Abstract
Fet3p from Saccharomyces cerevisiae is a multicopper oxidase (MCO) that contains 3 cupredoxin-like β-barrel domains and 4 copper ions located in 3 distinct metal sites (T1 in domain 3, T2, and the binuclear T3 at the interface between domains 1 and 3). To better understand how protein structure and stability is defined by cofactor coordination in MCO proteins, we assessed thermal unfolding of apo and metallated forms of Fet3p by using spectroscopic and calorimetric methods in vitro (pH 7). We find that unfolding reactions of apo and different holo forms of Fet3p are irreversible reactions that depend on the scan rate. The domains in apo-Fet3p unfold sequentially [thermal midpoint (Tm) of 45 °C, 62 °C, and 72 °C; 1 K/min]. Addition of T3 imposes strain in the apo structure that results in coupled domain unfolding and low stability (Tm of 50 °C; 1 K/min). Further inclusion of T2 (i.e., only T1 absent) increases overall stability by ≈5 °C but unfolding remains coupled in 1 step. Introduction of T1, producing fully-loaded holo-Fet3p (or in the absence of T2), results in stabilization of domain 3, which uncouples unfolding of the domains; unfolding of domain 2 occurs first along with Cu-site perturbations (Tm 50–55 °C; 1 K/min), followed by unfolding of domains 1 and 3 (≈65–70 °C; 1 K/min). Our results suggest that there is a metal-induced tradeoff between overall protein stability and metal coordination in members of the MCO family.
Keywords: spectroscopy, calorimetry, cupredoxin, copper-binding protein
The cupredoxin fold is one of the most common β-strand arrangements found in biology (1). This Greek-key, β-barrel structure typically has 7 or 8 β-strands forming 2 β-sheets that pack against each other (2, 3). Because this motif is found in small copper-containing electron-transfer proteins in bacteria, fungi, and plants termed blue-copper proteins, or cupredoxins (4), it has become known as the cupredoxin fold. Cupredoxins, e.g., azurin, plastocyanin, and stellacyanin, are excellent model systems for studies of the effects of metal cofactors on protein stability and folding because they are soluble and easily overexpressed and mutated. The best-characterized cupredoxin with respect to the role of metals in folding is Pseudomonas aeruginosa azurin (2). In P. aeruginosa azurin, the copper (Cu) is coordinated in a typical blue-copper site, designated as a T1 Cu site. It has been suggested that the cupredoxin fold defines the exact geometry of the Cu ligands, leading to the unusual Cu coordination in these proteins (5).
The cupredoxin fold also dominates another class of proteins, the multicopper oxidases (MCOs). Examples include laccases found in bacteria, fungi, and plants, and metallooxidases found in bacteria, algae, fungi, and mammals. These latter enzymes exhibit specificity toward low valent first-row transition metals, e.g., Mn(II), Cu(I) and Fe(II); MCOs with ferrous iron specificity are known as ferroxidases (6).
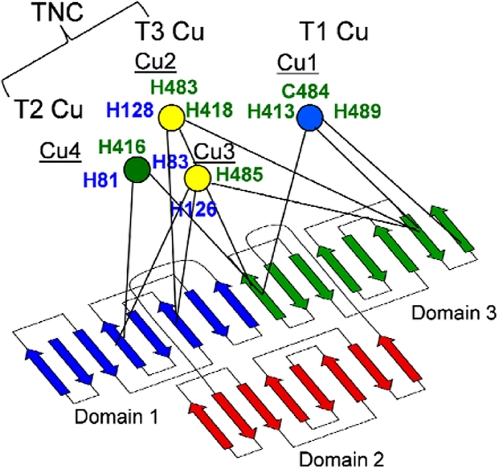
MCOs are composed of multiple cupredoxin domains, defined within these larger proteins as foldons of 125–175 residues. For example, an MCO from Bradyrhizobium japonicum (445 residues) contains 2 cupredoxin domains, the Fet3 protein from Saccharomyces cerevisiae (560 residues) contains 3, and the human ferroxidase, ceruloplasmin (Cp), has 1,065 residues and 6 domains. In all of these MCO proteins, there are 3 distinct copper sites arrayed within the domains in a conserved pattern. One [or 3 in the case of human Cp (hCp)] of the domains contains the typical T1 site found in single-domain cupredoxins. This site is most often composed of a coordination sphere of 2 His and 1 Cys; the Cys thiolate ligand provides strong charge transfer to Cu(II) that gives rise to the intense blue color of T1 Cu-containing proteins (ε600nm ≅ 5,000 M−1·cm−1). In the canonical organization of MCO proteins, T1 is found in the carboxyl-terminal domain, as illustrated in Fig. 1 for yeast Fet3p (7).
Fig. 1.
Schematic diagram of Fet3p topology and Cu sites. The β-strands of the 3 domains are shown in different colors to indicate boundaries. The 3 Cu sites [T1 (Cu1), blue; T2 (Cu4), green; T3 (Cu2 and Cu3), yellow; T2 and T3 form TNC] and the coordinating side chains are indicated. The side-chain labels are color-coded based on the domain origin.
The other 2 Cu sites in MCOs, designated as T2 and T3, are typically, but not always, found at the interface of the amino- and carboxyl-terminal domains. The protein coordination sphere at the T2 Cu is 2 His; a third ligand is water or hydroxide. The T3 site contains 2 Cu atoms bridged by a nonprotein oxygen atom; this ligand provides an electronic superexchange pathway that renders the T3 Cu(II) cluster diamagnetic. Each Cu in this cluster is coordinated by 3 histidine imidazoles. The T2 and T3 sites are collectively known as the trinuclear cluster (TNC); dioxygen binds at the TNC and is reduced to 2 H2O via inner-sphere transfer of 4 electrons. Dioxygen coordination and subsequent reduction depends on the fact that, with 3 Cu atoms and only 8 protein ligands, the TNC is coordinately undersaturated. The 8 ligands are distributed equally between the 2 cupredoxin domains that thus serve as a structural template for assembly of the TNC (domains 1 and 3 in Fet3p; Fig. 1).
More than 1,000 proteins have been identified as MCOs based on the multiples of the cupredoxin motifs they contain (6). Despite the interest in MCOs as catalysts in biofuel cells and in other biotechnological applications (8), very few have been characterized with respect to the role of the metal prosthetic groups in protein stability. Studies have indicated that the apo form of hCp adopts an extended structure (like beads on a string) because of the lack of the Cu-ligand bonds, the TNC, that connect the first and the last domain (9). In support of all-or-none Cu binding to hCp (10), metabolic-labeling experiments indicated that achieving native hCp required occupation of all 6 Cu sites (11). However, there have been reports that endorse partially-metallated forms of hCp (12). We recently reported chemical and thermal unfolding reactions of apo and holo forms of WT hCp (13, 14). We found that chemical or thermal perturbation resulted in temporary formation of an intermediate with less secondary structure and loss of 2 Cu. There have also been reports on chemical and thermal stability of ascorbate oxidase (which is a dimer of 2 MCOs) (15) and laccase (16) proteins.
Yeast ferroxidase, Fet3p, has proven to be a paradigm for structure–function studies because of the collection of demetallated forms that have allowed quantification of the role of each Cu site to the electronic properties of Fet3p and to its redox activity (6, 17–19). These forms are designated T1D, T2D, and T1D/T2D (where D is depleted), in which the T1, T2, or both the T1 and T2 Cu atoms are absent because of mutation of specific Cu ligands. Fet3p(Cys484Ser) lacks the T1 Cu, whereas Fet3p(His81Gln) lacks the T2 Cu; the double mutant lacks both Cu atoms. Here, we use these reagents to probe the roles of the 3 Cu sites in the stability and unfolding mechanism of Fet3p. We find that whereas the domains in apo-Fet3p unfold sequentially, unfolding of domains 1 and 3 is coupled in the holo form because of the T3 coppers “stitching” the domains together. In partially-metallated forms lacking T1, the 3 domains unfold as 1 cooperative unit with low stability. Our data demonstrate a metal-induced tradeoff between overall protein stability and metal coordination.
Results
Apo-Fet3p.
After core glycosylation in the endoplasmic reticulum, apo-Fet3p transfers to a post-Golgi compartment where Cu is added before delivery to the plasma membrane (20). Here, apo protein was prepared in vitro from purified holo protein. The absence of Cu in the apo protein was confirmed by a colorimetric assay developed by Felsenfeld (21). The apo form of Fet3p exhibits the same far-UV CD signal as the holo form [supporting information (SI) Fig. S1], implying that metal removal does not affect the content of secondary structure. Identification of the unfolding behavior and stability of apo-Fet3p provides the baseline for examination of how these characteristics are altered by coordination of Cu at 1 or more of Fet3p's Cu sites.
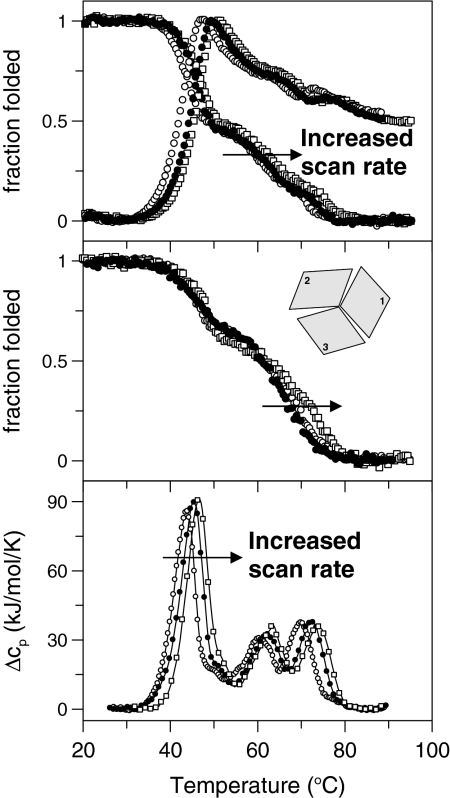
Thermal unfolding of apo-Fet3p was probed by far-UV CD (secondary structure), Trp fluorescence (environment near aromatics), anilino naphthalene-1-sulfonic acid (ANS) emission (exposure of hydrophobic surfaces), and differential scanning calorimetry (DSC; heat of reaction) as a function of scan rate (Fig. 2). Thermal perturbation of apo-Fet3p (and all other forms we have studied) is completely irreversible and, in accord, the reactions are scan rate-dependent: the faster the heating rate, the higher the apparent thermal stability. This behavior implies that the reactions are under kinetic control, and no thermodynamic parameters can be obtained. Based on far-UV CD spectra, Fet3p (in apo and metallated forms) adopts a nonnative structure at high temperatures that has a significant content of secondary structure (Fig. S1), which was found earlier upon heating of hCp and assigned to a molten-globule like denatured state (14). By CD detection there appears to be 2 detectable steps during heating of apo-Fet3p, whereas the 2 fluorescence detection methods and DSC reveal 3 sequential transitions as a function of increasing temperature for apo-Fet3p. Thermal midpoint (Tm) values for different steps obtained by different detection methods at 3 different scan rates are listed in Table 1. Each thermal step was analyzed by a 2-state irreversible model (22); the Tm values from different scan rates obtained by each detection methods were plotted as described in Materials and Methods to estimate activation energies (Ea) for each step (Table 1).
Fig. 2.
Thermal unfolding of WT apo-Fet3p probed by CD at 210 nm (Middle), Trp fluorescence (ratio F331/F355), ANS fluorescence (at 510 nm) (Top), and DSC (Bottom). Scan rates were 0.5, 1.0, and 1.5 K/min.
Table 1.
Thermal data for holo variants and apo form of Fet3p determined by different detection methods (50 mM phosphate buffer, pH 7)
| Fet3p form | Method | Midpoint, °C | Scan rate, K/min |
Ea, kJ/mol | ||
|---|---|---|---|---|---|---|
| 0.5 | 1.0 | 1.5 | ||||
| Holo | DSC | Tm,1 | 52.0 | 53.0 | 55.6 | 230 ± 100 |
| Tm,2 | 66.9 | 68.0 | 69.2 | 450 ± 80 | ||
| CD | Tm,1 | 54.4 | 56.0 | 58.5 | 230 ± 40 | |
| Tm,2 | 68.0 | 70.5 | 71.8 | 290 ± 10 | ||
| FlTrp | Tm,1 | 55.9 | 57.5 | 59.5 | 260 ± 60 | |
| Tm,2 | 70.4 | 72.2 | 73.8 | 320 ± 40 | ||
| FlANS | Tm,1 | 54.4 | 56.0 | 58.7 | 210 ± 70 | |
| Tm,2 | 69.3 | 71.7 | 73.4 | 260 ± 10 | ||
| Abs330 | Tm,1 | 52.0 | 54.5 | 55.9 | - | |
| T2D | DSC | Tm,1 | n.r. | n.r. | ∼52 | - |
| Tm,2 | n.r. | n.r. | ∼64 | - | ||
| CD | Tm,1 | - | - | - | - | |
| Tm,2 | 61.2 | 63.3 | 64.2 | 190 ± 20 | ||
| FlTrp | Tm,1 | - | - | - | - | |
| Tm,2 | 59.4 | 61.4 | 62.2 | 280 ± 10 | ||
| Abs330 | Tm,1 | 48.5 | 49.7 | 50.7 | - | |
| T1D | DSC | Tm,1 | 53.5 | 55.0 | 57.0 | - |
| CD | Tm,1 | 56.0 | 57.9 | 59.8 | 280 ± 10 | |
| FlTrp | Tm,1 | 53.6 | 55.2 | 57.4 | 320 ± 30 | |
| T1D/T2D | DSC | Tm,1 | 49.0 | 50.0 | 52.5 | - |
| CD | Tm,1 | 49.2 | 51.2 | 52.9 | 230 ± 20 | |
| FlTrp | Tm,1 | 48.1 | 50.9 | 51.4 | 250 ± 20 | |
| Apo | DSC | Tm,1 | 43.9 | 45.3 | 46.7 | 290 ± 30 |
| Tm,2 | 60.3 | 62.2 | 63.5 | 370 ± 10 | ||
| Tm,3 | 69.9 | 72.2 | 73.7 | 290 ± 10 | ||
| CD | Tm,1 | 45.0 | 47.9 | 50.0 | 180 ± 10 | |
| Tm,2 | - | - | - | - | ||
| Tm,3 | 69.4 | 71.0 | 74.5 | 190 ± 70 | ||
| FlTrp | Tm,1 | 43.4 | 45.7 | 46.6 | 280 ± 30 | |
| Tm,2 | 60.0 | 62.4 | 64.1 | 250 ± 10 | ||
| Tm,3 | 70.0 | 73.6 | 75.0 | 210 ± 20 | ||
| FlANS | Tm,1 | 42.3 | 44.5 | 45.9 | 210 ± 10 | |
| Tm,2 | 59.5 | 62.0 | 63.7 | 240 ± 10 | ||
| Tm,3 | 71.6 | 74.8 | 76.8 | 210 ± 10 | ||
All reactions are irreversible and each individual step (for variants with multiple steps) is analyzed as an apparent 2-state irreversible transition. Whereas apo-Fet3p exhibits 3 thermal transitions, the WT holo form and T2D Fet3p have 2 transitions and T1D and T1D/T2D Fet3p variants have single-step thermal transitions. n.r., not resolvable. Error in Tm is ± 0.5 °C.
Based on the presence of 3 distinct peaks in the DSC data (Tm values of 45 °C, 62 °C, and 72 °C; 1 K/min), it appears that the apo protein unfolds in 3 steps that likely correspond to independent unfolding of the 3 domains. The Ea values for the 3 transitions are similar, supporting sequential domain unfolding (Table 1). We propose that the first domain to unfold is domain 2, as the first DSC peak corresponds to the largest change in entropy. As domains 1 and 3 will have higher starting entropy because of terminal fraying, unfolding of the middle domain will likely increase entropy the most.
Fet3pT1D/T2D.
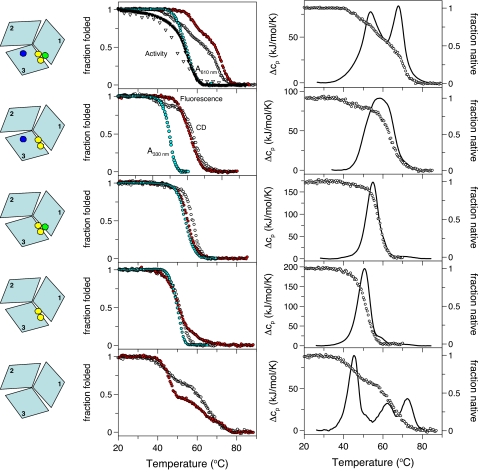
In this protein, the binuclear T3 cluster at the interface is present, but not T1 and T2. Absorbance and extended x-ray absorption fine structure spectra of Fet3pT1D/T2D indicate that T3 is native-like in its electronic characteristics (6, 17). In Fig. 3, we show CD, DSC, fluorescence, and 330-nm absorption (reporting on intact T3 site) data for heating of this mutant at 1 selected scan rate (1 K/min). A complete set of data (CD, DSC, Trp fluorescence, and ANS emission) at 3 scan rates are provided in Fig. S2. Tm and Ea values for different scan rates and detection methods are listed in Table 1.
Fig. 3.
Thermal unfolding of WT, T2D, T1D, T1D/T2D and apo-Fet3p variants (top to bottom) probed by CD at 210 nm (open circles), Trp fluorescence (ratio F331/F355; red), and absorption at 330 nm (yellow) (Center). For WT holo, oxidase activity (open triangles) and 610-nm absorption (filled triangles) are shown. CD (circles) and DSC (solid line) profiles are overlaid at Right. Scan rate in all experiments shown was 1.0 K/min. For additional mutant data see Fig. S2.
It is clear from the data that addition of T3 to the apo protein results in a relatively unstable molecule that unfolds as 1 cooperative unit (Tm of 50°C; 1 K/min). Note that if the T3 Cu atoms were to dissociate when the yellow color disappeared for Fet3pT1D/T2D (because of the T3 Cu absorbance at 330 nm), we would get apo protein that should have additional DSC peaks at higher temperatures; this is not the case and indicates Cu site perturbation but not Cu dissociation. In support, direct Cu content analysis (see Materials and Methods) revealed 1.5 ± 0.2 Cu ions (determined by colorimetric assay; ref. 21) per Fet3pT1D/T2D protein after heating to 80 °C.
Fet3pT1D.
In Fet3pT1D, both T3 and T2 sites are present, completing the TNC at the interface of domains 1 and 3. Again, spectral evidence on this variant shows this cluster to have native-like electronic properties (17). We find that addition of T2 to T3-containing Fet3p increases overall protein stability by ≈5° C (Tm of 55°C for Fet3pT1D; 1 K/min) but the thermal process remains coupled in a single step (Fig. 3 and Fig. S2). Tm and Ea values for different scan rates and detection methods are listed in Table 1. In support of Cu ions remaining bound to the perturbed protein, Cu content analysis (see Materials and Methods) revealed 2.4 ± 0.2 Cu ions per Fet3pT1D protein after heating to 80 °C.
Holo-Fet3p.
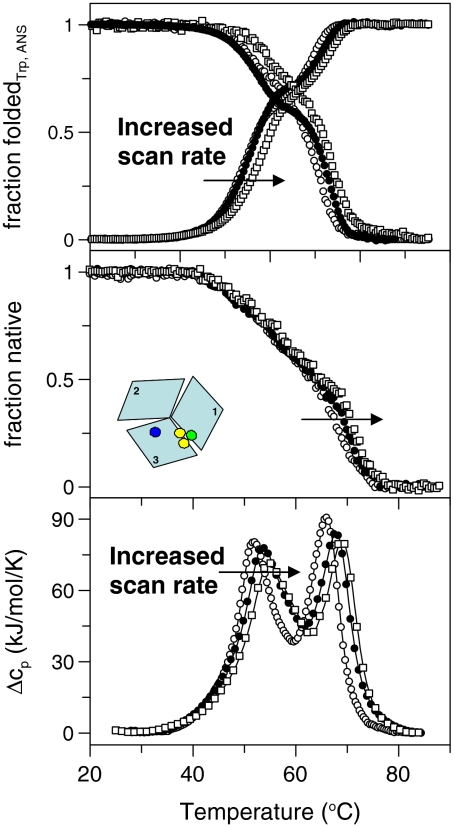
The addition of the final Cu (i.e., T1) results in the native holo protein. Thermal unfolding of WT holo-Fet3p (i.e., with T1, T2, and T3 intact) was probed by the same methods as the other variants as a function of scan rate (Figs. 3 and 4). Thermal denaturation of WT holo-Fet3p is irreversible and also exhibits a scan rate dependence of the unfolding transitions. Tm and Ea values for the different steps at different scan rates, analyzed as individual 2-step irreversible reactions, are listed in Table 1. For the WT holo form, absorption at 610 nm (reporting on an intact T1) and enzymatic activity (functional 4-Cu MCO) was monitored as a function of temperature as well (Fig. 3).
Fig. 4.
Thermal unfolding of WT holo-Fet3p probed by CD at 210 nm (Middle), Trp fluorescence (ratio F331/F355), ANS fluorescence (at 510 nm) (Top), and DSC (Bottom). Scan rates were 0.5, 1.0, and 1.5 K/min.
We find that holo-Fet3p unfolds in 2 transitions with Tm values of ≈55 °C and 68 °C (1 K/min) (Figs. 3 and 4). In the first step there is a change in far-UV CD of ≈30–40%, which suggests that 1 of the 3 domains unfolds; this conformational transition correlates with the disappearance of both the 610-nm (blue) and 330nm (yellow) transitions, indicating loss of S → Cu and O → Cu charge transfer at T1 and T3 sites, respectively. This does not mean the Cu ions dissociate in this step; if this were the case, the rest of the thermal traces should match those for the apo protein but they do not.
Loss of protein visible color caused by Cu site perturbations (but not dissociation) have been noted in single-domain cupredoxins (23). As mentioned, in the case of P. aeruginosa azurin, the Cu remained bound to the unfolded polypeptide but color was lost (5, 24–26). In nitrate reductase, a protein that evolved from the common ancestor of the MCOs that has 1 T1 site in each of its 3 domains, thermal perturbation was found to result in conversion of the T1 site to a colorless T2 site (27). In addition, estimates of total Cu content at different temperatures (see Materials and Methods) demonstrate that all Cu ions are retained with Fet3p up to at least 60 °C, which is after the first thermal transition. At 20 °C (i.e., before heating), as expected, we detect 3.9 ± 0.3 Cu per protein. After heating to 58 °C, the Cu content remains high (3.7 ± 0.3 Cu/protein), whereas heating to 80 °C corresponds to detection of 1.5 ± 0.5 Cu per protein, indicative of loss of some but not all Cu ions. The Ea values for the 2 holo-Fet3p transitions are consistently higher for the second step (Table 1), which agrees with 1 domain unfolding in the first step and 2 domains in the second step as also suggested by the CD data.
Fet3pT2D.
With this protein species we can determine the relationship between T1 and T2 Cu sites in defining Fet3p stability. Above, we quantified the effect of adding T2 to T3 in the absence of T1. With Fet3pT2D we can test the effect of T1 and T3 in the absence of T2. It appears reasonable to predict that removal of T2 will have less effect on Fet3p stability when T1 is present and stabilizes 1 of the 2 domains that contribute to the TNC interface. In agreement with this prediction, thermal unfolding of Fet3pT2D is similar to that of WT holo-Fet3p (with all Cu sites metallated) although the steps are shifted somewhat to lower temperatures (Fig. 3, Fig. S2, and Table 1). The DSC trace appears as a broad peak indicative of 2 underlying transitions; as with the WT holo form, 330-nm absorbance disappears together with ≈1/3 of the CD signal at a lower temperature (Tm ≈50 °C; 1 K/min); the remainder of the CD signal and most of the fluorescence is lost with midpoint Tm ≈63 °C (at 1 K/min). Cu content analysis revealed 2.4 ± 0.2 Cu ions per Fet3p T2D protein at 80 °C; thus, like the other Fet3p holo forms studied, most Cu ions remain bound to Fet3p T2D after structural perturbation.
We also collected DSC traces for each of the mutants (Fet3p T1D, T2D, and T1D/T2D) in the apo form. We find the apo variants to exhibit somewhat lower thermal stability than WT apo-Fet3p. Nonetheless, 3 separate DSC peaks are always detected (data not shown). The observation of lowered apo-protein stability is reasonable; many different point mutations in azurin were found to reduce apo-protein stability significantly (28–31).
Fet3p Glycan.
Fet3p is a glycoprotein; the glycan mass in the native protein has not been quantified. To examine the possible role of this carbohydrate in protein stability, we examined EndoH-treated WT Fet3p, which has been well-characterized by chemical and mass-spectral analyses and X-ray structural determination. This preparation retains ≈30% of the glycan of the native form (32). As indicated by diffraction data (7), this carbohydrate is concentrated on 6 of the 13 N-linked glycosylation sites identified in Fet3p. The same set of thermal experiments as above was repeated with apo and holo forms of this desglycan WT Fet3p form; the results were qualitatively and quantitatively equivalent to those presented for the native protein in apo and holo forms (Fig. S3 and Table S1). Of course, this result does not exclude the possibility that the remaining carbohydrate does contribute to the in vitro stability of Fet3p.
Discussion
MCO proteins are found in all kingdoms of life; a majority of MCOs archived in public databases contain 3 cupredoxin domains and 4 Cu ions distributed in 3 distinct sites that provide functionality and a connection between the amino- and carboxyl-terminal domains (6). Calorimetric profiles on apo and holo forms of Rhus laccase implied that the thermal reaction involved 2 steps that depended on Cu although little change in far-UV CD was observed during these steps (16). In contrast, thermal perturbation of dimeric ascorbate oxidase was found to involve 3 calorimetric steps that did not change upon T2 removal (33). Our recent biophysical work on hCp (composed of 6 cupredoxin motifs) (13, 14), revealed that thermal unfolding involved 2 irreversible steps: the first step involved unfolding of 1 of the 6 domains and perturbation of 1 of the 3 T1 sites. The intermediate species was partially active and, therefore, we speculated that the interface between domains 1 and 6 and thus T2 and T3 remained intact. In many ways, this is analogous to what we find here for Fet3p in which the T2 and T3 Cu atoms reside between domains 1 and 3. The important contribution of the current study is that we are able to identify the possible role of each Cu center via characterization of strategic Cu-site variants.
We find that on its own the presence of T3 in Fet3p results in a single cooperative structure with low thermal stability as compared with the more dynamic apo form where the domains likely can rotate freely with respect to each other. Addition of T2 to the T3-containing Fet3p increases the overall stability of the protein but the single-step mechanism is retained. This implies that T3 (and possibly also T2) “stitches” domains 1 and 3 together, which results in 1 cooperative unit. This large unit has low stability, as compared with the apo form, likely because of loss of entropy arising from the structural constraints of the T3/T2 coordination.
In relation to MCO evolution, it is significant that formation of the TNC raises the energy of the folded state of domains 1 and 3, thus reducing their stability. This energetic loss may be counterbalanced by the heat released upon formation of the ligand-Cu bonds. Assembling a TNC at an intermolecular interface as in dimers or higher order oligomers would be even more entropically costly. In this evolutionary context, domain 2 in Fet3p may be seen as a tether that exerts a “proximity” effect on domains 1 and 3 and thus provides stability by reducing the degrees of freedom of the protein's terminal domains.
Our data indicate that T1 markedly alters the Fet3p-unfolding landscape by introducing an intermediate species not present for the variants that lack T1. Incorporation of T1 to T3 only, or to the T3/T2 combination of Fet3p, adds significantly to the stability of domain 3; this results in uncoupling of domain 2 unfolding (a domain that has no Cu site) from that of domains 1 and 3 (which are held together by T3 and T2 and still unfolds as 1 unit). That a T1 Cu makes the structural domain more stable is in line with earlier work on azurin and other curpedoxins (34).
In summary, this study demonstrates the importance of T3 in holding the Fet3p trimeric structure together. This closed structural arrangement, required for ferroxidase function, however, results in loss of overall protein stability as compared with the nonfunctional apo form. The presence of T1 counters this by increasing the stability of domain 3, which is transmitted to domain 1 via the interfacial T3/T2 interactions. Fet3p, thus, is an excellent example of a biological situation in which trade-offs between stability and structure is mediated by metal prosthetic groups to obtain a desired function. We propose that this Fet3p unfolding landscape applies to most, if not all, MCO proteins.
Materials and Methods
Protein Preparation.
WT (with and without carbohydrates) and mutant Fet3p variants was prepared as described (17, 32). The variants used are as follows: T1D contains a Cys-484-to-Ser mutation at the T1 site; T2D includes a His-81-to-Gln mutation at the T2 site; T1D/T2D is a double mutant with both the above changes introduced (17). Apo protein was made by initial dialysis at 4 °C in 50 mM ascorbate and 0.1 M Tris·HCl, pH 7.2, followed by dialysis against the same Tris buffer including 50 mM NaCN and 10 mM EDTA, before the sample was dialyzed into 50 mM phosphate buffer, pH 7 (the condition used in all subsequent experiments). The apo forms prepared this way contained <0.5 copper per protein determined by the method of Felsenfeld (21). The metal contents of the holo forms of WT and the 3 variants were determined by flameless atomic absorption spectrophotometry. Contamination of Fe and Zn were below the detection limit in all proteins (i.e., <0.1 atoms per protein molecule). The Cu analyses showed that WT Fet3p has 3.8 +/− 0.2 Cu atoms per protein molecule; both T1D and T2D Fet3p have 2.7 +/− 0.3 Cu atoms per protein molecule; and T1D/T2D has 1.8 +/− 0.2 Cu atoms per Fet3p molecule. For this quantification, the Fet3p concentration was determined by the Bradford assay on protein samples dialyzed against Chelex-treated buffer (17).
DSC.
Thermal experiments were performed on a VP-DSC microcalorimeter (Microcal) at 0.5, 1, and 1.5 K/min scan rates. Protein concentration was 3–4 μM (50 mM phosphate buffer, pH 7). Before measurements, sample and reference solutions were degassed for 5 min and carefully loaded into the cells to avoid bubble formation. A pressure of 2 atm was kept in the cells throughout the heating cycles to prevent degassing. Background was subtracted from each protein trace. Reversibility of the transitions was assessed by performing second heating cycles after cooling. In all cases, the thermal reactions are 100% irreversible. Excess heat capacity curves were plotted with Origin software (MicroCal).
Spectroscopy.
Far-UV CD spectra were recorded on a Jasco-810. Thermal CD experiments were monitored at 210 nm (1-mm path), using heating rates of 0.5, 1, and 1.5 K/min in separate experiments (50 mM phosphate buffer, pH 7). For fluorescence, protein was mixed with 200 μM ANS and incubated for 1 h before thermal experiments. Tryptophan and ANS fluorescence at 335 nm (excitation at 295 nm) and 510 nm (excitation at 390 nm), respectively (1 cm × 1-cm cell), was monitored as a function of temperature (0.5, 1, and 1.5 K/min in separate experiments) in a Varian Eclipse. Fluorescence is reported as ratio of emission at 331 and 355 nm. Absorption at 330 and 610 nm was monitored on a Cary Bio-100 (1-cm path) with a scan rate of 1 K/min.
Activity and Cu Content Assays.
Oxidase activity of WT holo-Fet3p was tested by using o-dianisidine as a substrate (35). Activity as a function of temperature was determined as follows: a sample was heated with rate 1 K/min; after reaching desired temperature a 25 μL of aliquot of the solution was transferred into 100 μL of 100 mM acetate, pH 5.0 and cooled on ice. The activity was then measured at 23 °C. To probe Cu content in WT holo-Fet3p and the 3 variants at different temperatures (between 20 °C and 80 °C), protein samples were heated to the desired temperature (1 K/min) and an aliquot was removed. The protein aliquots were incubated on ice followed by dialysis against buffer to remove dissociated Cu. Then, the Cu content in the remaining protein sample was tested by Felsenfeld's assay (21). At 20 °C before before heating, all holo forms had the Cu content expected for each variant (±0.2) as determined by this assay.
Data Analysis.
The CD data were fit to 2-step reactions, whereas the fluorescence and DSC data were fit to 3-step reactions for the WT apo protein. For the WT holo form and T2D, 2-step reactions were observed, whereas T1D and T1D/T2D variants exhibited apparent single-step reactions. The Tm values for each step as a function of detection method and variant are reported in Table 1. Each step was analyzed as a separate 2-state irreversible process (22). For each step, the Tm values at different scan rates (as obtained by a particular detection method) were fit to the following equation, which allows estimation of apparent Ea values (v is scan rate, R is gas constant) (22):
Supplementary Material
Acknowledgments.
This work was supported by Robert A. Welch Foundation Grant C-1588 (to P.W.S.) and National Institutes of Health Grant DK53820 (to D.J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806431105/DCSupplemental.
References
- 1.Richardson JS. β-Sheet topology and the relatedness of proteins. Nature. 1977;268:495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- 2.Adman ET. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 3.Nar H, Messerschmidt A, Huber R, van de Kamp M, Canters GW. Crystal structure of Pseudomonas aeruginosa apo-azurin at 1.85-Å resolution. FEBS Lett. 1992;306:119–124. doi: 10.1016/0014-5793(92)80981-l. [DOI] [PubMed] [Google Scholar]
- 4.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. [Google Scholar]
- 5.Wittung-Stafshede P, et al. Reduction potentials of blue and purple copper proteins in their unfolded states: A closer lock at rack-induced coordination. J Biol Inorg Chem. 1998;3:367–370. [Google Scholar]
- 6.Kosman DJ. FET3P, ceruloplasmin, and the role of copper in iron metabolism. Adv Protein Chem. 2002;60:221–269. doi: 10.1016/s0065-3233(02)60055-5. [DOI] [PubMed] [Google Scholar]
- 7.Taylor AB, Stoj CS, Ziegler L, Kosman DJ, Hart PJ. The copper–iron connection in biology: Structure of the metallo-oxidase Fet3p. Proc Natl Acad Sci USA. 2005;102:15459–15464. doi: 10.1073/pnas.0506227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakurai T, Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem Rec. 2007;7:220–229. doi: 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- 9.Vachette P, et al. A key structural role for active site type 3 copper ions in human ceruloplasmin. J Biol Chem. 2002;277:40823–40831. doi: 10.1074/jbc.M207188200. [DOI] [PubMed] [Google Scholar]
- 10.Sato M, Gitlin JD. Mechanisms of copper incorporation during the biosynthesis of human ceruloplasmin. J Biol Chem. 1991;266:5128–5134. [PubMed] [Google Scholar]
- 11.Hellman NE, et al. Mechanisms of copper incorporation into human ceruloplasmin. J Biol Chem. 2002;277:46632–46638. doi: 10.1074/jbc.M206246200. [DOI] [PubMed] [Google Scholar]
- 12.Vassiliev VB, et al. Copper depletion/repletion of human ceruloplasmin is followed by the changes in its spectral features and functional properties. J Inorg Biochem. 1997;65:167–174. doi: 10.1016/s0162-0134(96)00119-5. [DOI] [PubMed] [Google Scholar]
- 13.Sedlak E, Wittung-Stafshede P. Discrete roles of copper ions in chemical unfolding of human ceruloplasmin. Biochemistry. 2007;46:9638–9644. doi: 10.1021/bi700715e. [DOI] [PubMed] [Google Scholar]
- 14.Sedlak E, Zoldak G, Wittung-Stafshede P. Role of copper in thermal stability of human ceruloplasmin. Biophys J. 2008;94:1384–1391. doi: 10.1529/biophysj.107.113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei G, et al. Role of quaternary structure in the stability of dimeric proteins: The case of ascorbate oxidase. Biochemistry. 1997;36:10917–10922. doi: 10.1021/bi970614p. [DOI] [PubMed] [Google Scholar]
- 16.Agostinelli E, Cervoni L, Giartosio A, Morpurgo L. Stability of Japanese-lacquer-tree (Rhus vernicifera) laccase to thermal and chemical denaturation: Comparison with ascorbate oxidase. Biochem J. 1995;306:697–702. doi: 10.1042/bj3060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackburn NJ, Ralle M, Hassett R, Kosman DJ. Spectroscopic analysis of the trinuclear cluster in the Fet3 protein from yeast, a multinuclear copper oxidase. Biochemistry. 2000;39:2316–2324. doi: 10.1021/bi992334a. [DOI] [PubMed] [Google Scholar]
- 18.Augustine AJ, Quintanar L, Stoj CS, Kosman DJ, Solomon EI. Spectroscopic and kinetic studies of perturbed trinuclear copper clusters: The role of protons in reductive cleavage of the O-O bond in the multicopper oxidase Fet3p. J Am Chem Soc. 2007;129:13118–13126. doi: 10.1021/ja073905m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer AE, et al. Spectroscopic characterization and O2 reactivity of the trinuclear Cu cluster of mutants of the multicopper oxidase Fet3p. Biochemistry. 2002;41:6438–6448. doi: 10.1021/bi011979j. [DOI] [PubMed] [Google Scholar]
- 20.Yuan DS, Dancis A, Klausner RD. Restriction of copper export in Saccharomyces cerevisiae to a late Golgi or post-Golgi compartment in the secretory pathway. J Biol Chem. 1997;272:25787–25793. doi: 10.1074/jbc.272.41.25787. [DOI] [PubMed] [Google Scholar]
- 21.Felsenfeld G. The determination of cuprous ion in copper proteins. Arch Biochem Biophys. 1960;87:247–251. doi: 10.1016/0003-9861(60)90168-5. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Ruiz JM, Lopez-Lacomba JL, Cortijo M, Mateo PL. Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry. 1988;27:1648–1652. doi: 10.1021/bi00405a039. [DOI] [PubMed] [Google Scholar]
- 23.Canters GW, Gilardi G. Engineering type 1 copper sites in proteins. FEBS Lett. 1993;325:39–48. doi: 10.1016/0014-5793(93)81410-2. [DOI] [PubMed] [Google Scholar]
- 24.Pozdnyakova I, Guidry J, Wittung-Stafshede P. Probing copper ligands in denatured Pseudomonas aeruginosa azurin: Unfolding His117Gly and His46Gly mutants. J Biol Inorg Chem. 2001;6:182–188. doi: 10.1007/s007750000189. [DOI] [PubMed] [Google Scholar]
- 25.Pozdnyakova I, Wittung-Stafshede P. Biological relevance of metal binding before protein folding. J Am Chem Soc. 2001;123:10135–10136. doi: 10.1021/ja016252x. [DOI] [PubMed] [Google Scholar]
- 26.Pozdnyakova I, Wittung-Stafshede P. Copper binding before polypeptide folding speeds up formation of active (holo) Pseudomonas aeruginosa azurin. Biochemistry. 2001;40:13728–13733. doi: 10.1021/bi011591o. [DOI] [PubMed] [Google Scholar]
- 27.Stirpe A, Sportelli L, Wijma H, Verbeet MP, Guzzi R. Thermal stability effects of removing the type-2 copper ligand His306 at the interface of nitrite reductase subunits. Eur Biophys J. 2007;36:805–813. doi: 10.1007/s00249-007-0151-5. [DOI] [PubMed] [Google Scholar]
- 28.Chen M, Wilson CJ, Wu Y, Wittung-Stafshede P, Ma J. Correlation between protein stability cores and protein folding kinetics: A case study on Pseudomonas aeruginosa apo-azurin. Structure (London) 2006;14:1401–1410. doi: 10.1016/j.str.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Wilson CJ, Apiyo D, Wittung-Stafshede P. Solvation of the folding-transition state in Pseudomonas aeruginosa azurin is modulated by metal: Solvation of azurin's folding nucleus. Protein Sci. 2006;15:843–852. doi: 10.1110/ps.051838206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson CJ, Wittung-Stafshede P. Role of structural determinants in folding of the sandwich-like protein Pseudomonas aeruginosa azurin. Proc Natl Acad Sci USA. 2005;102:3984–3987. doi: 10.1073/pnas.0501038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson CJ, Wittung-Stafshede P. Snapshots of a dynamic folding nucleus in zinc-substituted Pseudomonas aeruginosa azurin. Biochemistry. 2005;44:10054–10062. doi: 10.1021/bi050342n. [DOI] [PubMed] [Google Scholar]
- 32.Hassett RF, Yuan DS, Kosman DJ. Spectral and kinetic properties of the Fet3 protein from Saccharomyces cerevisiae, a multinuclear copper ferroxidase enzyme. J Biol Chem. 1998;273:23274–23282. doi: 10.1074/jbc.273.36.23274. [DOI] [PubMed] [Google Scholar]
- 33.Savini I, D'Alessio S, Giartosio A, Morpurgo L, Avigliano L. The role of copper in the stability of ascorbate oxidase toward denaturing agents. Eur J Biochem. 1990;190:491–495. doi: 10.1111/j.1432-1033.1990.tb15600.x. [DOI] [PubMed] [Google Scholar]
- 34.Wittung-Stafshede P. Role of cofactors in folding of the blue-copper protein azurin. Inorg Chem. 2004;43:7926–7933. doi: 10.1021/ic049398g. [DOI] [PubMed] [Google Scholar]
- 35.Schosinsky KH, Lehmann HP, Beeler MF. Measurement of ceruloplasmin from its oxidase activity in serum by use of o-dianisidine dihydrochloride. Clin Chem. 1974;20:1556–1563. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.