Abstract
Mutasynthesis in pikromycin PKS: The amenability of pikromycin polyketide synthase to mutational biosynthesis has been demonstrated. A natural triketide and its analogues, activated as N-acetyl-cysteamine thioesters, were synthesized and fed to a pikAI-deleted strain; this led to the production of new antibiotics. A vinyl analogue was found to have better antibacterial activity than pikromycin.

The pikromycin polyketide synthase (PKS) of S. venezuelae, which consists of one loading module and six extension modules, is responsible for the formation of the hexaketide narbonolide, a key intermediate ok? in the biosynthesis of the antibiotic pikromycin. S. venezuelae strains in which PikAI, which houses the loading domain and first two modules of the PKS, is either absent or catalytically inactive, produce no pikromycin product. When these strains are grown in the presence of a synthetically prepared triketide product, activated as the N-acetylcysteamine thioester, pikromycin yields are restored to as much as 11 % of that seen in the wild-type strain. Feeding analogues of the triketide intermediate provides pikromycin analogues bearing different alkyl substituents at C13 and C14. One of these analogues, Δ15,16-dehydropikromycin, exhibits improved antimicrobial activity relative to pikromycin.
Keywords: biosynthesis, natural products, pikromycin, polyketides, triketides
Introduction
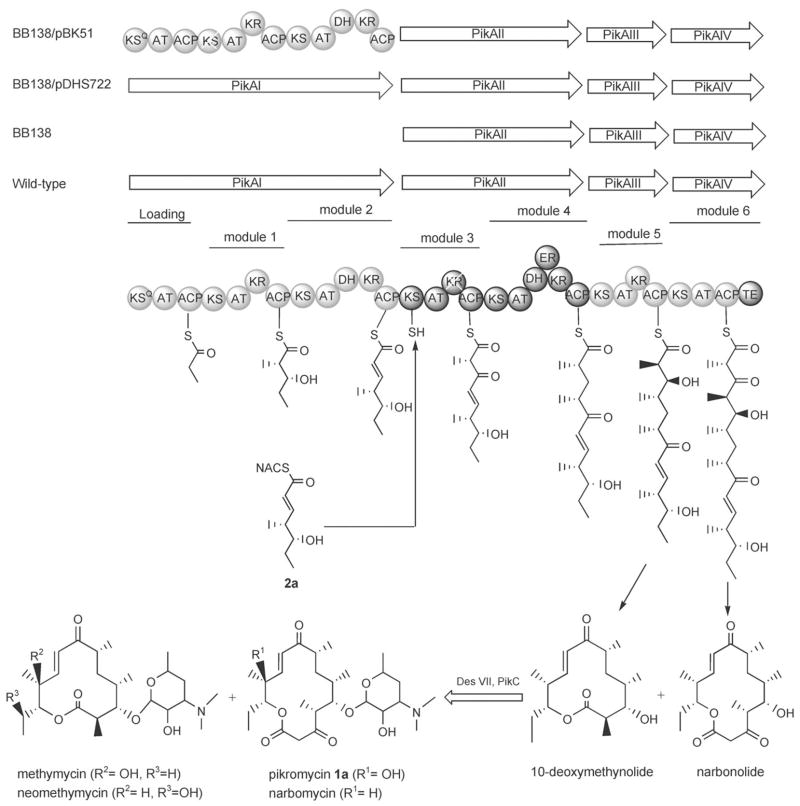
Polyketides constitute a large and diverse group of natural products that possess important anticancer, antibiotic, immunosuppressive, and other biological activities[1–3] Many of them are biosynthesized by modular type I polyketide synthases (PKSs), which are giant multifunctional enzymes consisting of several discrete modules, each responsible for one cycle of polyketide acyl chain elongation. Each module contains three basic domains for each biosynthetic step; an acyltransferase (AT), an acyl carrier protein (ACP), and a ketosynthase (KS). The extent of modification within each module of the β-ketoacyl thioester formed depends on the presence or absence of ketoreductase (KR), dehydratase (DH), or enoyl reductase (ER) domains.[4] The pikromycin (Pik) PKS of S. venezuelae comprises four separate polypeptides (PikAI-PikAIV) that house six such extension modules, a loading module, and a thioesterase domain (TE) responsible for priming and terminating the polyketide biosynthetic process, respectively (Figure 1).[5] Full extension through this Pik PKS gives rise to a heptaketide product, which is then cyclized to the 14-membered aglycone narbonolide. Modification by the DesVII–DesVIII glycosyltransferase and PikC (a P450 hydroxylase) gives the biologically active pikromycin product (1a).[5] Termination of the chain-elongation process at the hexaketide intermediate (without extension by module 6) has been shown to give rise to the 12-membered aglycone 10-deoxymethynolide, which is then converted by DesVII and PikC to methymycin.[5]
Figure 1.
Modular organization and function of the proteins (PikAI-PikAIV) of the pikromycin PKS. 2a is proposed to load onto the active-site cysteine of KS3 of PikAII and be processed by the downstream Pik PKS to form 12- and 14-membered macrolides.
It has been shown through both in vivo and in vitro experiments that PKS intermediates activated as N-acetylcysteamine (NAC) thioesters can be loaded onto the KS domains of elongation modules.[6–11] Such loading presumably accounts for the observation that polyketide production can be achieved by feeding diketide and triketide intermediates to bacterial strains in which either the priming or initial elongation steps have been abrogated.[6–11] Feeding of isomers of presumptive natural intermediates in such systems has proven to be a useful tool for determining unresolved stereochemical questions, as shown recently for diketides involved in ansamitocin,[9] rifamycin,[12] and phoslactomycin biosynthesis.[8] In all of these cases, feeding of the diketide intermediate led to only partial restoration of the final polyketide product. In the case of ansamitocin feeding, the presumed triketide intermediate gave no restoration.[9] These observations have led to the suggestion that loading polyketide intermediates onto the cognate PKS modules is inefficient.[9]
There are a few in vivo examples in which feeding analogues of polyketide intermediates leads to the generation of novel polyketide products. This mutasynthetic approach[13, 14] with type I PKSs has been used to generate new epothilones[11] and erythromycins.[6, 7, 15] In the latter case, a 6-deoxyerythonolide B synthase (DEBS) in which module 1 was made catalytically inactive has been used successfully to produce many new structures with unique physicochemical properties and new functional groups.[15] Yields of the new natural products in these cases were very low, presumably as a result of inefficient loading onto the cognate module, and slower conversion of the unnatural intermediate by the downstream PKS modules and post-PKS modification enzymes. A number of approaches have been taken to improve the efficiency of this process. In the case of erythromycin biosynthesis, it has been shown that natural product yields improve as much as tenfold if diketide intermediates are fed to strains in which the loading domain and module 1 of the PKS are deleted, rather than made catalytically inactive through mutation.[10]
To date there have been no reports of the generation of novel pikromycins by the same type of mutasynthetic approaches as for erythromycins. A number of observations suggest potentially significant barriers to the success of such an approach. Previous incorporation experiments with dual-labeled diketide and triketide intermediates in the wild-type Streptomyces venezuelae were shown initially to lead to no intact incorporation into a polyketide product.[16] Refinement of the fermentation conditions and timing of addition of the intermediates led to successful but extremely low incorporation levels (0.15–0.5 % of the polyketide product was generated from the intact intermediate), potentially indicating poor loading onto the cognate PKS modules. Recent in vivo and in vitro analyses of modules 3, 5, and 6 of the pikromycin PKS have also demonstrated that they have significant substrate specificity[17, 18] and thus might not efficiently process unnatural pathway intermediates.
We report here a series of mutasynthetic experiments with a pikAI deletion mutant of S. venezuelae (BB138) (Figure 1), in which PikAI, which houses the loading domain and first two modules, is absent. Feeding the N-acetyl cysteamine thioester of the triketide intermediate restores the yield of pikromycin to approximately 11% of production levels of the wild-type strain. Expression of a catalytically inactive PikAI in BB138 does not alter the efficiency of this process. A series of triketide analogues was synthesized and shown to be processed by the BB138 strain, and four were shown to be processed into new biologically active 14-membered macrolide products. The levels of production of these new macrolides varied, but in all cases were at least tenfold lower than seen for pikromycin production from the natural triketide. Preliminary analysis of one new product, Δ15,16-dehydropikromycin, indicated slightly improved antibacterial activity. These data show for the first time that the Pik PKS system is amenable to mutasynthesis and can be used to generate new and potentially useful antibiotics. However, we observed incorporation inefficiencies similar to those encountered in other type I PKSs.
Results and Discussion
Synthesis of natural triketide and its analogues
The natural triketide 2a and its analogues 2b–f were prepared as shown in Scheme 1. The aldol intermediates 3a–f were generated by stereoselective aldol coupling reaction between an Evans’ acyl oxazolidinone and various commercially available aldehydes. Silylation of the hydroxyl groups of 3a–f followed by replacement of the chiral auxiliary with a benzyl group gave intermediates 4a–f. Subsequent reduction of the benzyl ester with DIBAL-H gave alcohols 5a–f. (Attempts to generate alcohols 5a–f by direct reductive removal of the chiral auxiliary by using various reducing agents resulted in poor yields and decomposition products.) The alcohols were oxidized by using the Dess–Martin periodinane, and the resulting crude aldehydes were immediately treated with the respective Wittig salts. The resulting unsaturated esters were hydrolyzed under mild basic conditions to generate the corresponding carboxylic acids 6a–f. Treatment of the acids with diphenylphosphoryl azide and triethylamine followed by addition of N-acetylcysteamine afforded the protected NAC-thioesters, which were treated with aqueous HF to afford the final triketide NAC thioesters 2a–f.
Scheme 1.
Synthesis of triketides 2a–f. Reagents and conditions: a) DBBT, DIEA, CH2Cl2, R1CHO; b) TBSOTf, 2,6-lutidine, CH2Cl2; c) nBuLi, BnOH, THF; d) DIBAL-H, CH2Cl2; e) Dess–Martin periodinane, CH2Cl2; f) Wittig salt, THF; g) K2CO3, MeOH/H2O; h) (PhO)2P(O)N3, Et3N, AcHN(CH2)2SH, DMF; i) 48% HF, CH3CN, H2O. DBBT = dibutylboron triflate, DIEA =N,N-diisopropylethylamine.
The synthesis of the triketide analogues 2g–i was troublesome due to difficulty in removing the chiral auxiliary. Thus, for these triketides, a different route, based on modifications of a literature procedure,[19] was used to prepare the aldehyde intermediate (Scheme 2). Briefly, treatment of the aldol products 3g–i with N,O-dimethylhydroxyl amine and trimethyl aluminum gave the corresponding Weinreb amides, which were protected to give the silyl ethers. Reduction with DIBAL-H in THF gave the corresponding aldehydes in a much higher yield and purity compared to the strategy employed in Scheme 1. The aldehydes were converted to the desired triketide NAC thioesters 2g–i by using the same approach described above (steps f–i, Scheme 1).
Scheme 2.
Synthesis of triketides 2g–i. Reagents and conditions: a) DBBT, DIEA, CH2Cl2, R3CHO; b) AlMe3, MeO(-Me)NH·HCl, CH2Cl2; c) TBSOTf, 2,6-lutidine, CH2Cl2; d) DIBAL-H, CH2Cl2.
Feeding of unsaturated triketide 2a to S. venezuelae BB138 mutant
Fermentations of the BB138 in 10 mL please define SGGP (SGGP) media[20] were supplemented after 8 h of incubation with 1 mM of the unsaturated triketide 2a. After incubation for additional 3 days at 30 °C and 220 rpm, the culture was centrifuged, and the broth was extracted twice with ethyl acetate. The extract was concentrated, redissolved in a small volume of methanol, and analyzed by LC/MS. Feeding of the natural triketide 2a to BB138 resulted in the production of natural pikromycin in 10–12 % relative yield compared to wild-type S. venezuelae (Table 1). We also observed the formation of the 12-membered macrolides methymycin and neomethymycin in BB138 supplemented with 2a, also at about 10–12 % of that seen for the wild-type strain under the same growth conditions. There was no significant difference in the production level of pikromycin when the triketide concentration was increased to 2 or 5 mM (Figure 2). However, the levels of pikromycin decreased dramatically when lower (<0.5 mM) concentrations were used; this suggests that, under these conditions, priming of module 3 of the Pik PKS in BB138 was limited by the availability of 2a.
Table 1.
Production of natural pikromycin and its analogues by feeding substrates 2a–i to BB138 mutant.[a] Production yield relative to wild type S. venezuelae.
| Substrate | Feeding product | Production [%][a] |
|---|---|---|
| 2a | Pikromycin (1a) | 10–12 |
| 2b | 1b | <1 |
| 2c | 1c | <1 |
| 2h | 1d | <1 |
| 2i | 1e | ~1 |
Figure 2.
Pikromycin yields from growing S. venezuelae BB138 in the presence of varying concentrations of the triketide 2a. Results are average (±SEM) of triplicate assays.
The efficiency of precursor-directed biosynthesis of erythromycins by using diketides has been shown to be improved by removal of the loading domain and module 1 of DEBS.[10] While the reasons for this observation remain undetermined, it seems likely that these preceding modules can interfere with the transfer of the diketide onto the active-site cysteine of module 2 of DEBS. Experiments were carried out to determine if the modules of PikAI interfered with loading of the natural triketide 2a onto module 3. A plasmid (pBK51) that expresses the entire PikAI with a Cys–Ala mutation in the active site of module 1, and pDHS722 (which expresses the natural PikAI) were introduced into BB138 (Figure 1). Pikromycin production was observed in BB138/pDHS722, but not in BB138/pBK51 or BB138/pSG1 (plasmid control). These observations were consistent with expression of an active PikAI from pDHS722 and an inactive PikAI from pBK51. The levels of pikromycin production obtained by growing BB138/pBK51 and BB138/pSG1 in the presence of a range of concentrations of the natural triketide 2a were determined (Figure 3). In both cases the same significant drop in pikromycin production was observed when triketide concentrations were less than 0.5 mM. However, there was no significant difference in pikromycin production for BB138/pBK51 and BB138/pSG1 at each triketide concentration. A feeding study with the natural diketide resulted in pikromycin production in BB138/pBK51, thus unequivocally demonstrating the presence of soluble PikAI with an active module 2 and inactive module 1 (data not shown). These experiments suggest that the PikAI protein does not affect loading of the triketide onto module 3 of the Pik PKS, and contrast the observations made with DEBS.[10] In the DEBS case, however, the experiments were carried out with diketides, and the preceding modules (loading domain and module 1) were housed on the same polypeptide. In the case of triketide feeding for the Pik PKS, the preceding modules are housed on a separate polypeptide and are therefore arguably less likely to present a significant barrier to priming of the module (Table 1).
Figure 3.
Pikromycin yields from growing S. venezuelae BB138/pBK51 and BB138pSG1 in the presence of varying concentrations of the triketide 2a. Results are average (±SEM) of triplicate assays.
A number of factors could account for the observation that maximal yields of pikromycin production from feeding triketide to BB138 were only about 10 % of that seen in the wild-type strain. One potential limiting factor is the loading of 2a onto module 3 of the Pik PKS. Indeed, previous in vitro work with individual modules of DEBS has shown that the catalytic efficiency for formation of a triketide lactone increases more than 100-fold if the priming diketide substrate is presented attached to the cognate ACP rather than as an N-acetylcysteamine.[21] In the normal Pik PKS process, the docking domains of PikAI and PikAII presumably facilitate efficient passage of the triketide intermediate from ACP3 of PikAI to the KS4 of PikAII.[22] The efficiency of this process coupled with the poor loading of 2a on to KS3 likely accounts for previous observations of extremely low intact incorporation of labeled triketide into pikromycin in the wild-type S. venezuelae.[16]
Production of novel pikromycin analogues by mutational biosynthesis in strain BB138
NAC thioester analogues of the natural triketide 2a were added to fermentations of BB138 to determine which could be processed into novel pikroymcin-related products. All triketide analogues were fed at a final concentration of 1 mM, and the resulting fermentation products were analyzed by LC-MS. As detailed in Figure 4, four of these triketides (2b, 2c, 2h, and 2i) were processed to the corresponding 14-membered analogues of pikromycin. In none of the triketide experiments were 12-membered products (analogues of methymycin, neomethymycin, and 10-deoxymethynolide) observed. The reason for this observation was unclear, but might simply reflect the fact that they were produced at levels harder to detect (the 14-membered pikromycin predominated in BB138 grown in the presence of 2a).
Figure 4.
Left: Analogues of pikromycin (1a) and narbomycin (7a) generated by growing S. venezuelae BB138 in the presence of analogues 2b–i of the natural triketide 2a. Right: ESI mass and HPLC retention times for 1a–e and 7a–e.
Replacement of the ethyl group of the natural triketide with propyl (2b), isopropyl (2c), or ethylene (2i) resulted in detectable levels of fully elongated polyketide products (1b, 1c, and 1e, respectively). Replacement of the ethyl group with the cyclohexyl group in 2d, did not give the corresponding product, potentially indicating an inability of the Pik PKS to process a bulky substituent at the end of the polyketide chain. Surprisingly, the triketide analogue 2e, in which the ethyl group was shortened to a methyl was also not processed. Thus, there appears to be an upper and lower limit to the size of the substituent used to replace the ethyl group.
Replacement of the methyl substituent at the γ-position of the triketide with an ethyl in 2h successfully led to the new fully elongated product 1d, while replacement with a hydrogen in 2g did not. The same negative result was obtained with 2 f, when the α-hydrogen in the natural triketide was replaced with a methyl group.
These experiments demonstrate clearly that the subunits PikAII–PikAIV of the Pik PKS have sufficient flexibility to process some analogues of the natural triketide in which the structural components introduced by the loading domain and module 1 of PikAI have been varied. In all cases the yields were extremely poor. In the case of triketides 2b, 2c, and 2h, production of the pikromycin analogue was <1 % of the normal pikromycin production (and thus more than a tenfold decrease over yields of pikromycin made by feeding the natural triketide 2a to strain BB138, Table 1). The vinyl triketide analogue 2i was utilized more efficiently than other unnatural analogues (~1 % relative yield, calculated by using a standard curve for pikromycin, Table 1). We cannot discount the possibility that the triketide analogues are either taken up more poorly or degraded faster than the natural triketide. Nonetheless, the compounds are very similar in structure, and we hypothesize that the low yields seen with all triketide analogues relative to the natural triketide are consistent with slower loading and elongation of these analogues due to the specificity of the Pik PKS modules. Presumably triketide analogues that did not result in detectable production of new products were either not loaded and processed by the Pik PKS, or did so very slowly.
In the cases of the triketides 2a–c, h, and i, analogues 7a–e (Figure 4) of narbomycin (pikromycin lacking the C-13 hydroxyl substituent) were also observed in levels higher than their hydroxylated counterparts, as indicated by LC-MS analyses (the low titers precluded an accurate determination of this ratio). Compounds 7b, c, and e were purified, and accurate mass analyses (Table 2) were shown to be consistent with the proposed structures (Figure 4). These observations clearly demonstrate that PikC is able to process analogues of the natural substrate, but likely does so with reduced catalytic efficiency. For none of these four triketide analogues were there detectable levels of the 14-membered aglycone products. This observation demonstrated that DesVII–DesVIII,[23] a protein complex that catalyzes attachment of D-desosamine, can tolerate structural variations in the C13–C14 region of the aglycone structure. The lack of aglycone products also indicates that the low levels of pikromycin products made from these triketides is not a result of poor hydroxylation and glycosylation, and more likely poor processing by the Pik PKS.
Table 2.
HRMS data of compounds 7b, 7c, and 7e.
| Compound | Calculated [M+H] | Observed [M+H] |
|---|---|---|
| 7b | 524.3587 | 524.3563 |
| 7c | 524.3587 | 524.3560 |
| 7e | 508.3274 | 508.3250 |
Antibacterial activity of Δ15,16-dehydropikromycin
A small quantity of Δ15,16-dehydropikromycin (1e) was isolated and its antibacterial activity against three different bacteria was compared to that of natural pikromycin (Table 3). In this preliminary analysis a twofold improvement in the MIC99 was observed. Directed biosynthesis of erythromycin analogues bearing an azido group at C15 has recently been reported.[15] In this case, a twofold improvement in activity against the Gram-negative respiratory pathogen H. influenzae was observed relative to erythromycin (this difference was not observed with other bacteria). These two studies show that directed-biosynthesis can be used to generate pikromycin and erythromycin analogues bearing new C14 substituents that either do not impact biological activity, or lead to a modest improvement. The method also allows vinyl and ethylamino groups to be added, thereby providing chemical handles for additional modification steps.
Table 3.
Antibacterial activity of 1a and vinyl analogue 1e.[a] MIC99 is the minimum concentration at which no visible growth was detected.
| Test organism ok? | MIC99 [μm][a] | |
|---|---|---|
| 1a | 1e | |
| E. coli To1C | 50–60 | 20–30 |
| S. aureus NorA | 90–100 | 40–50 |
| Bacillus subtilis | 25–30 | 10–15 |
In conclusion, the data show for the first time that the Pik PKS system is amenable to directed biosynthesis and that analogues of the natural triketide intermediate can be used to generate new and potentially useful antibiotics.
Experimental Section
All reactions were carried out with dry solvents under anhydrous conditions (under nitrogen), ok? unless otherwise noted. All solvents and reagents were purchased from Aldrich. Normal-phase flash column chromatography was carried out on Davisil® silica gel (100–200 mesh, Fisher). Preparative thin-layer chromatography (PTLC) separations were carried out on 1 or 2 mm Merck silica gel plates (60F–254). 1H NMR spectra were recorded on Tecmag Libra-modified NM-500 MHz or Bruker AC-F 300 MHz spectrometers and calibrated by using residual undeuterated solvent as an internal reference. 13C NMR spectra were recorded on Bruker AMX-400 MHz or Bruker AC-F 300 MHz NMR spectrometers. High-resolution mass spectra were recorded on a Micromass LCT Electrospray mass spectrometer at the Mass Spectrometry & Proteomics Facility (Ohio State University).
HPLC and LC-MS analysis
HPLC was performed on a system equipped with Waters 600 pump connected to Waters 2487 Dual Absorbance. Products were analyzed by using a 5 μm Discovery HS C18 reversed-phase column (4.6 × 250 mm, Supelco) with an elution gradient from 20 to 80 % acetonitrile in ammonium acetate buffer (10 mM) at flow rate of 1 mL min−1. LC/MS analysis was performed by using the same gradient solvent system as described for HPLC analysis at a flow rate of 0.3 mLmin−1 on a Surveyor HPLC system (Thermofinnigan) connected to diode ray detector equipped with a 2.1 μm Discovery HS C18 reversed-phase column (4.6 × 250 mm, Supelco). Mass spectra were collected on an LCQ quadrupole ion trap (Thermofinnigan) mass spectrometer equipped with an electrospray ion source operating in positive mode.
Strains
The construction of PikAI deletion mutant (BB138) used for the feeding studies has been described previously.[24] The plasmid SG1 was constructed from pDHS702[5] by digestion with BamHI and Bg1II and religation by using a Roche Ligation kit. For the construction of pBK51, a BamHI–Bg1II fragment of the pikAI complementation plasmid pDHS722[24] was subcloned, and the desired mutation was introduced by using a QuickChange kit and the primers 5′-GTGGACACGGCCGCTAGCTCGTCGCTG-3′ and GTCCAC 5′CAGCGACGAGCTAGCGGCCGT. The resulting mutated fragment was then subcloned back into pDHS722 to give pBK51 according the methodologies previously described.[25] S. venezuelae transformants were selected on R2YE agar plates by overlaying with thiostrepton (1 mL, 500 μgmL−1). SGGP liquid medium was used for propagation of all the mutants of Streptomyces.
Feeding experiments
A loop full of spores of BB138 was inoculated in SGGP medium (10 mL) in a 50 mL flask and grown for 16 h at 30°C and 220 rpm. This seed culture (100 μL) was then used to inoculate fresh SGGP medium (10 mL) in a 50 mL flask. The culture was grown for 8 h at 30 °C and 220 rpm, after which triketide SNACs (1 mM) were added to it, and the culture was grown for an additional 3 days. At the end of the fermentation period the mycelia were removed by centrifugation, the pH of the supernatant was adjusted to 9.5 (1 N NaOH)), and the supernatant ? was extracted with ethyl acetate (2 × 20 mL). The organic extract was concentrated by rotary evaporation, redissolved in methanol, and analyzed by HPLC and LC/MS. The feeding of compounds 2 a–i was performed in triplicate.
Quantification of pikromycin analogues
Pikromycin was purified from S. venezuelae (wild-type) by HPLC on a C-18 column. Specific amounts of pikromycin were injected into the chromatograph, and its UV absorption was determined. A standard plot of concentration versus area under the peak (UV absorption) was obtained and used to quantify the production levels of other pikromycin analogues.
Determination of MIC99
Minimum inhibitory concentrations were determined in triplicate according to the broth microdilution method.[26] E. coli TolC, S. aureus NorA, and Bacillus subtilis (stock solutions in glycerol) were grown overnight at 37°C in LB, tryptic broth, and nutrient broth media respectively. The cells were diluted to ~0.001 OD600 with the respective media. The assay was performed in a 96-well plate. Stock solutions of compounds 1a and 1e in ethanol introduced into a 96-well plate were diluted with medium to produce the desired concentration. The plate was incubated for 6 h at 37°C for E. coli TolC and S. aureus NorA, and over-night for Bacillus subtilis. MIC99 values are reported as the minimum concentration at which there was no visible growth. The OD600 was used as a measure of cell growth.
Chemical synthesis
General procedure for aldol coupling
Diisopropylethylamine (25.8 mmol) and dibutylboron triflate (1.0 M in CH2Cl2, 25.8 mmol) were added to a stirred solution of Evans’ acyl-oxazolidinone (25.8 mmol) in CH2Cl2 (0.3 M) at 0 °C. The resulting reaction mixture was stirred at 0°C for 30 min and then cooled to −78°C. Propionaldehyde (17.2 mmol) in CH2Cl2 (0.5 M) was added, and the mixture was stirred at −78 °C for 1 h and then allowed to warm to 0 °C. After the mixture had been stirred for 2 h at this temperature, the reaction was quenched by addition of phosphate buffer pH 7 (20 mL). The reaction mixture was poured into a flask containing MeOH (85 mL) at 0°C, treated with precooled 30% H2O2 (107 mL), and stirred at 0 °C for 1 h. MeOH was removed by rotary evaporation, saturated aqueous NaHCO3 was added, and the resultant aqueous layer was extracted with CH2Cl2 (3 × 100 mL) and purified by flash silica gel chromatography to afford aldols 3.
General procedure for protection of aldol 3
2,6-Lutidine (14.9 mmol) was added to a solution of aldols 3 (4.9 mmol) in CH2Cl2 (0.2 M) at 0 °C. After the mixture had been stirred for 5 min at that temperature, tert-butyldimethylsilyltrifluoromethane sulfonate (7.5 mmol) was added dropwise, and the reaction mixture was stirred at 0°C for 20 min, after which time no starting material was detected by TLC. Saturated aqueous NH4Cl was then added. The organic phase was separated, and the aqueous layer was extracted with CH2Cl2 (3 × 30 mL). The combined organic extracts were dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography.
General procedure for benzylation
nBuLi (1.6 M in hexanes, 6.4 mmol) was added to a solution of benzyl alcohol (10.4 mmol) in THF (0.5 M) at 0 °C. The resulting solution was stirred at 0 °C for 30 min, and a solution of the protected aldols 3 (4.9 mmol) in THF (0.8 M) was added. The reaction mixture was stirred at this temperature for 5 h and then quenched by addition of saturated aqueous NH4Cl. The organic phase was separated, and the aqueous layer was extracted with diethyl ether ok? (3 × 20 mL). The combined organic extracts were washed with water, dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography to afford the benzyl esters 4.
General procedure for DIBAL-H reduction
DIBAL-H (1 M in hexanes, 8.9 mmol) was added to a solution of 4 (4.1 mmol) in CH2Cl2 (0.6 M) at −78 °C. The mixture was stirred at −78°C for 30 min and then warmed up to 0°C. After the mixture had been stirred at 0°C for 30 min, the reaction was quenched with MeOH (0.2 mL), and the mixture was diluted with CH2Cl2 (14 mL). A saturated aqueous solution of Rochelle salt (14 mL) was added, and the mixture was stirred at room temperature until there were two clear layers. The organic layer was separated, and the aqueous layer was extracted with diethyl ether (3 × 20 mL). The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography to afford alcohols 5.
General procedure for Wittig olefination
Dess–Martin periodinane (3.1 mmol) was added to a solution of 5 (2.6 mmol) in CH2Cl2 (0.2 M) at 0 °C. The resulting mixture was stirred at room temperature for 30 min, after which time it was subjected to silica gel column chromatography. The column was eluted with 10 % EtOAc in hexane to obtain the corresponding aldehyde as colorless oil, which was used directly in the next step. Methyl (triphenylphosphoranylidene) acetate was added to a solution of aldehyde (2.6 mmol) in THF (0.06 M). The mixture was heated under reflux for 24 h, after which time no starting material was detected by TLC. The reaction mixture was applied to a silica gel column, and the unsaturated esters were eluted with 1% EtOAc in hexane.
General procedure for hydrolysis of unsaturated esters
Potassium carbonate (3.5 mmol) was added to a solution of the unsaturated esters (0.69 mmol) in methanol (0.08 M) and water (0.25 M). The reaction mixture was heated under reflux for 3 h, after which methanol was removed by rotary evaporation, and the aqueous layer was acidified to pH 2 with concentrated HCl. The mixture was saturated with solid NaCl and extracted with diethyl ether (3 × 15 mL). The combined organic extracts were washed with saturated aqueous NaHCO3, dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography to afford the unsaturated acid 6.
General procedure for the preparation of protected NAC
Thioesters: Triethylamine (1.1 mmol) and diphenylphosphorylazide (0.8 mmol) were added to a solution of 6 (0.5 mmol) in DMF (0.2 M) at 0 °C. The reaction mixture was stirred at 0 °C for 2 h. N-acetylcysteamine (0.6 mmol) was added, and the mixture was stirred at room temperature for 2 h after which water (2.6 mL) was added. The organic layer was separated, and the aqueous layer was extracted with EtOAc (3 × 2.6 mL). The combined organic extracts were dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography.
General procedure for the deprotection and preparation of final NAC thioesters
Hydrofluoric acid (48 % wt in H2O) was added to a solution of the protected NAC thioester (0.4 mmol) in acetonitrile (0.1 M) and water (0.6 M). After being stirred for 2 h at room temperature the reaction mixture was cooled to 0°C and the pH was adjusted to 7.5 with saturated aqueous NaHCO3. Acetonitrile was removed by rotary evaporation, and the resultant aqueous layer was extracted with EtOAc (3 × 10 mL). The combined organic extracts were dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography to afford the final Triketide NAC thioesters 2a–f.
General procedure for Weinreb amides
AlMe3 (2.0M in heptane, 0.9 mmol) was added to a suspension of N,O-dimethylhydroxylamine hydrochloride (0.9 mmol) in THF (1.9 M) at 0°C. The solution was stirred at room temperature for 30 min and then cooled to −15°C. A solution of 3g–i (0.3 mmol) in THF (0.7 M) was added, and the mixture was allowed to warm to 0 °C. After stirring at 0°C for 2.5 h, CH2Cl2 (0.1 M) and HCl (0.5 N, 0.06 M) were added to the mixture, and it was stirred at 0°C for an additional 1 h. The organic layer was separated, and the aqueous layer was extracted with CH2Cl2 (3 × 5 mL). The combined organic extracts were washed with saturated aqueous NaHCO3, dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography.
General procedure for protection of Weinreb amides
2,6-Lutidine (15.3 mmol) was added to a solution of alcohol (5.1 mmol) in CH2Cl2 (0.2 M) at 0 °C. After stirring for 5 min at that temperature, tert-butyldimethylsilyltrifluoromethane sulfonate (7.7 mmol) was added dropwise to the reaction mixture, and it was stirred at 0°C for 20 min, after which time no starting material was detected by TLC. Saturated aqueous NH4Cl was added. The organic phase was separated, and the aqueous layer was extracted with CH2Cl2 (3 × 30 mL). The combined organic extracts were dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography.
General procedure for DIBAL-H reduction of Weinreb amides
To a solution of the protected Weinreb amide (3.3 mmol) in THF (0.08M) at −78°C was added DIBAL-H (1 M in hexanes, 6.6 mmol). The mixture was stirred at −78°C for 1 h, after which the reaction was quenched by the addition of acetone (0.3 mL). The solution was immediately poured into a vigorously stirring mixture of hexane (64 mL) and tartaric acid (0.5 M, 64 mL) at 0°C. The mixture was stirred at 0 °C for 30 min. The organic phase was separated, and the aqueous layer was extracted with dichloromethane (3 × 100 mL). The combined organic extracts were washed with water and brine, dried over anhydrous Na2SO4, concentrated, and purified by flash silica gel chromatography.
Analytical data for final triketide NAC thioesters 2a–i
Compound 2a
Rf = 0.18 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.94 (dd, J = 7.5, 15.5 Hz, 1H), 6.18 (dd, J = 1.0, 16.0 Hz, 1 H), 5.87 (br s ok?, 1H), 3.53 (m, 1 H), 3.48 (q, J =6.0 Hz, 2 H), 3.11 (t, J =6.0 Hz, 2 H), 2.45 (m, 1H), 1.98 (s, 3H), 1.63–1.53 (m, 1 H), 1.46–1.38 (m, 1 H), 1.12 (d, J =7.0 Hz, 3H), 0.99 (t, J =8.0 Hz, 3 H); 13C NMR (100 MHz, CDCl3): δ = 190.5, 170.5, 148.6, 128.4, 76.0, 42.4, 39.9, 28.6, 27.6, 23.4, 14.0, 10.5; HRMS calcd for C12H21NO3S + Na: 282.1140; found 282.1118 [M+Na].
Compound 2b
Rf = 0.19 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.93 (dd, J = 7.5, 15.5 Hz, 1 H), 6.15 (m, 2 H), 3.60 (br s, 1 H), 3.44 (q, J =6.0 Hz, 2 H), 3.08 (t, J =6.5 Hz, 2 H), 2.41 (m, 1 H), 2.06 (s, 1 H), 1.95 (s, 3 H), 1.41 (m, 4 H), 1.08 (d, J =6.5 Hz, 3 H), 0.92 (t, J =6.5 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ = 190.5, 170.6, 148.7, 128.4, 74.3, 42.8, 39.9, 36.8, 28.6, 23.4, 19.4, 14.2, 14.0; HRMS calcd for C13H23NO3S + Na: 296.1296; found 296.1278 [M+Na].
Compound 2c
Rf = 0.22 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.90 (dd, J = 8.0, 15.5 Hz, 1H), 6.16 (d, J = 16.0 Hz, 1 H), 6.01 (br s, 1 H), 3.46 (q, J =6.0 Hz, 2 H), 3.28 (m, 1 H), 3.09 (t, J = 6.5 Hz, 2H), 2.52 (m, 1 H), 1.97 (s, 3 H), 1.70 (m, 2 H), 1.11 (d, J = 6.0 Hz, 3H), 0.93 (d, J =7.0 Hz, 6H); 13C NMR (100 MHz, CDCl3): δ = 194.2, 170.2, 143.6, 135.5, 39.8, 39.4, 28.6, 27.9, 23.3, 15.1, 12.8, 10.2; HRMS calcd for C13H23NO3S + Na: 296.1296; found 296.1296 [M+Na].
Compound 2d
Rf = 0.15 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.94 (dd, J = 8.5, 16.5 Hz, 1H), 6.16 (d, J = 16.0 Hz, 1 H), 5.92 (br s, 1 H), 3.46 (q, J =6.0 Hz, 2H), 3.32 (m, 1H), 3.11 (t, J = 6.5 Hz, 2H), 2.57 (m, 1H), 1.98 (s, 3 H), 1.88 (m, 1 H), 1.10 (d, J = 7.5 Hz, 3H), 0.93–1.79 (m, 1 H); 13C NMR (100 MHz, CDCl3): δ= 190.6, 170.4, 149.6, 128.1, 78.6, 41.1, 40.0, 39.3, 29.9, 28.6, 27.9, 26.5, 26.4, 26.1, 23.5, 13.3; HRMS calcd for C16H27NO3S + Na: 336.1609; found 336.1605 [M+Na].
Compound 2e
Rf = 0.21 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.78 (dd, J = 8.0, 16.0 Hz, 1H), 6.19 (dd, J = 1.0, 15.5 Hz, 1 H), 5.87 (br s, 1H), 3.83 (m, 1 H), 3.48 (q, J =6.0 Hz, 2H), 3.12 (t, J =6.5 Hz, 2 H), 2.42 (m, 1H), 1.44 (d, J =5.0 Hz, 1 H), 1.20 (d, J = 6.5 Hz, 3H), 1.12 (d, J =6.5 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ = 190.3, 189.9, 147.6, 128.6, 70.6, 43.7, 39.8, 28.4, 23.3, 20.6, 14.3; HRMS calcd for C11H19NO3S + Na: 268.0983; found 268.0989 [M+Na].
Compound 2f
Rf = 0.12 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.64 (d, J = 10 Hz, 1 H), 5.89 (br s, 1H), 3.45 (m, 3 H), 3.08 (t, J =6.5 Hz, 2 H), 2.67 (m, 1H), 1.98 (s, 3 H), 1.92 (s, 3 H), 1.56 (m, 1 H), 1.38 (m, 1H), 1.09 (m, J =6.5 Hz, 3H), 0.98 (t, J =7.5 Hz, 3 H); 13C NMR (100 MHz, CDCl3): δ = 194.2, 170.2, 143.6, 135.5, 76.5, 39.8, 39.4, 28., 27.9, 23.3, 15.1, 12.8, 10.2; HRMS calcd for C13H23NO3S + Na: 296.1296; found 296.1296 [M+Na].
Compound 2g
Rf = 0.08 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.97 (m, 1H), 6.22 (d, J = 15.5 Hz, 1 H), 5.91 (br s, 1 H), 3.73 (m, 1H), 3.48 (q, J =6.0 Hz, 2H), 3.11 (t, J =6.0 Hz), 2.43 (m, 1H), 2.34 (m, 1H), 1.98 (s, 3 H), 1.55 (m, 2 H), 0.98 (t, J =7.5 Hz); 13C NMR (100 MHz, CDCl3): δ = 190.1, 170.2, 142.6, 130.5, 71.9, 39.8, 39.6, 30.1, 28.4, 23.2, 9.8; HRMS calcd for C11H19NO3S + Na: 268.0983; found 268.0989 [M+Na].
Compound 2h
Rf = 0.16 (5 % MeOH/CH2Cl2); 1H NMR (500 MHz, CDCl3): δ = 6.75 (dd, J = 9.5, 15.0 Hz, 1H), 6.16 (d, J = 15.5 Hz, 1 H), 6.0 (br s, 1H), 3.51 (m, 1H), 3.46 (q, J =6.0 Hz, 2 H), 3.10 (t, J = 6.5 Hz, 2H), 2.16 (m, 1 H), 1.97 (s, 3 H), 1.76 (m, 2H), 1.55 (m, 1 H), 1.36 (m, 2 H), 0.97 (t, J =8.0 Hz, 3 H), 0.87 (t, J =7.5 Hz, 3 H); 13C NMR (100 MHz, CDCl3): δ = 190.2, 170.4, 146.9, 129.9, 75.3, 51.0, 39.8, 28.5, 27.7, 23.3, 22.6, 12.0, 10.3; HRMS calcd for C13H23NO3S + Na: 296.1296; found 296.1299 [M+Na].
Compound 2i
Rf = 0.10 (5 % MeOH/CH2Cl2); 1H NMR (300 MHz, CDCl3): δ = 6.94 (dd, J = 15, 7.5 Hz, 1H), 6.16 (dd, J = 18, 1.2 Hz, 1 H), 5.95 (br s, 1 H), 5.83 (ddd, J =18, 9, 4.5 Hz, 1 H), 5.17–5.32 (m, 2 H), 4.13 (dt, J =9, 1.2 Hz, 1H) 3.46 (q, J =15, 6 Hz, 2H), 3.09 (t, J =6 Hz, 2 H), 2.54 (dsext, J =6, 1.2 Hz, 1 H), 1.96 (s, 3 H), 1.79 (br d, J = 18.9 Hz, 1H), 1.09 (d, J =6.3 Hz, 3 H); 13C NMR (75 MHz, CDCl3): δ = 190.1, 170.4, 147.4, 137.9, 128.4, 116.6, 75.6, 42.2, 39.7, 28.4, 23.2, 14.2; HRMS calcd for C12H19NO3S + Na: 280.0978; found 280.0990 [M+Na].
Acknowledgments
This work was supported by a grant from the NIH (R01 GM0876477).
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 2.O’Hagan D. The Polyketide Metabolites. Ellis Horwood; town?: 1991. [Google Scholar]
- 3.Staunton J, Weissman KJ. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 4.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 5.Xue Y, Sherman DH. Nature. 2000;403:571–574. doi: 10.1038/35000624. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen JR, Keatinge-Clay AT, Cane DE, Khosla C. Bioorg Med Chem. 1998;6:1171–1177. doi: 10.1016/s0968-0896(98)00081-9. [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen JR, Hutchinson CR, Cane DE, Khosla C. Science. 1997;277:367–369. doi: 10.1126/science.277.5324.367. [DOI] [PubMed] [Google Scholar]
- 8.Alhamadsheh MM, Palaniappan N, Daschouduri S, Reynolds KA. J Am Chem Soc. 2007;129:1910–1911. doi: 10.1021/ja068818t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota T, Brunjes M, Frenzel T, Xu J, Kirschning A, Floss HG. Chem-BioChem. 2006;7:1221–1225. doi: 10.1002/cbic.200500506. [DOI] [PubMed] [Google Scholar]
- 10.Ward SL, Desai RP, Hu Z, Gramajo H, Katz L. J Ind Microbiol Biotechnol. 2007;34:9–15. doi: 10.1007/s10295-006-0156-6. [DOI] [PubMed] [Google Scholar]
- 11.Boddy CN, Hotta K, Tse ML, Watts RE, Khosla C. J Am Chem Soc. 2004;126:7436–7437. doi: 10.1021/ja048108s. [DOI] [PubMed] [Google Scholar]
- 12.Hartung IV, Rude MA, Schnarr NA, Hunziker D, Khosla C. J Am Chem Soc. 2005;127:11202–11203. doi: 10.1021/ja051430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weist S, Sussmuth RD. Appl Microbiol Biotechnol. 2005;68:141–150. doi: 10.1007/s00253-005-1891-8. [DOI] [PubMed] [Google Scholar]
- 14.Thiericke R, Rohr J. Nat Prod Rep. 1993;10:265–289. doi: 10.1039/np9931000265. [DOI] [PubMed] [Google Scholar]
- 15.Ashley GW, Burlingame M, Desai R, Fu H, Leaf T, Licari PJ, Tran C, Abbanat D, Bush K, Macielag M. J Antibiot. 2006;59:392–401. doi: 10.1038/ja.2006.56. [DOI] [PubMed] [Google Scholar]
- 16.Cane DE, Lambalot RH, Prabhakaran PC, Ott WR. J Am Chem Soc. 1993;115:522–526. [Google Scholar]
- 17.Beck BJ, Aldrich CC, Fecik RA, Reynolds KA, Sherman DH. J Am Chem Soc. 2003;125:4682–4683. doi: 10.1021/ja029974c. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe K, Wang CC, Boddy CN, Cane DE, Khosla C. J Biol Chem. 2003;278:42020–42026. doi: 10.1074/jbc.M305339200. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson AL, (née Cutter), Hanefeld U, Wilkinson B, Leadlay PF, Staunton J. Tetrahedron Lett. 1998;39:9827–9830. [Google Scholar]
- 20.Lambalot RH, Cane DE. J Antibiot. 1992;45:1981–1982. doi: 10.7164/antibiotics.45.1981. [DOI] [PubMed] [Google Scholar]
- 21.Wu N, Tsuji SY, Cane DE, Khosla C. J Am Chem Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 22.Kim BS, Cropp TA, Florova G, Lindsay Y, Sherman DH, Reynolds KA. Biochemistry. 2002;41:10827–10833. doi: 10.1021/bi0256779. [DOI] [PubMed] [Google Scholar]
- 23.Borisova SA, Zhang C, Takahashi H, Zhang H, Wong AW, Thorson JS, Liu HW. Angew Chem. 2006;118:2814–2819. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:2748–2753. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]
- 24.Yoon YJ, Beck BJ, Kim BS, Kang HY, Reynolds KA, Sherman DH. Chem Biol. 2002;9:203–214. doi: 10.1016/s1074-5521(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 25.Kim BS, Sherman DH, Reynolds KA. Protein Eng Des Sel. 2004;17:277–284. doi: 10.1093/protein/gzh032. [DOI] [PubMed] [Google Scholar]
- 26.Domenech P, Reed MB, Barry CE., 3rd Infect Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]