Allaby et al. (1) investigate the phylogenetic consequences of crop domestication using forward simulations to mimic the domestication process. Their results suggest that monophyly of crop accessions is not a reasonable criterion for differentiating between multiple and single domestication events. Their simulations also produce the nonintuitive result that multiple domestication events more often result in monophyly than do single domestication events. We believe that these results are a function of their population-genetic model, which employs arbitrary population frequencies, excludes mutation, assumes unrealistically small effective population sizes, and incorporates neither their own protracted domestication model nor evolution of the wild population.
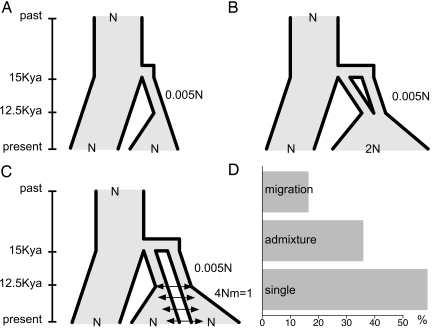
Here we present coalescent simulations (2) with entirely contrary results. We simulate codominant data with recombination, convert the data into dominant genotypes, and follow the phylogenetic methods outlined in ref. 1. We investigate a single domestication event, independent domestications with admixture (akin to ref. 1), and independent domestications with migration (Fig. 1). Under these models, single domestications are much more likely to appear as monophyletic (Fig. 1). Moreover, additional simulations (data not shown) suggest that our results are independent of the size of the domestication bottleneck and the model of population growth after domestication. Like Allaby et al. (1), we find that multiple domestication events appear monophyletic more often over time, but this process occurs much more slowly than claimed: To double the proportion of monophyletic phylogenies under the admixture model requires increasing the age of domestication by an order of magnitude (from 4N to 40N generations following the nonstandard scaling of ref. 1).
Fig. 1.
Coalescent models and observed monophyly. Simulations assume θ = 0.017, equivalent to N = 625,000 and μ = 6.8 × 10−9. Time is shown on the y-axis as thousands of years (Kya) before present. (A) Single domestication. (B) Two domestications with admixture. (C) Two domestications with migration. (D) Proportion of 1,000 simulations in which all domesticated taxa appear monophyletic.
Acknowledgments.
This work was partially supported by National Science Foundation Grant DBI0321467.
Footnotes
The authors declare no conflict of interest.
References
- 1.Allaby RG, Fuller DQ, Brown TA. The genetic expectations of a protracted model for the origins of domesticated crops. Proc Natl Acad Sci USA. 2008;105:13982–13986. doi: 10.1073/pnas.0803780105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]