Abstract
Cell proliferation and cell type specification are coordinately regulated during normal development. Cyclin E, a key G1/S cell cycle regulator, is regulated by multiple tissue-specific enhancers resulting in dynamic expression during Drosophila development. Here we further characterized the enhancer that regulates cyclin E expression in the developing peripheral nervous system (PNS) and show that multiple sequence elements are required for the full cyclin E PNS enhancer activity. We further show that Wg signaling is important for the expression of cyclin E in the sensory organ precursor (SOP) cells through two conserved TCF binding sites. Blocking Wg signaling does not completely block SOP cell formation but does completely block SOP cell proliferation as well as the subsequent differentiation.
Keywords: Cyclin E, Wingless (Wg), Peripheral Nervous System (PNS), Sensory Organ Precursor (SOP), Drosophila, TCF
1. Introduction
Cell proliferation is coordinated with patterning and cell differentiation during the development of multicellular organisms. There is increasing evidence suggesting that this coordination is mediated through the control of key cell cycle regulators by developmental mechanisms (Cayuso et al., 2006; Coffman, 2004; Edgar and Lehner, 1996; Gutierrez, 2005; Luo and Kessel, 2004; Sukhanova et al., 2007). In Drosophila, dissection of the enhancers that regulate the transcription of string (encoding a G2-M phase regulator), cyclin E (encoding a key G1-S phase regulator, that binds to and activates the Cdk2 protein kinase) and dacapo (encoding the p21/p27/p57 Cyclin E/Cdk2 inhibitor) during embryogenesis all revealed a complex cis-regulatory region containing tissue and stage specific enhancers (Jones et al., 2000; Lehman et al., 1999; Liu et al., 2002; Meyer et al., 2002; Sukhanova et al., 2007). These observations indicate that transcriptional control of key cell cycle regulators by developmental cues is likely to be an important mechanism that coordinates cell proliferation and patterning. However, the precise developmental mechanism that regulates the expression of these cell cycle regulators is still largely unknown.
Drosophila embryogenesis starts with 13 synchronous, syncytial division cycles, followed by three cycles of G2-regulated cell divisions. These early cell cycles have no G1 phase and Cyclin E activity is not limiting during these cycles. Consistent with this, maternal cyclin E mRNA was found throughout the embryo during cleavage cycles and was degraded in all cells except germ cells at cycle 14 while the zygotic expression of cyclin E was ubiquitously observed in all somatic cells of the epidermis (Richardson et al., 1993). Later, zygotic expression of cyclin E was abruptly down regulated in most of the cells at stage 11 but was dynamically expressed in the developing central nervous system (CNS), peripheral nervous system (PNS), and the endocycle domains (Knoblich et al., 1994; Richardson et al., 1993). The expression of cyclin E in the proliferating embryonic PNS cells precedes S phase in these cells, (Richardson et al., 1993) and Cyclin E is required for DNA replication in PNS cells, as revealed by the analysis of cyclin E mutant embryos (Knoblich et al., 1994). Furthermore, precise inactivation of Cyclin E/Cdk2 activity by Dacapo (Dap) is required for normal cell cycle exit (de Nooij et al., 1996; Lane et al., 1996), and ectopic expression of Cyclin E leads to precocious S phase entry and developmental defects (Li et al., 1999; Richardson et al., 1995). Thus, appropriate regulation of Cyclin E expression and of Cyclin E/Cdk2 activity is important for limiting cell proliferation.
The Drosophila PNS contains several distinct types of sensory organs, including the external sensory (es) organs and the chordotonal (ch) organs (for a review, see (Jan and Jan, 1993). A typical es or ch organ is composed of four components: a neuron and three supporting cells. The different components of the es and ch organs are derived from divisions of the sensory organ precursor (SOP) cells, which can be identified by the expression of the protein Senseless (Sens) (Nolo et al., 2000). The SOPs undergo two to three rounds of asymmetric cell division to give rise to a neuron, a sheath cell, which expresses the protein Prospero (Pros), and two accessory cells to form the es organs and the ch organs (Bodmer et al., 1989; Doe et al., 1991; Gho et al., 1999; Hartenstein and Posakony, 1989; Reddy and Rodrigues, 1999; Roegiers and Jan, 2004; Wang and Chia, 2005).
In this report, we characterize the enhancer that regulates zygotic cyclin E expression in the embryonic PNS precursor cells and show that multiple sequences elements are required for full PNS enhancer activity. We show that Wingless (Wg) signaling is required for cyclin E expression in the developing embryonic PNS precursor cells and identify two TCF binding sites that are required for full PNS enhancer activity. These results indicate that Wg signaling directly regulates cyclin E expression in the developing embryonic PNS precursor cells.
2. Results and discussion
2.1. Deletion analysis of the PNS enhancer of cyclin E
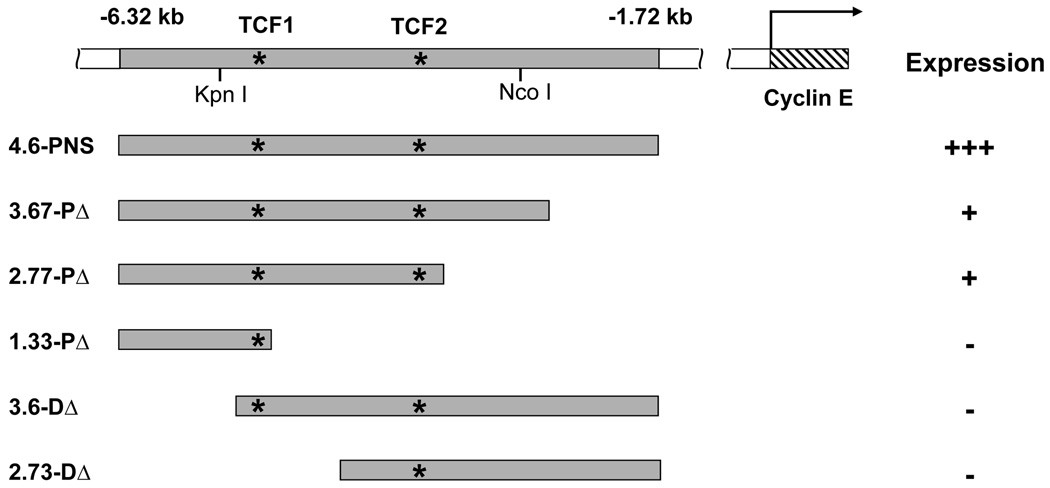
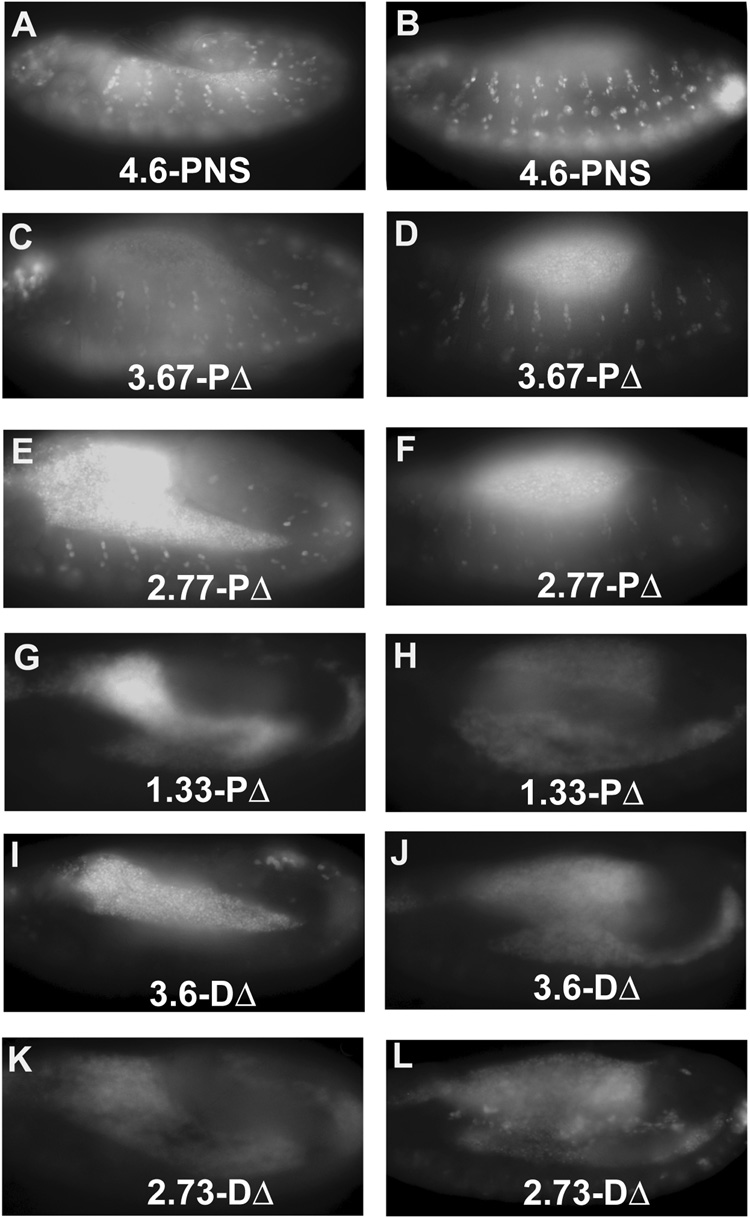
To begin dissection of the promoter region required for the regulation of cyclin E gene expression in the developing embryonic PNS, a DNA fragment (4.6-PNS, Fig. 1), containing the PNS enhancer defined to be within a 2.6 kb KpnI-NcoI fragment (Jones et al., 2000), was cloned into the GFP-expressing pH-Stinger vector (Barolo et al., 2000) and transgenic flies were generated. As expected, expression of the GFP reporter was observed in the PNS region of stage 12 embryos (Fig. 1 and 2A). Antibody staining revealed that all the GFP-positive cells also expressed Sens (Nolo et al., 2000), confirming that the GFP reporter is indeed expressed in PNS SOP cells (Supplemental Fig. 1). The GFP reporter was also observed in stage 13 embryos (Fig. 1 and Fig. 2B) as well as in later stage embryos when the cyclin E message was no longer detected. This is likely due to the extended stability of the nuclear GFP protein as reported previously (Sukhanova et al., 2007). Deletion of the PNS enhancer from the proximal end leads to a gradual decrease in the expression of GFP reporter (Fig. 1, 3.67-PΔ, 2.77-PΔ, 1.33-PΔ and Fig. 2C–H). In contrast, deletions from the distal end completely abolished GFP reporter expression (Fig. 1, 3.6-DΔ, 2.73-DΔ, Fig. 2I–L). These results indicate that both the proximal and the distal end sequences of the 4.6-PNS enhancer contribute to the enhancer activity, with the distal end sequences playing an essential role while the proximal end sequence augments the enhancer activity.
Fig 1. A diagram of the cyclin E genomic region and of the different cyclin E PNS enhancer constructs.
cyclin E genomic fragments were used to generate a series of nuclear GFP (nGFP) reporter constructs. The numbers on top represent the locations of the cyclin E genomic sequence relative to the translation start of the type I cyclin E transcript. The location of the Kpn I and Nco I restriction sites, which defines the PNS enhancer in the work of Jones et al. (2000) are indicated. Asterisks indicate the locations of the two TCF-binding sites in the cyclin E PNS enhancer. The presence or absence of GFP expression is indicated by plus or minus signs, respectively. The number of plus signs indicates the level of GFP expression relative to the full-length 4.6-PNS enhancer.
Fig. 2. GFP reporter expression from different cyclin E PNS enhancer constructs.
GFP reporter expression of the indicated cyclin E transgenic constructs at embryonic stages 11/12 (A, C, E, G, I, and K) or stages 12/13 (B, D, F, H, J, and L) are shown. Multiple insertions of each deletion construct were examined and no significant differences were observed between insertions of the same constructs. The level of GFP expression is summarized in Fig. 1.
2.2. Wg regulates cyclin E PNS expression and the cyclin E 4.6-PNS enhancer activity
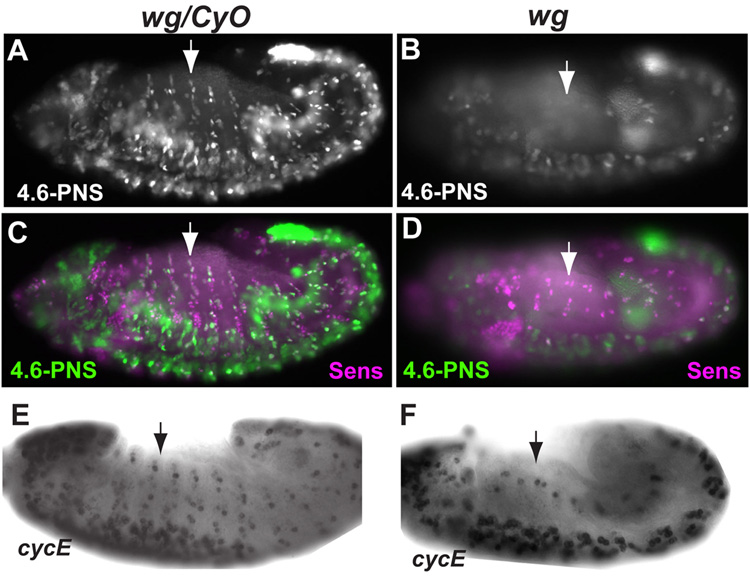
To determine the developmental mechanisms that regulate cyclin E expression in PNS, we examined the activity of cyclin E PNS enhancer in several mutant backgrounds. Interestingly, 4.6-PNS GFP reporter expression in PNS precursor cells were mostly absent in the wg mutants, even though 4.6-PNS GFP reporter expression in the CNS cells can still be observed (Figs 3A and B). Double staining with anti Sens antibody showed that a large number of the Sens positive cells were expressing GFP reporter from the 4.6-PNS enhancer in WT embryos, while only a few Sens positive cells express 4.6-PNS GFP reporter in wg mutant embryos (Fig. 3C and D). These results suggest that Wg signaling regulates cyclin E expression in the developing PNS. To further test this idea, in situ hybridization of cyclin E message was carried out. As shown in Figs. 3E and F, cyclin E expression in the developing PNS but not CNS was significantly decreased in wg mutants (see also supplemental Fig. 2). We conclude from these observations that Wg signaling plays an important role in the expression of cyclin E in the developing PNS cells.
Fig. 3. The effect of wg mutation on cyclin E expression in the embryonic PNS.
(A, B) Expression of the GFP reporter from the 4.6-PNS constructs was significantly reduced in the wg mutant embryos (B) as compared to that in the control (wg/CyO) embryos (A). (C, D) Double labeling of 4.6-PNS GFP reporter expression and Sens revealed large number of Sens positive cells express GFP reporter in wg/CyO embryos (C) while very few Sens positive cells express GFP reporter in wg mutant embryos (D). (E, F) Whole-mount in situ hybridization showing expression of zygotic cyclin E in the developing PNS. (E), wg/CyO embryos show normal cyclin E expression in the PNS. (F), wg mutant embryos exhibit significantly reduced PNS cyclin E expression.
2.3. Wg signaling does not block PNS progenitor cell determination but does block PNS progenitor cell proliferation and differentiation
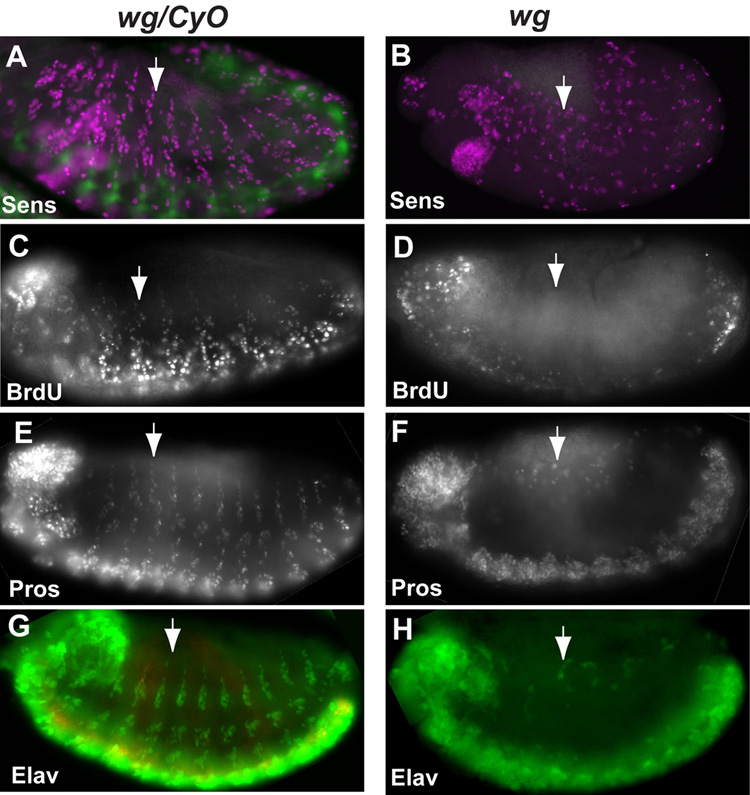
As described above, the 4.6-PNS reporter specifically targets GFP reporter expression in the developing PNS precursors, which can be identified by Sens antibody. To determine if the effect of wg mutation on the expression of cyclin E was due to its effect on SOP determination, we examined the embryonic PNS SOP cells by Sens staining. As shown in Figs. 4A and B, significant number of Sens positive cells were still observed in wg mutants, suggesting that the failure to express cyclin E from the 4.6-PNS enhancer in wg mutant embryos is not likely simply due to a failure in SOP determination.
Fig. 4. Wg signaling is required for cell proliferation in the developing PNS and for the development of neurons and Pros expressing cells in the PNS.
(A, B) Significant numbers of SOP cells were observed in wg mutant (B) embryos. PNS SOP cells were identified by Senseless staining shown in magenta. Homozygous wg mutant embryos were identified by the absence of β-gal expression from the ftz-lacZ construct on the CyO balancer shown in green. (C, D) wg mutation blocks cell proliferation in the developing PNS. BrdU incorporation was not observed in the PNS region of the wg mutant embryos (D) but was observed in the wg/CyO embryos (C). It should be noted that BrdU incorporation in the head and the ventral CNS region was still observed in wg mutant embryos. (E–H) wg mutation blocks the differentiation of neurons and Pros expression cells in the developing PNS. Sheath cells in the es and scolopale cells in the ch organs are the PNS cell types that accumulate high levels of Pros. Very few Pros-expressing cells were observed in wg mutants (F) as compared to wg/CyO embryos (E). Similarly, very few neurons identified by Elav staining were observed in wg mutant (H) as compared with that of the wg/CyO embryos (G). Homozygous wg mutant embryos were identified by the absence of β-gal expression from the ftz-lacZ construct on the CyO balancer shown in red in G and H. Arrows point to a PNS segment in the embryos.
To test if decreased cyclin E expression in wg mutants blocks cell proliferation in developing PNS cells, BrdU incorporation in the developing PNS cells was determined. As shown in Fig. 4C, D, BrdU incorporation in the developing PNS cells were completely blocked in wg mutants while BrdU incorporation in the developing CNS and the head region can still be observed in wg mutant embryos (Fig. 4C, D). These observations are consistent with the observation that wg mutation does not affect cyclin E expression in these regions (Fig. 3E, F).
To further characterize the effect of wg mutation on PNS development, we examine the expression of Pros and Elav in developing PNS. Expression of Pros in the PNS, which labels the sheath cell of the es organ and the scolopale cell of the ch organ (Doe et al., 1991; Vaessin et al., 1991), was significantly decreased in wg mutant embryos as compared with those in WT embryos (Figs. 4E and F). Similarly, expression of Elav in PNS, which labels the PNS neurons, was also largely absent in the PNS region (Fig. 4G and H). These observations showed that Wg signaling is required for the proliferation of PNS progenitor cells and is required for the proper differentiation of multiple cell types in the developing PNS.
2.4. Two conserved TCF-binding sites identified in the 4.6-PNS enhancer are required for PNS enhancer activity
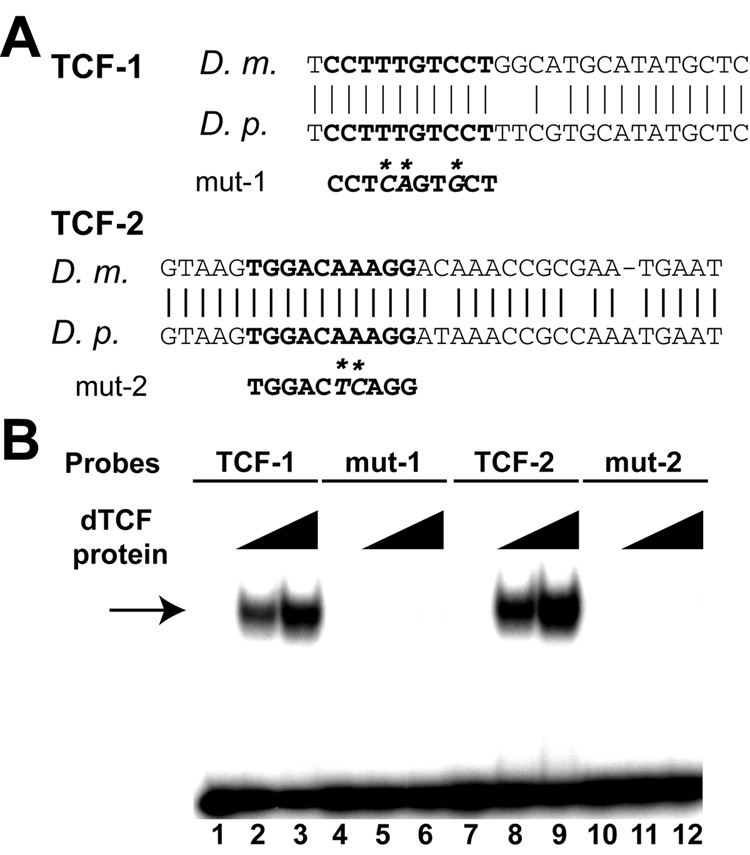
To examine the possibility that Wg signaling regulates cyclin E PNS expression directly, we used bioinformatics tools to identify putative binding sites for the Wg pathway transcription factor TCF in the 4.6-PNS enhancer that are conserved in different Drosophila species. Two TCF-binding sites (−5101 to −5110 and −3768 to −3777) were identified that are conserved between different species of Drosophila (Fig. 5A). To determine if dTCF protein can bind to the observed TCF-binding sites, gel shift assays were carried out using purified His-tagged dTCF protein. As shown in Fig. 5B, probes containing either binding site TCF-1 or TCF-2 can bind strongly to dTCF protein. Furthermore, mutations in the conserved TCF-binding sites completely abolished dTCF binding, indicating that dTCF protein bound specifically to TCF-1 and TCF-2 sites.
Fig. 5. Two conserved TCF-binding sites in the 4.6-PNS enhancer.
(A) Alignment of sequences around the conserved TCF-binding sites between D. melanogaster and D. pseudoobscura. The bold letters represent the TCF-binding sites in the 4.6-PNS enhancer. The sequences of the mutant TCF sites are shown below the binding sites and base pair changes are indicated by “*”. (B) Gel shift assay showing that the Drosophila TCF protein bound strongly and specifically to the wild type TCF1 (lanes 2 and 3) and TCF-2 (lanes 8, 9). In contrast, no binding was observed when the TCF sites were mutated (mut-1 and mut-2, lanes 5, 6 and 11, 12). No dTCF protein were added in lanes 1, 4, 7, and 10; half the amount of dTCF protein was added to lanes 2, 5, 8, 11 compared to lanes 3, 6, 9, and 12. An arrow points to the dTCF-DNA complex. The sequences of TCF and mutated TCF probes are shown in Materials and Methods.
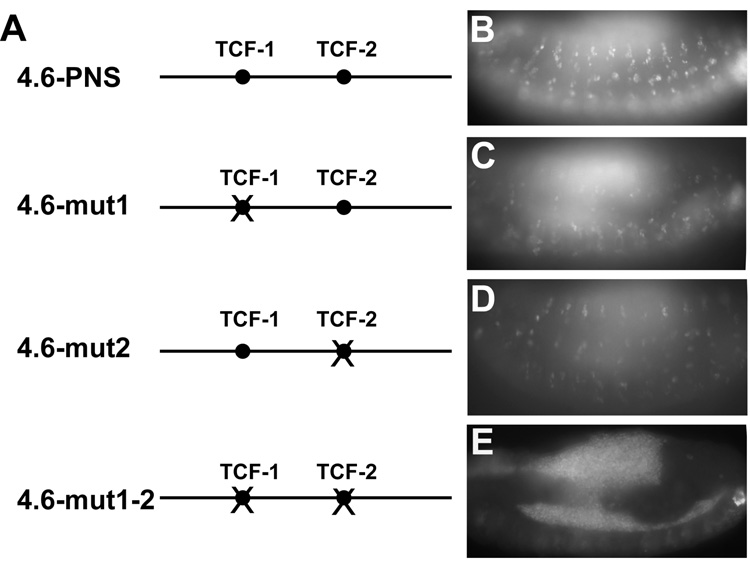
To determine if the observed TCF-binding sites are important for regulating cyclin E 4.6-PNS enhancer activity, the effect of mutating the TCF-binding sites on the activity of the 4.6-PNS enhancer was determined. As shown in Fig. 6, mutations of either the TCF-1 (4.6-mut1) or the TCF-2 sites (4.6-mut2) partially reduced 4.6-PNS enhancer activity (Figs. 6A–D). Furthermore, mutation of both TCF-binding sites (4.6-mut1-2) completely abolished cyclin E 4.6-PNS enhancer activity (Fig. 6E). Taken together, these results indicate that Wg signaling directly regulates the expression of cyclin E in the developing embryonic PNS cells through these two TCF-binding sites.
Fig 6.
(A) A diagram of the 4.6-PNS enhancer and the 4.6-PNS enhancers with each of the TCF binding sites mutated either alone or in combination. Filled circles represent the TCF binding sites, and crossed (x) circles indicate mutated TCF binding sites in the PNS enhancer. (B–E) Both TCF binding sites contribute to the full activity of the 4.6-PNS enhancer. Transgenic lines carrying the 4.6-PNS enhancer with mutated TCF-1 binding site (C) or the TCF-2 binding site alone (D) show decreased GFP reporter expression when compared to WT 4.6-PNS enhancer (B). Transgenic lines carrying the 4.6-PNS enhancer with mutations of both TCF binding sites only show background levels of GFP reporter expression (E).
2.5. Conclusions and Discussion
Taken together, our results indicate that Wg signaling is required for the expression of cyclin E in proliferating PNS precursor cells. In the absence of Wg signaling, the expression of cyclin E is dramatically reduced, resulting in an inhibition of PNS precursor cell proliferation and the determination of different cell types in the developing PNS.
Our results also revealed that cyclin E expression in developing PNS precursor cells is regulated by a large enhancer containing multiple sequence elements, including two TCF-binding sites that mediate the regulation by Wg signaling. While these TCF-binding elements are essential for the activity of the PNS enhancer, proximal and distal elements in the 4.6-PNS sequence appear to be important for full activity. The importance of Wg in the regulation of the PNS expression of cyclin E is supported by the fact that wg mutant embryos displayed decreased cyclin E expression in the developing PNS cells. This reduction in cyclin E expression in wg mutant embryos was accompanied by an inhibition of BrdU incorporation in the developing PNS, and an inhibition of the determination of the Pros and Elav expression cells in the developing PNS. It is possible that the block in differentiation into the Pros and Elav positive cells is a consequence of the inhibition of cyclin E expression or perturbations to the cell proliferation as has been previously observed (reviewed by Chia and Prokopenko, 2005). However it is also possible that the observed differentiation block in PNS is due to a function of Wg that is independent of PNS cell proliferation. Further studies will be needed to resolve this issue.
In addition to wg, a number of other mutations such as achaete/scute (ac/sc) complex and da have also been reported to block PNS precursor proliferation and affect the expression of several cell cycle genes (Bodmer et al., 1989; Hassan and Vaessin, 1997). Ac/Sc complex proteins and Da are bHLH proteins that are important in all aspects of es-PNS precursor differentiation while bHLH protein Atonal (ato) and Da are required for all aspects of ch-PNS precursor development. Recent studies of the expression of the Cdk inhibitor Dap during cell type specification revealed that Dap expression is directly regulated by the same developmental mechanisms that control the differentiation of these cell types (Sukhanova et al., 2007). Therefore it will be interesting to test if bHLH proteins such as Da also directly regulate cyclin E expression in the developing PNS cells.
3. Experimental procedures
3.1. Fly stocks
wg1–17 mutant flies, a molecular null mutant described previously (van den Heuvel et al., 1993), was obtained from the Bloomington Stock center and used.
3.2. Gel shift assay
The HMG (High Mobality group) DNA binding domain of dTCF was amplified by PCR (van de Wetering et al., 1997), and cloned into the pET15b vector. The pET15b-dTCF plasmid was transformed into BL21(DE3) to produce His-tagged dTCF protein. His-tagged dTCF protein was purified by Ni-NTA column and was used in gel shift experiments as described previously (Duman-Scheel et al., 2002). WT or TCF-binding site-mutated nucleotides were used as probes in gel shift experiments, and the sequences of wild type (WT) and TCF-binding site mutated probes are:
WT TCF1: 5’ GCATGCCAGGACAAAGGAGCCGGACCAG 3’
Mut TCF1: 5' GCATGCCAGCACTGAGGAGCCGGACCAG 3’
WT TCF2: 5’ GGT TTG TCCTTTGTCCACTTACCCTCC 3’
Mut TCF2: 5’ GGT TTG TCCTGAGTCCACTTACCCTCC 3’
The dTCF-binding sites are underlined and the mutated bases are shown in italics.
3.3. Generation of transgenic flies
The 4.6 kb enhancer (4.6-PNS) and the different deletions of the cyclin E PNS enhancer region were amplified by PCR from the P1 clone DS07108 (BDGP Resources) and were cloned into the BamH1 site of the pH-Stinger vector (Barolo et al., 2000) to generate transgenic lines using standard P-element mediated germ line transformation (Spradling and Rubin, 1982). To generate the 4.6-PNS enhancer with mutated dTCF binding sites, the same TCF-binding site mutations used in the gel shift experiments were introduced into the 4.6-PNS enhancer. The WT and the TCF-binding site-mutated constructs were confirmed by DNA sequencing and cloned into the pH-Stinger vector to generate transgenic lines. At least two different transgenic lines were examined for each construct with enhancer deletions and mutated TCF-binding sites. No significant variations were observed between different insertions of the same constructs.
3.4. in situ hybridization, BrdU incorporation, and antibody staining
The antibodies used in this paper are: rabbit anti β-gal antibody (Cappel), rat anti Elav (7E8A10), 22C10, and mouse anti Prospero antibodies (Hybridoma Bank), mouse anti BrdU (Becton Dickinson), and guinea pig anti Senseless antibody (Nolo et al., 2000). wg mutant embryos were identified by the lack of β-gal antibody staining. All embryos were mounted in 80% glycerol in 1xPBS.
For in situ hybridization, Digoxigenin-labeled DmcycE antisense RNA probe was prepared by in vitro transcription reaction. Properly aged embryos were fixed with 4% Paraformaldehyde over heptane and devitellinized with methanol. Hybridization was carried out at 65°C as described (Du and Dyson, 1999).
For BrdU incorporation, properly aged embryos were pulse-labeled for 15 min in Schneider’s medium containing 1 mg/ml BrdU as described previously (Du and Dyson, 1999). BrdU was incorporated into replicating nuclei and the pulse was subsequently fixed and detected by anti-BrdU mouse monoclonal antibody (Becton Dickinson, 1:50 dilution).
Supplementary Material
The 4.6-PNS cyclin E enhancer drives expression in a subset of SOP cells in the PNS during embryonic development. Low (A) and high (B–D) magnification views of embryos carrying the 4.6-PNS transgene stained with the anti-Sens antibody. All of the GFP expressing cells (green in A, B, and white in C) correspond to the SOP cells that express Senseless (magenta in A, B, and white in D). Arrowheads in B–D point to GFP-expressing cells that also express Sens while arrows point to Sens expression cells that do not have GFP expression. The rectangle in A indicates the area shown in B–D.
In situ hybridization detecting cyclin E message (in blue) and antibody staining detecting β-gal protein (in brown). Significantly reduced cyclin E expression was observed in wg mutant embryos (B) as compared to wg/CyO control embryos (A). Homozygous wg mutant embryos were identified by the absence of β-gal staining from the ftz-lacZ construct on the CyO balancer.
Acknowledgements
We would like to thank Dr. James Posakony for the pH-Stinger vector, Dr. Hugo J. Bellen for providing the anti-Senseless antibody, Dr. H.C. Clevers for the dTCF cDNA, and the Developmental Studies Hybridoma Bank for providing Elav, 22C10, and Prospero antibodies. We also want to thank the Berkeley Drosophila Genome Project for providing the cyclin E P1 genomic clone, the Bloomington fly stock center for fly stocks and Gabe Gordon for reading the manuscript. This work was supported by grants from the American Cancer Society and National Institute of Health to W. Du. W. Du is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–728. 730–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Carretto R, Jan YN. Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron. 1989;3:21–32. doi: 10.1016/0896-6273(89)90112-8. [DOI] [PubMed] [Google Scholar]
- Cayuso J, Ulloa F, Cox B, Briscoe J, Marti E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- Chia W, Prokopenko SN. Cyclin E at the centre of an identity crisis. Nat Cell Biol. 2005;7:3–5. doi: 10.1038/ncb0105-3. [DOI] [PubMed] [Google Scholar]
- Coffman JA. Cell cycle development. Dev Cell. 2004;6:321–327. doi: 10.1016/s1534-5807(04)00067-x. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Du W, Dyson N. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. Embo J. 1999;18:916–925. doi: 10.1093/emboj/18.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman-Scheel M, Weng L, Xin S, Du W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature. 2002;417:299–304. doi: 10.1038/417299a. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Lehner CF. Developmental control of cell cycle regulators: a fly's perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- Gho M, Bellaiche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573–3584. doi: 10.1242/dev.126.16.3573. [DOI] [PubMed] [Google Scholar]
- Gutierrez C. Coupling cell proliferation and development in plants. Nat Cell Biol. 2005;7:535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- Hassan B, Vaessin H. Daughterless is required for the expression of cell cycle genes in peripheral nervous system precursors of Drosophila embryos. Dev Genet. 1997;21:117–122. doi: 10.1002/(SICI)1520-6408(1997)21:2<117::AID-DVG1>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Jan Y, Jan L. The Peripheral Nervous System. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Vol. 2. Cold Spring Harbor Laboratory Press; 1993. pp. 1207–1244. [Google Scholar]
- Jones L, Richardson H, Saint R. Tissue-specific regulation of cyclin E transcription during Drosophila melanogaster embryogenesis. Development. 2000;127:4619–4630. doi: 10.1242/dev.127.21.4619. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- Lehman DA, Patterson B, Johnston LA, Balzer T, Britton JS, Saint R, Edgar BA. Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development. 1999;126:1793–1803. doi: 10.1242/dev.126.9.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Pazdera TM, Minden JS. Drosophila embryonic pattern repair: how embryos respond to cyclin E-induced ectopic division. Development. 1999;126:2299–2307. doi: 10.1242/dev.126.10.2299. [DOI] [PubMed] [Google Scholar]
- Liu TH, Li L, Vaessin H. Transcription of the Drosophila CKI gene dacapo is regulated by a modular array of cis-regulatory sequences. Mech Dev. 2002;112:25–36. doi: 10.1016/s0925-4773(01)00626-8. [DOI] [PubMed] [Google Scholar]
- Luo L, Kessel M. Geminin coordinates cell cycle and developmental control. Cell Cycle. 2004;3:711–714. [PubMed] [Google Scholar]
- Meyer CA, Kramer I, Dittrich R, Marzodko S, Emmerich J, Lehner CF. Drosophila p27Dacapo expression during embryogenesis is controlled by a complex regulatory region independent of cell cycle progression. Development. 2002;129:319–328. doi: 10.1242/dev.129.2.319. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Rodrigues V. A glial cell arises from an additional division within the mechanosensory lineage during development of the microchaete on the Drosophila notum. Development. 1999;126:4617–4622. doi: 10.1242/dev.126.20.4617. [DOI] [PubMed] [Google Scholar]
- Richardson H, O'Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Richardson HE, O'Keefe LV, Reed SI, Saint R. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development. 1993;119:673–690. doi: 10.1242/dev.119.3.673. [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Sukhanova MJ, Deb DK, Gordon GM, Matakatsu MT, Du W. Proneural basic helix-loop-helix proteins and epidermal growth factor receptor signaling coordinately regulate cell type specification and cdk inhibitor expression during development. Mol Cell Biol. 2007;27:2987–2996. doi: 10.1128/MCB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Klingensmith J, Perrimon N, Nusse R. Cell patterning in the Drosophila segment: engrailed and wingless antigen distributions in segment polarity mutant embryos. Dev Suppl. 1993:105–114. [PubMed] [Google Scholar]
- Wang H, Chia W. Drosophila neural progenitor polarity and asymmetric division. Biol Cell. 2005;97:63–74. doi: 10.1042/BC20040064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 4.6-PNS cyclin E enhancer drives expression in a subset of SOP cells in the PNS during embryonic development. Low (A) and high (B–D) magnification views of embryos carrying the 4.6-PNS transgene stained with the anti-Sens antibody. All of the GFP expressing cells (green in A, B, and white in C) correspond to the SOP cells that express Senseless (magenta in A, B, and white in D). Arrowheads in B–D point to GFP-expressing cells that also express Sens while arrows point to Sens expression cells that do not have GFP expression. The rectangle in A indicates the area shown in B–D.
In situ hybridization detecting cyclin E message (in blue) and antibody staining detecting β-gal protein (in brown). Significantly reduced cyclin E expression was observed in wg mutant embryos (B) as compared to wg/CyO control embryos (A). Homozygous wg mutant embryos were identified by the absence of β-gal staining from the ftz-lacZ construct on the CyO balancer.