Summary
The correct annotation of genes encoding the smallest proteins is one of the biggest challenges of genome annotation, and perhaps more importantly, few annotated short ORFs have been confirmed to correspond to synthesized proteins. We used sequence conservation and ribosome binding site models to predict genes encoding small proteins, defined as having 16-50 amino acids, in the intergenic regions of the Escherichia coli genome. We tested expression of these predicted as well as previously-annotated genes by integrating the sequential peptide affinity tag directly upstream of the stop codon on the chromosome and assaying for synthesis using immunoblot assays. This approach confirmed that 20 previously-annotated and 18 newly-discovered proteins of 16-50 amino acids are synthesized. We summarize the properties of these small proteins; remarkably more than half of the proteins are predicted to be single transmembrane proteins, nine of which we show co-fractionate with cell membranes.
Keywords: Information theory, genome annotation, membrane proteins
Introduction
One of the challenges inherent in characterizing the proteome of an organism is the isolation and identification of small proteins. It is difficult to reliably annotate genes encoding small proteins computationally, since these genes frequently lack sufficient sequence for domain and homology determination (Basrai et al., 1997; Blattner et al., 1997; Cliften et al., 2001; Consortium, 2004; Rudd et al., 1998). The small size of these genes also limits the frequency in which they are disrupted in random genetic screens (Basrai et al., 1997; Kastenmayer et al., 2006). The problem of identifying small polypeptides is further compounded by difficulties involved in using standard proteomic techniques to isolate and identify proteins less than 10 kDa in size (Garbis et al., 2005).
Although proteins of 16-50 amino acids (herein referred to as small proteins) are difficult to predict, isolate and characterize, an increasing body of evidence shows that these polypeptides have important cellular and intercellular functions. For example, in the bacterium Bacillus subtilis, the Sda protein (46 amino acids) represses aberrant sporulation by inhibiting the activity of the KinA kinase (Burkholder et al., 2001; Rowland et al., 2004). In eukaryotes, small proteins play important roles at both a cellular and organismal level. Recent work has shown that three previously unannotated essential small proteins in yeast are members of the kinetochore complex and are necessary for proper chromosome segregation (Miranda et al., 2005). Small proteins are important components of photosystem II in plants (Shi and Schröder, 2004). In animals, cationic antimicrobial peptides are a first-line defense against pathogen attack (Gallo and Nizet, 2003), and many hormones are peptides derived from larger proteins (Canaff et al., 1999). These examples illustrate the diverse functions of small proteins across species, and suggest that future studies of small proteins will provide new biological insights.
Relatively few E. coli proteins of 16-50 amino acids have been characterized. Most of the characterized small proteins are members of three different categories: leader peptides, ribosomal proteins, or toxic proteins. Leader peptides have been identified upstream of 11 genes which primarily encode proteins involved in amino acid metabolism. In these cases, translation of the short open reading frame (ORF) regulates transcription and/or translation of the downstream genes during periods of amino acid starvation (reviewed in (Yanofsky, 2000)). It is still unknown whether these leader peptides have independent functions after they are translated, although the peptides can accumulate upon overexpression (Gong et al., 2006). The E. coli ribosome also contains a number of relatively small proteins, and two components of the 50S subunit, L36 (encoded by rpmJ) and L34 (encoded by rpmH) are proteins of less than 50 amino acids. One ribosome-associated protein, Sra (also denoted S22 and RpsV) also is only 45 amino acids in length.
The small proteins in the third category can be toxic to cells, especially when overexpressed. Their toxicity can be mitigated by co-expression of a corresponding antitoxin protein blocking activity or antisense small RNA blocking expression. Included in this group are members of the Hok family. This toxic gene family was originally identified on plasmids, but intact hok genes are also encoded on some E. coli genomes (Gerdes et al., 1997; Rudd et al., 1998). In plasmids, the Hok system insures that cells retain the plasmid during replication, however the function of the chromosomally-encoded toxic genes is still unclear. The 35-amino acid protein LdrD expressed from one of the long direct repeat sequences in E. coli K-12 has been shown to be toxic when overproduced, and it is likely that the homologous LdrA, LdrB and LdrC proteins expressed from the other three copies of the LDR sequences are also toxic at high levels (Kawano et al., 2002). Overexpression of the 18 or 19 amino acid Ibs proteins encoded by the five copies of the E. coli K-12 SIB repeats is similarly toxic (Fozo et al., 2008). Three other small proteins shown to be toxic at elevated levels are the 48-amino acid entericidin B protein (Bishop et al., 1998), the 29-amino acid TisB protein (Vogel et al., 2004) and the 26-amino acid ShoB protein (Fozo et al., 2008). The mechanisms by which elevated levels of these proteins kill cells or inhibit growth are not known though all are predicted to be membrane proteins.
Aside from the three classes of proteins listed above, only a few small proteins have been identified in E. coli. As already mentioned, it has been difficult to reliably annotate small proteins (Ochman, 2002). Automated annotation methods usually either under-annotate or over-annotate small proteins. Among the sequenced E. coli strains, the number of annotated short ORFs ranges from 0 in strain APEC 01 to 323 in strain E24377A. A simple way to improve the accuracy of protein annotation is to adopt a threshold for hypothetical ORF translations and not annotate any proteins less than 40 or 50 amino acids in length. This minimizes false positives, but knowingly under-annotates, excluding many small proteins likely to be synthesized. In cases where less stringent criteria are used, a large number of short proteins are annotated, but many of these are unsupported by conservation or experimentation. The net result is that a significant number of highly unlikely short proteins are annotated in all major protein databases. This over-annotation can self-propagate when new closely related genomes are annotated using automated methods that score these as valid protein hits due to their previous annotation. Ultimately, experimental validation of protein synthesis is necessary to confirm small protein predictions. To more accurately define the E. coli proteome, we set out to systematically re-annotate, identify and test the chromosomal expression of the predicted small proteins in E. coli K-12 MG1655.
Results
Previously-annotated genes encoding small proteins
As a starting point for our analysis, we re-examined the existing annotation for proteins of less than 50 amino acids. In the initial U00096.1 annotation of the E. coli K-12 MG1655 genome, 42 proteins of 14 to 50 amino acids, including 11 leader peptides, were annotated (Blattner et al., 1997). For this initial annotation, the difficulty of accurately predicting small genes was noted. However, the presence of gene remnants corresponding to fragments of full-length genes found in other organisms was not fully recognized. In a subsequent EcoGene 10 reevaluation (Rudd, 1998) of the U00096.1 annotation, 13 of the 42 small proteins were rejected as unlikely genes (the annotation history is given in Table S1). In addition, homology analysis led to the prediction of additional small protein genes. These analyses together with published experimental documentation of new small proteins led to the annotation of 33 small proteins and 11 leader peptides in EcoGene 10 (Rudd, 1998).
The annotation of small protein genes was improved with the availability of an increasing number of genomic sequences for bacteria closely related to E. coli. Comparisons between the related sequences allowed for the identification of additional regions of conservation in intergenic regions. Some potential small protein genes were found during searches for small, regulatory RNAs. The patterns of conservation, with higher sequence variation every third position, indicated that these regions encode small proteins; YpfM, YneM, YncL (Wassarman et al., 2001), SgrS (protein named SgrT, (Wadler and Vanderpool, 2007)) and ISO92/IsrB (protein named AzuC, Miranda-Rios, unpublished observation). The results of the comparative analyses together with expression data and biochemical identification of additional small proteins led to a list of 40 proteins for which we had reasonable confidence [annotated in EcoGene 19 (December 2006) and listed in Table 1]. In addition, we noted one region, ydfW, for which there were overlapping candidate ORFs of 49 and 75 amino acids in different frames.
Table 1.
Previously annotated small proteins
|
Protein detected previouslya |
Potentially toxic proteinsb |
Detected in current study |
Not detected in current study |
||||
|---|---|---|---|---|---|---|---|
| Name | Lengthc | Name | Lengthc | Name | Lengthc | Name | Lengthc |
| KdpF | 29 | IbsE | 18 | YjeV | 17 | Tpr | 29 |
| RpmJ | 38 | IbsA | 19 | YpfM | 19 | YlcH | 33 |
| Blr | 41 | IbsB | 19 | AzuC | 28 | YoaI | 34 |
| EcnA | 41 | IbsC | 19 | YccB | 30 | DinQ | 42 |
| Sra | 45 | IbsD | 19 | YncL | 31 | YjjY | 46 |
| RpmH | 46 | ShoB | 26 | YneM | 31 | ||
| TisB | 29 | YniD | 35 | ||||
| LdrA | 35 | YohO | 35 | ||||
| LdrB | 35 | YbgT | 37 | ||||
| LdrC | 35 | YdfB | 42 | ||||
| LdrD | 35 | YmiA | 42 | ||||
| EcnB | 48 | SgrT | 43 | ||||
| HokB | 49 | YqgB | 43 | ||||
| HokA | 50 | YdaG | 44 | ||||
| HokC | 50 | YceO | 46 | ||||
| HokE | 50 | YlcG | 46 | ||||
| YkgO | 46 | ||||||
| YobF | 47 | ||||||
| MgrB | 47 | ||||||
| YbhT | 49 | ||||||
These proteins are toxic upon overexpression. None of the proteins have been detected directly, though genetic evidence is consistent with synthesized proteins (Bishop et al., 1998; Fozo et al., 2008; Gerdes et al., 1997; Kawano et al., 2002; Vogel et al., 2004).
Number of amino acids in unprocessed size.
Synthesis of the majority of annotated small proteins in rich medium
To determine whether the annotated small proteins were synthesized, we integrated the sequential peptide affinity (SPA) tag (Zeghouf et al., 2004) into the MG1655 chromosome adjacent to the carboxy-terminal amino acid of the predicted short ORFs. The SPA tag is a dual epitope tag, consisting of a calmodulin binding protein epitope sequence followed by a 3xFLAG tag. The SPA tag provided two advantages. First, sensitive and specific antibodies are available for detecting the 3xFLAG epitope tag. Second, the tag itself is around 8 kDa, and this increase in size allowed for easier detection of small proteins that normally are difficult to visualize by gel electrophoresis. For example, although the YpfM-SPA protein is highly expressed (see below), we were unable to detect the 2.4 kDa YpfM protein in its native form even when overexpressed from a multicopy plasmid (data not shown).
We tested expression of three short ORFs (Tpr, YbgT and SgrT) for which proteins reportedly had been detected, seven annotated short ORFs (YpfM, AzuC, YncL, YneM, DinQ, YobF and MgrB) for which transcripts have been detected by S1 or Northern analysis, as well as 15 annotated short ORFs for which direct tests of expression were not available (YjeV, YccB, YlcH, YoaI, YniD, YohO, YdfB, YmiA, YqgB, YdaG, YjjY, YceO, YlcG, YkgO and YbhT) (Tables S1 and S2). In addition, we generated two tagged strains corresponding to the predicted 49 and 75 amino acid ORFs in the ydfW region.
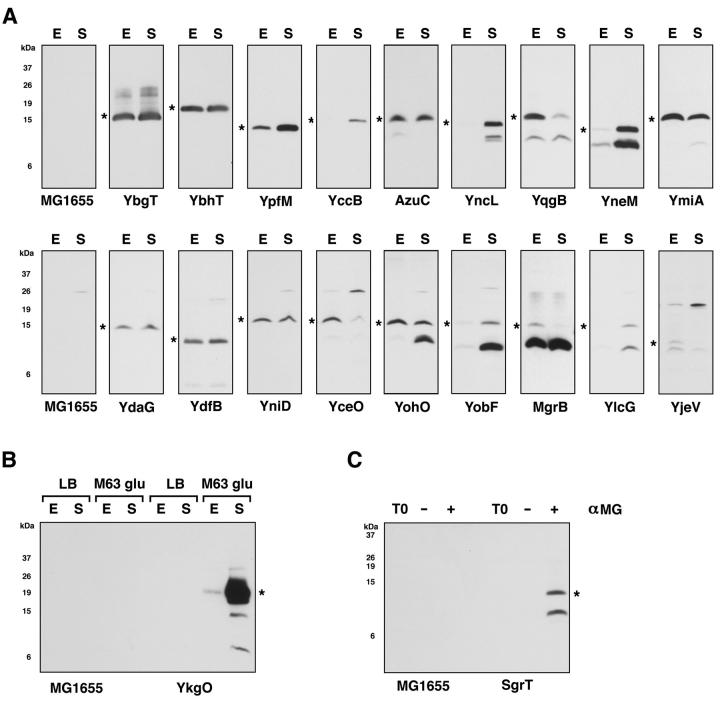
Each strain encoding a tagged protein was grown to exponential (OD600 = 0.3-0.4) or early stationary phase (OD600 = 2.0–3.0) in LB rich medium. Cells were harvested and the SPA-tagged proteins detected by immunoblot analysis by comparison with the MG1655 control lanes included on each blot. As shown in Fig. 1A, we could detect 18 of the 25 tagged 16-50 amino acid proteins in at least one of the two growth conditions. There was a wide range in the levels of the tagged proteins. Some, such as YbgT, appear to be extremely abundant and were easily detected. Others, such as YobF, were present at much lower levels. For a number of tagged proteins, we noted an 8 kDa product which may be due to cleavage between the small protein and the SPA tag, giving rise to a stable fragment corresponding to the epitope tag. It is also possible the fragment could arise from translation of just the tag if the in frame ATG in the SPA coding sequence happens to be preceded by a putative Shine-Dalgarno sequence within the small ORF. We observed only the small 8 kDa fragment for YjjY and a barely discernable 8 kDa band for Tpr (Fig. S1). For the ydfW region, we detected expression for the predicted 75 amino acid protein but not the predicted 49 amino acid protein (data not shown).
Fig. 1.
Immunoblot analysis of previously-annotated short ORFs tagged with SPA. Immunoblot analysis using anti-FLAG, alkaline phosphatase-conjugated antibodies was carried out with whole cell extracts harvested from MG1655 cultures.
A. Cells were grown to (E) exponential and (S) stationary phase in LB medium. Immunoblots shown on the top row are of highly expressed small proteins, whereas immunoblots on the bottom row are of proteins with lower expression. MG1655 control lanes were run for each blot and representative lanes are shown. For those proteins with lower expression (bottom row) an additional cross-reacting band of approximate 26 kDa can also be observed for MG1655 grown to stationary phase.
B. Cells were grown to (E) exponential and (S) stationary phase in LB and M63 medium containing 0.2% glucose.
C. Cells were grown to (T0) exponential phase in LB medium and then treated with (−) water or (+) 1% α-methylglucoside. In all cases, a fraction equivalent to the cells in OD600 = 0.057 was loaded in each lane. The star (*) indicates the band corresponding the fusion protein. The positions of the markers for one blot are shown. This only provides the approximate sizes of the proteins because there was slight variation in the running of gels. Exposure times were optimized for each panel for visualization here, therefore the signal intensity shown does not indicate relative abundance between proteins. Given the need for longer exposure times, some background bands were detected for the immunoblots in the second row of (A).
For five annotated short ORFs (SgrT, DinQ, YlcH, YoaI and YkgO) we did not detect any synthesis under the LB growth conditions (data not shown and Fig. S1). Thus we also examined expression of these fusions in strains grown to exponential (OD600 = 0.3-0.4) and stationary (OD600 = 1.8-2.0) phase in M63 glucose minimal medium. Interestingly, synthesis of the YkgO-SPA fusion was very strongly induced in minimal medium, particularly in stationary phase (Fig. 1B). We again did not detect any bands for SgrT, DinQ, YlcH and YoaI (data not shown). The levels of the dinQ and sgrS/sgrT RNAs were reported to be induced by exposure to the DNA damaging agent mitomycin C and the nonmetabolizable glucose analog α-methylglycoside, respectively (Fernández De Henestrosa et al., 2000; Wadler and Vanderpool, 2007). Thus we also examined expression under these conditions. We still did not detect DinQ, but observed high levels of the SgrT-SPA protein (Fig. 1C). All together, our detection of 20 tagged proteins, together with 6 small proteins detected previously (KdpF, RpmJ, EcnA, Blr, Sra, RpmH, (Bishop et al., 1998; Gaßel et al., 1999; Wong et al., 2000)) and 16 potential small toxin proteins of 50 amino acids or less (listed in the introduction), indicate that at least 42 proteins of 16-50 amino acids are encoded by E. coli K-12 (Table 1). We did not include Tpr and YjjY in this list but given that we did detect the lower 8 kDa band for these short ORFs it is possible that they are bona fide protein coding genes.
Homology-based searches using intergenic DNA sequences as input
Since a number of small protein genes were previously identified on the basis of homology, we sought to systematically search for additional conserved small proteins. To streamline our analysis, we batch processed all of the intergenic regions of the E. coli K-12 genome of 40 basepairs or greater (from EcoGene 19) through a series of blastx and tblastx searches. For the blastx analysis, the intergenic regions were screened against the nonredundant protein database to identify potential homologues that had been annotated in other genomes. For the tblastx analysis, the intergenic sequences were compared separately to a DNA database consisting of 300 finished and unfinished microbial genomes and to a DNA database with the genomes from 22 Enteric bacteria. The blastx and tblastx search results for each intergenic region were saved and then sorted according to output file size (Table S3).
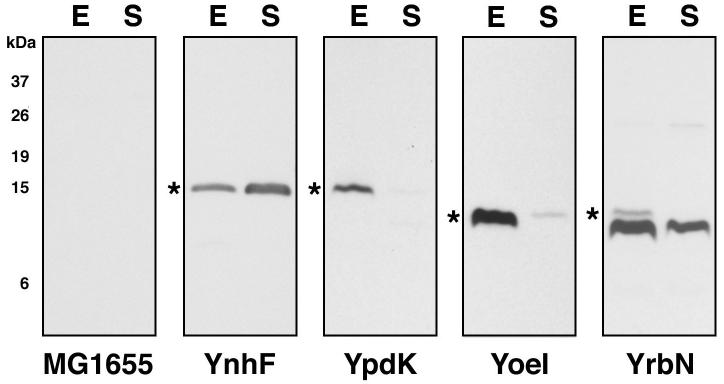
We carefully analyzed the 200 intergenic regions that gave the largest file size in the tblastx search of the 22 Enteric species (summarized in Fig. 2A). The alignments in each file were searched for small ORFs that were conserved in at least two genera and contained examples of conservative amino acid replacements between putative homologues. 522 ORFs fit these criteria, and out of these, 126 were found to encode proteins between 16 and 50 amino acids. We chose this arbitrary range because it corresponded to the size range of previously-annotated small proteins we had tested. Of the 126 short ORFs, 58 were identified as having potential homologs with conserved start and stop codons in at least two different genera of bacteria. These sequences were considered the most likely candidates for new short genes and were used in DNA and protein alignments to evaluate the sequences for the presence of synonymous codons and conservative replacements of amino acids. For most of the protein alignments, we observed that a portion of the short ORF was absolutely conserved with no conservative amino acid replacements, while the surrounding sequences are highly divergent. Many of such short ORFs overlap known transcription factor binding sites. We suggest that the conservation of these regions is driven by the need to conserve DNA regulatory features. Four short ORFs contained an abundance of conservative amino acid replacements in other organisms (Fig. 3). We chose to SPA tag these four potential proteins, YnhF (29 amino acids), YpdK (23 amino acids), YoeI (20 amino acids) and YrbN (26 amino acids), as well as one potential small protein in the fabG-acpP intergenic region (26 amino acids) which showed stretches of continuous conservation and fewer conservative amino acid replacements, to test for their synthesis during growth in LB medium (Fig. 4). We observed high levels of YnhF in both the exponential and stationary-phase cultures. Interestingly this protein was also found as a small chloroform-soluble protein (Z. Guan, X. Wang and C. R. H. Raetz, personal communication). In addition we detected high levels of tagged YpdK and YoeI, especially in exponential phase. For YrbN, we observed high levels of the 8 kDa SPA fragment. We also observed a higher band though the mobility was faster than expected from the predicted molecular weight of YrbN-SPA. No protein was detected for the ORF in the fabG-acpP intergenic region (data not shown).
Fig. 2.

Summary of approaches used to predict genes encoding small proteins.
A. Homology-based searches using intergenic DNA sequences as input.
B. Searches for ribosome binding sites using intergenic DNA sequences as input.
C. Homology-based searching using protein sequences predicted from ribosome binding sites as input.
aa = amino acids
Fig. 3.
Alignments for short ORFs identified on the basis of DNA homology.
Gene sequences identified in the DNA-as-input search were translated and the predicted protein was used to search for homologs using tblastn. Alignments were generated using ClustalW (Fig. 1http://align.genome.jp). “*” indicates the residues are identical in all sequences and “:” and “.” respectively indicate that conserved and semi-conserved substitutions as defined by ClustalW. Swiss-Prot organism codes are from EcoGene (www.ecogene.org/modules.php?name=NEWT).
Fig. 4.
Immunoblot analysis of small proteins predicted on the basis of DNA homology.
Whole cell extracts of MG1655 cells grown to (E) exponential and (S) stationary phase in LB medium were analyzed as in Fig. 1. Again the star (*) indicates the band corresponding the fusion protein. The caveats of the marker lane and exposure times are as for Fig. 1.
Upon closer examination of the genes for which we only observed the 8 kDa SPA fragment (tpr and yjjY) as well as for most genes or regions (dinQ, yoaI and fabG-acpP) for which no expression was detected, we noted that their potential Shine-Dalgarno sequences diverged substantially from the canonical sequence E. coli ribosome binding site (RBS) sequence of “GGAGG”. In contrast, the potential Shine-Dalgarno sequences for the three highly expressed small proteins above, YnhF, YpdK, and YoeI were closer to a canonical RBS.
Searches for ribosome binding sites
To test the possibility that genes encoding small proteins could be identified by the presence of a Shine-Dalgarno sequence, we screened the intergenic regions ≥ 40 basepairs for a canonical “GGAGG” sequence followed by a short ORF. Two sequences were identified that had potential RBSs but were not found in our search above. One potential gene, ydgU, is preceded by a putative Shine Dalgano sequence of “GGAGGG”. Although YdgU can be found in a number of bacteria, it did not fall within the group analyzed in the conservation-based search. The other potential gene, ythA, is preceded by the sequence “AGGAGG”. This intergenic region along with the adjacent genes are likely of prophage origin and no homologs are found outside of E. coli. The putative short ORFs were tagged with the SPA epitope tag and analyzed for expression. We detected synthesis of both proteins in cells grown in rich medium (Fig. 5A). The YthA-SPA protein was present at a high level in exponential cells and to a lesser extent in stationary-phase cells, whereas YdgU-SPA was detected only in stationary-phase cultures.
Fig. 5.
Immunoblot analysis of small proteins predicted on the basis of potential RBSs with high information content.
A. Small proteins predicted in an initial search for genes with canonical RBSs.
B. Small proteins predicted using an information theory-based RBS search for open reading frames. For both (A) and (B), whole cell extracts of MG1655 cells grown to (E) exponential and (S) stationary phase in LB medium were analyzed as in Fig. 1. Again the star (*) indicates the band corresponding the fusion protein. The caveats of the marker lane and exposure times are as for Fig. 1. Given the need for longer exposure times, some background bands were detected for the immunoblots in second row of (B).
To more comprehensively search for RBSs followed by short ORFs, we utilized a RBS model previously constructed using information theory and confirmed E. coli RBSs (Shultzaberger et al., 2001). This model evaluates the information content of the Shine-Dalgarno sequence, the start codon and the distance between these two elements. Based on this model, the information content for the RBSs for the confirmed small proteins ranges from 1 to 14.2 bits of information (Table S2). Those previously-annotated short ORFs for which we did not detect expression generally had lower information content. We used the RBS model to search the intergenic regions for potential RBSs followed by short ORFs (Fig. 2B). The data set used for this screen included the intergenic regions (given in EcoGene 19) as well as 30 basepairs on each 5′ side, so potential hits could contain short overlaps with adjacent genes. The hits were ordered according to the information content, with the maximum of 17.1 bits and a minimum cutoff of 5 bits (Table S4). To test this method of predicting short genes, the 13 predicted short proteins of 16-50 amino acids located downstream of the sites with the highest information content (14.1 to 17.1 bits) were tagged with SPA. Six of these putative genes had predicted σ70 −10 and −35 binding sites upstream of the short ORF (Shultzaberger et al., 2007), while five were in the same orientation as the upstream gene suggesting that they could be transcribed in a polycistronic message.
As with the other short ORFs predicted to encode proteins, the strains encoding the SPA-tagged short ORFs were grown to exponential or early stationary phase in LB, and synthesis of the SPA-tagged proteins was detected by immunoblot analysis. As shown in Fig. 5B, we could detect synthesis of nine of the tagged proteins, IlvX (16 amino acids), YobI (21 amino acids), YmjD (21 amino acids), YnbG (21 amino acids), YqeL (26 amino acids), YohP (27 amino acids), YkgR (33 amino acids), YqfG (41 amino acids), YqcG (46 amino acids) in at least one of the two growth conditions. For one ORF of 34 amino acids encoded in the gmr-rnb intergenic region, we only detected the 8 kDa band corresponding to the SPA tag (Fig. S1). The only intergenic regions for which we did not detect the predicted protein are the ileY-ygaQ, ykgDykgE, and ycgI-minE intervals (data not shown). These regions may not be transcribed. The short ORF in the ileY-ygaQ intergenic region is not likely to be part of an operon and the potential σ70-binding site has low information content. It is also not clear that the ykgD, ykgE and ycgI transcripts are long enough to include the short ORFs in the ykgD-ykgE and ycgI-minE intergenic regions.
In general, most of the genes identified on the basis of the RBSs were not conserved, consistent with the fact that they were not identified in the screen for conserved genes. An exception is the highly-expressed YohP protein for which there are conservative replacements in the gene and synonymous amino acid replacements in the protein sequence. While YohP is conserved, it was missed in the tblast search because it also fell outside of the range that was analyzed in detail.
Homology-based searches using predicted protein sequences as input
The average information content of the RBSs of characterized E. coli genes is 10 bits. Thus short ORFs downstream of translation initiation regions of less than our initial cutoff of 14.1 bits are likely to be translated if they are transcribed. To identify additional candidates among the remaining 1908 ORFs identified downstream of predicted RBSs, we searched this data set for conserved proteins (Fig. 2C). To do this, the protein sequences from the predicted short genes were used as inputs for homology searches against microbial genome DNA sequences that were translated by tblastn. Again the tblastn results from these searches were ordered on the basis of output file size (Table S5). Out of 1921 putative small proteins identified by the RBS search, only 217 gave non-self tblastn hits. These 217 files were screened for conservation of the short ORF including conservation of the start and stop codon positions. While a number of regions showed conservation in one or two other species, ten short ORFs showed strong conservation in at least four species. Of the ten sequences, three potential short ORFs contained an abundance of conservative amino acid replacements in other organisms (Fig. S2). We again tagged these ORFs, YshB (36 amino acids), YoaJ (24 amino acids) and YoaK (32 amino acids) as well as one short ORF in the ydjA-sppA intergenic region (ydjA_sppA_0, 32 amino acids), which showed fewer conservative amino acid replacements. We observed high levels of YoaJ-SPA and YoaKSPA and lower levels of YshB-SPA (Fig. 6). Only the 8 kDa band was detected for the tagged ORF in the ydjA-sppA intergenic region which had lower conservation across the putative homologs (Fig. S1).
Fig. 6.
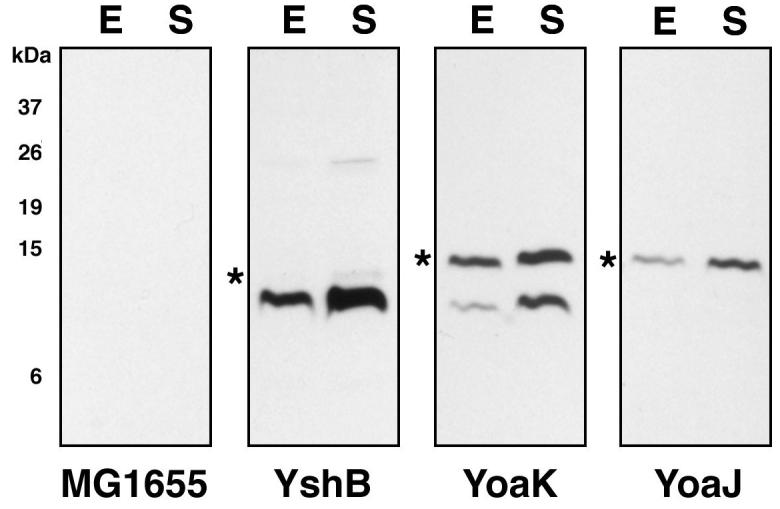
Immunoblot analysis of small proteins predicted on the basis of the presence of a RBS and protein homology.
Whole cell extracts of MG1655 cells grown to (E) exponential and (S) stationary phase in LB medium were analyzed as in Fig. 1. Again the star (*) indicates the band corresponding the fusion protein. The caveats of the marker lane and exposure times are as for Fig. 1.
Abundance of transmembrane proteins
In total, the searches for conservation and translation initiation regions led to the identification of 18 new proteins of less than 50 amino acids (Table 2). Interestingly, 10 of the 18 novel proteins and 65% of all proteins of less than 50 amino acids were predicted to contain a transmembrane segment by at least two out of three prediction programs. This abundance of predicted transmembrane segments prompted us to test whether the tagged proteins could be detected in the membrane fraction. Cell-free extracts were generated from eight strains encoding tagged proteins predicted to have transmembrane segments (YohP, YnhF, YneM, YoaJ, YoaK, YbgT, YbhT and YpdK) and three strains encoding tagged proteins predicted not to have transmembrane segments (YpfM, AzuC and YqgB). In addition, cell-free extracts were prepared from control strains in which the membrane-localized OmpA protein and the cytosolic protein Pgm were tagged with SPA. The extracts were fractionated into supernatant and pellet fractions by ultracentrifugation, and, as predicted, the tagged YohP, YnhF, YneM, YoaK, YbgT, YbhT and YpdK proteins showed fractionation similar to OmpA-SPA while the tagged YpfM and YqgB proteins showed fractionation similar to Pgm-SPA (Fig. 7). The 8 kDa SPA fragment detected for the YneM-SPA and YoaK-SPA extracts also fractionated with the supernatant fraction as expected. Although some of the YoaJ-SPA protein was detected in the supernatant, the majority of this protein was in the pellet fraction. The tagged AzuC, which was not predicted to have a transmembrane segment, also fractionated with the pellet. Closer examination of the AzuC amino acid sequence suggests it could form an amphipathic helix that could be associated with the membrane or a membrane-bound protein. In general, the results from the fractionation are consistent with the majority of the small proteins being localized to membranes.
Table 2.
Newly identified small proteins
|
Protein Name |
Lengtha |
Flanking Genes |
Strandb |
Information Contentc |
Predicted TM domain |
How Predicted | Amino Acid Sequence |
|---|---|---|---|---|---|---|---|
| IlvX | 16 | ilvL/ilvG | >>> | 15.3 | no | RBS | MNNSTKFCFS RFRTGN |
| YoeI | 20 | yeeF/yeeY | <<< | 8.6 | no | DNA query | MGQFFAYATV ITVKENDHVA |
| YobI | 21 | ruvA/yebB | <<> | 16.0 | no | RBS | MYIFITHFFT EYVILKYLLP I |
| YmjD | 21 | ymjC/ymjY | <<< | 14.8 | no | RBS | MKHIQIRNSD MDWHIAANNL G |
| YnbG | 21 | paaY/ydbA | ><> | 14.6 | no | RBS | MKYINCVYNI NYKLKPHSHY K |
| YpdK | 23 | lpxP/yfdZ | >>< | 8.3 | yes | DNA query | VKYFFMGISF MVIVWAGTFA LMI |
| YoaJ | 24 | yoaK/yeaQ | >><d | 11.5 | yes | RBS + protein query | MKKTTIIMMG VAIIVVLGTE LGWW |
| YqeL | 26 | yqeJ/yqeK | ><< | 15.9 | no | RBS | MKDVDQIFDA LDCHILREYL ILLFYD |
| YrbN | 26 | deaD/nlpI | <<< | 5.3 | no | DNA query | MKIADQFHDE LCRLAAINFE AHVLHG |
| YohP | 27 | mdtQ/dusC | <>< | 15.8 | yes | RBS | MKIILWAVLI IFLIGLLVVT GVFKMIF |
| YdgU | 27 | asr/ydgD | >>> | 4.2 | yes | RBS | MVGRYRFEFI LIILILCALI TARFYLS |
| YnhF | 29 | ydhP/purR | <<> | 5.4 | yes | DNA query | MSTDLKFSLV TTIIVLGLIV AVGLTAALH |
| YoaK | 32 | yeaP/yoaJ | >>>d | 6.9 | yes | RBS + protein query | MRIGIIFPVV IFITAVVFLA WFFIGGYAAP GA |
| YkgR | 33 | ykgM/ykgP | <<< | 15.4 | yes | RBS | MKENKVQQIS HKLINIVVFV AIVEYAYLFL HFY |
| YshB | 36 | hemN/glnG | ><< | 10.6 | yes | RBS + protein query | MLESIINLVS SGAVDSHTPQ TAVAAVLCAA MIGLFS |
| YqfG | 41 | ygfU/idi | >>> | 14.6 | yes | RBS | MNFLMRAIFS LLLLFTLSIP VISDCVAMAI ESRFKYMMLL F |
| YthA | 41 | yjhC/yjhD | >>< | 11.7 | yes | RBS | MIKNFIFDNL IILAVPFMIK TSLKTNLIFF FLCVFVPHMA S |
| YqcG | 46 | ygcF/ygcG | <>> | 16.0 | no | RBS | MSEENKENGF NHVKTFTKII FIFSVLVFND NEYKITDAAV NLFIQI |
Unprocessed size.
Orientation of flanking genes. > and < denote genes present on the clockwise (Watson) or counterclockwise (Crick) strand of the E. coli chromosome, respectively.
Information content for ribosome binding site in bits.
yoaJ and yoaK are in the same intergenic interval flanked by yeaP and yeaQ
Fig. 7.
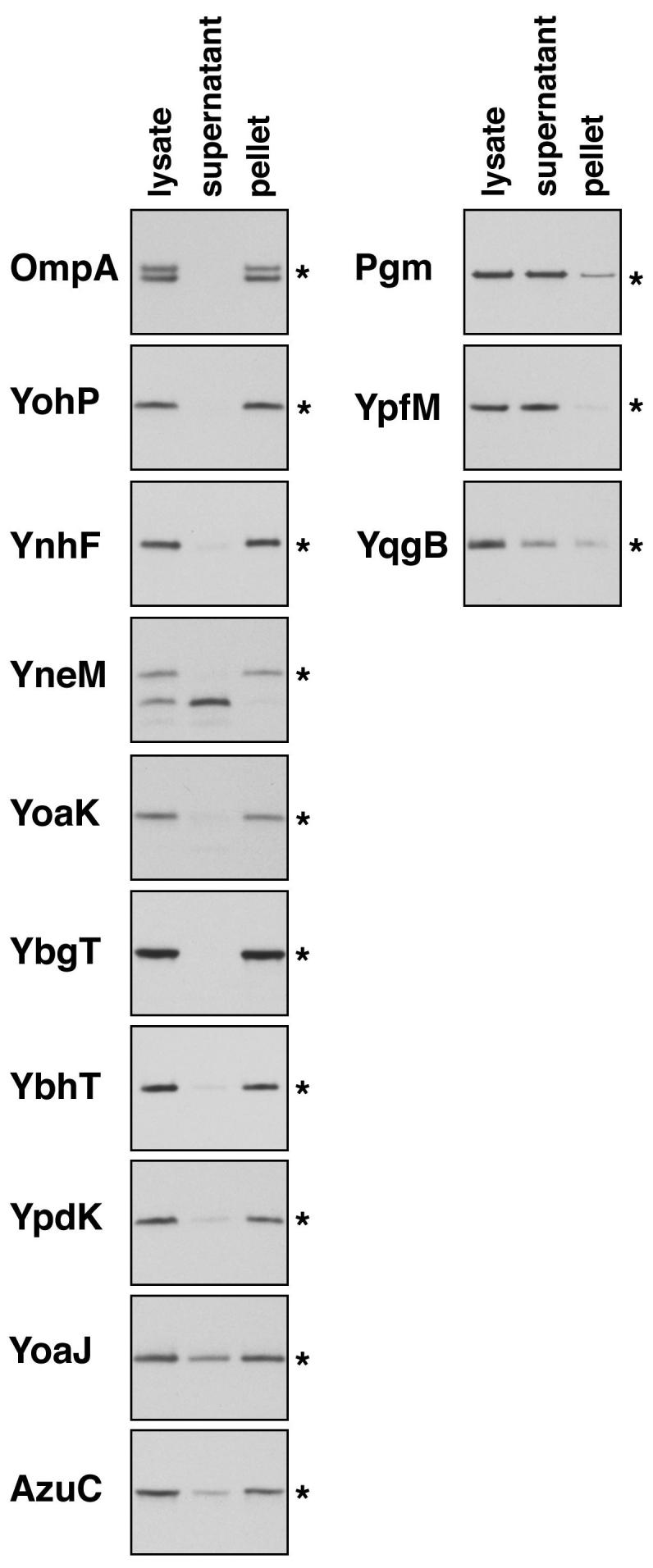
Subcellular fractionation of small proteins predicted to have transmembrane segments.
Whole cell lysates were generated from the SPA-tag strains and fractionated into cytoplasmic supernatant and membrane pellet fractions. Immunoblot analysis of the lysates and fractions was carried out as in Fig. 1. The star (*) indicates the band corresponding the fusion protein.
Discussion
In summary, we validated the translation of 20 previously annotated short genes with epitope-tagged protein detection, more than doubling the number of short genes demonstrated to encode proteins in E. coli K-12. Bioinformatic searches for undiscovered short proteins encoded in E. coli K-12 intergenic regions identified almost two thousand short ORFs that are candidates to encode small proteins. Of these, 24 were selected for experimental validation. Translated products were detected for 18 of these short ORFs. This raises the number of small proteins with validated expression in E. coli K-12 to 44 (not including the 16 potentially toxic small proteins described in the introduction), and suggests that additional small protein-coding genes remain to be identified. The observation that many of the small proteins showed differential expression under the conditions tested is consistent with the supposition that these proteins have specific functions in the cell. Interestingly, the majority of newly identified (10 of 18) and previously annotated (29 of 42) short ORFs are predicted to contain single transmembrane α helices, suggesting that many of these proteins could be functioning at the cell membranes.
Identification of novel small proteins
The processes of identifying statistically significant homologs and defining protein families using multiple sequence alignments present novel challenges when applied to small proteins. The short length and limited number of homologous sequences can preclude a statistically compelling alignment. The small proteins also do not seem to include enzymes or DNA binding proteins with recognizable motifs. In addition, short unrelated hydrophobic regions might align to each other, and short highly-conserved DNA motifs can falsely align at the protein level. Thus the existing annotation of small protein sequences for all organisms is highly variable and unreliable. We mitigated these limitations by (1) computational identification and elimination of gene remnants, dubious Genbank annotations and known DNA motifs, (2) assessment of the conservation of the start and stop codon positions of a short ORF, in addition to its amino acid sequence conservation, (3) RBS predictions, and (4) experimental verification of protein expression. The identification of protein sequence conservation and RBS predictions are complementary search strategies. There are known E. coli genes with good RBSs that do not appear to be conserved and other genes with convincing homologs that do not have discernible RBSs, so both approaches are needed. We found that candidates with either strong evidence of protein sequence conservation (conservative amino acid replacements and aligned synonymous codons) or predicted highly efficient RBSs were generally expressed (9 of 13) whereas candidates with no convincing homologs and moderately or weakly predicted RBSs generally showed very low or no detectable expression under the conditions tested.
In the initial DNA-as-input homology searches, we used the intergenic regions that were cleaned of all known intergenic repeat families. Blastx translates all six frames of the input into protein sequence (ignoring stops and starts) and searches the protein databases for protein similarity. These blastx searches are limited by the fact that many small genes have not been recognized and annotated and thus are not in the protein databases. We also utilized tblastx, which also translates the input DNA sequence but searches against a six-frame translation of the DNA sequence databases. These searches did uncover several new short genes, however the screening of the complex output files was inefficient. Thus we took all intergenic short ORFs preceded by predicted RBSs and used these predicted protein sequences as input for blastp and tblastn searches of protein and translated DNA databases, respectively. Several additional conserved proteins were identified in the more limited output of the tblastn searches.
A number of programs are available for predicting genes in microbial sequences (Bocs et al., 2003; Borodovsky et al., 2003; Delcher et al., 2007; Kosuge et al., 2006). Perhaps the most commonly employed program is Glimmer, which uses a hidden Markov model for identifying protein coding genes in a DNA sequence (Delcher et al., 2007). To be able to compare the predictive capability of Glimmer for identifying short genes to the methods we used, we analyzed the E. coli K-12 MG1655 genome using Glimmer 3.02 (www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi). 244 genes encoding proteins of ≤50 amino acids were predicted by Glimmer. Surprisingly, there was little overlap between our predictions and those of Glimmer. Of the 244 genes identified by Glimmer, only 46 were among the ORFs identified in our information theory-based RBS screen. Of the 64 genes identified as having good RBSs (13 bits or greater) in our information theory-based RBS screen, only six were identified by Glimmer. In addition, Glimmer identified only five (YoaJ, YqeL, YnhF, YoaK, and YqfG) of the 18 newly identified genes (30%) for which we detected protein expression. Programs other than Glimmer have also been used to identify potential bacterial genes, although protein synthesis from genes predicted by these programs has not been demonstrated. We noted that three (YnhF, YshB, and YqfG) of the short genes validated in our study were annotated in a screen with the program Gene Trek in Prokaryotic Space which also combines filtering for the presence of RBSs and homology, though with different parameters (Kosuge et al., 2006). The short genes we confirmed could be used as a teaching set to improve predictions and programs such as Glimmer.
The intrinsic limitations in the ability to predict small protein genes underline the continued need to experimentally validate predictions. We chose to integrate an epitope tag into the chromosome since it allowed the testing of a large number of predictions. However other approaches such as mass spectrometric analysis of the small protein pool can also provide evidence for small protein synthesis. Ultimately, knowledge of the complete proteome of an organism will only be obtained by the iterative application of both computational and experimental approaches.
Small proteins not identified in this screen
Based on the nature of our screen, there are several categories of small proteins that would not have been identified in this study. Most obviously, additional proteins of less than 16 and greater than 50 amino acids are likely to be encoded by the E. coli genome. In fact 170 putative small proteins ranging between 51 and 147 amino acids were predicted in the RBS screen, many with RBSs of high information content. A few of these putative proteins are widely conserved (Table S5). Our searches also did not include proteins encoded opposite previously annotated genes. We recently found that five potentially toxic proteins of 18-19 amino acids are encoded opposite the Sib small RNA genes (Fozo et al., 2008), and it is possible that other small proteins are encoded opposite known genes.
In general we did not test the expression of apparent gene remnants (pseudogenes) of less than 50 amino acids, and removed them from the list of previously-annotated short ORFs (see Table S1). Five of these gene remnants correspond to the amino terminal sequence of a larger protein usually with an intact allele in another E. coli strain. Since the RBS sequences are not deleted, these remnants could be expressed as small proteins. We note however, one of the newly-discovered short ORFs, ymjD, encodes a highly expressed 21 amino acid protein that is almost identical to the amino terminus of a 310 amino acid hydrolase found in other E. coli and Shigella strains and thus may be a pseudogene.
Another group of small proteins that would be missed in our analysis are peptides processed from larger proteins. There are many examples of these proteins in bacteria, and proteolytic processing of larger proteins to give active peptides is prevalent in eukaryotes (Canaff et al., 1999; Grossman, 1995). We would also miss proteins that are only expressed under very specific conditions or are quickly degraded and do not accumulate in the cell. In a number of cases, we detected an 8 kDa band corresponding to the SPA tag while no full-length protein product was apparent. A possible explanation for this observation is that after translation of the tagged protein, the protein was cleaved from the SPA tag and degraded, while the stable SPA tag accumulated in the cell. The presence of the SPA tag could be considered evidence that the short ORF does correspond to a labile translated protein, but for the purposes of this study, we used the more stringent requirement of accumulation of a protein > 8 kDa as evidence of a new small protein.
Possible roles of small proteins
More than half of the E. coli K-12 proteins of less than 50 amino acids are predicted to be single transmembrane segment proteins. Genes encoding a family of short (20-23 amino acids) hydrophobic proteins has also been predicted in streptococci (Ibrahim et al., 2007). The prevalence of the small single transmembrane segment proteins suggests that these proteins are playing roles at the cell membranes. It is tempting to speculate that some of the highly expressed transmembrane proteins serve to modulate the character of the membrane. This could be the role of very small proteins such as YohP, which have only four amino acids outside of the predicted transmembrane helix. Alternatively, oligomeric complexes of the single transmembrane proteins could form a pore as has been proposed for the toxic Hok proteins (Gerdes et al., 1997). The somewhat larger single transmembrane proteins may be part of multiprotein complexes. Two such proteins, YbgT and YccB, are encoded in cytochrome oxidase operons and are in fact paralogs of each other. Small transmembrane proteins, such as PsbE and PsbF in the plant photosystem II complex, have been shown to act as cofactors that coordinate heme and other molecules (Shi and Schröder, 2004). Both YbgT and YccB have conserved cysteines in the transmembrane helix, so it is intriguing to speculate that they could coordinate the heme for the cytochrome oxidase. The sequences of a few of the hydrophobic small proteins also are reminiscent of the pheromone peptides produced by Enterococcus faecalis raising the possibility that some of the E. coli proteins might be cleaved and secreted as signals.
Although not a potential transmembrane protein, another interesting protein characterized in this study is YkgO. This protein was originally identified as a potential paralog of the ribosomal protein RpmJ/L36 (Blattner et al., 1997), although to our knowledge this is the first evidence that E. coli K-12 YkgO is expressed. The intriguing characteristic of YkgO is that it is undetectable in cells growing at high rates in rich medium, a condition in which most ribosomal proteins are synthesized at high levels (Kaczanowska and Rydén-Aulin, 2007). The levels of YkgO are highest in stationary-phase cells grown in minimal medium where synthesis of other ribosomal proteins is reduced. Comparative genomics has suggested that ykgO is repressed by zinc in various bacteria (Panina et al., 2003). Unlike RpmJ, YkgO lacks a zinc-binding motif. It has been proposed that ribosomal protein paralogs lacking zinc binding motifs are induced to provide zinc to the cell during zinc starvation (Akanuma et al., 2006; Moore and Helmann, 2005; Panina et al., 2003) and/or allow ribosome function under zinc-limiting conditions (Natori et al., 2007).
Density of E. coli genes
Ultimately, the results of this analysis suggest a reevaluation of the size of intergenic regions in bacteria is warranted. During the original annotation of the E. coli K-12 genome, the 70 intergenic regions of larger than 600 nt were reanalyzed for the presence of ORFs, from which 15 regions were found to contain potential genes (Blattner et al., 1997). Our results suggest that such a reanalysis could have been performed on much smaller intergenic regions. The finding of new small protein genes, coupled with the discovery of nearly one hundred small RNA genes, raises the question of how much sequence in the E. coli genome is truly “intergenic”. As small RNA and small protein discovery continues, we expect that the average intergenic size will continue to shrink.
Experimental procedures
Construction of SPA-tagged strains
The sequences of all oligonucleotides used in the study are given in Table S6. Oligonucleotides were designed such that the sequential peptide affinity (SPA) tag [containing the 3xFLAG and the calmodulin binding peptide (CBP) sequences separated by a TEV protease cleavage site] together with the adjacent kanamycin resistance genes surrounded by FRT sites could be amplified from the plasmid pJL148 (Zeghouf et al., 2004) and be flanked by 40-45 nt of sequence homologous to the region of insertion. All PCR reactions were carried out using either Accuprime Pfx or Platinum Taq High Fidelity DNA polymerase (Invitrogen). The gel-purified PCR product was used to transform NM400 (MG1655 mini-λ cam) to introduce the SPA tag at the end of the putative short ORF using mini-λ Red recombination (Yu et al., 2000). All the short ORF-SPA-kan alleles were moved into MG1655 by P1 transduction. All insertions were confirmed by sequencing.
Immunoblot assays
Whole cells grown in Luria Broth (LB, Invitrogen) or M63 medium (KD Medical) supplemented with 10 μg/ml vitamin B1 and 0.2% glucose, were resuspended in 50 mM sodium phosphate buffer (pH 8). Resuspended whole cells were mixed with 4X sample buffer (1X stacking buffer, 2% SDS, 0.025 mg bromophenol blue, 52% glycerol), heated at 95°C for 10 min and centrifuged for 10 min. A fraction equivalent to the cells in OD600 = 0.057 was separated on a Novex 16% Tricine gel (Invitrogen), and transferred to a nitrocellulose membrane (Invitrogen). The membranes were blocked with 3% milk, and probed with anti-FLAG M2-AP monoclonal antibody (Sigma-Aldrich) in 2% milk. Signals were visualized using the Lumi-Phos WB (Pierce) for anti-FLAG M2-AP antibody detection.
Cell fractionation
The cells in 20 ml of culture, grown to OD600 = 0.6-0.7 at 37°C in LB medium, were collected by centrifugation at 4,000 rpm for 10 min at 4°C and resuspended in 0.9 ml of cold 20% sucrose, 100 mM NaCl, 50 mM Tris pH 8 and EDTA-free protease inhibitor (Roche). After the addition of 50 μl of a solution of 10 mg/ml lysozyme and 10 mM EDTA, the samples were incubated at 25°C for 1 h and then pipetted 10X through a 30 Gauge needle at 4°C. The lysates were centrifuged 3X at 12 000 g for 5 min at 4°C to remove unlysed cells. Supernatant fractions were taken for the whole cell lysate sample (100 μl) and for further centrifugation at 56,000 rpm (130,000 × g) for 2 h at 4°C in a Beckman TLA100.3 rotor (500 μl). Aliquots of the supernatant from the last ultracentrifugation step were taken for the cytoplasmic fraction (500 μl). The membrane pellet was resuspended in 500 μl of 20% sucrose 50 mM Tris pH 8 by pipetting followed by the addition of SDS (1% final concentration). SDS (1% final concentration) was also added to the whole cell lysate and cytoplasmic fractions and all samples were incubated overnight at room temperature. Subsequently, the samples were mixed with 4X sample buffer, incubated at 95°C for 10 min, centrifuged for 5 min and then analyzed by immunoblots as described above.
blastx and tblastx searches
The details of all databases used in the searches are given in Table S7. For the blastx searches, the eg19_ig_clean intergenic sequence file (2579 intervals) was searched against a combined translated non-redundant and environmental protein sequence database denoted as the nr_env database. For the tblastx searches, two different sets of microbial genomes were downloaded from NCBI and formatted for use as the 300_unfin_micro and 22_entero databases. The various output files were formatted as HTML files that could be accessed using a spreadsheet with HTML hyperlinks. The HTML outputs were then sorted by file size. After sorting, the tblastx outputs were screened for conserved sequences that contained amino acids potentially encoded by E. coli start codons (Met for “ATG”, Val for “GTG” and Leu for “TTG”), followed by at least two examples of conservative replacement. In most cases, a single intergenic region yielded multiple potential hits. 522 hits were identified based on these criteria. The predicted protein sequence of these hits was then analyzed by tblastn against the E. coli K-12 genome in order to confirm start codons, identify stop codons and examine ORF conservation. 126 hits contained a short ORF that could encode a small protein of 16 to 50 amino acids. One hit that was identified as a newly annotated probable gene remnant and was not considered further. Of the 126 hits, 58 ORFs were conserved in bacterial species of two or more genera. DNA and protein alignments were generated for these 58 genes and inspected for conservative replacement and synonymous codon usage.
RBS search
The same 2579 intergenic regions subjected to blastx and tblastx searches above were searched for short ORFs ≥ 9 amino acids and with at least 5.0 bits of information in the translation initiation region as follows. Programs of the Delila system (dbbk, catal, delila)were used to extract the intergenic sequences (Schneider et al., 1982; Schneider, 1996) from the E. coli K-12 genome (GenBank Accession NC_000913). For each intergenic interval and its complementary sequence, 30 bases were included at the 5′ side to ensure locating RBSs that intersect the 5′ end. Likewise, 30 bases on the 3′ side were removed to avoid picking up the known gene starts. These sequences were searched (multiscan) for RBSs with a minimum of 5 bits using the rbseg12 ribosome model (Shultzaberger et al., 2001). A new program (orf) identified open reading frames that began with identified RBSs and were at least 10 codons and at most 150 codons long (including the stop codon). These short ORFs were sorted by the individual information content of the initiating RBS (Schneider, 1997). The sorted short ORF features, along with the identified RBSs, annotated gene ends from the GenBank entry and potential σ70 promoters (Shultzaberger et al., 2007) were used to generate a genetic map in a PostScript file using the lister program. These features, displayed from 30 bases upstream to 20 bases downstream of the intergenic region, are given at http://www.ccrnp.ncifcrf.gov/~toms/papers/smallproteins/.
tblastn search
Again the details of all databases used in the searches are given in Table S7. The eg19_RBS_9-50 protein sequence library (1921 entries) was used to search the microbial genome sequences for conservation with tblastn with the low complexity filter off against the all_micro and coli_only databases and with blastp against the eg19_RBS_9-50 input database and the nr_env database. Again the various output files were formatted as HTML files that could be accessed using a spreadsheet with HTML hyperlinks. 217 of these sequences returned non-null results files, and the degree of conservation of these genes was analyzed by tblastn of the predicted amino acid sequence against the microbial genome database. 27 of these genes were found to be conserved in bacteria in three of more genera based on DNA and protein alignments, and these were considered as candidates for experimental testing of protein expression.
Acknowledgments
We thank J. Miranda-Rios for pointing out the ORF in the ISO92/IsrB RNA, R. Hegde for advice on cell fractionation, M. W. Stone and J. Zhou for computational assistance, C. Raetz for communicating unpublished data, and S. Gottesman, E. Koonin and members of the Storz lab for helpful discussions and comments. This research was supported by NIH grant number R01-GM58560 (K.E.R.), the Intramural Research Programs of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (M.R.H., B.J.P. and G.S.) and the National Cancer Institute (T.D.S.) and by a postdoctoral fellowship from the Life Sciences Foundation (M.R.H.).
Abbreviations
- ORF
open reading frame
- RBS
ribosome binding site
References
- Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J. Bacteriol. 2006;188:2715–2720. doi: 10.1128/JB.188.7.2715-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J. Mol. Biol. 1998;280:583–596. doi: 10.1006/jmbi.1998.1894. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bocs S, Cruveiller S, Vallenet D, Nuel G, Médigue C. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res. 2003;31:3723–3726. doi: 10.1093/nar/gkg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M, Mills R, Besemer J, Lomsadze A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr. Protoc. Bioinformatics. 2003 doi: 10.1002/0471250953.bi0405s01. Chapter 4: Unit 4.5. [DOI] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Canaff L, Bennett HP, Hendy GN. Peptide hormone precursor processing: getting sorted? Mol Cell Endocrinol. 1999;156:1–6. doi: 10.1016/s0303-7207(99)00129-x. [DOI] [PubMed] [Google Scholar]
- Cliften PF, Hillier LW, Fulton L, Graves T, Miner T, Gish WR, Waterston RH, Johnston M. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RL, Nizet V. Endogenous production of antimicrobial peptides in innate immunity and human disease. Curr. Allergy Asthma Rep. 2003;3:402–409. doi: 10.1007/s11882-003-0074-x. [DOI] [PubMed] [Google Scholar]
- Garbis S, Lubec G, Fountoulakis M. Limitations of current proteomics technologies. J. Chromatogr. A. 2005;1077:1–18. doi: 10.1016/j.chroma.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Gaßel M, Möllenkamp T, Puppe W, Altendorf K. The KdpF subunit is part of the K(+)-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 1999;274:37901–37907. doi: 10.1074/jbc.274.53.37901. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Gultyaev AP, Franch T, Pedersen K, Mikkelsen ND. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- Gong M, Gong F, Yanofsky C. Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2Pro availability. J. Bacteriol. 2006;188:1892–1898. doi: 10.1128/JB.188.5.1892-1898.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- Ibrahim M, Nicolas P, Bessières P, Bolotin A, Monnet V, Gardan R. A genome-wide survey of short coding sequences in streptococci. Microbiology. 2007;153:3631–3644. doi: 10.1099/mic.0.2007/006205-0. [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis RW, Boeke JD, Snyder MA, Basrai MA. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16:365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- Kosuge T, Abe T, Okido T, Tanaka N, Hirahata M, Maruyama Y, Mashima J, Tomiki A, Kurokawa M, Himeno R, Fukuchi S, Miyazaki S, Gojobori T, Tateno Y, Sugawara H. Exploration and grading of possible genes from 183 bacterial strains by a common protocol to identification of new genes: Gene Trek in Prokaryote Space (GTPS) DNA Res. 2006;13:245–254. doi: 10.1093/dnares/dsl014. [DOI] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 2005;8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol. Microbiol. 2007;63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- Ochman H. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet. 2002;18:335–337. doi: 10.1016/s0168-9525(02)02668-9. [DOI] [PubMed] [Google Scholar]
- Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland SL, Burkholder WF, Cunningham KA, Maciejewski MW, Grossman AD, King GF. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell. 2004;13:689–701. doi: 10.1016/s1097-2765(04)00084-x. [DOI] [PubMed] [Google Scholar]
- Rudd KE. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol. Mol. Biol. Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd KE, Humphery-Smith I, Wasinger VC, Bairoch A. Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis. 1998;19:536–544. doi: 10.1002/elps.1150190413. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stormo GD, Haemer JS, Gold L. A design for computer nucleic-acid sequence storage, retrieval and manipulation. Nucleic Acids Res. 1982;10:3013–3024. doi: 10.1093/nar/10.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider >TD. Reading of DNA sequence logos: Prediction of major groove binding by information theory. Meth. Enzym. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- Schneider >TD. Information content of individual genetic sequences. J. Theor. Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- Shi LX, Schröder WP. The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta. 2004;1608:75–96. doi: 10.1016/j.bbabio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EGH, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RS, McMurry LM, Levy SB. 'Intergenic' blr gene in Escherichia coli encodes a 41-residue membrane protein affecting intrinsic susceptibility to certain inhibitors of peptidoglycan synthesis. Mol. Microbiol. 2000;37:364–370. doi: 10.1046/j.1365-2958.2000.01998.x. [DOI] [PubMed] [Google Scholar]
- Yanofsky >C. Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]