Abstract
Homeobox genes encode transcription factors whose expression organizes programs of development. A number of homeobox genes expressed in reproductive tissues have been identified recently, including a colinear cluster on the X chromosome in mice. This has led to an increased interest in understanding the role(s) of homeobox genes in regulating development of reproductive tissues including the testis, ovary, and placenta. Here we report the identification and characterization of a novel homeobox gene of the paired-like class on the X chromosome distal to the reproductive homeobox (Rhox) cluster in mice. Transcripts are found in the testis and ovary as early as 13.5 days post-coitum (dpc). Transcription ceases in the ovary by 3 days post-partum (dpp), but continues in the testis through adulthood. The Rhox13 gene encodes a 25.3 kDa protein expressed in the adult testis in germ cells at the basal aspect of the seminiferous epithelium.
1. Introduction
At its most basic level, the process of development is regulated by changes in gene expression. Homeobox genes encode transcription factors that play an important role in the regulation of various developmental events including embryo patterning, limb formation, and organogenesis. The members of this diverse group are characterized by a well-conserved 60 amino acid homeodomain containing 3 alpha helices involved in DNA-binding (Gehring et al., 1994). Efforts to classify homeobox genes have focused not only on peptide sequence similarities within the homeodomain, but motifs outside the homeodomain (Burglin, 1994). One such class, the paired class of homeobox genes, is characterized by a 130 amino acid DNA binding “paired domain” in the N-terminal portion of the protein and a conserved serine residue at position 50 in the third, or “recognition helix” of the homeodomain (Burglin, 1994). Changes at this position alter the DNA sequence binding specificity (Treisman et al., 1989). Genes with homeodomains similar to the paired class that lack a paired domain are grouped into the paired-like class.
A large cluster of reproductive homeobox genes (Rhox) of the paired-like class was identified recently on the X chromosome in mice (MacLean et al., 2005) containing all of the described reproductive homeobox genes except Esx1 (Li et al., 1997; Branford et al., 1997) and Arx (Miura et al., 1997). Since the initial report of 12 Rhox genes, another 20 have been identified within the cluster, all of which are apparent duplications of Rhox2, Rhox3, and Rhox4 in mouse and rat (Morris et al., 2006; MacLean et al., 2006; Wang and Zhang, 2006, Jackson et al., 2006). The genomic organization of the Rhox gene subfamily is conserved, with the homeodomain sequence being interrupted by two introns between residues 31/32 and 46/47. In the mouse, all of the genes characterized to date are expressed to some degree in placenta, and to varying degrees in testis, ovary, and other tissues (MacLean et al., 2005). Human reproductive homeobox genes are not organized in a cluster as in mice and rats, and only three genes have been described to date: ESX1L (Fohn and Behringer, 2001), RHOXF1 (previously called PEPP1 or OTEX) (Wayne et al., 2002; Geserick et al., 2002) and RHOXF2 (previously called PEPP2) (Wayne et al., 2002). ESX1L is expressed in the testis and the placenta, while RHOXF2 is restricted to the testis. RHOXF1 is expressed in testis, ovary, and prostate, and at much lower levels in epididymis, mammary gland, and uterus. Neither RHOXF1 nor RHOXF2 was expressed in full-term placenta (Wayne et al., 2002; Geserick et al., 2002).
Three of the reproductive homeobox genes (Esx1, Rhox5, and Rhox9) have been disrupted in mice (Li and Behringer, 1998; Pitman et al., 1998; Takasaki et al., 2001). All of the mutant mice are viable and fertile, although male fertility in Rhox5 null mice is reduced (MacLean et al., 2005) and Esx1 null mice have low birth weights (Li and Behringer, 1998). The overlapping expression patterns of reproductive homeobox genes (MacLean et al., 2005) and the viability and fertility of the mutant models listed above suggest a degree of functional redundancy in the reproductive homeobox genes. Therefore, characterization of additional homeobox genes in this region of the X chromosome will be necessary to provide a foundation to dissect the interactions of these genes in the development of reproductive tissues. To this end, we have identified and characterized a novel X-linked homeobox gene transcribed in prenatal and neonatal ovary and in prenatal, neonatal, and postnatal testis in the mouse and translated in cells at the periphery of the seminiferous epithelium in the adult testis. The presence of a homeodomain, lack of a paired domain, location on the X chromosome, and expression pattern indicate this gene is a germ cell-line specific member of the Rhox cluster.
2. Materials and methods
2.1 Identification and sequence determination
OmniViz software (Version 3.9.0, OmniViz, Inc) was used to cluster and mine a database of expressed sequence tags (ESTs) from seven mouse spermatogenic cell cDNA libraries and one Sertoli cell cDNA library (McCarrey et al., 1999) to identify putative transcription factors. An EST of interest from a cDNA library (dbEST Library ID. 11285) prepared with mRNA from a highly enriched population of primitive type A spermatogonia isolated from testes of 6 day-old mice was found to have high identity with a region of a previously uncharacterized cDNA, 1700123J19Rik.
Verification of the coding and promoter sequences of Rhox13 was performed by RT-PCR using 100 ng of adult mouse testis cDNA (Clontech) and 30 ng of mouse genomic DNA amplified for 35 cycles in a reaction containing: 10 mM Gene Amp PCR buffer II, 2 mM MgCl2, 1.5 mM dNTPs, 1.25 µM each primer, and 1.25 U Taq Gold (Perkin-Elmer). Primer sequences used were: 1) 5’-TGAAGGATAGGCAGCCCATG, 2) 5’-CAGTCCAGACGGAAGCCAG, 3) 5’-CTGCACAGTCTACTGGTCG, 4) 5’-CTAGATGTTCACACAAGAATTCTG.
Sequencing of amplified PCR products was performed on an ABI 3100 Avant (Applied Biosystems) with Big Dye Ver 1.1 terminator chemistry (Applied Biosystems) according to the manufacturer’s instructions.
2.2 Northern blot and RT-PCR analysis
All animal procedures were approved by the National Institute of Environmental Health Sciences (NIEHS) Animal Care and Use Committee. Tissues were dissected from CD-1 mice and immersed immediately in RNAlater (Ambion). RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Northern analysis was performed using standard methods (10 µg total RNA/lane) and the blot was probed with a radiolabeled (α-32P-dCTP) 582 bp fragment of Rhox13 cDNA (nt 472–1054, Fig. 1) generated by PCR (primers: 5’- ACCACAATTACTACATTGTGGAG and 5’-CGCTCTCCGATTGGTAAACC).
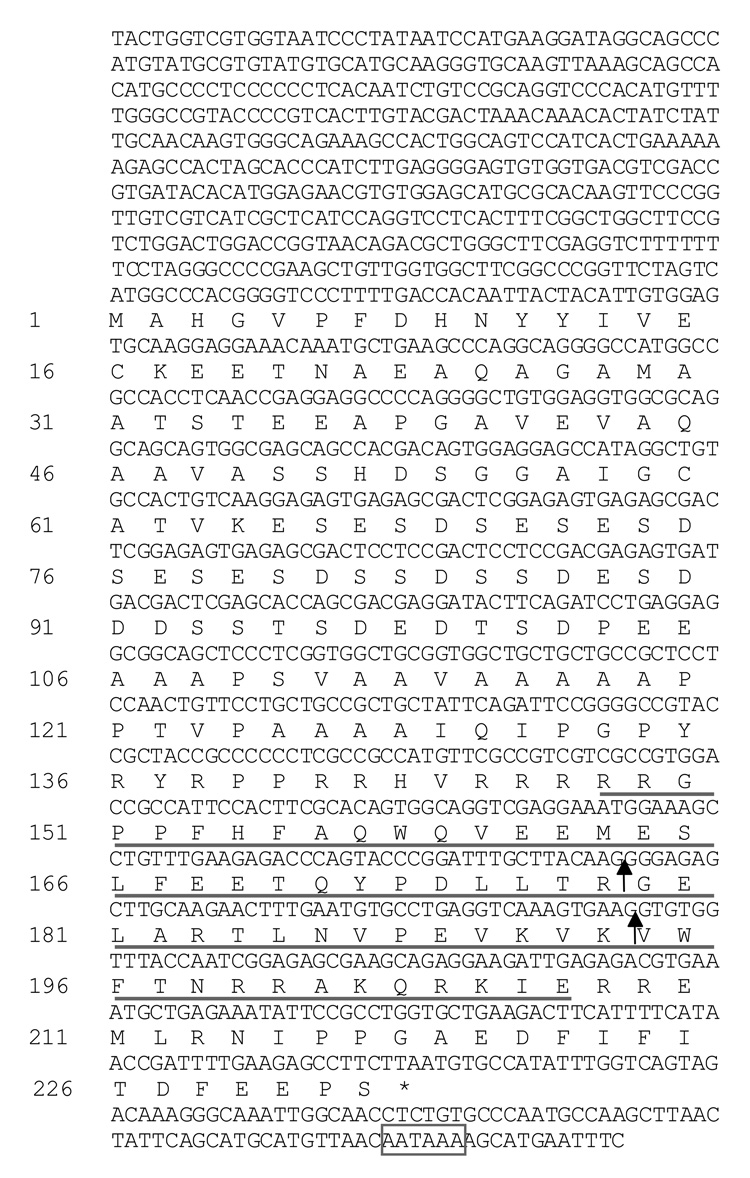
Figure 1. The nucleotide and deduced amino acid sequences of the Rhox13 cDNA.
The approximate 5’ end of the cDNA was determined by RT-PCR and northern blotting. The positions of the two introns are marked by solid arrows. The homeodomain is underlined, an asterisk indicates the stop codon, and the putative polyadenylation signal is boxed.
For RT-PCR, 1 µg of total RNA from each tissue sample was reverse transcribed using oligo dT primers and MuLV reverse transcriptase (Applied Biosystems). 50 ng of cDNA was then amplified for 35 cycles using the conditions described above with primers (5’-ACCGCCATTCCACTTCGCAC and 5’-ATTGGGCACAGAGGTTGC) that span both introns of Rhox13 to control for amplification of genomic DNA. Equal loading was confirmed by coincident amplification of the cytoplasmic beta actin (Actb) housekeeping gene (primers: 5’-TCCGATGCCCTGAGGCTCTTTTC and 5’-CTTGCTGATCCACATCTGCTGGAA).
2.3 Antiserum production, western blot analysis, and immunoprecipitation
A synthetic peptide was generated (Sigma Genosys) corresponding to the first 24 amino acids (C-MAHGVPFDHNYYIVEKEETNAEAQ) of the putative coding sequence of Rhox13 along with a N-terminal cysteine that was added to enable conjugation with keyhole limpet hemocyanin (KLH). Polyclonal antisera (anti-RHOX13) were collected from immunized rabbits (Harlan Bioproducts), and the final bleeds were used for the experiments described here.
Protein was extracted from adult CD-1 mice by homogenization as previously described (Goto and Eddy, 2004). Fifteen µg of the supernatant fraction from each of the whole testis, brain, and liver extracts were boiled in SDS buffer for 10 minutes and separated on a 10% Tris-glycine gel (BioRad), transferred, and western analysis done using standard procedures. Antiserum was used at a 1:1,500 dilution, and a HRP-conjugated goat anti-rabbit secondary antibody (Sigma) was used at a 1:20,000 dilution. Detection was done using the ECL Plus Detection System substrate (Amersham).
Five hundred µg of soluble testis protein was pre-cleared with 100 µl of protein G-agarose (Upstate Biotechnology), and immunoprecipitations were performed overnight at 4°C by adding 1 µl of preimmune serum or anti-RHOX13 to the pre-cleared lysates. The following day, 100 µl of protein G-agarose was used to capture the antigen-antibody complexes. Following three washes in 1X PBS, the pellet was resuspended and boiled in SDS-PAGE loading buffer. Western blots were done as described above.
2.4 Expression of recombinant RHOX13
The coding sequence of Rhox13 was amplified by PCR (primers: 5’-CACCATGGCCCACGGGGTCCCTTTG and 3’-CACCTTAAGAAGGCTCTTCAAAATCGTTATG) and directionally cloned into pENTR/D/TOPO/TEV (Invitrogen). For expression in E. coli [Rosetta 2(DE3) pLacI (Novagen)], the Rhox13 coding sequence was then subcloned into pDEST 566 (a gift from D. Espsito, NCI/SAIC, Frederick, Maryland) as an N-terminal His-MBP fusion protein. Cells were grown in LB-Ampicillin media, induced with 0.2 mM IPTG (Molecula) for 4 hours at 30°C, and then harvested by centrifugation and stored at −80°C.
Cells were resuspended in bacterial protein extraction reagent B-PER (Pierce). After 15 minutes of incubation, the mixture was cooled on ice and then sonicated 3 times with 30 second bursts at maximum power using a Bronson model 450 sonicator. Insoluble material was removed by centrifugation at 27,000 × g for 30 minutes. The supernatant was passed through an equilibrated (50 mM sodium phosphate, pH 7.3, 300 mM NaCl at 1 ml/minute) Talon resin column (Clontech). The His-MBP-RHOX13 fusion protein was eluted using a 0–150 mM imidazole step gradient; the 75–150 mM imidazole fractions were combined and concentrated by ultracentrifugation through a 10 kDa MWCO filter (Millipore). The protein was purified through an equilibrated (25 mM HEPES, 150 mM NaCl, pH 8) Superdex (16/60) 75 sizing column (Amersham). The N-terminal His-MBP fusion tags were removed by overnight digestion with tobacco etch virus (TEV) protease at 4°C. The cleavage proceeded to completion, as assessed by SDS-PAGE (data not shown). The identity of recombinant RHOX13 (rRHOX13) was confirmed by the unambiguous sequencing of the 12 C-terminal amino acids by reverse-phase liquid chromatography electrospray ionization tandem mass spectrometry on an Agilent SCT Ultra Ion trap mass spectrometer in line with an Agilent 1100 series HPLC system.
2.5 Immunohistochemistry
Tissues were collected from CD-1 mice, fixed overnight in Bouins solution (Polysciences, Inc.), dehydrated through an ethanol series, and embedded in paraffin using standard procedures. Antiserum was diluted 1:400 and applied to 6 µm sections without antigen retrieval followed by a biotinylated secondary goat anti-rabbit antibody (Vector Laboratories). Detection was done using a Vectastain Elite ABC avidin-biotin-peroxidase kit (Vector Laboratories) according to manufacturer’s instructions. Sections were then counterstained with hematoxylin, mounted with Permount (Sigma), visualized using a Zeiss Axioplan microscope, and images captured using a QImaging QICAM digital camera with QCapture 2 software. Periodic-Acid Schiff staining was performed using standard methods.
3. Results and discussion
3.1 Sequence analysis
An in silico search for novel putative transcription factors expressed during spermatogenesis identified an EST that mapped to the homeobox domain-containing hypothetical gene 1700123J19Rik (GenBank AK007253), a putative “extraembryonic, spermatogenesis, homeobox 1 gene”. Four additional EST sequences in UniGene (Mm.50505, Build 159) map to 1700123J19Rik (CF356891, AV210275, AV265101, AV269723). All of these ESTs are expressed in testis. This gene is predicted to contain a 699 bp open reading frame (ORF) encoding a 232 amino acid protein with a mass of 25.3 kDa (Fig. 1). The locus symbol Rhox13 was assigned to this gene by the Mouse Genome Informatics database.
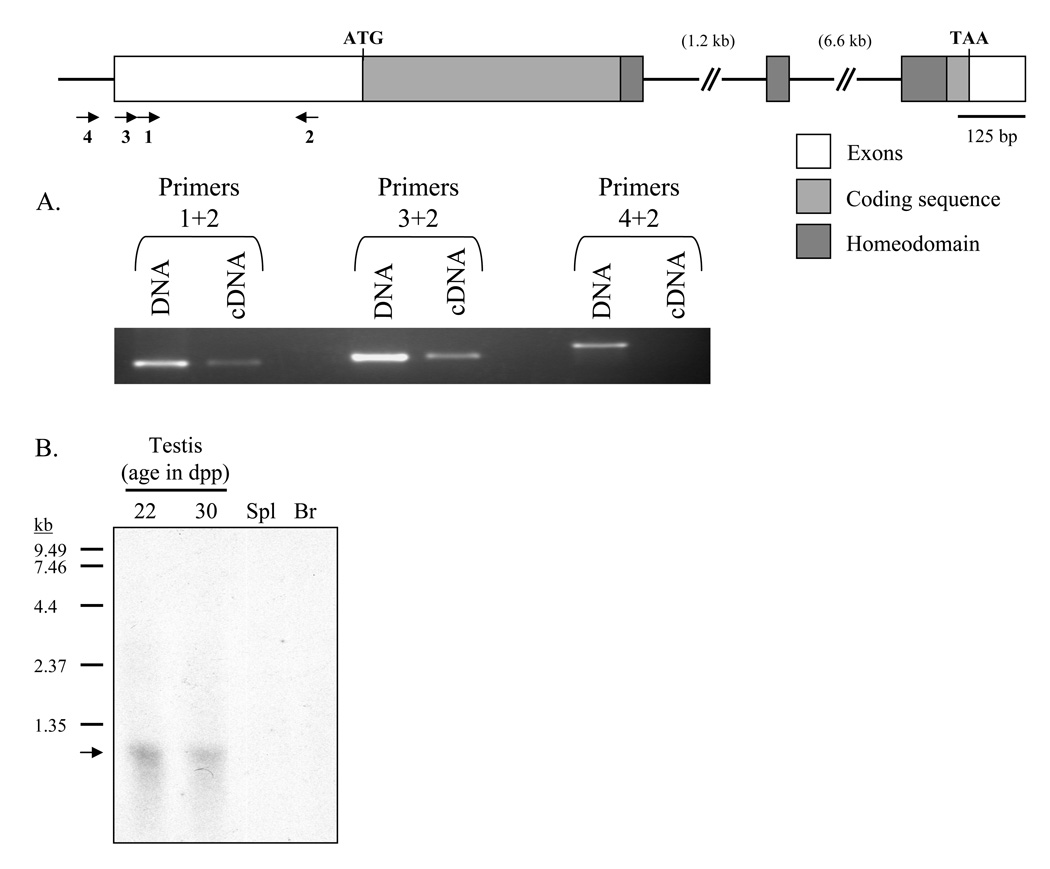
The predicted coding sequence and 3’ untranslated region (UTR) were confirmed by RT-PCR followed by sequencing of the amplified products. A consensus polyadenylation signal was found in the 3’ UTR, 86 bp from the stop codon. Five-prime rapid amplification of cDNA ends (RACE) was employed initially to obtain the sequence of the 5’ UTR, but this technique failed to yield specific products despite repeated attempts. Therefore, primer sets were designed to progressively amplify cDNA and genomic DNA sequences upstream of the start codon (Fig. 2A) to characterize the 5’ UTR. Primers 1+2 and 3+2 were able to amplify both genomic DNA and cDNA, while primers 4+2 could only amplify genomic DNA, suggesting that the 5’ UTR is approximately 460 bp in length. Northern analysis revealed a single 1.2 kb band, comparable to the transcript size estimated from the cDNA and PCR analyses (Fig. 2B).
Figure 2. Gene structure of Rhox13.
Boxes represent exonic sequence; coding sequence is in light gray, and the homeodomain is in dark gray. A: Identifying the 5’ end of the mRNA. Primer sets (indicated by arrows and numbered in panel A) are designed to amplify progressively upstream regions of the Rhox13 5’ UTR from genomic DNA and cDNA. Primer sets 1+2 and 3+2 are able to amplify both genomic DNA and cDNA, while primer set 4+2 is only capable of amplifying genomic DNA, indicating that the upstream primer is located outside of the 5’ UTR. B: Northern blot analysis of Rhox13. 10 µg of total RNA from whole testis (22 and 30 dpp, respectively), spleen (Spl), and brain (Br) were separated and subjected to northern blot analysis. A ~1.2 kb band representing Rhox13 mRNA is present in whole testis (indicated by an arrow) but not in spleen or brain. Sizes of the RNA ladder (in kb) are to the left of the blot.
3.2 Genomic Organization
BLAST analysis indicated that the 1700123J19Rik gene was located on the X chromosome at XA3.3, distal to and near the Rhox cluster (MacLean et al., 2005). The start codon of Rhox12 resides 13 kb upstream of 1700123J19Rik, and the two genes are oriented head-to-head. The deduced mRNA of 1700123J19Rik is encoded by three exons, with exon-intron boundaries that match homeobox genes of the paired-like class, with the second exon containing residues 32–46 of the homeodomain (Burglin, 1994). The location and structural features of this gene are consistent with it being designated Rhox13, in accordance with the current mouse reproductive homeobox nomenclature (MacLean et al., 2005; Gallardo et al., 2007).
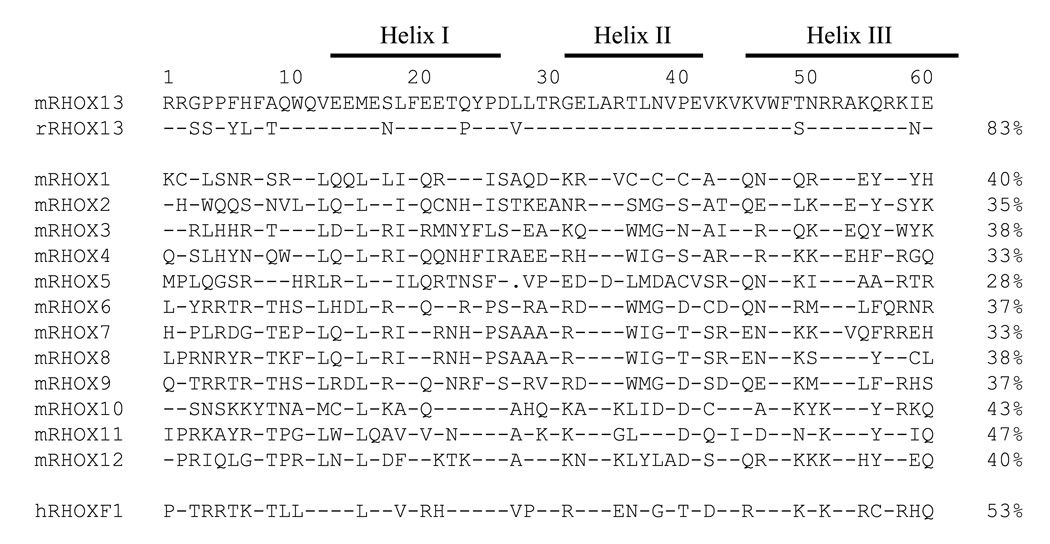
The open reading frame of Rhox13 encodes a 232 amino acid protein with a calculated mass of 25.3 kDa and a consensus homeodomain of amino acids 148–207 (Fig. 3). The syntenic rat Rhox13 ortholog (LOC683136) was identified by BLAST analysis using the mouse cDNA sequence. It encodes a 234 amino acid protein that is 67% identical and 74% similar to the mouse Rhox13 sequence (data not shown). No other characterized domains were found in either the mouse or rat RHOX13 coding sequences. It is important to note that RHOX13 has either a threonine (mouse) or a serine residue (rat) at position 50 in the third, or recognition helix of the homeodomain (Fig. 3). This residue has been implicated in DNA binding specificity (Treisman et al., 1989) and is not shared by any of the other Rhox genes, but is rather a hallmark of the paired class of homeobox proteins (Burglin, 1994). The N-terminus of the RHOX13 protein contains a long acidic region, a characteristic of activation domains in transcription factors (Mitchell and Tjian, 1989), suggesting a role in transcriptional activation. This acidic region contains numerous glutamic acid residues, a feature shared by several of the reproductive homeobox genes (Wayne et al., 2002).
Figure 3. Comparison of the RHOX13 homeodomain with those of other reproductive homeobox proteins.
Amino acids 1–60 of the homeodomains from various proteins are shown, with hyphens indicating residues that match RHOX13 at each position, and periods representing gaps. The percent identity to RHOX13 is represented to the right of each sequence. The three solid lines above indicate those stretches of amino acids involved in the formation of helices I, II, and III, respectively. The small letter in front of each gene name designates the species of origin (m=mouse, r=rat, h=human).
3.3 RNA analysis in prenatal and adult gonads
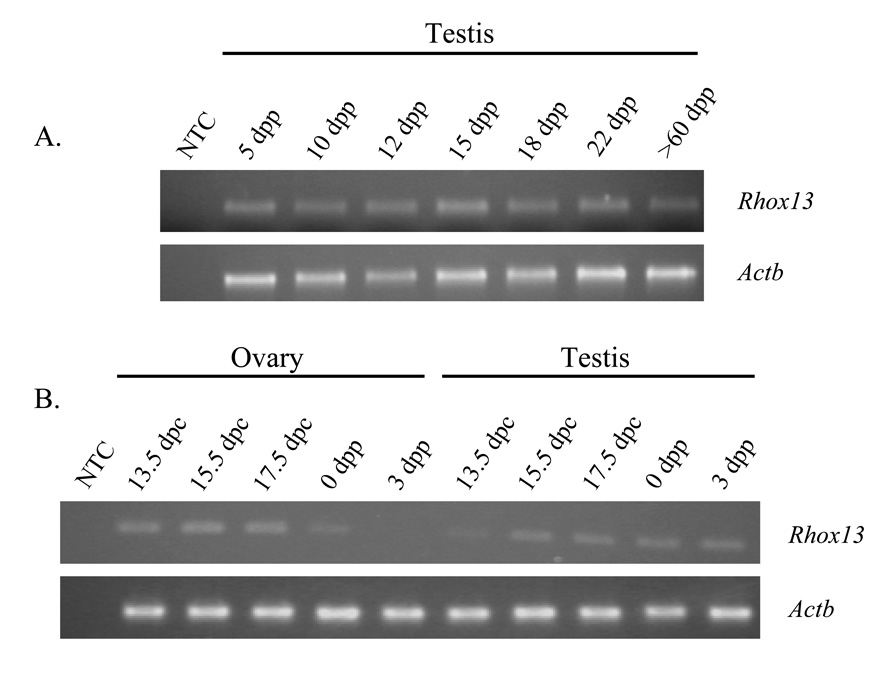
Transcripts for Rhox13 were initially detected by northern analysis of total testis RNA (Fig. 2B). The developmental expression of Rhox13 transcripts was determined by RT-PCR using Rhox13-specific primers and total testis RNA from mice ranging in age from 13.5 dpc to adulthood. The relative levels of Rhox13 transcripts in fetal testis and ovary were determined using semi-quantitative RT-PCR. Transcripts for Rhox13 were barely detectable in the testis at 13.5 dpc, but then increased substantially at 15.5 dpc. This increase in steady-state transcript levels in the testis between 13.5–15.5 dpc correlates with the proliferation of prospermatogonia during that interval, after which they stop dividing and remain quiescent until after birth (reviewed by McCarrey, 1993). Transcript levels in the testis remained at a constant level throughout the remainder of fetal life and in all of the postnatal ages examined (Fig. 4A).
Figure 4. Rhox13 gonadal expression analysis by semi-quantitative RT-PCR.
A: Rhox13 transcript levels are consistent in whole testis from 5 dpp to adult (>60 dpp). B: In the prenatal and neonatal ovary, transcripts for Rhox13 are present from 13.5 dpc-17.5 dpc, and become undetectable by 3 dpp. In the prenatal testis, however, transcript levels increase from 13.5 – 15.5 dpc and remain consistent through 3 dpp. NTC = no template control. The amplicons from the Actb and Rhox13 primer sets span one and two introns, respectively, in order to eliminate potential amplification of genomic DNA.
In addition, Rhox13 transcripts were found in the prenatal ovary and appeared to be more abundant in ovary than in testis at 13.5 dpc. However, transcript levels declined over the next few days and by 3 dpp no signal was detectable (Fig. 4B). There was no detectable expression of Rhox13 in the adult ovary by RT-PCR (data not shown).
The majority of reproductive homeobox genes characterized thus far are transcriptionally active to some extent in extraembryonic tissues (Li et al., 1997; Han et al., 1998; Han et al., 2000; MacLean et al., 2005). However, Rhox13 transcripts were not detected by RT-PCR (35 cycles) in placenta at 12.5 or 15.5 dpc (data not shown).
3.4 Protein analysis in prepuberal and adult testis
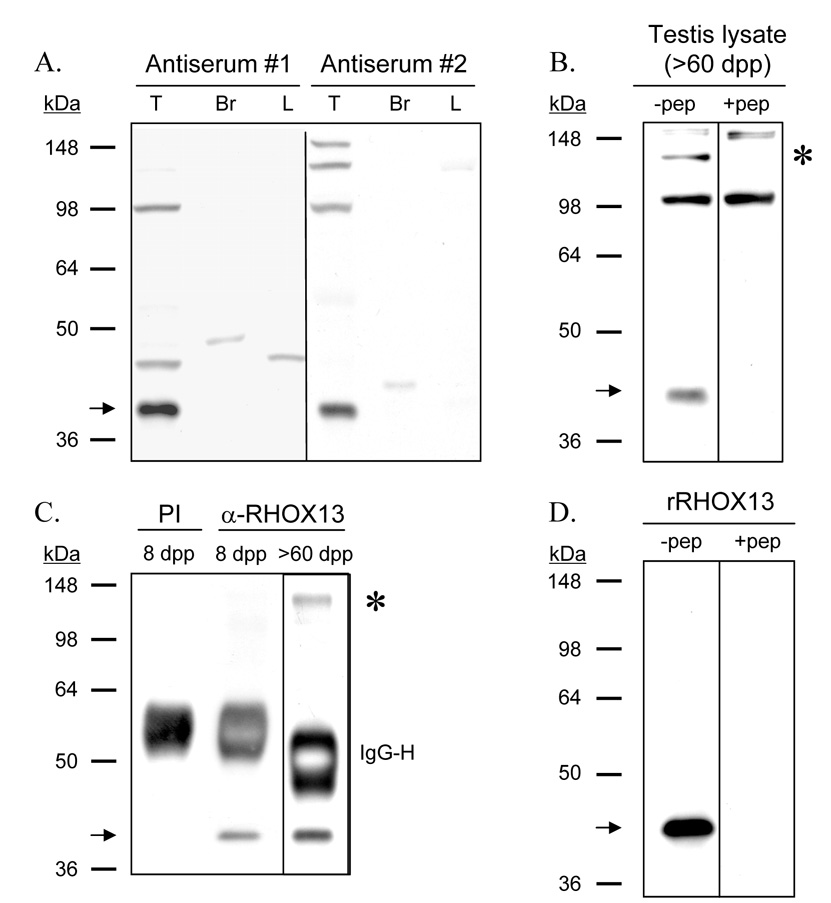
Western blot analysis with antisera to the N-terminus of RHOX13 generated in two rabbits identified a ~45 kDa band in supernatants of adult testis homogenates, but not in those of adult brain or liver (Fig. 5A). No bands were present when preimmune serum was used as the primary antibody (data not shown). Antiserum #2 was used for subsequent analyses, although results were comparable using both antisera. The ~45 kDa band was present in testis homogenates from adult mice, and this band was not recognized when antiserum was preincubated with the immunizing peptide (Fig. 5B). A ~130 kDa band recognized in supernatants of adult testis was also not detected after the antiserum was preincubated with the immunizing peptide; however, it was not detected by either antiserum in prepuberal testis (data not shown). This band presumably represents interaction of antiserum #2 with an epitope on a ~130 kDa protein similar to one on RHOX13. The ~45 kDa protein was immunoprecipitated from supernatants of 8 dpp and adult testis using anti-RHOX13, but the ~130 kDa protein was immunoprecipitated only from adult testis supernatant (Fig. 5C).
Figure 5. Immunoblots using anti-RHOX13.
A: A ~45 kDa protein corresponding to RHOX13 (indicated by an arrow) is present in soluble extracts from adult testis but not brain or liver. B: The RHOX13 band is present in soluble protein from whole testes of adult (>60 dpp) mice. The ~45 kDa RHOX13 band (indicated by an arrow) is not recognized when anti-RHOX13 was preincubated with 0.01 µg of the N-terminal immunizing peptide. In addition, a ~130 kDa band seen only in adult testis using antiserum #2 is not recognized when anti-RHOX13 is preincubated with the immunizing peptide (indicated by an asterisk). C: IP using anti-RHOX13 on 8 dpp and adult testis lysates (2 separate gels). The ~45 kDa RHOX13 band (indicated by an arrow) is pulled down from both 8 dpp and adult testis lysates using anti-RHOX13, but not pre-immune serum (PI). The ~130 kDa band (indicated by an asterisk) seen above in A in adult testis lysate is also pulled down by anti-RHOX13. The dark bands at approximately 55 kDa represent the heavy chain of IgG (IgG-H). D: Recombinant RHOX13 (rRHOX13) migrates at ~45 kDa (left lane, −pep) and is not recognized when anti-RHOX13 is preincubated with the immunizing peptide (right lane, +pep).
The ~45 kDa protein band observed on western blots migrated considerably slower than the 25.3 kDa mass of RHOX13 predicted from the primary sequence. We considered 3 possible explanations for this discrepancy: 1) there was additional exonic sequence unaccounted for in the predicted coding sequence, 2) the highly acidic stretches of amino acids in the primary sequence (amino acids 65–105, Fig. 1) may have caused the protein to migrate aberrantly on the gel, and 3) our antiserum recognized a protein distinct from RHOX13. To address these possibilities, we generated a recombinant RHOX13 (rRHOX13) fusion protein in E. coli with an N-terminal His-MBP tag. Purified rRHOX13 (minus the His-MBP tag) was separated by electrophoresis on a 10% polyacrylamide gel and assayed by western blotting. It migrated at ~45 kDa, the same size as in the prepuberal and adult testis lysates, and this band was not recognized when antiserum was preincubated with the immunizing peptide (Fig. 5D). It is not uncommon for proteins with charged regions to migrate aberrantly on polyacrylamide gels (Query et al., 1989; Renz et al., 1996; Iakoucheva et al., 2001), and we conclude that the primary sequence of RHOX13 was responsible for its aberrant migration.
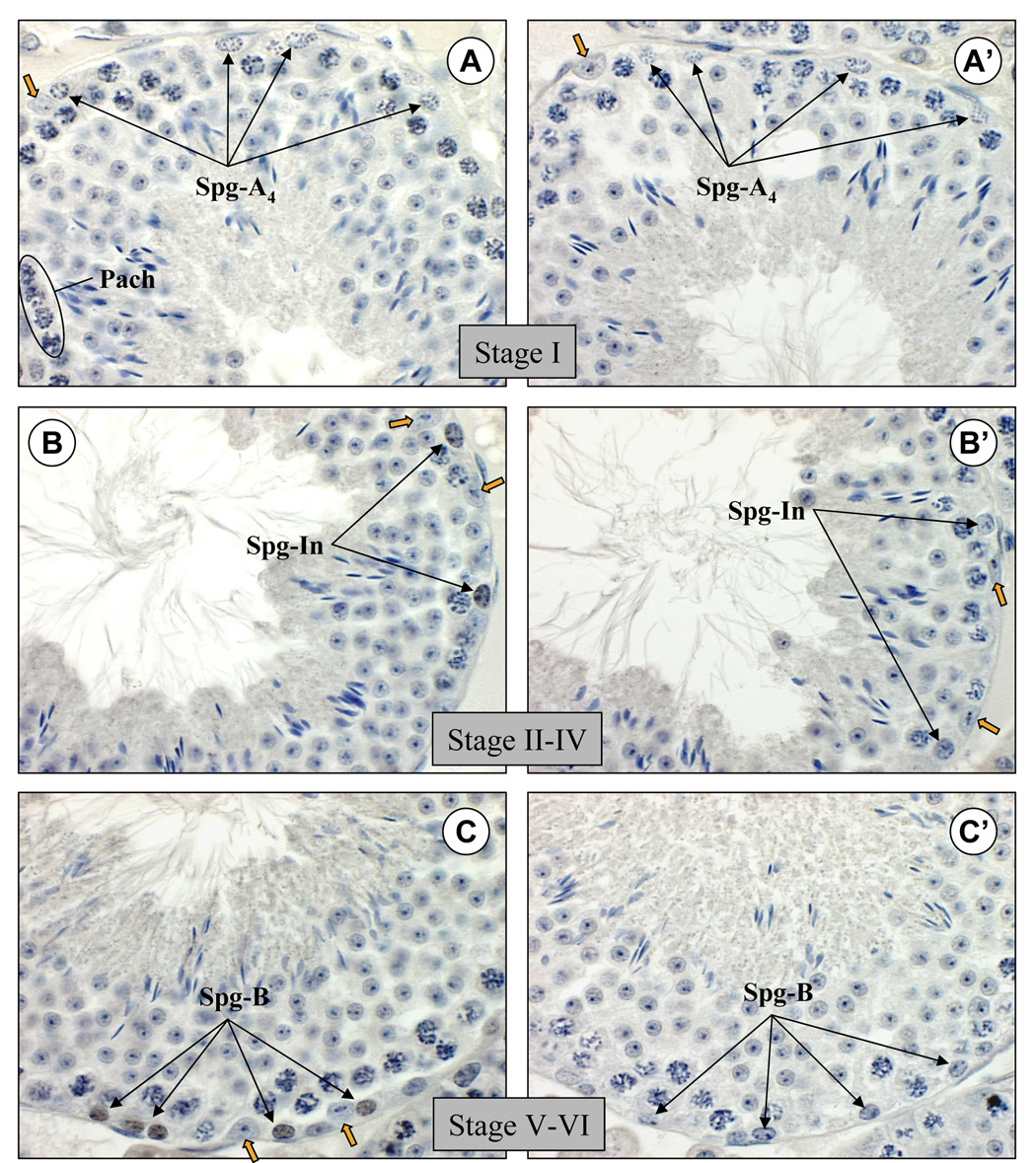
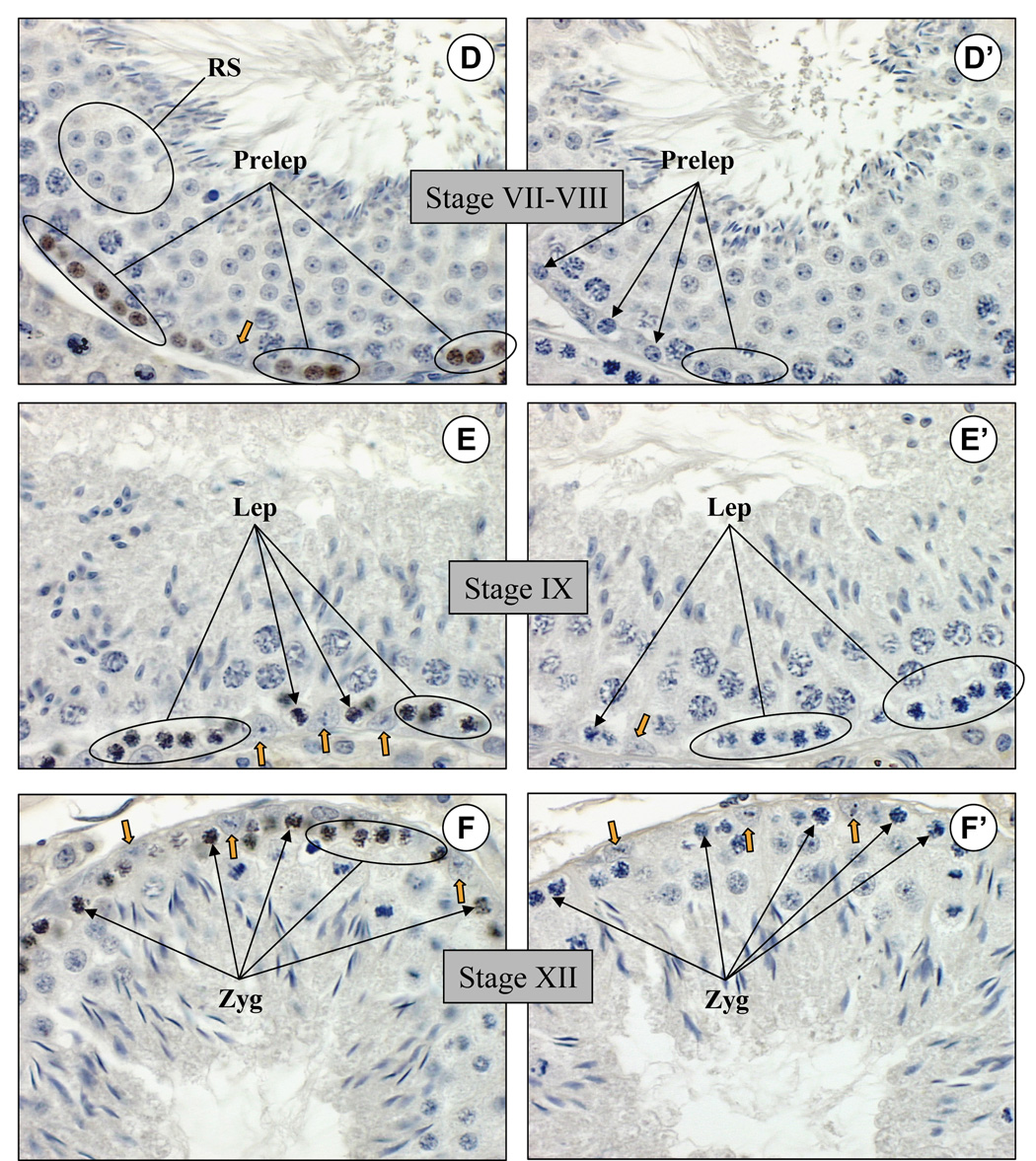
Spermatogenesis proceeds in the adult seminiferous epithelium in 12 well-defined developmental stages. Spermatogonia sequentially differentiate from type A4 to intermediate and then to type B spermatogonia, which then become preleptotene spermatocytes to begin meiosis and progress to leptotene, zygotene, and then pachytene spermatocytes (Bellvé et al., 1977; de Rooij, 2001). Postmeiotic haploid round spermatids undergo remodeling during spermiogenesis to become spermatozoa. Tubules were staged by determining the specific cohort of spermatogenic cell types present in the testis section along with the extent of development of the acrosome by PAS staining of adjacent sections (data not shown). Spermatogonia in the basal compartment of the seminiferous epithelium were identified based on their morphology as well as the stage of the tubule in which they were present, and this allowed for determination of which specific spermatogonial cell types expressed RHOX13. No detectable nuclear staining was observed in type A4 spermatogonia of stage I tubules (Fig. 6A). Nuclear staining was first evident in intermediate spermatogonia (Fig. 6B), which increased in type B spermatogonia (Fig. 6C) and preleptotene spermatocytes (Fig. 6D). Nuclear staining decreased in leptotene spermatocytes (Fig. 6E) and zygotene spermatocytes (Fig. 6F), and was not seen in pachytene spermatocytes (Fig. 6A–F) or round spermatids (Fig. 6A–D). In addition, RHOX13 protein was not detectable in Sertoli, Leydig, or peritubular myoid cells (Fig. 6A–F, and data not shown). No staining was seen when anti-RHOX13 that had been preincubated with the immunizing peptide was applied to adjacent sections (Fig. 6A’–F’).
Figure 6. RHOX13 protein is expressed in a stage-specific manner in the seminiferous epithelium of adult testis.
A, B: RHOX13 is not detectable in type A4 spermatogonia (Spg-A4) in stage I tubules, but becomes detectable in intermediate spermatogonia (Spg-In). C–D: Expression increases in type B spermatogonia (Spg-B) and in preleptotene spermatocytes (Prelep). E–F: RHOX13 expression decreases in leptotene (Lep) and zygotene (Zyg) spermatocytes, respectively. No expression is seen in pachytene (Pach) spermatocytes or round spermatids (RS) at any of the stages examined. Additionally, no signal is evident in Sertoli cells (indicated by orange arrowheads in A–F). A’–F’: Adjacent sections are stained with anti-RHOX13 preincubated with the immunizing peptide. Scale bar = 25 µm.
Conclusions
We have identified a novel X-linked reproductive homeobox gene and characterized its expression pattern in the prenatal and postnatal gonads in the mouse. Like other reproductive homeobox genes, Rhox13 is expressed in reproductive tissues, including the prenatal ovary and the prenatal and postnatal testis; however, unlike other characterized Rhox genes, Rhox13 does not appear to be expressed in Sertoli cells or in extraembryonic tissues. In the adult testis, RHOX13 protein is found in differentiating spermatogonia and preleptotene spermatocytes, but diminishes and then disappears as these cells enter meiosis. The unique expression pattern and homeobox sequence of Rhox13 suggest that this novel gene may regulate a subset of genes in the prenatal ovary and prenatal and postnatal testis distinct from other Rhox genes.
Acknowledgements
We would like to thank Bob Petrovich and Jason Williams of the protein expression and microcharacterization core facilities, respectively, at the NIEHS for their help in expression and identification of recombinant RHOX13. We wish to thank Paula Brown for help with searching the EST database, Katherine Burns and Daniel Gilchrist for critical reading of the manuscript, and members of the Eddy lab for technical advice.
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences
abbreviations
- dpp
days post partum
- dpc
days post coitum
- Rhox
reproductive homeobox
- kDa
kilodalton
- bp
base pair
- nt
nucleotide
- KLH
keyhole limpet hemocyanin
- HRP
horseradish peroxidase
- MWCO
molecular weight cutoff
- TEV
tobacco etch virus
- MBP
maltose binding protein
- EST
expressed sequence tag
- ORF
open reading frame
- UTR
untranslated region
- RACE
rapid amplification of cDNA ends
- BLAST
Basic local alignment and search tool
- E. coli
Escherichia coli.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bellvé AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. J. Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford WW, Zhao GQ, Valerius MT, Weinstein M, Birkenmeier EH, Rowe LB, Potter SS. Spx1, a novel X-linked homeobox gene expressed during spermatogenesis. Mech. Dev. 1997;65:87–98. doi: 10.1016/s0925-4773(97)00058-0. [DOI] [PubMed] [Google Scholar]
- Burglin TR. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the Homeobox Genes. New York: Oxford University Press; 1994. pp. 25–71. [Google Scholar]
- De Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- Fohn LE, Behringer RR. ESX1L, a novel X chromosome-linked human homeobox gene expressed in the placenta and testis. Genomics. 2001;74:105–108. doi: 10.1006/geno.2001.6532. [DOI] [PubMed] [Google Scholar]
- Gallardo TD, John GB, Shirley L, Contreras DM, Akbay EA, Haynie JM, Ward SE, Shidler MJ, Castrillon DH. Genomewide discovery and classification of candidate ovarian fertility genes in the mouse. Genetics. 2007;177(1):179–194. doi: 10.1534/genetics.107.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu. Rev. Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Geserick C, Weiss B, Schleuning W, Haendler B. OTEX, an androgen-regulated human member of the paired-like class of homeobox genes. Biochem. J. 2002;366:367–375. doi: 10.1042/BJ20020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Eddy EM. Speriolin is a novel spermatogenic cell-specific centrosomal protein associated with the seventh WD motif of Cdc20. J. Biol. Chem. 2004;279(40):42128–42138. doi: 10.1074/jbc.M403190200. [DOI] [PubMed] [Google Scholar]
- Han YJ, Park AR, Sung DY, Chun JY. Psx, a novel murine homeobox gene expressed in placenta. Gene. 1998;207:159–166. doi: 10.1016/s0378-1119(97)00620-3. [DOI] [PubMed] [Google Scholar]
- Han YJ, Lee YH, Chun JY. Identification and characterization of Psx-2, a novel member of the Psx (placenta-specific homeobox) family. Gene. 2000;241:149–155. doi: 10.1016/s0378-1119(99)00453-9. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ. Aberrant mobility phenomena of the DNA repair protein XPA. Prot. Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Watt AJ, Gautier P, Gilchrist D, Driehaus GJ, Graham GJ, Keebler J, Prugnolle F, Awadalla P, Forrester LM. A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genomics. 2006;7:212. doi: 10.1186/1471-2164-7-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lemaire P, Behringer RR. Esx1, a novel X chromosome-linked homeobox gene expressed in mouse extraembryonic tissues and male germ cells. Dev. Biol. 1997;188:85–95. doi: 10.1006/dbio.1997.8640. [DOI] [PubMed] [Google Scholar]
- Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat. Genet. 1998;20:309–311. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: A new homeobox gene cluster. Cell. 2005;130:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Lorenzetti D, Hum Z, Salerno WJ, Miller J, Wilkinson MF. Rhox homeobox gene cluster: recent duplication of three family members. Genesis. 2006;44:122–129. doi: 10.1002/gene.20193. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Development of the germ cell. In: Desjardins C, Ewing LL, editors. Cell and Molecular Biology of the Testis. New York: Oxford University Press; 1993. pp. 58–89. [Google Scholar]
- McCarrey JR, O’Brien DA, Skinner MK. Construction and preliminary characterization of a series of mouse and rat testis cDNA libraries. J. Androl. 1999;20:635–639. [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Miura H, Yanazawa M, Kato K, Kitamura K. Expression of a novel aristaless related homeobox gene Arx in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 1997;25:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Morris L, Gordon J, Blackburn CC. Identification of a tandem duplicated array in the Rhox alpha locus on mouse chromosome X. Mamm. Genome. 2006;17:178–187. doi: 10.1007/s00335-005-0138-4. [DOI] [PubMed] [Google Scholar]
- Pitman JL, Lin TP, Kleeman JE, Erickson GF, MacLeod CL. Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Dev. Biol. 1998;202:196–214. doi: 10.1006/dbio.1998.8978. [DOI] [PubMed] [Google Scholar]
- Query CC, Bentley RC, Keene JD. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Renz A, Fackelmayer FO. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 1996;24(5):843–849. doi: 10.1093/nar/24.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki N, Rankin T, Dean J. Normal gonadal development in mice lacking GPBOX, a homeobox protein expressed in germ cells at the onset of sexual dimorphism. Mol. Cell. Biol. 2001;21:8197–8202. doi: 10.1128/MCB.21.23.8197-8202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J. Remarkable expansions of an X-linked reproductive homeobox gene cluster in rodent evolution. Genomics. 2006;88:34–43. doi: 10.1016/j.ygeno.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Wayne CM, MacLean JA, II, Cornwall G, Wilkinson MF. Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene. 2002;301:1–11. doi: 10.1016/s0378-1119(02)01087-9. [DOI] [PubMed] [Google Scholar]