Abstract
The chloroplast NAD(P)H dehydrogenase complex, a homologue of mitochondrial complex I, consists of >15 subunits, of which 11 are encoded by the chloroplast genome (ndhA-K). The ndhC and ndhK genes are partially overlapped and cotranscribed in many land plants. The downstream ndhK mRNA possesses 4 possible AUG initiation codons in many dicot plants. By using an efficient in vitro translation system from tobacco chloroplasts, we defined that the major initiation site of tobacco ndhK mRNAs is the third AUG that is located 4 nt upstream from the ndhC stop codon. Mutation of the ndhC stop codon (UAG) arrested translation of the ndhK cistron. Frameshift of the ndhC coding strand inhibited also translation of the distal cistron. The results indicated that ndhK translation depends on termination of the preceding cistron, namely translational coupling. Surprisingly, removal of the ndhC 5′-UTR and its AUG still supported substantial translation of the ndhK cistron. This translation was abolished again by removing the ndhC stop codon. Although translation of the downstream cistron of an overlapping mRNA is generally very low, we found that the ndhC/K mRNA produces NdhK and NdhC in similar amounts. Based on subunit compositions of the bacterial complex I, the stoichiometry of NdhK and NdhC is suggested to be 1:1 in chloroplasts. To meet this stoichiometry, the ndhC/K mRNA is translated not only by a translational coupling event but also by a termination codon-dependent pathway.
Keywords: mRNA, cistron, initiation codon, translational coupling
Chloroplasts are photosynthetic organelles that contain their own genetic system. The chloroplast genome of higher plants contains ≈80 protein-coding genes (1). Many of these genes are transcribed as polycistronic pre-mRNAs by multiple RNA polymerases (2). These pre-mRNAs are generally processed into complex sets of overlapping transcripts including mono-, di-, and poly-cistronic mRNAs (3). Many nuclear-encoded proteins are involved in the mRNA processing as well as translation and mRNA stability (4–6). Translational control is the major step of chloroplast gene expression, and it is especially important for the stoichiometric production of individual subunits in photosynthetic complexes (7, 8). The existence of a NAD(P)H dehydrogenase (NDH) complex was first assumed based on chloroplast DNA sequences whose predicted amino acid sequences resemble those of human mitochondrial complex I subunits (9, 10). The chloroplast NDH complex consists of >15 subunits (11, 12). Eleven ndh genes (A to K) that encode subunits of the NDH complex are found in the chloroplast genome of most land plants except pines (13) and some parasitic plants (14). These genes are clustered and organized in 4 transcription units: ndhC/K/J in the large single-copy region, ndhB in the inverted repeat, and ndhH/A/I/G/E/D and ndhF in the small single-copy region (15). The ndhC and ndhK genes overlap in part in the chloroplast DNAs of many higher plants (www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plastids_tax.html), but are separated by a spacer in some legumes (16). The ndhC/K overlapping genes are cotranscribed with the downstream ndhJ gene, and their major transcripts are long enough to include all 3 cistrons (15), suggesting that the overlapping ndhC and ndhK are translated by polycistronic mRNAs but not by monocistronic mRNAs.
The initiation process represents a crucial point for the synthesis of correct proteins. There are often multiple possible initiation codons in chloroplast mRNAs (17, 18). A striking case is the ndhK mRNA from tobacco and many other dicot plants, and the mRNA contains 4 possible AUG codons (15). Three in-frame AUG triplets are present in many monocot plants (19). There are several techniques to determine which is the real start codon. The N termini of proteins are generally determined, but this method requires protein isolation and is not always conclusive when nascent products are processed at the N terminus. The site-specific disruption of candidate codons by chloroplast transformation was successfully applied to study some of the Chlamydomonas chloroplast genes (20, 21). However, this method cannot be applied to genes essential with viability. In contrast, in vitro translation systems that support accurate translation initiation allow us to determine in principle the initiation site of any mRNAs. By using our chloroplast in vitro translation system, we found that translation of tobacco ndhD mRNAs starts only at the edited AUG from ACG but not at the upstream in-frame AUG and GUG (22). We recently improved the original in vitro system, and the refined system is highly active enough to measure the relative rate of translation by means of the fluorescence intensity of fused green fluorescent protein (23). We used the modified (m)GFP, in which 3 amino acids were replaced to enhance fluorescence (23), so that one can detect minor translation products that are undetectable by using 35S-Met. This system has allowed us to analyze the effect of mRNA processing on translation (23), the translation efficiency of several synonymous codons (24), and the translation initiation site of psbC mRNAs (18).
Based on in vitro translation analyses, we here report that the major initiation site of the tobacco chloroplast ndhK cistron is the third AUG within the upstream ndhC cistron. We then show that the ndhK translation depends exclusively on translational termination codon (UAG) of the ndhC cistron. This result indicates that the ribosome translated from the ndhC cistron moves upstream after translation and reinitiates at the third AUG, translational coupling. Surprisingly, removal of the 5′-UTR and its following AUG from the ndhC/K mRNA still supported substantial translation of the ndhK cistron, which is also termination codon-dependent. Although translation of the downstream cistron of overlapping genes is generally very low, NdhK is produced as efficiently as NdhC from ndhC/K mRNAs. Therefore, in addition to translational coupling, the downstream ndhK cistron should be translated by an additional mechanism, also in a termination codon-dependent manner.
Results
Translation Initiation Sites of Tobacco ndhK mRNAs.
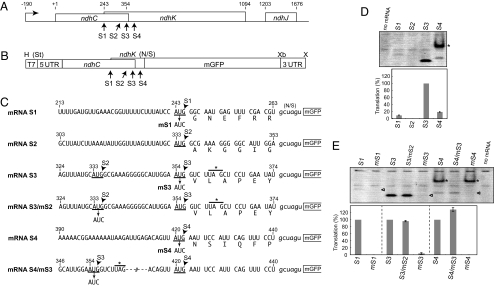
Fig. 1A shows the ndhC/K/J gene cluster from tobacco chloroplasts (25). In the ndhK coding frame, there are 4 ATG triplets (named S1 to S4), of which S1 to S3 are within the ndhC coding region. To determine which is the translation initiation codon, we performed in vitro translation assays. Transcription of the tobacco ndhC/K/J cluster starts at position −190 relative to the ndhC ATG (A as +1) initiation codon (34). We constructed chimeric mRNAs consisting of the full 5′ UTR, ndhC and ndhK coding parts, the mGFP coding region, and the 3′ UTR from tobacco chloroplast rps16 mRNA as described (23). Because sequences downstream of the initiation codon were reported to be important for translation efficiency (26), we fused the mGFP coding region at 6 codons after each of the 4 AUGs, so that mGFP is in-frame with ndhK, but not with ndhC (mRNAs S1 to S4 in Fig. 1C). Translation in vitro of the chimeric mRNAs was performed by the standard procedure, and translation products were separated on native gels and monitored by mGFP fluorescence. Because the fluorescent signal was very low, we first reexamined reaction conditions to improve translation activity. The optimized reaction mixture included 40 mM KOAc, 8 mM MgOAc, 24 A280 units per mL S30, and 200 fmol/mL mRNA, which led to >2-fold increase in activity (data not shown). Next, we increased the reaction volume from 20 μL (standard mixtures) to 60 μL, and native gels 2-mm thick instead of 1-mm thick were used, resulting in ≈10-fold enhancement.
Fig. 1.
Determination of the translation initiation site of ndhK mRNAs from the tobacco chloroplast ndhC/K/J cluster. (A) Schematic representation of the ndhC/K/J. Positions relative to the ndhC ATG start codon (A as +1) are shown above. Bent-arrow indicates the transcription initiation site. S1 to S4 indicate potential ATG start sites for ndhK translation. (B) Schematic representation of the plasmid construct, which contains the T7 promoter (19 bp, T7), the ndhC 5′UTR (190 bp), and its coding region (360 bp), a 5′ ndhK part (21–198 bp), the mGFP coding region (no ATG, 714 bp), and the rps16 3′UTR (199 bp). Restriction sites are H (HindIII), Xb (XbaI), and X (XhoI). St shows the StuI site removed during plasmid construction. N/S indicates the junction to mGFP (“gctagt” derived from ligation of NheI- and SpeI- cut fragments). The construct in a pUC18 derivative was linearized with XhoI and transcribed with T7 RNA polymerase. (C) Parts of test mRNA sequences (mRNAs S1 to S4). Positions are as in A; mRNA S1 has S1, mRNA S2 has S1 and S2, mRNA S3 has S1 to S3, and mRNA S4 has S1 to S4. The middle AUG in sequences is 1 of the 4 AUGs (S1 to S4, underlined). The ndhC stop codon UAG is lined with an asterisk. Each AUG is replaced with AUC (mS1 to mS4). Predicted amino acid sequences are shown below. (D) Gel pattern of translation products (mGFP fluorescence). Translation efficiencies are shown relative to mRNA S3 as 100%. SEs were obtained from 3 independent assays. Asterisk indicates the translation from S3. (E) Gel pattern of products from mutated mRNAs mS1 mS3, S3/mS2, mS4 and S4/mS3. Translation efficiencies are relative to each of the mRNAs S1 to S4 as 100%. Triangles indicate expected positions of products.
Fig. 1D shows a native gel pattern of mGFP products synthesized from the 4 test mRNAs. Translation of mRNA S3 was clearly observed, whereas that of mRNA S2 was not detected. A faint signal was observed from mRNA S1 and from mRNA S4 (the upper dense band was from the S3 site, indicated by an asterisk). The size of translation products was slightly different, probably due to differences in amino acid compositions of N-terminal extensions in mGFP products. Translation of mRNA S3 increased linearly for 1 h and gradually up to at least 3 h, whereas that of the other mRNA was too low to calibrate (data not shown). We then exchanged each of the 4 AUGs for AUC (mS1 to mS4, Fig. 1C). This alteration arrested translation completely from S1, S3, and S4 sites (Fig. 1E, indicated by arrowheads). Mutation at S2 did not affect translation from S3. Change of S3 in mRNA S4 slightly enhanced translation from S4. These results indicate that the major site of translation initiation of tobacco ndhK cistron is S3.
The ndhC Termination Codon Is Essential for Translation of the ndhK Cistron.
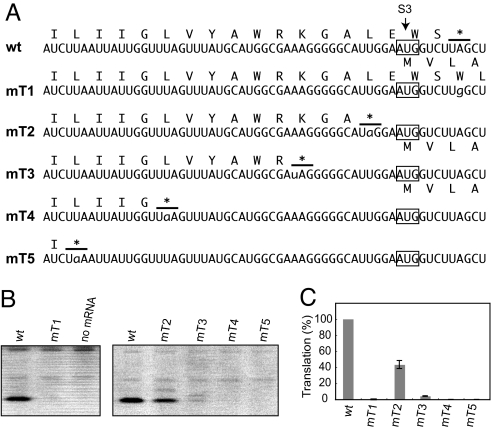
The start codon S3 of tobacco ndhK cistron is located 4 nt upstream from the ndhC UAG termination codon (Fig. 2A, WT). To examine whether the ndhC termination codon affects translation of the ndhK cistron, we first altered the UAG to UGG, which resulted in no amino acid change in NdhK (UUA to UUG, both coding for Leu) but in shifting the ndhC termination codon located 22 codons further downstream (Fig. 2A, mT1). Surprisingly, no translation of the ndhK cistron was observed with mRNA mT1 (Fig. 2B). This observation strongly suggests that the presence of the ndhC termination codon is crucial for translation of the downstream ndhK cistron, namely translational coupling. We then created premature termination codons, located upstream from S3. As shown in Fig. 2B (lines mT2–5), translation of the mRNA with a termination codon 2 nt upstream (mT2) decreased, whereas slight or no translation was observed from the mRNAs with termination codons further upstream (mT3–5). These results indicate that translation of the ndhK cistron requires the ndhC termination codon located in close vicinity of the start codon. Unexpectedly, ndhC translation was higher when the ndhC termination codon was situated after S3 (WT) than before S3 (mT2).
Fig. 2.
Effect of removing the ndhC termination codon and introducing premature termination codons on ndhK translation. (A) Test mRNA sequences around S3. Predicted amino acid sequences from ndhC and ndhK are shown above and below, respectively. Lowercase nt represents altered nt. Asterisks with lines indicate the ndhC termination codons. The termination codon in mT1 is located 22 codons further downstream (data not shown). (B) Gel patterns of products. (C) Translation efficiencies are shown as in Fig. 1D.
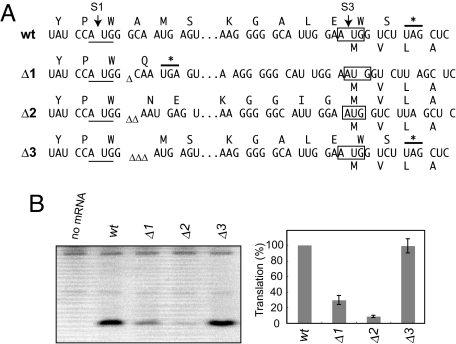
As the above mRNAs contain point mutations close to S3, these mutations may affect intrinsic translation of the ndhK cistron. Therefore, translation assays were performed by using frameshift mRNAs. We deleted 1 to 3 nt in the immediate downstream of S1, 107 nt upstream from S3, so that sequences surrounding S3 are identical with the wild-type mRNA (Fig. 3A). Deletion of 1 nt (Δ1) creates a premature stop codon (UGA) 4 nt after S1 in the ndhC cistron, 2 nt deletion (Δ2) leads ndhC and ndhK into 1 long reading frame, and 3 nt deletion (Δ3) results in 1 codon (GCA for Ala) shorter than the wild-type ndhC cistron. As shown in Fig. 3B, translation of the ndhK cistron was markedly reduced by mRNA Δ1, and that from mRNA Δ2 was hardly detected. However, mRNA Δ3 produced its ndhK product similar in amount to that from the original mRNA. The results obtained from mRNAs Δ2 and Δ3 further confirmed that translation of the ndhK cistron depends on translational termination of the ndhC cistron. However, translation from mRNA Δ1 was clearly observed (≈30% of WT) suggesting that there is an unknown mechanism to translate in part the ndhK cistron (see the next section).
Fig. 3.
Effect of frameshift of the ndhC coding strand on ndhK translation. (A) Test mRNA sequences around S1 and S3. Triplets are in-frame with the ndhC start codon. Asterisks with lines indicate termination codons. Triangles indicate deleted nt. Predicted amino acid sequences from ndhC and ndhK are shown above and below, respectively. (B) Gel pattern of products. Translation efficiencies are shown as in Fig. 1D.
Effect of 5′UTRs on Translation of the ndhK Cistron.
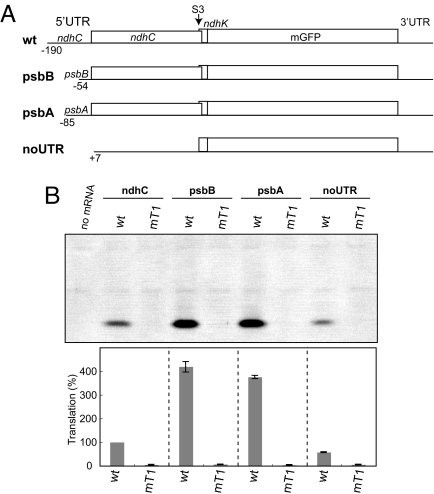
As described, translation activity of the 5′UTR from the ndhC/K cluster was very low. A preliminary assay showed that translation from the ndhC 5′UTR-mGFP mRNA is <1/10th of that from the psbA 5′UTR counterpart (data not shown). To examine the effect of 5′UTRs on translation of the ndhK cistron, we replaced the authentic 5′UTR with that of psbB or psbA mRNA (Fig. 4A), because these 5′UTRs have high activity in vitro (23). The both chimeric mRNAs produced ≈4-fold more mGFPs from the ndhK cistron than the original ndhC mRNA (Fig. 4B). Also, translation of the ndhK cistron again arrested when the ndhC termination codon was removed (UAG to UGG as before, lanes mT1s).
Fig. 4.
Effect of replacing the ndhC 5′UTR on ndhK translation. (A) Schematic representation of test mRNAs. As a negative control mRNA, the 5′UTR and the following AUG and UUG codons were removed (noUTR). The ndhC termination codon of respective test mRNAs was removed as mT1 in Fig. 2A. (B) Gel pattern of products. Translation efficiencies are shown as in Fig. 1D.
In the above assays, we used the test mRNA lacking the 5′UTR and the following AUG codon (noUTR) as a negative control, with which no translation was expected. Quite unexpectedly, substantial translation (≈58% of WT) was observed from the control mRNA (lane noUTR, WT). However, removing the ndhC termination codon abolished this translation (lane noUTR, mT1). These observations demonstrated the existence of an additional mechanism to translate the ndhK cistron. Translation from mRNA Δ1 (see Fig. 3B) was due probably to the additional pathway.
Efficient Translation of the Downstream ndhK Cistron.
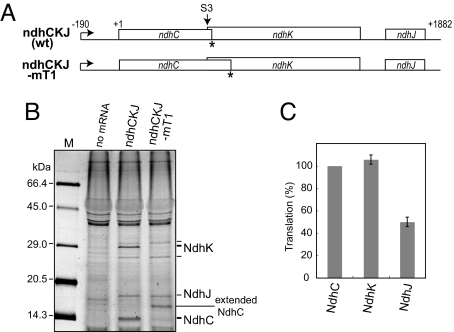
Then, translation efficiencies were compared between ndhC and ndhK cistrons. Because the tobacco ndhC/K/J cluster was cotranscribed to produce a major 2 kb mRNA (15), we prepared its tricistronic mRNA of 2,097 nt (Fig. 5A). We translated the tricistonic mRNA in the presence of fluorescent-labeled fMet-tRNAfmet. Reaction mixtures included RNase and proteinase inhibitors from S30, and mRNA templates were stable during 2-h incubation and translation products were stable after 12-h incubation (23). The products from ndhC/K/J mRNAs were resolved by denatured gel and the fluorescence intensity from fMet (or Met) incorporated at the N terminus was detected and quantified. As shown in Fig. 5B, the major NdhK band of the expected size from S3 (28.0 kDa) was clearly observed together with faint upper and lower bands that correspond to translation products from S1 and S4, respectively, based on their sizes. The translation product from ndhC was also detected as a band of the expected size (13.9 kDa). Translation of the ndhK (S3) cistron occurred as efficiently as, or slightly higher than, that of the preceding ndhC cistron (Fig. 5C). By contrast, the last ndhJ cistron was translated less than the upstream 2 cistrons (≈50% of NdhC).
Fig. 5.
Translation of full-length ndhC/K/J mRNAs. (A) Schematic representation of the tricistronic mRNAs. Asterisks indicate ndhC termination sites. (B) Gel pattern of products. Translation was performed with fluorescent fMet-labeled tRNAfmet. Products were separated on denatured gels. The calculated kDa of NdhC, NdhK (S3), and NdhJ are 13.9, 28.0, and 18.6, respectively. Faint bands above and below NdhK are those from S1 (32.3 kDa) and S4 (25.5 kDa), respectively. A band between NdhJ and NdhC in lane mT1 is an extended NdhC (16.4 kDa). Size markers are fluorescent (FITC)-labeled molecular mass marker (APRO Life Science Institute). (C) Translation efficiencies are as in Fig. 1D. Only the major NdhK from S3 was used.
We then replaced the ndhC termination codon UAG with UGG as mT1 in Fig. 2A, which shifts its termination 20 codon further downstream (Fig. 5A). Removal of the authentic ndhC stop codon arrested translation from the ndhK S3 site, but hardly affected translation from the ndhK S1 and S4 sites and that from the ndhC cistron (to produce an extended NdhC of the expected size of 16.4 kDa). Using mRNA noUTR (see Fig. 4A) and fluorescent-fMet tRNAfmet, no NdhC was detected, as expected, whereas translation of the ndhK -mGFP cistron was observed (data not shown). These results support that an additional pathway operates to translate the ndhK cistron.
Discussion
The chloroplast genome in higher plants is tightly packed, and protein-coding genes sometimes overlap each other to increase the number of proteins encoded by the size-limited genome. The ndhC/K cluster is such an example. Our studies showed that the major translation initiation site of ndhK mRNAs is the third AUG among 4 in-frame AUGs, which is situated 4 nt upstream from the ndhC stop codon. We then demonstrated that location of the ndhC termination codon has a crucial role in directing translation of the ndhK cistron, namely translational coupling or coupled translation reinitiation.
In eukaryotes, cellular mRNAs are generally monocistronic, and their translation initiation depends on the 5′cap structure (27). However, many virus transcripts are bicistronic or polycistronic. Translational initiation of a downstream cistron requires specialized mechanisms. Translation reinitiation, leaky scanning, ribosome jumping, and internal ribosome entry are possible mechanisms for translation of downstream cistrons (27, 28). For example, the M2 mRNA of human respiratory syncytial virus contains 2 ORFs that overlap partially. Translation of the second ORF initiates at 1 of the 3 AUGs located upstream of the termination codon for the first ORF, and translation of these AUGs requires the termination of the first ORF translation (29). However, unlike the case observed in ndhK translation, all of the 3 AUGs can function as initiators, although different in efficiency, and the distance between the AUG and the UGA in the virus mRNA is much longer (up to 26 nt) than that in the ndhC/K mRNAs (4 nt). Also, the frequency of such translation reinitiation was suggested to be very low. This indication would result in low levels of expression of the second ORF protein in viruses.
In prokaryotes, many genes are organized in operons and hence cotranscribed as polycistronic mRNAs. The start codon of each cistron is usually distinguished by its own Shine–Dalgarno sequence that pairs with the 3′ end of 16S rRNA, and translation can occur at multiple sites, in principle, independently throughout a polycistronic mRNA (27). However, translational coupling has been reported for some dicistronic mRNAs through mainly an overlapping termination and initiation codon (27, 28, 30). For example, expression of the upstream coat and downstream lysis genes in Escherichia coli RNA phage GA is translationally coupled by means of an overlapping termination and initiation codon, UAAUG (30). In this case, only 25–30% of the ribosome participated in translating the downstream cistron and the major portion of the ribosome was released at the UAAUG. Translational coupling was also reported for the maize chloroplast atpB/E gene cluster, which has overlapping translation initiation and termination codons (AUGA), introduced into heterologous systems E. coli and Synecochocystis sp. PCC6803 (31). The chloroplast ATP synthase includes 3 β subunits and 1 ε subunit, suggesting that translation efficiency of the downstream atpE cistron is lower than that of its upstream atpB cistron. Together, if the ndhC/K mRNA is translated only by means of translational coupling, translation efficiency of the ndhK cistron should be low.
We demonstrated that translation efficiency of the downstream ndhK cistron is similar to, or slightly higher than, that of the upstream ndhC cistron (see Fig. 5C). Current models of translational coupling cannot explain this finding. Although the stoichiometry of the chloroplast NDH complex is not elucidated yet due to very low amounts and its low functional stability, coomassie blue-stained SDS/PAGE of a tobacco chloroplast NDH complex showed that NdhK is not a minor component but its amount looks comparable with several other subunits (NdhC could not be detected) (11). Based on the subunit compositions of cyanobacterial NDH-1 complexes (32), the stoichiometry of NdhK and NdhC in chloroplasts is likely to be 1:1. Because ndhC and ndhK are single copies in the chloroplast genome, the ndhK cistron should be translated at least as efficiently as the upstream cistron from the overlapping mRNA. If the ribosome participated in ndhC translation, moves 4 nt upstream after termination and resumes translation from the ndhK cistron, synthesis of NdhK should be no more than that of NdhC. It was suggested that the ribosome could undergo bidirectional diffusion after termination but reinitiation was very inefficient (33).
To meet the proper stoichiometry, translation of the ndhK cistron would need an additional pathway besides translational coupling. Surprisingly, removal of the ndhC 5′UTR and its AUG start codon still supported substantial translation of the ndhK cistron. This pathway is also termination codon-dependent. We favor the hypothesis that free ribosomes are loaded on somewhere within the ndhC coding region, migrate to the ndhC stop codon and start to translate the ndhK cistron. In conclusion, we propose that the downstream ndhK cistron is translated by 2 ways, a translational coupling event and a termination codon-dependent mechanism.
Materials and Methods
Plasmids.
The plasmids (S1-S4) for synthesizing mRNAs S1-S4 were constructed as described (23) by using fragments from the transcription start site of ndhCKJ to 18 nt after each of the 4 ndhK AUGs. Site-directed mutants of the 4 ndhK AUGs and the ndhC stop codon were prepared by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). To construct deletion mutants, fragments were amplified from plasmid S3 by PCR by using 1 of the upstream primers including the NcoI site adjacent to 1–3 nt deletion and the downstream primer including the NdeI site, and then the sequence between the NcoI and NdeI sites in plasmid S3 was replaced with 1 of these fragments. Plasmids psbA and psbB were made by replacing the ndhC 5′UTR of plasmid S3 with the psbA or psbB 5′UTR. Plasmid noUTR was constructed as plasmid S3 except the sequence from the third codon (+7) of ndhC cistron was used as the upstream primer. Plasmid ndhCKJ was prepared by replacing the StuI/EcoRV region of pHK309 with the PCR-amplified fragments from the transcription start site to a predicted termination site of ndhCKJ (203 nt downstream from the ndhJ termination codon). All constructs were verified by sequence analysis.
Messenger RNA Templates and in Vitro Translation.
Messenger RNA templates and tobacco chloroplast S30 extracts were prepared as described (23). Translation reaction was carried out at 28 °C for 2 h in a 60-μL reaction mixture with modifications as described in Results. Products were separated by 12.5% native-PAGE and the fluorescence intensity of mGFP was quantified as described (23). For fluorescent-fMet tRNAfmet labeling, 0.5-μL F-Detector tRNA solution (iNtRON Biotechnology) was added to 20-μL reaction mixture. After incubation, products were resolved by 15% denatured SDS/PAGE (23), and the fluorescent intensity was quantifies by a Typhoon9400 with 488-nm light and a 520BP40 filter (GE Healthcare).
Acknowledgments.
We thank Drs. Y. Yukawa and H. Kuroda for advice and discussion. This work was supported by the New Energy and Industrial Technology Development Organization (the Green Biotechnology Program) and Ministry of Education, Culture, Sports, Science, and Technology Grants-in-aids 19370021 and 19657072.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 2.Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- 3.Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts: The psbB operon from spinach. Eur J Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- 4.Robida MD, Merhige PM, Hollingsworth MJ. Proteins are shared among RNA-protein complexes that form in the 5′ untranslated regions of spinach chloroplast mRNAs. Curr Genet. 2002;41:53–62. doi: 10.1007/s00294-002-0283-1. [DOI] [PubMed] [Google Scholar]
- 5.Manuell A, Beligni MV, Yamaguchi K, Mayfield SP. Regulation of chloroplast translation: Interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem Soc Trans. 2004;32:601–605. doi: 10.1042/BST0320601. [DOI] [PubMed] [Google Scholar]
- 6.Baginsky S, Grossmann J, Gruissem W. Proteome analysis of chloroplast mRNA processing and degradation. J Proteome Res. 2007;6:809–820. doi: 10.1021/pr060473q. [DOI] [PubMed] [Google Scholar]
- 7.Marin-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynth Res. 2007;94:359–374. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- 8.Wobbe L, Schwarz C, Nickelsen J, Kruse O. Translational control of photosynthetic gene expression in phototrophic eukaryotes. Physiol Plant. 2008;133:507–515. doi: 10.1111/j.1399-3054.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 9.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohyama K, et al. Chloroplast gene organization deduced from complete seqeuence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. [Google Scholar]
- 11.Rumeau D, et al. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell. 2005;17:219–232. doi: 10.1105/tpc.104.028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu H, et al. CRR23/NdhL is a subunit of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 2008;49:835–842. doi: 10.1093/pcp/pcn058. [DOI] [PubMed] [Google Scholar]
- 13.Wakasugi T, et al. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci USA. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsubayashi T, et al. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: Determination of the splice sites. in ndhA and ndhB pre-mRNAs. Mol Gen Genet. 1987;210:385–393. doi: 10.1007/BF00327187. [DOI] [PubMed] [Google Scholar]
- 16.Whelan J, Young S, Day DA. Cloning of ndhK from soybean chloroplasts using antibodies raised to mitochondrial complex I. Plant Mol Biol. 1992;20:887–895. doi: 10.1007/BF00027160. [DOI] [PubMed] [Google Scholar]
- 17.Sugiura M, Hirose T, Sugita M. Evolution and mechanism of translation in chloroplasts. Annu Rev Genet. 1998;32:437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda H, et al. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 2007;48:1374–1378. doi: 10.1093/pcp/pcm097. [DOI] [PubMed] [Google Scholar]
- 19.Hiratsuka J, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Kindle KL, Stern DB. The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell. 1995;7:1295–1305. doi: 10.1105/tpc.7.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rimbault B, et al. Identification of the initiation codon for the atpB gene in Chlamydomonas chloroplasts excludes translation of a precursor form of the beta subunit of the ATP synthase. Mol Gen Genet. 2000;264:486–491. doi: 10.1007/s004380000332. [DOI] [PubMed] [Google Scholar]
- 22.Hirose T, Sugiura M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: A possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yukawa M, Kuroda H, Sugiura M. A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: Effect of pre-mRNA processing on translation in vitro. Plant J. 2007;49:367–376. doi: 10.1111/j.1365-313X.2006.02948.x. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura M, Sugiura M. Translation efficiencies of synonymous codons are not always correlated with codon usage in tobacco chloroplasts. Plant J. 2007;49:128–134. doi: 10.1111/j.1365-313X.2006.02945.x. [DOI] [PubMed] [Google Scholar]
- 25.Yukawa M, Tsudzuki T, Sugiura M. The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum) Plant Mol Biol. 2005;23:359–365. [Google Scholar]
- 26.Kuroda H, Maliga P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 2001;125:430–436. doi: 10.1104/pp.125.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 28.Jackson RJ, Kaminski A, Poyry TAA. Translational Control in Biology and Medicine. Plainview, NY: Cold Spring Harbor Lab Press; 2007. pp. 197–223. [Google Scholar]
- 29.Ahmadian G, Randhawa JS, Easton AJ. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 2000;19:2681–2689. doi: 10.1093/emboj/19.11.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inokuchi Y, Hirashima A, Sekine Y, Janosi L, Kaji A. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 2000;19:3788–3798. doi: 10.1093/emboj/19.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatenby AA, Rothstein SJ, Nomura M. Translational coupling of the maize chloroplast atpB and atpE genes. Proc Natl Acad Sci USA. 1989;86:4066–4070. doi: 10.1073/pnas.86.11.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, et al. Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem J. 2005;390:513–520. doi: 10.1042/BJ20050390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 34.Matsubayashi T. Nagayo, Japan: Nagoya University; 1990. Structure and expression of chloroplast ndh gene clusters (in Japanese) PhD dissertation. [Google Scholar]