Abstract
Activating signal cointegrator-2 (ASC-2), a transcriptional coactivator of multiple transcription factors that include the adipogenic factors peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα, is associated with histone H3-Lys-4-methyltransferase (H3K4MT) MLL3 or its paralogue MLL4 in a complex named ASCOM (ASC-2 complex). Indeed, ASC-2-null mouse embryonic fibroblasts (MEFs) have been demonstrated to be refractory to PPARγ-stimulated adipogenesis and fail to express the PPARγ-responsive adipogenic marker gene aP2. However, the specific roles for MLL3 and MLL4 in adipogenesis remain undefined. Here, we provide evidence that MLL3 plays crucial roles in adipogenesis. First, MLL3Δ/Δ mice expressing a H3K4MT-inactivated mutant of MLL3 have significantly less white fat. Second, MLL3Δ/Δ MEFs are mildly but consistently less responsive to inducers of adipogenesis than WT MEFs. Third, ASC-2, MLL3, and MLL4 are recruited to the PPARγ-activated aP2 gene during adipogenesis, and PPARγ is shown to interact directly with the purified ASCOM. Moreover, although H3K4 methylation of aP2 is readily induced in WT MEFs, it is not induced in ASC-2−/− MEFs and only partially induced in MLL3Δ/Δ MEFs. These results suggest that ASCOM-MLL3 and ASCOM-MLL4 likely function as crucial but redundant H3K4MT complexes for PPARγ-dependent adipogenesis.
Keywords: peroxisome proliferator-activated receptor γ, transcription, activating signal cointegrator-2, coactivator, ASCOM
In higher eukaryotes, histone H3-Lys-4 (H3K4) trimethylation, an evolutionarily conserved mark for transcriptionally active chromatin, is closely associated with promoters and early transcribed regions of active genes (1, 2) and counters the generally repressive chromatin environment imposed by H3-K9/K27-methylation (3). H3K4-methyltransferases (H3K4MTs) include yeast and human Set1, MLL1, MLL2, MLL3/HALR, MLL4/ALR, Ash1, and Set7/9 (4). These proteins contain a SET domain, which is associated with an intrinsic histone lysine-specific methyltransferase activity (3). Mammalian Set1 and MLL complexes belong to a highly conserved family of Set1-like complexes (4), which also contain complex-specific subunits and a common core subcomplex consisting of RbBP5, ASH2L and WDR5 (5–7). In particular, WDR5 mediates interactions of the H3K4MT unit with the histone substrate and also plays crucial roles in maintaining the integrity of the complex (6–8).
Activating signal cointegrator-2 (ASC-2; also named NCOA6, AIB3, TRBP, TRAP250, NRC, and PRIP) is a coactivator of numerous nuclear receptors and transcription factors (9). Importantly, ASC-2 is an integral and unique component of a Set1-like complex named ASCOM (for ASC-2 complex), which contains MLL3 or MLL4 (5, 10). ASCOM indeed possesses H3K4MT activity (5, 10–12). More recent studies identified additional components of ASCOM, including UTX (11, 12), a protein subsequently shown to be a H3K27-demethyase enzyme (13–16). Thus, ASCOM, unlike other Set1-like complexes, contains 2 distinct histone-modifying enzymes linked to transcriptional activation.
The importance of ASC-2 as a key coactivator of multiple nuclear receptors has been reported from studies with various ASC-2 mouse models (9, 17). In particular, ASC-2-null mice display at least 2 major phenotypes comparable with those described for mice with targeted inactivation of the peroxisome proliferator-activated receptor γ (PPARγ) and its dimerization partner retinoid X receptor (18, 19). First, the labyrinthine layer is less vascularized in placentas from ASC-2-null embryos than in those from their WT littermates (20–22). Second, ASC-2-null embryo hearts have unusually thin cardiac ventricular walls and trabecular hypoplasia (20–22). In further support for ASC-2 as a physiological coactivator of PPARγ is the observation that the transcriptional activity of PPARγ is impaired in ASC-2-null mouse embryonic fibroblasts (MEFs) (20–22). In addition, ASC-2 plays essential roles for the adipogenic program directed by PPARγ, a master regulator of adipogenesis (23), as demonstrated by the finding that ASC-2-null MEFs are refractory to PPARγ-stimulated adipogenesis and fail to express the PPARγ-responsive, adipogenic marker gene aP2 (24). Interestingly, ASC-2 has been reported to play crucial roles in granulocyte differentiation as a coactivator of C/EBPα (25), which also functions as a key adipogenic factor through its ability to trigger expression of PPARγ during adipogenesis (23). Taken together, these results suggest that ASC-2 may exert its adipogenic function as a coactivator of at least 2 key adipogenic transcription factors, PPARγ and C/EBPα.
To elucidate the physiological role of ASCOM, we have established a homozygous mouse line MLL3Δ/Δ (10). In these mice, WT MLL3 is replaced by a mutant MLL3 that bears an in-frame deletion of a 61-aa catalytic core region in the MLL3 SET domain and is expressed and incorporated into ASCOM (10). Unlike ASC-2-null mice, which die at approximately embryonic day (E) 9.5–E13.5 (19–21, 25), MLL3Δ/Δ mice in C57BL/6–129S6 background show only a partial embryonic lethality (10). Interestingly, MLL3Δ/Δ mice provide genetic evidence for complex formation between MLL3 and ASC-2, as these animals display at least 3 phenotypes that are shared with isogenic 129S6 ASC-2+/− mice (26), i.e., stunted growth, decreased cellular doubling rate, and lower fertility (10). MLL3Δ/Δ mice also show decreased white adipose tissue (WAT), as described here, and kidney ureter urothelium tumors (unpublished results) and resistance to high-fat diet-induced fatty liver formation (27). The early embryonic lethality of ASC-2-null mice (20–22, 26) versus the milder phenotypes of MLL3Δ/Δ mice, along with the demonstrated association of ASC-2 with MLL3 or MLL4 in ASCOM (5, 10) and the overlapping phenotypes of isogenic ASC-2+/− mice (26) and MLL3Δ/Δ mice (10), suggest that many ASC-2-target genes are likely to be regulated by MLL3 and MLL4 in a redundant manner. Indeed, our recent results have shown that ASCOM-MLL3 and ASCOM-MLL4 appear to be redundant coactivators both for the tumor suppressor p53 (unpublished results) and the liver X receptor (LXR) and retinoic acid receptor (RAR) (10, 27).
Despite the critical role for H3K4 trimethylation in transcriptional activation (3), the potential role for H3K4MTs in adipogenesis has been poorly understood. In this article, we present evidence that MLL3 and MLL4 function as important, but redundant, H3K4MTs for PPARγ-dependent adipogenesis. Our results also reveal that MLL3Δ/Δ mice display not only decreased WAT content but also enhanced insulin sensitivity, although the underlying target tissues and mechanisms for the insulin sensitivity need further investigation. Importantly, our results provide a potentially novel therapeutic venue to control obesity and insulin resistance, given the fact that MLL3 and MLL4 are enzymes amenable to chemical modulation.
Results
Decreased WAT in MLL3Δ/Δ Mice.
MLL3+/Δ mice showed no apparent phenotype but MLL3Δ/Δ mice weighed ≈30–40% less at birth and remained ≈20% smaller through adulthood (10). This difference in body weight, which persisted even after 4 months of high-fat diet (Fig. 1A), was correlated with a significantly decreased amount of WAT in MLL3Δ/Δ mice (Fig. 1 A and B). Interestingly, this WAT phenotype, observed with 100% of MLL3Δ/Δ mice, was evident from birth and became progressively worse with age (data not shown). In contrast, MLL3Δ/Δ mice had a relatively normal amount of brown adipose tissue (BAT) (Fig. 1B). From our histological analysis, WAT cells of MLL3Δ/Δ mice were found to be smaller than those of WT mice both before and after 2 months on a high-fat diet (Fig. 1C). In addition, despite the unaltered overall level of BAT in MLL3Δ/Δ mice (Fig. 1B), these animals accumulated less lipid in their BAT than did WT controls after 2 months on a high-fat diet (Fig. 1D). Compared with WT mice, MLL3Δ/Δ mice also appeared to have lower fasting blood glucose and insulin levels (≈55–78% of WT) and displayed improvement in both glucose and insulin tolerance tests on either normal chow or high-fat diets (Fig. 2 A–C). Interestingly, insulin tolerance tests with 4 h of fasting carried out for WT and MLL3Δ/Δ mice fed 2 months of high-fat diet resulted in the death of MLL3Δ/Δ mice from hypoglycemia [supporting information (SI) Fig. S1]. Thus, we carried out subsequent insulin tolerance tests without fasting. Even under this mild condition, at 120 min, all MLL3Δ/Δ mice on normal chow diet died because of hypoglycemia (Fig. 2C). In contrast, MLL3Δ/Δ mice showed normal blood triglyceride levels and unaltered circulating levels of leptin, adiponectin, and resistin (data not shown). Importantly, and despite their lower body weight, MLL3Δ/Δ mice consumed similar amounts of food as did WT controls on a normal chow diet or more food than WT on a high-fat diet (Fig. 2D). As an explanation for the improved insulin sensitivity (Fig. 2 A and C) despite increased food consumption per body weight (Fig. 2D), compared with control animals, MLL3Δ/Δ mice showed significantly greater energy expenditure and appeared to have higher overall activity (Fig. 2E). Overall, these results suggest that MLL3Δ/Δ mice eat more per body weight but are hyperactive and more efficient in energy expenditure, which may contribute to their improved sensitivity to insulin.
Fig. 1.
Decreased WAT in MLL3Δ/Δ mice. (A) Even after high-fat diet, MLL3Δ/Δ mice maintain lower weight and WAT than control WT mice and, unlike WT animals, MLL3Δ/Δ mice do not appear to develop hepatic steatosis (arrows). Six-month-old male mice fed 4 months of high-fat diet are shown as representative images. (B) BAT and WAT weights of 4-month-old WT and MLL3Δ/Δ male mice. (C and D) Histology of WAT (C) and BAT (D) of 4-month-old WT and MLL3Δ/Δ male mice fed chow diet (ND) or high-fat diet (HD) for 2 months. Female mice show identical phenotypes (data not shown). (Scale bars: 50 micrometer.)
Fig. 2.
Improved insulin sensitivity and increased energy expenditure in MLL3Δ/Δ mice. (A) Results are shown for glucose tolerance tests (GTTs) after 4 h of fasting for WT and MLL3Δ/Δ mice upon 2 months of normal chow (ND) or high-fat diet (HD) (n = 3∼5). (B) Blood glucose level was independently measured after 16 h fasting for WT (n = 19) and MLL3Δ/Δ (n = 17) mice fed ND. (C) Because MLL3Δ/Δ mice fed 2 months of HD developed hypogylcemia in insulin tolerance tests (ITTs) with 0.75 units of insulin after 4 h fasting (Fig. S1), ITTs for WT and MLL3Δ/Δ mice upon 2 months of ND or HD (n = ≈3–5) were carried out with 0.75 units of insulin and no fasting. Even under this mild condition, at 120 min, all MLL3Δ/Δ mice fed ND (#) died because of hypoglycemia. (D) Food intake of mice fed ND or HD was measured for 3 days, and the average daily food intake was as shown. (E) For WT and MLL3Δ/Δ mice fed 2 months of ND or HD (n = 3∼5), mean oxygen consumption, carbon dioxide production, and spontaneous locomotor activity were measured through a 12-h light/dark cycle as described (40).
Importantly, these phenotypes are much more complex than those observed in mice with adipose-specific inactivation of PPARγ (28, 29). For instance, the latter animals are known to have a decreased amount of both WAT and BAT and to accumulate more lipids in the liver than WT mice (28, 29). In contrast, the livers of MLL3Δ/Δ mice hardly accumulated lipid even after 2–4 months on a high-fat diet (arrows in Fig. 1A; ref. 27). These results are consistent with the notion that, because ASCOM is a multifunctional coactivator, inactivation of MLL3Δ/Δ is expected to result in impaired transactivation not only by PPARγ but also by other transcription factors targeted by ASCOM.
Partially Impaired Adipogenic Potential of MLL3Δ/Δ Cells.
To gain insight into the molecular basis underlying the complex metabolic phenotypes observed with MLL3Δ/Δ mice, we isolated total RNA from WAT and BAT of WT and MLL3Δ/Δ mice and carried out DNA microarray analyses. These experiments identified a total of 72 and 563 genes whose expression is significantly altered between WT and MLL3Δ/Δ mice (P < 0.0005), from WAT and BAT, respectively (Tables S1 and S2). Strikingly, >46% of the 563 affected BAT genes are involved with diverse aspects of metabolism, strongly suggesting selective roles for MLL3 in regulating metabolic genes. In further support of this idea, our DNA microarray analyses also identified 353 genes that are differentially expressed between the ureters of WT and MLL3Δ/Δ mice, and ≈40% of these genes are involved in metabolism (unpublished results). Similarly, among the 72 affected WAT genes, we identified a number of metabolic genes whose altered expression results in phenotypes with significant similarities to the metabolic phenotypes observed with MLL3Δ/Δ mice. These genes include down-regulated Rbp4, Lipin1, Ctsl, and Vldlr and up-regulated Igf1r and Apoc1 in MLL3Δ/Δ WAT (Table S1). For instance, insulin resistance is ameliorated by overexpressed ApoC1 (30) but facilitated by Rbp4 (31). Our quantitative PCR (Q-PCR) analysis appeared to support the DNA microarray results for ApoC1, Rbp4, and other genes (Fig. 3A and data not shown). Further analyses of these identified ureter and BAT/WAT genes may provide crucial insights into metabolic functions of MLL3. Overall, these results reinforce the idea that MLL3Δ/Δ mice are likely perturbed in signaling for multiple transcription factors and suggest that ASCOM (at least ASCOM-MLL3) is an important regulator of metabolic genes.
Fig. 3.
Decreased expression of adipogenic genes in MLL3Δ/Δ mice and partially impaired adipogenic potential of E13.5 MLL3Δ/Δ MEFs. (A–C) Expression of ApoC1 and RBP4 (A) and MLL3/4 (B) and aP2, adiponectin, C/EBPα and PPARγ (C) in E13.5 WT and MLL3Δ/Δ WAT or BAT was measured by Q-PCR (n = ≈3–5). We also measured expression of aP2 as positive controls for WAT and BAT (B). (D) E13.5 WT and MLL3Δ/Δ MEFs were induced to differentiate to adipocytes. Experiments were repeated 3 times, and MLL3Δ/Δ MEFs reproducibly showed ≈80% efficiency of WT MEFs in adipogenesis. (Magnification: ×4.)
Among the complex metabolic phenotypes of MLL3Δ/Δ mice, we decided to further focus on the prominent decrease in WAT and examined the potential role for MLL3 in adipogenesis. In WT mice, the expression level of MLL3 and MLL4 was ≈75% and ≈60% of aP2 in WAT and not significantly different from that of aP2 in BAT (Fig. 3B). NIH3T3-L1 cells efficiently carry out an adipogenic program in response to the PPARγ ligand rosiglitazone. In our hands, 80–90% of these cells efficiently underwent adipogenesis over 8 days (data not shown). MLL3 and MLL4 were also expressed through adipogenesis of NIH 3T3-L1 cells (data not shown). Interestingly, expression of aP2 and adiponectin, 2 target genes of PPARγ, was significantly down-regulated in WAT of MLL3Δ/Δ mice (Fig. 3C), although the circulating level of adiponectin was not altered in these animals (data not shown). These results are consistent with the coactivator function of ASC-2 for the key adipogenic factors PPARγ and C/EBPα (23). In addition, as in ASC-2-null cells (24), PPARγ was up-regulated in MLL3Δ/Δ mice, despite the fact that expression of C/EBPα, which induces PPARγ expression (23), was significantly down-regulated (Fig. 3C). These results further emphasize the complexity in the metabolic phenotypes of MLL3Δ/Δ mice. Nonetheless, it is important to note that expression of aP2 was completely ablated in ASC-2-null cells (24). Consistent with the significant down-regulation of aP2 and adiponectin expression, E13.5 MEFs isolated from MLL3Δ/Δ mice were ≈20% less efficient than the E13.5 WT MEFs in adipogenesis (Fig. 3D). These results are in sharp contrast to the complete inability of ASC-2-null MEFs to undergo adipogenesis under the same condition (24). Taken together, these results raise the interesting possibility that ASCOM-MLL3 and ASCOM-MLL4 have redundant functions in adipogenesis and that ASC-2 plays crucial roles for the adipogenic function of these 2 related complexes, likely as a key adaptor to recruit ASCOM to PPARγ and C/EBPα during adipogenesis (20–22, 25). Notably, we have recently shown that ASC-2 functions in this manner to tether LXRs and RARs to both ASCOM-MLL3 and ASCOM-MLL4 (10, 27).
Redundant Functions for ASCOM-MLL3 and ASCOM-MLL4 in H3K Trimethylation on aP2.
To directly test a role for ASCOM in regulating aP2 expression during adipogenesis and to assess a potential redundancy between ASCOM-MLL3 and ASCOM-MLL4, we carried out ChIP assays with NIH 3T3-L1 cells. Adipogenesis of these cells was accompanied by a timely recruitment of PPARγ to aP2 (Fig. 4A). In strong support of redundant roles for ASCOM-MLL3 and ASCOM-MLL4 in adipogenesis, both MLL3 and MLL4, along with ASC-2, were recruited to aP2 in a time-dependent manner, with peak recruitment at 4 days after treatment with rosiglitazone (Fig. 4A). Consistent with these results, H3K4 trimethylation was induced with similar kinetics (Fig. 4B). In addition, H3 and H4 acetylation were concomitantly induced (Fig. 4B and data not shown), as expected from the reported cross-talk between these 2 modifications (32, 33) and between ASCOM and histone acetyltransferases CBP and p300 (34–36).
Fig. 4.
Crucial roles for ASC-2 and MLL3 in H3K4 trimethylation of aP2. (A and B) ChIPs for the PPAR-responsive element in the aP2 promoter, using antibodies against ASC-2, MLL3, MLL4, and PPARγ (A) or trimethylated H3K4 and acetylated H4 (B), were carried out for NIH 3T3-L1 cells, which were induced to differentiate to adipocytes. (C and D) ChIPs using antibody against trimethylated H3K4 were also carried out for cells derived from E9.5 WT and ASC-2−/− MEFs transfected with PPARγ (C) and E13.5 WT and MLL3Δ/Δ MEFs induced to differentiate to adipocytes (D). All ChIP experiments, repeated >3 times, produced similar results. IgG was used as negative control.
Next, we wanted to test the involvement of ASC-2 in these histone modifications on aP2. Because the early embryonic lethality of ASC-2-null mice (9) makes it difficult to carry out ChIP experiments with primary ASC-2-null MEFs, we used the previously described MEF cell lines established from E9.5 ASC-2-null embryos and their WT littermates (10). We expressed PPARγ in these cell lines and examined the induction of H3K4 trimethylation on aP2 in response to the PPARγ ligand rosiglitazone. In direct support of crucial roles for ASC-2 in triggering H3K4 trimethylation on aP2, the robust rosiglitazone-induced H3K4 trimethylation observed in WT cells was abolished in ASC-2-null cells (Fig. 4C). Remarkably, H3K4 trimethylation of aP2 was significantly attenuated but not ablated in E13.5 MEFs isolated from MLL3Δ/Δ mice (Fig. 4D), suggesting that another H3K4MT, most likely MLL4, still functions in these cells. Importantly, these results parallel the partially impaired adipogenic potential of E13.5 MLL3Δ/Δ MEFs (Fig. 3D) and the completely ablated adipogenic potential of ASC-2-null MEFs (24). In control experiments, both ASC-2-null cells and E13.5 MLL3Δ/Δ MEFs fully supported H3K4 trimethylation of Hoxa9 (data not shown), which is a known target for the Set1-like complex containing MLL1 (37). Although the participation of MLL4 in adipogenesis remains to be directly tested, through establishment of either MLL4 mutant mice or preadipogenic cells expressing siRNAs against MLL3 and MLL4, our results suggest that ASCOM-MLL3 and ASCOM-MLL4 likely play crucial, but redundant, roles in adipogenesis.
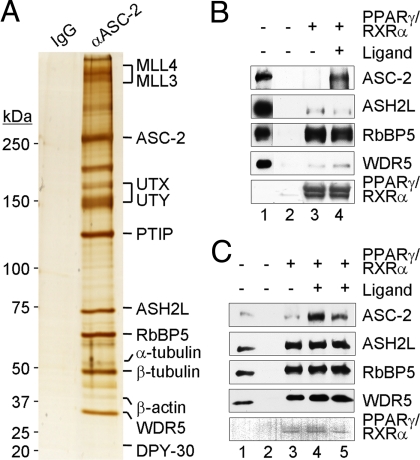
To further investigate the basis for ASCOM recruitment to PPARγ target genes, the complex was immuno-affinity-purified by using an ASC-2-specific mAb (5). Consistent with previous studies (5, 11, 12), the complex contained ASC-2, MLL3, MLL4, PTIP, and UTX and components (WDR5, RbBP5/RBQ-3, ASH2L and DPY30) common to other Set1-like complexes (Fig. 5A). This purified complex showed a strong interaction with M2 agarose-immobilized PPARγ·RXRα, but not with M2 agarose alone (Fig. 5B). Intriguingly, the common core subunits (ASH2L, RbBP5, and WDR5) showed ligand-independent interactions, whereas ASC-2 showed a strong ligand-enhanced interaction. In an immobilized template assay with bound PPARγ·RXRα, the purified complex also showed a strong PPARγ·RXRα-dependent recruitment (Fig. 5C). Again, core subunits showed ligand-independent binding, whereas ASC-2 showed ligand-enhanced binding. These results argue strongly for recruitment of ASCOM-MLL3 and ASCOM-MLL4 complexes based on direct interactions with PPARγ·RXRα, although we do not yet understand the basis for the ligand-dependent versus ligand-independent interactions of different components of the ASCOM complex (see Discussion).
Fig. 5.
Direct interaction of ASCOM with promoter-free and promoter-bound PPARγ·RXRα. (A) Purified ASCOM complex. The immuno-affinity purified complex was analyzed by SDS/PAGE (4–20% gradient) with silver staining. Polypeptides identified by mass spectrometry are indicated. (B) ASCOM binding to promoter-free PPARγ·RXRα. The purified complex was incubated with M2 agarose (lane 2) or M2 agarose-bound FLAG-PPARγ·RXRα in the absence (lane 3) or presence (lane 4) of 5.0 μM 15d-deoxy-Δ12,14-Prostagladin J2. (C) ASCOM binding to promoter-bound PPARγ·RXRα. The purified complex was incubated with a Streptavidin agarose-immobilized biotinylated DNA fragment (containing 3 DR1 sites) in the absence (lane 2) or presence (lanes 3–5) of PPARγ and RXRα and either with no ligand (lanes 2 and 3) or with 5.0 μM 15d-deoxy-Δ12,14-prostagladin J2 (lane 4) or 5.0 μM rosiglitazone (lane 5). Bound proteins in B and C were detected by immunoblot (ASC-2, ASH2, RbBP5, and WDR5) or staining (PPARγ and RXRα). Lanes 1 in B and C contained 10% of the input samples.
Discussion
H3K4 trimethylation is an evolutionarily conserved mark for transcriptionally active chromatin (1, 2). Interestingly, whereas yeast has a single enzyme responsible for this modification (H3K4MT), higher eukaryotes carry a number of H3K4MTs (4), which raises the possibility that individual H3K4MTs in higher eukaryotes may have distinct target genes. However, the physiological roles for higher eukaryotic H3K4MTs and their target genes remain poorly understood. The results presented here indicate that MLL3, and possibly MLL4, may be specialized for regulating genes involved in metabolic homeostasis, thus revealing key roles for specific H3K4MTs in metabolism. We have found that MLL3Δ/Δ mice have a significantly decreased amount of WAT with a favorable overall metabolic profile, including improved insulin sensitivity and increased energy expenditure, although these animals also develop ureter urothelium tumors that likely result from an ASCOM coactivator function for the tumor suppressor p53 (unpublished results). These animals are also resistant to high-fat diet-induced fatty liver formation (27). These results raise the interesting possibility that novel antidiabetic and/or antisteatohepatitis therapeutics might result from modulating the MLL3/4-H3K4MT activity in proper target tissues.
Set1-like complexes contain not only a shared core subcomplex of ASH2L, RbBP5/RBQ-3 and WDR5 but also proteins unique to each complex. ASC-2, a distinct subunit in ASCOM, has been proposed as a key adaptor used by many members of the nuclear receptor superfamily to recruit ASCOM (9). Indeed, we have shown that ASC-2 is an essential adaptor for recruitment of ASCOM by RARs and LXRs (10, 27). Consistent with earlier demonstrations that ASC-2 binds directly to PPARγ and C/EBPα, 2 factors important for adipogenesis (10, 25), we show here a direct interaction of ASCOM with both free and promoter-bound PPARγ·RXRα heterodimers. Thus, it is possible that ASC-2 plays a similar role in facilitating ASCOM function through PPARγ and C/EBPα during adipogenesis, which may explain the functional redundancy for ASCOM-MLL3 and ASCOM-MLL4 in adipogenesis. These results are also consistent with our earlier proposal that unique subunits of Set1-like complexes determine the target specificity of these H3K4MTs through direct protein–protein interactions with target transcription factors (10). Finally, as we observe some distinct phenotypes in MLL3Δ/Δ mice, we expect that ASCOM-MLL3 and ASCOM-MLL4 are not completely redundant and that each complex may have its own specialized targets. To fully understand the physiological roles for ASCOM, it will be important to elucidate not only common targets for both complexes but also distinct targets for each complex.
Our observation of both ligand-dependent and ligand-independent interactions of different components of the ASCOM complex uncovers a previously unappreciated complexity in recruiting ASCOM to nuclear receptors. This could reflect, for example, the presence of a dynamic equilibrium distribution of ASC-2-containing and ASC-2 free (38) complexes and an associated ligand-independent recruitment of the core complex, possibly through interactions with a PPARγ domain other than AF2, followed by a ligand- and AF2-dependent recruitment or stabilization of ASC-2. As this implies that ASC-2-PPARγ interactions could require a preassembled PPARγ–ASCOM complex, the availability of ASCOM may serve as a switch that selects whether PPARγ acts via ASC-2 or other ASCOM-independent coactivators. Another possibility is the presence in our purified PPARγ of an active AF2 conformation, caused either by an endogenous ligand or a natural equilibrium between active and inactive AF2 states (39), and an AF2-dependent recruitment of ASCOM through LXXLL motifs in MLL3/4 (27, 38) or ASC-2 with further stabilization of MLL3/4 or ASC-2 by the potent ectopic ligands. Whereas these and other possibilities remain to be further examined, the present observations are clearly indicative of ASCOM recruitment, possibly after other PPARγ-dependent chromatin remodeling events, by direct interactions with PPARγ.
In summary, we have presented evidence that MLL3 and MLL4 function as crucial, but redundant, H3K4MTs for adipogenesis, uncovering an interesting connection between H3K4 trimethylation and adipogenesis. Our results also reveal that targeted removal of MLL3 H3K4MT activity results in complex metabolic phenotypes in the mouse and, consistently, altered expression of many metabolic genes. Thus, our results raise an interesting possibility that specific agonists/antagonists of MLL3/4 H3K4MT activity could be useful for treating diverse metabolic disorders.
Materials and Methods
Mice, Histochemistry, and Metabolic Profiling.
As we found that 100% of MLL3Δ/Δ animals are embryonic lethal in pure C57BL/6 background, we maintained them in C57BL/6–129S6 background. BAT and WAT were isolated, fixed in 4% paraformaldehyde, and paraffin-embedded. Sections were subjected to standard hematoxylin/eosin staining. Two-month-old male or female MLL3Δ/Δ mice in C57BL/6–129S6 background were fed a standard rodent chow (Purina) or a high-fat diet (40% calories from fat; Bioserv) for 2 months on a 12-h light/dark cycle before being killed for glucose and insulin tolerance tests and energy expenditure and activity measurements, as described (40). Blood samples were drawn at 15, 30, 60, and 120 min after the injection. No difference between males and females was observed.
RT-/Q-PCR.
Total RNA was isolated from cells after lysis in TRIzol reagent according to the manufacturer's protocol (Invitrogen), and RT- and SYBR Green Q-PCRs were performed as described (10). The primer sequences are available on request.
Adipogenesis of MEFs.
Primary MEFs isolated from E13.5 WT and MLL3Δ/Δ embryos were induced to differentiate to adipocytes by Preadipocyte Growth Medium and Adipocyte Differentiation Medium according to the manufacturer's protocol (Cell Applications). These cells were subjected to standard oil red O staining.
ChIP Assays.
Soluble chromatin was prepared and immunoprecipitated with the indicated antibodies, as described (10). The final DNA extractions were amplified by using primers that encompass the PPAR-responsive element in the aP2 promoter. The primer sequences are available on request.
Statistical Analysis.
All results are expressed as mean ± SE. Statistical significance was determined by using Student's t test; *, P < 0.05; **, P < 0.01.
Antibodies.
Polyclonal ASC-2, ASH2L, and RbBP5 antibodies were from Bethyl Laboratories, and polyclonal WDR5 antibody was from David Allis (Rockefeller, New York). MLL3 and MLL4 antibodies were as described (5, 10).
Purification of ASCOM.
Affinity-purified anti-ASC-2 mAb from cultured A3C1 hybridoma cells (provided by Paul S. Meltzer, National Institutes of Health, Bethesda, MD; ref. 5) was covalently cross-linked to agarose beads. Clarified nuclear extract (400 mg) from U-937 cells then was incubated with anti-ASC-2 agarose beads (300 μL) for 7 h at 4 °C in BC300 [40 mM Tris (pH 7.9), 300 mM KCl, 20% glycerol, 0.05% Tween 20, 0.05% CHAPS, 0.5 mM PMSF, 1.0 μM leupeptin, 1.0 μM pepstatin, and 2.0 μM MG132]. After extensive washing with BC300, anti-ASC-2 agarose beads were transferred to a mini-column, and bound proteins were eluted with 300 μg/ml of ASC-2 epitope peptide. As a control, an identical amount of nuclear extract was incubated with normal mouse IgG agarose beads and processed in a similar manner. Eluted ASC-2 and its associated proteins (ASCOM) were resolved by SDS/PAGE and visualized by Coomassie blue staining. All visible bands were subjected to MALDI mass spectrometry analysis (performed at the Rockefeller University Proteomics Resource Center).
ASCOM Binding Assays.
FLAG-tagged PPARγ·RXRα heterodimer (≈5.0 μg) was expressed via baculovirus vectors in Sf9 cells, coupled to M2 agarose beads (20 μL), and incubated with the purified ASCOM (1.0 μg) for 5 h at 4 °C in BC200 [40 mM Tris (pH 7.9), 200 mM KCl, 20% glycerol, 0.05% Tween 20, 0.5 mM PMSF, 1.0 μM leupeptin, 1.0 μM pepstatin, and 2.0 μM MG132], either in the presence or in the absence of a PPARγ ligand. After extensive washing of the beads in BC200, bound proteins were eluted by boiling the beads in 2% SDS and analyzed by SDS/PAGE and immunoblot. For immobilized template binding assays, a 266-bp DNA fragment containing 3 DR1 sites was end-labeled with biotin and immobilized on Streptavidin-agarose beads (Invitrogen). The immobilized DNA (350 ng) was incubated sequentially with PPARγ·RXRα (500 ng) for 1 h and with ASCOM (1.0 μg) for 2 h at 4 °C in BC200 (above) in the presence or the absence of a PPARγ ligand. Beads were washed 5 times with 1 mL of BC200. Bound proteins were eluted by boiling the beads in SDS sample buffer and analyzed by SDS/PAGE and immunoblot.
Supplementary Material
Acknowledgments.
We thank David Allis and Paul Meltzer for antibodies. This work was supported by National Institutes of Health Grants DK064678 (to J.L.), HL51586 (to L.C.), and DK071900 (to R.G.R.) and Baylor Diabetes Endocrinology Research Center Grant DK-079638 (to L.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810100105/DCSupplemental.
References
- 1.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 2.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 3.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Goo YH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 7.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 8.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 9.Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: Role in hormone action, metabolism, growth, and development. Endocr Rev. 2005;26:583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci USA. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 14.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 15.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 16.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Chu MJ, Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol Cell Biol. 2007;27:8073–8086. doi: 10.1128/MCB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barak Y, et al. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 19.Kastner P, et al. Genetic analysis of RXRα developmental function: Convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 20.Kuang SQ, et al. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem. 2002;277:45356–45360. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- 21.Antonson P, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu YJ, et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- 23.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 24.Qi C, et al. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor γ (PPARγ)-interacting protein, is required for PPARγ-mediated adipogenesis. J Biol Chem. 2003;278:25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- 25.Hong S, Lee MY, Cheong J. Functional interaction of transcriptional coactivator ASC-2 and C/EBPα in granulocyte differentiation of HL-60 promyelocytic cell. Biochem Biophys Res Commun. 2001;282:1257–1262. doi: 10.1006/bbrc.2001.4727. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan MA, Das S, Zhu H, Tomic-Canic M, Samuels HH. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol. 2008;22:1312–1319. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He W, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones JR, et al. Deletion of PPARγ in adipose tissues of mice protects against high-fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jong MC, et al. Protection from obesity and insulin resistance in mice overexpressing human apolipoprotein C1. Diabetes. 2001;50:2779–2785. doi: 10.2337/diabetes.50.12.2779. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 32.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SK, et al. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol Endocrinol. 2001;15:241–254. doi: 10.1210/mend.15.2.0595. [DOI] [PubMed] [Google Scholar]
- 35.Ko L, Cardona GR, Chin WW. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci USA. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahajan MA, Samuels HH. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048–5063. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 38.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281:15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 39.Nolte RT, et al. Ligand binding and coactivator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 40.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10:625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.