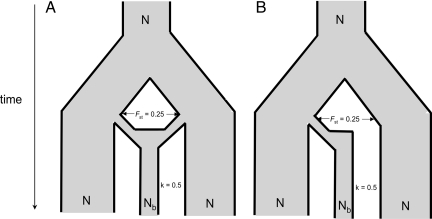
Ross-Ibarra and Gaut (1) use an inappropriate model with a single wild population, a single genetic locus system, and poor accountability for biological and archaeological data. Their results do not conflict with ours (2) and are easily explained. Our model (Fig. 1) has two structured wild populations, consistent with the current archaeological view of a multiregional agricultural transition (3) and genetic data (4). Our bottleneck uses established biological dimensions (5). In their model the same wild population is domesticated twice, and the resultant populations are kept separate for 2,500 years, a scenario that has no archaeological basis. Their single-locus system achieves monophyly in ways different from the multilocus systems used in real studies of crop origins and modeled by us. The 60% monophyly they observe results from allele extinction in the wild population. Further monophyly under this system is caused by fixation of derived mutations, this process of allele turnover equal to the mutation rate, in their case taking 2.4–24 million years. Mutation is obviously not a major influence over the relevant timescales, which is why we excluded it from our model. The differentiation of allele frequencies between populations, which causes monophyly in multilocus systems, is absent from their approach. Their artificially small wild population does not reflect the barley metapopulation that stretches from western Turkey to Pakistan but has a dramatic effect on the outcome of their simulation by making wild allele extinction an influential process when it should be negligible; hence, they observe monophyly by artifact.
Fig. 1.
Description of population simulation used by Allaby and colleagues (3). Simulations assume that N is very large, Nb is the bottleneck size, and k is the bottleneck severity expressed by Nb/d where d is the number of bottleneck generations.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ross-Ibarra J, Gaut BS. Multiple domestications do not appear monophyletic. Proc Natl Acad Sci USA. 2008;105:E105. doi: 10.1073/pnas.0809042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaby RG, Fuller DQ, Brown TA. The genetic expectations of a protracted model for the origins of domesticated crops. Proc Natl Acad Sci USA. 2008;105:13982–13986. doi: 10.1073/pnas.0803780105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown TA, Jones MK, Powell W, Allaby RG. The complex origins of domesticated crops. Trends Ecol Evol. 2008 doi: 10.1016/j.tree.2008.09.008. in press. [DOI] [PubMed] [Google Scholar]
- 4.Morrell PL, Clegg MT. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA. 2007;104:3289–3294. doi: 10.1073/pnas.0611377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Q, Zheng X, Luo J, Gaut B, Ge S. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: Severe bottleneck during domestication of rice. Mol Biol Evol. 2007;24:875–888. doi: 10.1093/molbev/msm005. [DOI] [PubMed] [Google Scholar]