The accuracy with which the genetic information contained in protein-coding genes is faithfully translated into the corresponding sequence of amino acids has long fascinated biologists. Before the mechanisms of transcription and protein synthesis had been uncovered in the exquisite molecular detail we know today, some of the inherent problems of faithful gene expression were obvious. Crick's seminal adaptor hypothesis (1) predicted the existence of many then-unknown components of translation, including the aminoacyl-tRNA synthetases. The aminoacyl-tRNA synthetases in effect define the genetic code by catalyzing a 2-step reaction that pairs amino acids with their cognate tRNAs to provide substrates for ribosomal protein synthesis. In the first step, an amino acid is condensed with ATP to form an aminoacyl-adenylate. In the second reaction, the aminoacyl group is transferred to the 3′ end of the tRNA. The aminoacyl-tRNA synthetases also provide a critical safeguard to maintain fidelity during translation of the genetic code by discriminating against and, when necessary, editing noncognate amino acids. Crick was quick to point out that specificity would be of paramount importance to the synthetases, because their function in protein synthesis would require them to precisely distinguish similar amino acids such as isoleucine and valine. Linus Pauling (2), who reasoned that small differences in binding energy between aliphatic amino acids would not provide the level of discrimination necessary for faithful protein synthesis, had also noted this particular problem in molecular recognition. This discrepancy, between the specificity achievable during recognition and the accuracy required for translation, was resolved with the discovery of editing.
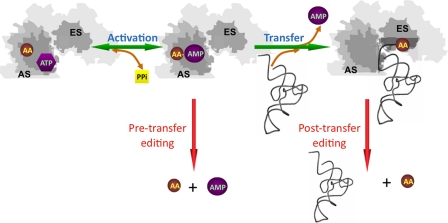
It was observed that although isoleucyl-tRNA synthetase did indeed recognize and activate valine, the intermediate valyl-adenylate was subsequently hydrolyzed in a tRNA-dependent reaction (3). The net result is that although isoleucyl-tRNA synthetase can use both isoleucine and valine, only the cognate product isoleucyl-tRNAIle accumulates. Numerous studies of isoleucyl-tRNA and other synthetases have provided a general picture of the structure and mechanism of editing (Fig. 1). It had long been known that editing could occur either before (pretransfer) or after (posttransfer) amino acids are covalently attached to tRNA, an essential component of the protein synthesis machinery. In both cases the net result is the same, synthesis and release of noncognate aminoacyl-tRNA is prevented and translational accuracy is maintained. In recent years the biological relevance of the pretransfer route had come under question, and the prevailing dogma was that, with a few notable exceptions, editing was essentially a posttransfer process. In this issue of PNAS, Martinis and coworkers (4) now show that posttransfer editing can mask pretransfer editing activity, a finding with far-reaching implications for both quality control and the evolution of protein synthesis.
Fig. 1.
Pretransfer and posttransfer editing of noncognate amino acids by aminoacyl-tRNA synthetases. The amino acid (AA) is activated in an ATP-dependent reaction at the active site (AS) to form an enzyme-bound aminoacyl-adenylate (AA-AMP). In pretransfer editing, AA-AMP is hydrolyzed directly, thereby preventing the synthesis of a noncognate aminoacyl-tRNA. In posttransfer editing, AA-AMP is a substrate for esterification of the 3′ end of the tRNA, which then translocates to the editing site (ES) where the AA is removed. PPi, pyrophosphate.
Posttransfer editing can mask pretransfer editing activity.
The posttransfer reaction either occurs in cis, before the noncognate aminoacyl-tRNA is released, or in trans catalyzed by synthetases or specific hydrolases (5). A large number of biochemical and structural studies have clearly shown that this reaction occurs in a second active site that is distinct from the ancient catalytic core. Less frequently, the noncognate aminoacyl-adenylate will be hydrolyzed before tRNA esterifcation in a so-called pretransfer reaction (6). However, the relationship between these 2 pretransfer and posttransfer editing pathways has remained unclear in most systems. In particular, the mechanism and physiological relevance of pretransfer editing has continued to be contentious. Many of the points of dispute arise from the inherent instability of aminoacyl-adenylates, which makes their study difficult in vitro and almost impossible in vivo. In addition, much of the controversy surrounding the pretransfer pathway had come from difficulties in envisioning the molecular mechanism by which an unstable aminoacyl-adenylate could translocate some 30 Å from the active site before hydrolysis. Indeed, several examples of pretransfer editing by synthetases that lack a separate editing domain have been reported (7–9).
Leucyl-tRNA synthetase (LeuRS) presents an ideal model system for studying the relationship between, and relative significance of, pretransfer and posttransfer editing. In addition to the well-documented modularity of the conserved CP1 editing domain (10–12), different LeuRSs show widely divergent editing mechanisms. For example, Escherichia coli LeuRS is known to use a posttransfer editing mechanism to deacylate tRNAs charged with a variety of noncognate amino acids, including isoleucine, valine, norvaline, and methionine. Most notably for the present study, although it has been proposed that the yeast enzyme predominantly relies on pretransfer editing, the E. coli enzyme shows no such activity and solely uses posttransfer editing (13). In their study, Martinis and coworkers (4) set out to exploit the known properties of the E. coli enzyme to probe the relationship between pretransfer and posttransfer editing in more detail.
Previous studies had suggested that particular residues are needed for the effective transfer of editing substrates from the synthetic active site to the hydrolytic editing site in the CP1 module of LeuRS (14). To study this postulated translocation pathway Martinis and coworkers (4) used a combination of LeuRS mutants defective in posttransfer editing and CP1 deletion mutants of both E. coli (ecΔCP1) and yeast mitochondrial (ymΔCP1) LeuRSs. The ΔCP1 deletion mutants were found to retain significant cognate aminoacylation activity (Leu-tRNALeu synthesis), although they did show some loss compared with the wild type. This modest loss in leucylation by the ΔCP1 mutants was not unexpected and was consistent with suggestions that the CP1 domain stabilizes enzyme–tRNA interactions. When tested for hydrolysis in trans of noncognate Ile-tRNALeu, both of the ΔCP1 deletion mutants display negligible levels of deacylation, confirming that, as expected, these mutants retain little or no posttransfer editing activity. Martinis and coworkers next performed a final control and tested the ability of the mutants to synthesize mischarged tRNAs, a typical property of enzymes in which posttransfer editing has been disrupted. Whereas the editing-site mutant T252Y showed robust Ile-tRNAIle synthesis as predicted, the ecΔCP1 and ymΔCP1 mutants were incapable of mischarging tRNALeu with Ile. Pyrophosphate exchange assays performed to test the misactivation of Ile and other noncognate substrates showed that the ecΔCP1 mutant misactivated Ile to a level comparable with that of wild type. The ability of ecΔCP1 to misactivate Ile, together with its inability to form Ile-tRNA, suggested the existence of a dormant pretransfer editing activity, which is only apparent when the posttransfer editing CP1 domain is removed. Finally, by using inactive tRNAs modified to lack the site for amino acid attachment, pretransfer editing activity was shown to be strongly stimulated by tRNA despite the absence of the CP1 domain.
In unveiling pretransfer editing activity in E. coli LeuRS, an enzyme believed to solely depend on posttransfer editing for quality control, Martinis and coworkers (4) raise a number of questions about the origin and function of editing. The most immediate result of their study is to force a reassessment of tRNA-dependent pretransfer editing. Their work clearly shows that translocation to a distinct editing domain is not a prerequisite for tRNA-dependent pretransfer editing as had previously been proposed, which, in turn, raises the question as to how tRNA facilitates editing without itself being aminoacylated. Presumably this would involve increasing the rate of hydrolysis within the active site itself and/or accelerating adenylate dissociation as a prelude to spontaneous hydrolysis, both of which were recently shown to contribute to tRNA-independent editing by prolyl-tRNA synthetase (6). However this particular pretransfer editing mechanism is eventually resolved, the observation that the extant E. coli LeuRS:tRNALeu pair contains the functional remnants of a rudimentary ribonucleoprotein has some interesting evolutionary implications. The tRNA dependence of this hidden pretransfer editing activity is consistent with the coevolution of tRNAs and quality-control mechanisms, helping to explain how structurally similar amino acids were added to the genetic code without compromising the fidelity of translation.
The biggest remaining mystery of this study is why the E. coli LeuRS has retained a “hidden” capacity for pretransfer editing that would appear to be redundant given that it already has robust posttransfer activity. Editing is not ubiquitous to synthetases as illustrated by the fact that some enzymes, including human mitochondrial LeuRS, have lost their proofreading capacity and rely instead on accurate substrate recognition (15–17). In this context, the retention of 2 distinct editing pathways is all the more surprising and would seem to suggest that the pretransfer reaction of E. coli LeuRS may actually be required for an activity other than translational quality control. No matter what this activity might turn out to be, to paraphrase Mark Twain, reports of the death of pretransfer editing are greatly exaggerated.
Footnotes
The authors declare no conflict of interest.
See companion article on page 19223.
References
- 1.Crick F. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 2.Pauling L. Festschrift für Prof Dr Arthur Stoll. Basel, Switzerland: Birkhauser; 1958. [Google Scholar]
- 3.Baldwin AN, Berg P. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J Biol Chem. 1966;241:839–845. [PubMed] [Google Scholar]
- 4.Boniecki MT, Vu MT, Betha AK, Martinis SA. CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc Natl Acad Sci USA. 2008;105:19223–19227. doi: 10.1073/pnas.0809336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Splan KE, Ignatov ME, Musier-Forsyth K. Transfer RNA modulates the editing mechanism used by class II prolyl-tRNA synthetase. J Biol Chem. 2008;283:7128–7134. doi: 10.1074/jbc.M709902200. [DOI] [PubMed] [Google Scholar]
- 7.Gruic-Sovulj I, Uter N, Bullock T, Perona JJ. tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase. J Biol Chem. 2005;280:23978–23986. doi: 10.1074/jbc.M414260200. [DOI] [PubMed] [Google Scholar]
- 8.Hati S, et al. Pretransfer editing by class II prolyl-tRNA synthetase: Role of aminoacylation active site in “selective release” of noncognate amino acids. J Biol Chem. 2006;281:27862–27872. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 9.Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. Hydrolysis of noncognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett. 2007;581:5110–5114. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 10.Lin L, Hale SP, Schimmel P. Aminoacylation error correction. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 11.Nureki O, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 12.Lincecum TL, et al. Structural and mechanistic basis of pretransfer and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 13.Englisch S, Englisch U, von der HF, Cramer F. The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res. 1986;14:7529–7539. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams AM, Martinis SA. Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity. Proc Natl Acad Sci USA. 2006;103:3586–3591. doi: 10.1073/pnas.0507362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lue SW, Kelley SO. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 16.Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 17.SternJohn J, Hati S, Siliciano PG, Musier-Forsyth K. Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc Natl Acad Sci USA. 2007;104:2127–2132. doi: 10.1073/pnas.0611110104. [DOI] [PMC free article] [PubMed] [Google Scholar]