Abstract
Sulfotransferases are a versatile class of enzymes involved in numerous physiological processes. In mammals, adenosine 3′-phosphate-5′-phosphosulfate (PAPS) is the universal sulfuryl donor, and PAPS-dependent sulfurylation of small molecules, including hormones, sugars, and antibiotics, is a critical step in hepatic detoxification and extracellular signaling. In contrast, little is known about sulfotransferases in bacteria, which make use of sulfurylated molecules as mediators of cell–cell interactions and host–pathogen interactions. Bacterial arylsulfate sulfotransferases (also termed aryl sulfotransferases), in contrast to PAPS-dependent sulfotransferases, transfer sulfuryl groups exclusively among phenolic compounds in a PAPS-independent manner. Here, we report the crystal structure of the virulence factor arylsulfate sulfotransferase (ASST) from the prototypic, pyelonephritogenic Escherichia coli strain CFT073 at 2.0-Å resolution, and 2 catalytic intermediates, at 2.1-Å and 2.4-Å resolution, with substrates bound in the active site. ASST is one of the largest periplasmic enzymes and its 3D structure differs fundamentally from all other structurally characterized sulfotransferases. Each 63.8-kDa subunit of the ASST homodimer comprises a 6-bladed β-propeller domain and a C-terminal β-sandwich domain. The active sites of the dimer are situated at the center of the channel formed by each β-propeller and are defined by the side chains of His-252, His-356, Arg-374, and His-436. We show that ASST follows a ping-pong bi–bi reaction mechanism, in which the catalytic residue His-436 undergoes transient sulfurylation, a previously unreported covalent protein modification. The data provide a framework for understanding PAPS-independent sulfotransfer and a basis for drug design targeting this bacterial virulence factor.
Keywords: beta propeller, crystal structure, pyelonephritis, uropathogenic Escherichia coli CFT073 periplasm
Sulfotransferases catalyze the transfer of a sulfuryl group from an activated donor to an acceptor and are essential for numerous physiological processes, such as sulfur metabolism, liver detoxification, signal transduction, hormone regulation, viral entry, and molecular recognition. A variety of small molecules, including hormones, sugars, and antibiotics, have been shown to undergo 3′-phosphate-5′-phosphosulfate (PAPS)-dependent sulfurylation (1, 2), and sulfotransferases are recognized as modulators of prokaryote–eukaryote interactions (3).
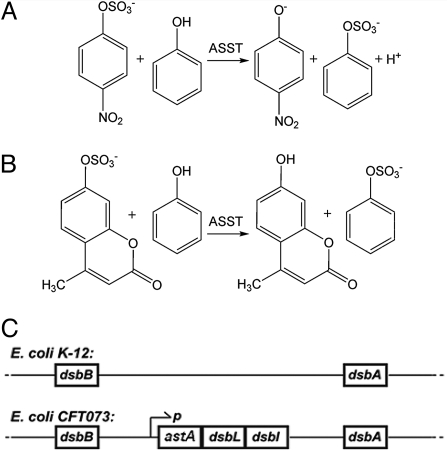
In mammals, sulfoconjugation and glucuronidation represent the dominant mechanisms for detoxification of endogenous and exogenous compounds bearing phenolic groups (4, 5). Therefore, hepatic sulfotransferases are of considerable toxicological and pharmacological interest. In addition to mammalian sulfotransferases, arylsulfate sulfotransferases of commensal intestinal bacteria have been proposed to play a role in the detoxification of phenolic compounds (6, 7). Although a number of eukaryotic sulfotransferases have been extensively studied, much less is known about bacterial sulfotransferases, with periplasmic arylsulfate sulfotransferases comprising a virtually uncharacterized class. In contrast to other sulfotransferases that exclusively use PAPS as a sulfuryl donor, bacterial arylsulfate sulfotransferases (ASSTs) catalyze sulfuryl transfer from a phenolic sulfate to a phenol (Fig. 1 A and B) and cannot use PAPS as the donor (1).
Fig. 1.
Substrates of the ASST-catalyzed reaction analyzed in this study and the genomic context of ASST. (A and B) ASST catalyzes the transfer of a sulfuryl group from an aromatic sulfuryl donor to acceptors with an aromatic hydroxyl group. The corresponding model substrates used in this study were PNS and phenol (A) and MUS and phenol (B), respectively. (C) Location of the dithiol oxidation pathway genes in E. coli K-12 and operon organization of the genes encoding ASST, DsbL, and DsbI in E. coli CFT073.
The genes encoding bacterial periplasmic ASSTs are often found clustered with the genes coding for the proteins DsbL and DsbI of the DsbA/DsbB-independent disulfide bond formation pathway. These DsbA/DsbB homologs form a specific dithiol oxidase system generating the single disulfide bond in each subunit of the ASST homodimer (see below) (8). Most commensal Escherichia coli strains do not contain a gene encoding ASST, but the prototypic uropathogenic E. coli strain CFT073 (9) as well as other uropathogenic E. coli strains (10) do contain the genes encoding ASST, DsbL, and DsbI as a tricistronic operon on the genome (Fig. 1C).
We here report complementary structural and biochemical studies on ASST, shedding light on the unusual, PAPS-independent sulfotransfer reaction catalyzed by this enzyme. The X-ray structure of ASST shows that the subunits of the homodimeric enzyme consist of a 6-bladed β-propeller domain fused to an N-terminal β-sandwich domain. In addition, we have also determined the structure of 2 sulfo-ASST intermediates formed upon incubation of ASST with different sulfuryl donors that provide snapshots of the sulfuryl transfer reaction. Together with biochemical data, these intermediates demonstrate catalysis of sulfuryl transfer to proceed via a ping-pong reaction mechanism involving transient sulfurylation of His-436, a previously unknown covalent protein modification. The residues His-252, His-356, Arg-374, and His-436 are shown to be essential to function, and together, form the active site in the central substrate binding pocket formed by the β-propeller domain. The structural and functional studies presented here provide a unique framework for understanding PAPS-independent sulfuryl transfer.
Results
Crystallization and Overview of the Crystal Structure of ASST.
Arylsulfate sulfotransferase from E. coli CFT073 is a homodimeric protein (Fig. S1), and at 63,763 Da (571 residues) per monomer it is one of the largest proteins in the bacterial periplasm.* Monomers of ASST are secreted into the periplasm with an N-terminal signal sequence of 27 aa,† as confirmed by Edman sequencing of the mature protein. ASST was overproduced by coexpression with DsbL and DsbI, purified as described (8), and obtained as a mixture of unmodified ASST and a sulfuryl-adduct (+81 ± 2 kDa). This mass difference (corresponding to +SO3−; i.e., 80.1 Da) provided the first clue that ASST becomes transiently sulfurylated during its catalytic cycle (see below). ASST was crystallized by the sitting drop vapor diffusion method from 1.8 M Li2SO4 in 100 mM cacodylic acid/NaOH pH 6.5. Hexagonal ASST crystals, belonging to the space group P3212 with 1 dimer of unmodified ASST in the asymmetric unit, were obtained, and X-ray diffraction data were collected to a resolution of 2.0 Å (Table S1).
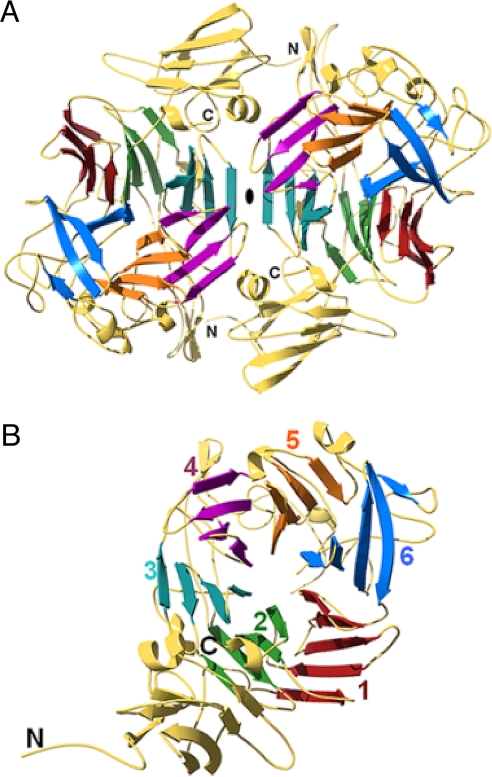
Each ASST monomer consists of 2 readily distinguishable domains: a small N-terminal domain (residues 1–116) forming a 7-stranded β-sandwich and a large C-terminal domain (residues 117–571) (Fig. 2 and Fig. S2). The C-terminal domain adopts a 6-bladed β-propeller fold formed by the packing of 6 four-stranded β-sheets in a circular array. The small (≈410 Å2) contact area between the 2 domains in the monomer involves residues from loop regions and β-strands in the N-terminal domain and the second and third blade of the propeller. The contact area between the 2 ASST monomers (≈6,200 Å2) comprises residues from the N-terminal domain, the third and fourth blade of the propeller, and the loop regions within them.
Fig. 2.
Ribbon diagrams of the ASST structure. (A) Top view of the ASST homodimer along the 2-fold noncrystallographic symmetry (NCS) axis, which relates the 2 monomers. (B) Each ASST monomer consists of a small, N-terminal 7-stranded β-sandwich domain, and a larger C-terminal, 6-bladed β-propeller domain.
As with most other β-propeller structures, the channel formed by the blades of the propeller is conical in shape (11). In the case of ASST, the channel narrows in diameter from ≈21–10 Å in the region of the active site, is solvent accessible, containing 11 ordered water molecules, and is suitable for substrate sequestration. The loop containing residues Val-32–Leu-327 flanks the narrower channel entrance. Its poorly defined electron density suggests a high degree of flexibility, facilitating free passage of substrates into the channel.
ASST contains 3 cysteine residues; 2 of these residues (Cys-418 and Cys-424) are located in a loop in the outer part of the sixth propeller blade and are linked by a disulfide bond of very unusual geometry (Fig. S3). The distance between the Cα atoms of the 2 cysteines is only 3.8 Å, which is extremely short in comparison to the mean of 5.63 Å for all protein disulfides (12). The dihedral angles of this disulfide classify it as being of a −RHStaple geometry and, together with the high dihedral strain energy of this bond [estimated at ≈30 kJ/mole (12)], these parameters help to explain the requirement for the ASST-specific dithiol oxidase DsbL for ASST activity (8).
Architecture of the Active Site in the Substrate-Free ASST.
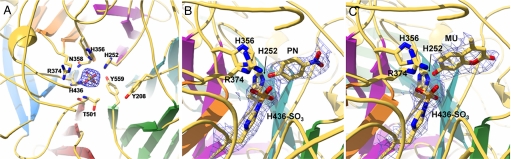
Subsequent to the building of the ASST model several sulfate groups were found. The majority of these sulfate groups are situated in close proximity to a positively charged amino acid side chain at the protein surface, acting as counter ions and believed to originate from the crystallization conditions. The sulfate group adjacent to His-436 is, however, bound in a very specific and complex manner, situated in the central channel of the β-propeller, and is involved in an extensive hydrogen bond network with the nitrogen atoms of 5 neighboring side chains (His-252, His-356, Asn-358, Arg-374, and His-436) and the backbone nitrogen of Thr-501 (Fig. 3A and Fig. S4A). Two further tyrosine residues, Tyr-208 and Tyr-559, are in close proximity to this sulfate (Fig. 3A). This complex network of hydrogen bonds coupled with a comparison of the location of the active site in other enzymes exhibiting a β-propeller fold (11) suggest the central channel of ASST's β-propeller fold contains the active site.
Fig. 3.
Active site geometry in ASST. (A) The structure of substrate-free ASST exhibits a sulfate dianion that is coordinated by nitrogen atoms from the active-site residues His-252, His-356, Asn-358, Arg-374, and His-436 and the backbone nitrogen of Thr-501. (B and C) Both PNS (B) and MUS (C) soaked ASST show His-436 to be sulfurylated at its Nε2 atom, whereas the desulfurylated donors [p-nitrophenol (PN) and MU, respectively] are located in the central channel of the β-propeller. Electron density is shown and contoured at 1.5 σ.
To clarify the catalytic role of residues surrounding the proposed active site we carried out mass spectrometry of sulfo-ASST, characterized the catalytic properties of ASST point variants, and determined the crystal structure of 2 catalytic intermediates.
Crystal Structures of Catalytic Intermediates of ASST and Substrate Binding.
To trap sulfurylated intermediates of ASST, crystals of substrate-free ASST were soaked with the sulfuryl donor substrates p-nitrophenylsulfate (PNS) and 4-methylumbelliferylsulfate (MUS) before being flash-cooled in liquid nitrogen. The crystal structure of the PNS and MUS adducts, solved to 2.1- and 2.4-Å resolution, respectively, revealed His-436 to be covalently sulfurylated on the Nε2 atom in both cases, whereas the desulfurylated products remained bound in the active site cleft (Fig. 3 B and C). The sulfuryl moiety of sulfohistidine is stabilized by extensive hydrogen bonding to the side chains of His-252, Asn-358, Arg-374, and His-356 and backbone nitrogen of Thr-501 (Fig. S4B), whereas the hydroxyl groups of the desulfurylated donors form a hydrogen bond with the sulfo-His-436 and a hydrophobic contact with the residue Phe-171. The only disordered segment in the ASST structure without detectable electron density is comprised by residues Val-321–Leu-327, which are relatively close to the active site and may contribute to the recognition of natural ASST substrates.
Active Site Residues and the Mechanism of PAPS-Independent Sulfotransfer by ASST.
The residues forming the active site of ASST are consistent with a mechanism in which first a His and then a Tyr side chain in the active site are sulfurylated, as proposed previously for the homologous enzyme from Eubacterium A-44 (7). To identify which residues in ASST are essential for catalysis, we characterized the catalytic properties of several ASST variants and analyzed residue conservation among its homologs.
We individually replaced the active site residues and compared the sulfotransferase activity of these variants to that of the wild-type variants (Table S2). Replacement of Arg-374, His-252, or His-356 by Leu dramatically decreased ASST activity to 0.1, 4.0, and 0.06%, respectively, suggesting that these residues are critically important for catalysis (the activity measurements of the His436Leu variant were precluded because this variant could not be expressed). In contrast, the activity of the point variants Tyr208Phe and Tyr559Phe and of the double variant Tyr208Phe/Tyr559Phe was comparable to that of the wild-type variant, indicating that these tyrosines do not participate in catalysis. Similarly, replacement of Tyr-96, a previously proposed catalytic ASST residue (13), had no effect on activity (Table S2). These data do not support the previously proposed model of sulfuryl transfer from a sulfo-His to a Tyr residue in ASST before sulfurylation of the acceptor substrate, instead favoring a mechanism in which a sulfurylated active-site histidine directly delivers the sulfuryl group to the aromatic hydroxyl group of an acceptor substrate. This reaction is a simple inverse of ASST sulfurylation by a donor (Fig. S4). The active-site residues His-252, His-356, and Arg-374 provide a means for coordination of the sulfuryl moiety during the catalytic cycle (Fig. S4) (14, 15). The essential role of the histidines in the active site of ASST was further supported by inactivation of ASST through treatment with diethylpyrocarbonate (DEPC), a reagent that specifically modifies histidine side chains (16). In contrast, reagents such as iodoacetamide and PMSF, targeting thiol and nucleophilic hydroxyl groups, respectively, had no influence on ASST activity (Fig. S5).
Sequence alignment of ASST homologs (Fig. S6) revealed all active-site residues (His-252, His-356, Arg-374, and His-436) to be invariant. In addition, the only invariant segment of >3 consecutive residues in all homologs corresponded to the pentapeptide sequence Tyr-434–Ala-438, centered at His-436. Furthermore, the residues around His-436 in the 3D structure of ASST undergo purifying selection [e.g., across-species mutations of the corresponding codons are less likely to be residue-changing than silent (17)], in agreement with a critical role in catalysis.
Biochemical Properties of ASST.
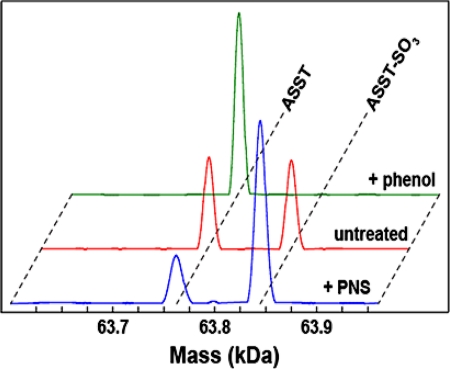
As previously mentioned, the initial indication of transient covalent sulfurylation of ASST during its catalytic cycle was obtained from electrospray ionization mass spectra (ESI MS). The spectra of freshly purified ASST exhibited a peak at 63,764.5 ± 2 Da (the mass expected for the monomer) and a second peak of similar intensity at 63,845.5 ± 2 Da for the sulfurylated enzyme (Fig. 4). Accordingly, incubation of ASST with an excess of the sulfuryl acceptor phenol resulted in the complete disappearance of the peak corresponding to the sulfoenzyme, whereas incubation of desulfurylated ASST with the sulfuryl donor PNS returned the peak corresponding to sulfo-ASST for ≈75% of the ASST molecules (Fig. 4). It was not possible to obtain mass spectra of uniformly sulfurylated ASST subsequent to treatment with PNS, suggesting that the sulfo-ASST reaction intermediate is prone to hydrolysis. This observation may also explain the initial occurrence of “sulfo-depleted” crystals even though the starting material for crystallization contained sulfo-ASST.
Fig. 4.
ESI MS highlighting the sulfurylation states of ASST. Spectra for ASST (red, as prepared), sulfo-ASST (blue, treated with PNS), and desulfo-ASST (green, treated with phenol) are shown. Desulfo-ASST was detected at a mass of 63,764.5 Da and the sulfo-form was detected at 63,845.5 Da. The calculated mass values were 63,763 Da and 63,843 Da, respectively.
Additional confirmation for the formation of sulfurylated His-436 was obtained by tryptic digestion of ASST pretreated with PNS, followed by mass spectrometry of tryptic ASST peptides, which was performed under basic conditions to minimize hydrolysis of sulfohistidine. Only the tryptic peptide Leu-405–Lys-443, bearing His-436, appeared as a double peak with a characteristic mass difference of 80 Da, confirming that the sulfurylated residue lies within this ASST fragment (Fig. S7). Furthermore, His-436 is the only histidine residue within this tryptic ASST peptide. Together with the crystal structures of the catalytic intermediates of ASST, these observations clearly demonstrate that His-436 is the residue that undergoes transient covalent sulfurylation during the catalytic cycle.
Sulfo-ASST in fresh preparations after expression in E. coli (see SI Methods for details) might indicate the presence of an unknown aromatic sulfuryl donor in the periplasm in vivo. However, we were unable to efficiently extract ASST from the periplasm by standard periplasmic extraction protocols, presumably because of the large size of the ASST homodimer (data not shown) and thus had to purify ASST from total cell extracts. We could not, therefore, eliminate the possibility that a nonphysiological, cytoplasmic sulfuryl donor was responsible for the fraction of sulfurylated enzyme in our ASST preparations. We could, however, confirm that the cytoplasmic sulfuryl donor PAPS is not an ASST substrate (data not shown) (1, 18–24).
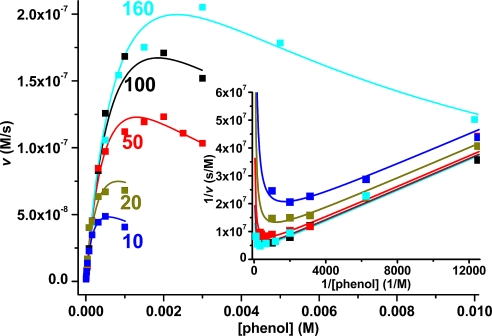
Ping-pong kinetics are common among transferases that are transiently modified by the chemical group to be transferred and possess only a single binding pocket for the donor and the acceptor substrate (25–27). Several sets of experimental data confirm that ASST follows this mechanism. Our mass spectrometry data (see above and Fig. 4) and the X-ray structures of the ASST intermediates (Fig. 3 B and C) demonstrate the covalent sulfurylation of His-436 upon reaction with donor substrates as well as regeneration of substrate-free ASST upon addition of the sulfuryl acceptor phenol to sulfo-ASST. In addition, we confirmed the ping-pong mechanism with steady state kinetics by using the sulfuryl donor MUS and the acceptor phenol. Fig. 5 shows the initial rate plot for the ASST-catalyzed reaction at pH 8.0, which yielded parallel lines in double reciprocal form, characteristic for ping-pong kinetics, and revealed substrate inhibition at high concentrations of phenol, which competes with MUS for the single substrate binding pocket at the enzyme (28). A global fit of the data according to the ping-pong model (28) yielded the following parameters at pH 8.0: KM,MUS = (4.45 ± 0.72) × 10−5 M, KM,phenol = (1.35 ± 0.19) × 10−3 M, KI,phenol = (1.13 ± 0.22) × 10−3 M, and kcat = (48.6 ± 0.5) s−1. The kinetic analysis of the reaction at different MUS and phenol concentrations in the range of pH 6–10 is depicted in Fig. S8. We found that the KM for MUS increased and the KM for phenol decreased with increasing pH, whereas kcat remained essentially constant in this pH range. Notably, phenol showed an increased substrate inhibition at pH 10 compared with pH 8 (KI,phenol 48 μM and 1.1 mM, respectively). This observation agrees with a higher affinity of ASST for the phenolate anion [pKa(phenol) = 9.95] that is expected to compete more efficiently with the negatively charged MUS at pH 10 compared with phenol at pH 8.
Fig. 5.
ASST-catalyzed sulfuryl transfer from MUS to phenol shows ping-pong kinetics. Initial velocities are plotted against the initial phenol concentration. The labeled curves for the reaction at pH 8.0 correspond to data sets at a constant initial MUS concentration of 10, 20, 50, 100, and 160 μM and are depicted in blue, green, red, black, and cyan, respectively. The solid lines correspond to a global fit of all data according to ping-pong kinetics with substrate inhibition by phenol. (Inset) Same data set (the data at lowest phenol concentrations not shown for clarity) in double reciprocal (Lineweaver–Burk) form (same color code). Parallel lines at various MUS concentrations (observed at low phenol concentrations) are diagnostic of the ping-pong mechanism. The upward curvature at high phenol concentrations (asymptotic toward the y axis) is a manifestation of substrate inhibition by phenol. The kinetic parameters obtained by global fitting of the entire dataset, spanning the range of 5–10 mM phenol and 10–160 μM MUS at pH 8.0 and an ASST concentration of 10 nM (monomer) are: Vmax = (4.86 ± 0.46) × 10−7 M·s−1, KM,MUS = (4.45 ± 0.72) × 10−5 M, KM,phenol = (1.35 ± 0.19) × 10−3 M, KI,phenol = (1.13 ± 0.22) × 10−3 M, and kcat = (48.6 ± 0.5) s−1. Details on data collection and evaluation are described in Methods.
Each ASST monomer contains 1 unpaired cysteine (Cys-322) located within the short loop preceding the fourth propeller blade. Treatment of native ASST with Ellman's reagent (29) yielded a mixed disulfide with thionitrobenzoic acid (TNB), which exhibited a similar catalytic activity to the native enzyme, as did ASST after treatment with the thiol-specific alkylating agent iodoacetamide, suggesting that Cys-322 is not involved in catalysis. This residue is also not conserved in ASST homologs (Fig. S6), showing ASST to be a protein in the oxidative environment of the bacterial periplasm, containing an unpaired cysteine residue with no catalytic function.
Discussion
The combined biochemical, crystallographic, and computational data presented here reveal that the periplasmic arylsulfate sulfotransferase from E. coli CFT073 is a homodimeric enzyme with a β-propeller fold that transfers sulfuryl groups specifically from phenolic sulfuryl donors to phenolic acceptors and, in contrast to the vast majority of characterized sulfotransferases, does not use PAPS as the sulfuryl group donor. Notably, none of the ≈50 structurally characterized sulfotransferases bear any resemblance to the β-propeller structure presented here (30), highlighting the difference in substrate specificity, cellular locations, and physiological roles between periplasmic ASSTs and other sulfotransferases. In common with other enzymes exhibiting a β-propeller fold, ASST's circular array of propeller blades forms a conical central channel that is well-suited to sequester either the donor or the acceptor substrate and contains the active site of the enzyme. Previous mechanistic studies of ASST suggested the catalysis to proceed by transient tyrosine sulfurylation (1, 13, 31). The residue Tyr-96 was proposed to undergo sulfurylation in ASST from Enterobacter amnigenus, which is 84% identical in sequence to the ASST of E. coli CFT073 (13, 31). The crystal structure presented here shows Tyr-96 to be located within the N-terminal β-sandwich domain far from the center of the β-propeller (Fig. 2), which, together with the identical catalytic activity of the Tyr96Phe variant to wild-type ASST, suggests that Tyr-96 is not involved in catalysis (Table S2).
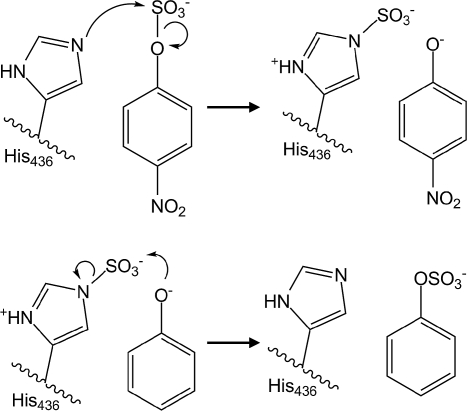
Whereas PAPS-dependent sulfotransferases catalyze direct sulfotransfer by binding both the sulfuryl donor PAPS and the acceptor simultaneously without becoming covalently modified (1, 32), ASST reacts first with a donor substrate and releases the first product (desulfurylated donor) before reacting with an acceptor substrate. Catalysis by ASST is therefore a 2-step process requiring transient covalent modification of the enzyme. Mass spectrometry and crystallography show that, in the first step of catalysis, ASST reacts with a sulfuryl donor and becomes covalently sulfurylated at the Nε2 atom of His-436 (Fig. 3). Subsequently, the sulfuryl group is transferred to an acceptor and the free enzyme is regenerated (Fig. 6). A cage of nitrogen atoms, from the side chains of His-252, His-356, Asn-358, and Arg-374 and the backbone nitrogen of Thr-501, define the active site, facilitating sulfotransfer most likely by coordinating the sulfuryl moiety during the reaction (Fig. S4). In the substrate-free crystal structure, solved from crystals obtained in the presence of sulfate, this site is occupied by an ordered sulfate dianion (Fig. 3). The 2 structures of ASST catalytic intermediates, with donor substrates PNS or MUS, exhibit His-436 covalently sulfurylated at Nε2, revealing sulfohistidine to form an analogous extensive hydrogen bond network with the same active site residues (Fig. 3 and Fig. S4). Despite being distant in the primary structure, these active site residues are all invariant between ASST homologs (Fig. S6).
Fig. 6.
Proposed reaction mechanism of arylsulfate sulfotransferase for the substrates PNS and phenol. During the first step of catalysis, the free electron pair of Ne2 from His-436 nucleophilically attacks the sulfur atom of PNS, yielding a covalent sulfohistidine intermediate and p-nitrophenylate. After dissociation of p-nitrophenylate and binding of phenol, the phenolate oxygen nucleophilically attacks the sulfur in the intermediate, and the sulfuryl group is transferred onto the acceptor phenol.
All currently available mechanistic studies on ASST homologs propose that the catalysis occurs by a covalent modification of a tyrosine residue in the active site of the enzyme, and some suggest that a tyrosine and a histidine form a catalytic pair in the active site (1, 20, 24, 31). Indeed, 2 tyrosines are within 10 Å of the sulfuryl group of sulfo-His-436. However, ASST variants lacking these tyrosines exhibit similar catalytic properties compared with the wild type, implying that tyrosine sulfurylation does not occur during catalysis (Table S2).
Taken together, our crystallographic and biochemical results show the mechanism of ASST to proceed via a ping-pong reaction mechanism and to involve transient formation of a sulfohistidine residue (Fig. 6). Furthermore, competitive substrate inhibition observed at high acceptor concentrations indicates that donor and acceptor bind at the same site, another typical feature of enzymes with ping-pong kinetics (28), and can easily be rationalized by ASST's substrate specificity, including phenolic compounds both as donors and acceptors. Additional support for the ping-pong catalytic mechanism is provided by the product inhibition patterns reported for analogous enzymes (6, 23, 25) as well as stereochemical studies of ASST-catalyzed sulfuryl transfer, which was found to occur with the retention of absolute stereochemical configuration of a chiral sulfuryl moiety (31). A direct transfer from the donor to the acceptor without formation of the sulfoenzyme would result in the inversion of absolute stereochemical configuration. The transient formation of sulfohistidine, a previously unknown covalent protein modification, avoids this inversion. Histidine sulfurylation is analogous to the formation of phosphohistidine as a covalent enzyme–substrate intermediate in histidine kinases (33, 34). This analogy highlights the striking structural and mechanistic resemblance of sulfotransfer, involved in extracellular signaling (35), and phosphotransfer, involved in intracellular signal transduction in bacteria (36, 37). It should be emphasized, however, that our results were obtained by using the phenolic substrates PNS and MUS, which bear electron withdrawing substituents. Therefore, it cannot be completely excluded that ASST's mechanism differs with its physiological substrates.
Disulfide bonds in proteins are traditionally classified as catalytic if they are formed and broken in each catalytic cycle or as structural if they stabilize the protein. A recent study proposed allosteric disulfide bonds as a third class of disulfide bonds that regulate catalytic activity and binding properties upon forming or breaking (12). These disulfide bonds are characterized by high dihedral strain, an unusually short distance between the Cα atoms of the 2 cysteines, and a −RHStaple geometry of the disulfide bond. The residues Cys-418 and Cys-424 of ASST are joined by such a disulfide bond (Fig. S3) with an extremely short Cα separation (3.8 Å) and with the dihedral angles corresponding to those of the −RHStaple class. The high steric strain energy of this bond (estimated to be 30 kJ/mol) could explain the requirement for the strong dithiol oxidase DsbL for its formation (8).
ASST's structure, unusual substrate specificity, periplasmic location, and genomic clustering with a specific dithiol oxidase system are suggestive of a number of physiological roles of ASST. Regulation of prokaryote–eukaryote interactions is known to involve sulfurylation of small molecules (35), and sulfotransferases play a significant role in modulating normal and pathogenic biological processes (26, 38, 39). ASST from the periplasm of uropathogenic E. coli strains is up-regulated in the uropathogenic habitat (10) and may therefore be involved in host–pathogen interactions. To our knowledge, the in vivo sulfuryl group donor for ASST has not yet been identified. It is also conceivable that there is no single, specific sulfuryl donor or acceptor, and that the physiological role of ASST may instead be to distribute the sulfuryl group within a defined set of biological recipients.
A second possible physiological role of ASST may be inferred from its genomic context. The gene that codes ASST is found clustered with genes coding for DsbL and DsbI. Because bacterial operons usually control gene clusters that code for functionally related proteins (37), it is probable that DsbL, DsbI, and ASST are functionally interrelated. Whereas DsbL and DsbI are thiol-disulfide oxidoreductases, ASST transfers sulfuryl groups between phenolic compounds. DsbL has been shown to be a specific oxidase for disulfide bond formation in ASST, and DsbI catalyzes the oxidation of the dithiol oxidase DsbL by ubiquinone-Q8 (8). Notably, ubiquinol-Q1, the soluble analog of ubiquinol-Q8, is also an acceptor substrate of ASST in vitro (data not shown). Therefore, ASST may sulfurylate membrane-bound quinones in vivo. Quinone sulfurylation has been described and identified as a dominant step following quinone reduction to hydroquinone in the mammalian liver (40, 41). Furthermore, it has been observed that sulfurylation of quinol rendered quinone a better oxidant (40). These observations raise the possibility that ASST contributes to quinone metabolism in uropathogenic bacteria.
In summary, the crystal structure of periplasmic ASST from E. coli strain CFT073 and those of its catalytic intermediates provide significant insight into the molecular mechanism of PAPS-independent sulfotransfer. This is a 2-step catalysis resulting in the formation of a transient high-energy sulfohistidine intermediate at the center of a β-propeller fold novel to sulfotransferases. The described crystallographic and biochemical experiments provide a basis for understanding PAPS-independent sulfotransfer and a basis for future work on the metabolism of phenolic compounds in uropathogenic bacteria and their physiological roles in pathogen–host interactions.
Methods
Molecular Cloning, Protein Expression and Purification, and Crystallization.
Cloning and expression of ASST was performed as described (8). ASST variants were constructed as described in SI Methods. See Table S3 for oligonucleotide primers used for site-directed mutagenesis of ASST.
Steady State Kinetics.
Initial rates of the ASST-catalyzed reaction were measured with MUS as donor substrate (concentration range 0.28–200 μM) and phenol as acceptor substrate (concentration range, 5 μM–10 mM) in the mixed buffer system prepared as described (20 mM final concentration of buffering species) (28), at pH values 6, 7, 8, 8.55, 9, and 10 at 25 °C by monitoring fluorescence of the reaction product 4-methylumbelliferone (MU) at 453 nm (excitation at 350 nm). For the data series at each pH, the instrument was calibrated with solutions of different MU concentrations enabling direct quantification of MU concentration changes. It was also verified that no species other than MU exhibited fluorescence at 453 nm or absorbed light at 350 nm. The concentration of ASST (monomer) was within the range of 10–30 nM. The ASST concentration was determined via its extinction coefficient at 280 nm (ε280 = 93,350 M−1·cm−1). Initial rates in the range of pH 8–10 were globally fitted with ORIGIN (Microcal) to the equation v = V[A][B]/(Ka[B](1+[B]/KIb)+Kb[A]+[A][B]), describing the initial velocity pattern for ping-pong kinetics (28), where [A] and [B] are the initial concentrations of MUS and phenol, respectively; Ka and Kb are the Michaelis constants (KM) for MUS and phenol, respectively; KIb is the inhibition constant of phenol; V is the maximum velocity; and v is the measured initial velocity. At pH 6.0 and 7.0, where the substrate inhibition by phenol was negligible, the equation used was v = V[A][B]/(Ka[B]+Kb[A]+[A][B]).
Protein Crystallization, Data Collection, and Structure Determination.
Details on crystallization of ASST, crystallographic data collection, processing, structure solution, and refinement are described in detail in SI Methods. Crystallographic data and refinement statistics are listed in Table S1. The structure of ASST was determined by using the single-wavelength anomalous dispersion technique with the anomalous signal from selenomethionine. The initial maps and models of the catalytic intermediates of ASST were obtained by refinement of substrate-free ASST against data collected from crystals soaked with donor substrates, as described in detail in SI Methods.
Supplementary Material
Acknowledgments.
We thank R. Aebersold, C. Carapito and B. Domon for help with the identification of sulfurylated peptide in ASST by MS; B. Blatmann for robotic crystallization screening; D. Sargent for help with initial diffraction experiments; C. Schulze-Briese for advice and assistance with experiments at the protein crystallography beam lines of the Swiss Light Source; the members of the Functional Genomics Center Zurich for protein analytics services; and the editor and 2 anonymous reviewers whose comments helped significantly strengthen the manuscript. G.M. acknowledges the instructors of the 2006 “X-ray Methods in Structural Biology” course at Cold Spring Harbor Laboratory for outstanding teaching and help with structure solution, E. Weber-Ban and N. Ban for helpful discussions, and the Huber-Kudlich Stiftung for a travel grant. This work was supported by the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland), Schweizerische Nationalfonds, and ETH Zurich in the framework of the Swiss Structural Biology National Center of Competence in Research program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3ELQ, 3ETS, and 3ETT).
Information was gathered from The E. coli Cell Envelope Protein Data Collection (ECCE), available at www.cf.ac.uk/biosi/staffinfo/ehrmann/tools/ecce/ecce.htm; accessed November 20, 2007.
The sequence of amino acids is MFDKYRKTLVAGTVAITLGLSASGVMA.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806997105/DCSupplemental.
References
- 1.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: Structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 2.Mougous JD, et al. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nat Struct Mol Biol. 2004;11:721–729. doi: 10.1038/nsmb802. [DOI] [PubMed] [Google Scholar]
- 3.Mougous JD, Green RE, Williams SJ, Brenner SE, Bertozzi CR. Sulfotransferases and sulfatases in mycobacteria. Chem Biol. 2002;9:767–776. doi: 10.1016/s1074-5521(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 4.Gamage N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 5.Jakoby WB, Ziegler DM. The enzymes of detoxication. J Biol Chem. 1990;265:20715–20718. [PubMed] [Google Scholar]
- 6.Kim DH, Kobashi K. The role of intestinal flora in metabolism of phenolic sulfate esters. Biochem pharmacol. 1986;35:3507–3510. doi: 10.1016/0006-2952(86)90619-2. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Konishi L, Kobashi K. Purification, characterization and reaction mechanism of novel arylsulfotransferase obtained from an anaerobic bacterium of human intestine. Biochim Biophys Acta. 1986;872:33–41. doi: 10.1016/0167-4838(86)90144-5. [DOI] [PubMed] [Google Scholar]
- 8.Grimshaw J, et al. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic E. coli. J Mol Biol. 2008;380:667–680. doi: 10.1016/j.jmb.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 9.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd AL, Rasko DA, Mobley HL. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. 2007;189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paoli M. Protein folds propelled by diversity. Prog Biophys Mol Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 13.Kwon AR, Yun HJ, Choi EC. Kinetic mechanism and identification of the active site tyrosine residue in Enterobacter amnigenus arylsulfate sulfotransferase. Biochem Biophys Res Commun. 2001;285:526–529. doi: 10.1006/bbrc.2001.5185. [DOI] [PubMed] [Google Scholar]
- 14.Cameron DR, Thatcher GRJ. Mechanisms of reaction of sulfate esters: A molecular orbital study of associative sulfuryl group transfer, intramolecular migration and pseudorotation. J Org Chem. 1996;61:5986–5997. [Google Scholar]
- 15.Graafland T, Wagenaar A, Kirby AJ, Engberts JBFN. Structure and reactivity in intramolecular catalysis. Catalysis of sulfonamide hydrolysis by the neighboring carboxyl group. J Am Chem Soc. 1979;101:6981–6991. [Google Scholar]
- 16.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 17.Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T. Selecton: A server for detecting evolutionary forces at a single amino-acid site. Bioinformatics. 2005;21:2101–2103. doi: 10.1093/bioinformatics/bti259. [DOI] [PubMed] [Google Scholar]
- 18.Kang JW, et al. Cloning, sequence analysis, and characterization of the astA gene encoding an arylsulfate sulfotransferase from Citrobacter freundii. Arch Pharm Res. 2001;24:316–322. doi: 10.1007/BF02975099. [DOI] [PubMed] [Google Scholar]
- 19.Kang JW, Kwon AR, Kim DH, Choi EC. Cloning and sequencing of the astA gene encoding arylsulfate sulfotransferase from Salmonella typhimurium. Biol Pharm Bull. 2001;24:570–574. doi: 10.1248/bpb.24.570. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Kim HS, Imamura L, Kobashi K. Kinetic studies on a sulfotransferase from Klebsiella K-36, a rat intestinal bacterium. Biol Pharm Bull. 1994;17:543–545. doi: 10.1248/bpb.17.543. [DOI] [PubMed] [Google Scholar]
- 21.Kobashi K, Kim D.-H, Morikawa T. A novel type of arylsulfotransferase. J Protein Chem. 1987;6:237–244. [Google Scholar]
- 22.Kwon AR, Oh TG, Kim DH, Choi EC. Molecular cloning of the arylsulfate sulfotransferase gene and characterization of its product from Enterobacter amnigenus AR-37. Protein Expr Purif. 1999;17:366–372. doi: 10.1006/prep.1999.1129. [DOI] [PubMed] [Google Scholar]
- 23.Lee NS, Kim BT, Kim DH, Kobashi K. Purification and reaction mechanism of arylsulfate sulfotransferase from Haemophilus K-12, a mouse intestinal bacterium. J Biochem. 1995;118:796–801. doi: 10.1093/oxfordjournals.jbchem.a124982. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, Kim HS, Kobashi K. Purification and characterization of novel sulfotransferase obtained from Klebsiella K-36, an intestinal bacterium of rat. J Biochem. 1992;112:456–460. doi: 10.1093/oxfordjournals.jbchem.a123921. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Kobashi K. Kinetic studies on a novel sulfotransferase from Eubacterium A-44, a human intestinal bacterium. J Biochem. 1991;109:45–48. [PubMed] [Google Scholar]
- 26.Pi N, Yu Y, Mougous JD, Leary JA. Observation of a hybrid random ping-pong mechanism of catalysis for NodST: A mass spectrometry approach. Protein Sci. 2004;13:903–912. doi: 10.1110/ps.03581904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voet D, Voet JG. Biochemistry. New York: Wiley; 2004. [Google Scholar]
- 28.Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. New York: Garland; 2007. [Google Scholar]
- 29.Ellman GL. A colorimetric method for determining low concentrations of mercaptans. Arch Biochem Biophys. 1958;74:443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 31.Chai CL, Lowe G. The mechanism and stereochemical course of sulfuryl transfer catalysed by the aryl sulfotransferase from Eubacterium A-44. Bioorg Chem. 1992;20:181–188. [Google Scholar]
- 32.Negishi M, et al. Structure and function of sulfotransferases. Arch Biochem Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- 33.Besant PG, Attwood PV. Mammalian histidine kinases. Biochim Biophys Acta. 2005;1754:281–290. doi: 10.1016/j.bbapap.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 34.Kennelly PJ. Archaeal protein kinases and protein phosphatases: Insights from genomics and biochemistry. Biochem J. 2003;370:373–389. doi: 10.1042/BJ20021547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Klumpp S, Krieglstein J. Phosphorylation and dephosphorylation of histidine residues in proteins. Eur J Biochem. 2002;269:1067–1071. doi: 10.1046/j.1432-1033.2002.02755.x. [DOI] [PubMed] [Google Scholar]
- 37.Lengeler JW, Drews G, Schlegel HG, editors. Biology of the Prokaryotes. New York: Thieme; 1999. [Google Scholar]
- 38.Armstrong JI, et al. Discovery of carbohydrate sulfotransferase inhibitors from a kinase-directed library. Angew Chem Int Ed Engl. 2000;39:1303–1306. doi: 10.1002/(sici)1521-3773(20000403)39:7<1303::aid-anie1303>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Bowman KG, Bertozzi CR. Carbohydrate sulfotransferases: Mediators of extracellular communication. Chem Biol. 1999;6:R9–R22. doi: 10.1016/S1074-5521(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 40.Chan TS, O'Brien PJ. Hepatocyte metabolism of coenzyme Q1 (ubiquinone-5) to its sulfate conjugate decreases its antioxidant activity. BioFactors. 2003;18:207–218. doi: 10.1002/biof.5520180223. [DOI] [PubMed] [Google Scholar]
- 41.Shangari N, Chan TS, O'Brien PJ. Sulfation and glucuronidation of phenols: implications in coenyzme Q metabolism. Methods Enzymol. 2005;400:342–359. doi: 10.1016/S0076-6879(05)00020-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.