10) mediates leukemic transformation in this setting.
Several approaches have been used to demonstrate that MLL fusion proteins are leukemogenic in mice. Using retroviral-mediated gene transfer to murine hematopoietic cells, followed by transplantation into irradiated mice, a number of MLL fusions, including MLL-AF9, MLL-GAS7, MLL-ENL, and MLL-CBP have been shown to be leukemogenic (13). Using ES cell gene targeting strategies, an MLL-AF9 fusion, under the regulatory control of endogenous MLL sequences, was leukemogenic (14). Interestingly, although the targeted ES cells expressed the MLL-AF9 fusion ubiquitously, the only malignancies noted were hematopoietic, suggesting that MLL fusions might only be oncogenic in hematopoietic cells (14).
Mechanisms of MLL gene rearrangement
Since MLL gene fusions lead to leukemia, it is important to understand the causes of MLL gene fusions. It should be noted that for any chromosomal rearrangement to be recognized clinically, two criteria must be fulfilled. First, the region involved must undergo a DNA double strand break (DSB) and re-ligation. Second, the break and re-ligation must in some way confer a clonal growth advantage to the cell. If both criteria are not fulfilled, then the translocation will not produce a clonal population, and not be recognized clinically. Proposed mechanisms to account for MLL translocations include recombination between Alu elements, recombination mediated by topo II poisons, and an error prone non-homologous end joining (NHEJ) of DNA DSB.
Although chromosomal translocations caused by interchromosomal recombination between Alu elements have been implicated in a case of an MLL-AF9 translocation, these seem to be relatively rare. However, the MLL partial tandem duplication (also known as MLL self-fusion), which leads to a duplication of MLL exons 2–6, is commonly mediated via inter or intra chromosomal recombination between Alu elements within the MLL locus, via a mechanism that is consistent with the single-strand annealing (SSA) repair pathway (15).
The association of t-AML with MLL translocations and topo II poisons has led to the hypothesis that these translocations are directly caused by topo II poisons (16). Topo II normally functions as a homodimeric enzyme that introduces a 4-bp staggered nick in double stranded DNA resulting in a short-lived intermediate in which the topo II monomer is covalently bonded to the DNA phosphodiester backbone. Topo II poisons stabilize this short-lived intermediate, which becomes recognized by the cell as a DNA DSB, which triggers an apoptotic cell death (16). Since topo II normally functions as a homo-dimer, it has been proposed that perfect, (ie, no net gain or loss of genetic material) or near-perfect (gain or loss of 4 or fewer nucleotides) reciprocal translocations could occur via an exchange of topo II subunits and covalently linked chromosomal DNA (17). Chromosomal translocations showing this type of perfect or near-perfect inter-chromosomal exchange have been identified in patients with t-AML following chemotherapy regimens that included topo II poisons. However, it should be noted that these cases may be the exception rather than the rule, as more extensive surveys of MLL translocation breakpoints, from de novo or t-AML patients, do not contain large numbers of samples with these near-perfect reciprocal interchromosomal exchanges (18).
Site-specific cleavage of the MLL bcr has been identified in leukemic patient samples and cell lines following treatment with topo II poisons (19). Although this cleavage site maps close to a consensus topo II cleavage site, it seems likely that this site encompasses a region that is generally susceptible to DNA DSB, as cleavage can be induced by DNAse I, or more generally by the high molecular weight (HMW) DNA fragmentation that occurs in cells undergoing apoptosis (19). Taken together, these observations have led to speculation that an “aborted” apoptosis program could lead to MLL gene rearrangement following abnormal repair at this cleavage site (19). Interestingly, PCR products consistent with MLL gene rearrangements produced by improper repair of DNA double strand cleavage at this site can be identified following treatment with genotoxic (etoposide) or non-genotoxic (fas ligand) triggers of apoptosis (20).
Breakpoint nucleotide sequence data suggests that a majority of the leukemogenic translocations involving MLL are mediated by inappropriate repair of DNA DSBs via an “error-prone” NHEJ pathway (18). However, the proximate cause(s) of the DNA DSBs within the MLL bcr remain unclear. Moreover, it remains unknown whether the MLL bcr is extraordinarily susceptible to DNA DSBs (ie, one of the most susceptible regions throughout the human genome), whether the MLL bcr is uniquely susceptible to aberrant, error-prone NHEJ repair of DNA DSBs, or whether neither of these are true, and the frequent presence of MLL translocations in leukemic samples is due to a remarkable growth/survival advantage conferred by oncogenic MLL fusion proteins.
Summary
Chromosomal translocations leading to MLL gene fusions are a common event in patients with acute leukemia, and are particularly common in infants with AML or ALL, and patients with t-AML. MLL is a “promiscuous” oncogene, with numerous partner genes involved in MLL fusions. The molecular mechanisms that lead to MLL gene rearrangements remain obscure, but seem to involve aberrant repair of DNA DSB breaks via NHEJ. The normal MLL protein undergoes proteolytic cleavage, and can function as a transcriptional repressor or activator; the activation function is associated with its ability to covalently modify histones. Expression of MLL gene fusions is oncogenic in mice, and seems to be due, at least in part, to upregulation of clustered homeobox genes such as Hoxa9. The presence of MLL fusions are currently used in risk-stratification schemes for childhood leukemia, and the observation that MLL fusion proteins modify histones suggests that this activity might be a target for modulation by small molecules.
Acknowledgments
We apologize in advance to authors whose work we have not cited due to space limitations. We thank Michael Kuehl, Ilan Kirsch, Rolf Marschalek, Shai Izraeli, Jay Hess, Matthew Strout, Michael Caligiuri, Clara Bloomfield, Martin Stanulla, Carolyn Felix, Terrance Rabbitts, Janet Rowley, Nancy Zeleznik-Le, and Michael Thirman for many thoughtful and stimulating discussions regarding MLL translocations. This work was supported by the Intramural Research Program of the NIH, NCI.
References
- 1.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Thirman MJ, Gill HJ, Burnett RC, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. New England Journal of Medicine. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 3.Eguchi M, Eguchi-Ishimae M, Greaves M. The role of the MLL gene in infant leukemia. Int J Hematol. 2003;78:390–401. doi: 10.1007/BF02983811. [DOI] [PubMed] [Google Scholar]
- 4.Tomizawa D, Koh K, Sato T, et al. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: a final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia. 2007;21:2258–2263. doi: 10.1038/sj.leu.2404903. [DOI] [PubMed] [Google Scholar]
- 5.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 6.Gale KB, Ford AM, Repp R, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci U S A. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pui CH, Behm FG, Raimondi SC, et al. Secondary acute myeloid leukemia in children treated for acute lymphoid leukemia. N Engl J Med. 1989;321:136–142. doi: 10.1056/NEJM198907203210302. [DOI] [PubMed] [Google Scholar]
- 8.Rowley JD, Reshmi S, Sobulo O, et al. All patients with the t(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997;90:535–541. [PubMed] [Google Scholar]
- 9.Felix CA, Megonigal MD, Chervinsky DS, et al. Association of germline p53 mutation with MLL segmental jumping translocation in treatment-related leukemia. Blood. 1998;91:4451–4456. [PubMed] [Google Scholar]
- 10.Pui CH, Chessells JM, Camitta B, et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia. 2003;17:700–706. doi: 10.1038/sj.leu.2402883. [DOI] [PubMed] [Google Scholar]
- 11.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 13.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 14.Corral J, Lavenir I, Impey H, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 15.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 17.Zhou RH, Wang P, Zou Y, Jackson-Cook CK, Povirk LF. A precise interchromosomal reciprocal exchange between hot spots for cleavable complex formation by topoisomerase II in amsacrine-treated Chinese hamster ovary cells. Cancer Research. 1997;57:4699–4702. [PubMed] [Google Scholar]
- 18.Reichel M, Gillert E, Angermuller S, et al. Biased distribution of chromosomal breakpoints involving the MLL gene in infants versus children and adults with t(4;11) ALL. Oncogene. 2001;20:2900–2907. doi: 10.1038/sj.onc.1204401. [DOI] [PubMed] [Google Scholar]
- 19.Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Molecular & Cellular Biology. 1997;17:4070–4079. doi: 10.1128/mcb.17.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]