Abstract
Surface modification by surface-mediated polymerization necessitates control of the grafted polymer film thicknesses to achieve the desired property changes. Here, a microarray format is used to assess a range of reaction conditions and formulations rapidly in regards to the film thicknesses achieved and the polymerization behavior. Monomer formulations initiated by eosin conjugates with varying concentrations of poly(ethylene glycol) diacrylate (PEGDA), N-methyldiethanolamine (MDEA), and 1-vinyl-2-pyrrolidone (VP) were evaluated. Acrylamide with MDEA or ascorbic acid as a coinitiator was also investigated. The best formulation was found to be 40 wt% acrylamide with MDEA which yielded four to eight fold thicker films (maximum polymer thickness increased from 180 nm to 1420 nm) and generated visible films from 5-fold lower eosin surface densities (2.8 vs. 14 eosins/µm2) compared to a corresponding PEGDA formulation. Using a microarray format to assess multiple initiator surface densities enabled facile identification of a monomer formulation that yields the desired polymer properties and polymerization behavior across the requisite range of initiator surface densities.
Keywords: surface-mediated polymerization, aqueous monomer formulations, visible light photopolymerization
1. Introduction
Surface-mediated polymerization is a capable and valuable method for modifying surface properties for a variety of applications including improved dispersion of nanoparticles [1,2], generation of antifouling coatings [3,4], cell encapsulation [5,6], drug delivery [7], microfluidic device fabrication [8] and biodetection [9–14]. An important parameter in determining the effectiveness of almost any surface graft in these applications is film thickness. Allowing for both spatial and temporal control, photoinitiation of surface polymerization is particularly advantageous for achieving the desired surface changes in defined regions [15–18]. Light intensity and surface density of photoinitiators are readily manipulated to control the polymerization extent. However, a critical, frequently overlooked factor is the monomer formulation itself, which obviously determines the range of thicknesses that are obtained by changing other factors.
Alternatively to photopolymerization, surface-mediated polymerizations are also achieved via thermal polymerization [1,19], atom transfer radical polymerization (ATRP) [13,20], reversible addition fragmentation chain transfer (RAFT) polymerization [14,21], and anionic or cationic polymerization [22,23]. Modifying surface densities of these types of initiating species also allows for control of film properties, but patterning of these surfaces is less straightforward than in photopolymerizable systems in which a simple photomask may be employed. Well known for their living polymerization characteristics, ATRP and RAFT are widely used for controlled surface modifications; however, these polymerizations proceed more slowly and tend to result in thinner films than non-living/controlled radical polymerization systems. Anionic and cationic polymerization can also have living characteristics, but are sensitive to the presence of water, a significant limitation in many biological applications.
This article compares the film thicknesses achieved from a range of eosin surface densities using several aqueous monomer formulations most commonly reported for use with eosin. To assess a broad range of initiator surface densities simultaneously, eosin is surface-immobilized in a microarray format in which each row of spots comprises a different initiator surface density. One specific motivation for the work presented here is to enhance the capabilities of the eosin-based biodetection platform presented by Hansen et al. [10] in which eosin is coupled to streptavidin, yielding a species that both participates in biorecognition and initiates polymerization from the test surface such that the presence of a polymer film indicates a positive biodetection response. Surface-mediated or interfacial polymerization with eosin has also been used for islet cell encapsulation from cell membrane-embedded eosin [5], to improve dispersion of silica nanoparticles [1] and for polymer surface coatings of arterial tissues [24]. For the proposed biodetection application it is desirable to achieve the highest polymer-based amplification, i.e. the thickest films possible from the fewest number of initiators on the surface and the formation of a visible polymer film from as few bioselectively coupled eosin moieties as possible. This outcome should be achieved while maintaining a dynamic response in thickness related to initiator surface density to facilitate analyte quantification.
Eosin is a visible light initiator which is typically used with a tertiary amine coinitiator such as methyldiethanolamine (MDEA). MDEA reacts with the photoexcited triplet state of eosin to undergo energy, charge, and electron transfer yielding a relatively stable eosin radical and a MDEA radical capable of initiating polymerization. Although the initiating radical is on the non-tethered coinitiator, the reaction is still considered surface-mediated since initiating radicals are only generated in the vicinity of eosin which is surface-tethered. Compared to UV initiators, visible light initiators are desirable in applications that require larger cure depths and in biological applications such as cell encapsulation where high doses of UV will damage proteins and DNA.
Aqueous monomer formulations are requisite in biological applications to maintain protein function, plus have the advantage of using a non-toxic solvent. A commonly used aqueous monomer formulation for use with eosin is PEGDA with a tertiary amine coinitiator and vinyl pyrrolidone (VP) as an accelerant [5,6,25–27]. Cruise and Kizilel have reported changes in film thickness by changing PEGDA, amine and VP concentrations; however, only up to three eosin surface densities were investigated [5,6,27]. We investigate changes to these factors across a wide range of eosin surface densities. Another common formulation used with eosin for the generation of holographic recording material is acrylamide with a tertiary amine coinitiator [28,29]. Acrylamide has also been used with ascorbic acid to initiate polymerization with eosin, and this reaction is reported to be either insensitive to [1] or favored by [30] oxygen. While not necessary, it would be beneficial for a biodetection platform to be resistant to oxygen inhibition. All of these formulations were evaluated in regards to both their bulk and surface-mediated polymer characteristics.
2. Experimental
2.1. Materials
Epoxy-functionalized glass slides (SuperEpoxy 2), 2X Protein Printing Buffer and solid printing pins were purchased from Telechem International, Inc. Biotin-functionalized slides were purchased from Xenopore Corp. Streptavidin, poly(ethylene glycol) diacrylate (PEGDA) (Mn = 575), 40% acrylamide/bis-acrylamide (19:1) in water, 1-vinyl-2-pyrrolidone (VP), N-methyldiethanolamine (MDEA), and ascorbic acid were purchased from Sigma-Aldrich. PEGDA was dehibited twice using columns from Scientific Polymer Products that remove MEHQ. Acrylamide was provided without inhibitor added. Eosin isothiocyanate was purchased from Invitrogen. A Cy3 Scanner Calibration slide was purchased from Full Moon BioSystems. Silicone gasket material was purchased from Grace Bio-Labs. Water was purified using a Milli-Q system.
2.2. Preparation of surfaces with eosin
2.2.1. Preparation and characterization of microarray surfaces
Streptavidin-eosin conjugate was prepared as described previously [10] by reacting eosin isothiocyanate with amines on the protein’s surface. The conjugate was printed onto epoxy-functionalized glass slides by a VersArray Chip Writer™ Pro (Bio-Rad) with a solid pin yielding spots of approximately 500 µm diameter. The surface epoxy groups form a covalent linkage with amine and thiol groups present on the protein. 9 ×5 arrays of spots were prepared containing five replicate spots of 8 decreasing eosin concentrations, and a 9th row with no eosin that served as a negative control suitable for evaluating non-specific polymerization. The highest print concentration was 0.27 mg/mL streptavidin-eosin conjugate (12 µM eosin, 1.3 eosin per streptavidin) in a final concentration of 1X printing buffer. Lower print concentrations were prepared by serial 1:3 dilutions into 1X print buffer with unmodified streptavidin such that all spots were printed with a total protein concentration of 0.27 mg/mL. Slides were printed with about 45% humidity and were allowed to sit in humidity for 10 minutes after spotting was completed, then were rinsed four times in water with rocking and finally were dried with nitrogen air flow and stored in conditions that protected from light exposure. Since eosin is a fluorophore, initiator surface density was characterized with an Agilent Technologies Microarray Scanner. A Cy3 Scanner Calibration slide was used to convert fluorescence readings into surface density of fluorophores.
2.2.2. Preparation of surfaces for IR studies
Streptavidin-eosin conjugate at a concentration of 30 µg/mL in PBS was incubated for 30 minutes with rocking on either biotin-functionalized glass slides or epoxy-functionalized glass slides, followed by four ten minute water rinses. Although streptavidin binds biotin non-covalently, it binds with extremely high affinity (Kd = 10−15), such that the reaction with biotin slides yields higher surface eosin densities than the reaction with epoxy slides.
2.3. Polymerization procedures
2.3.1. Polymerization from microarrays
The Chip Clip System from Whatman was used to form wells around each of the 2 arrays per slide, and 300 µL of monomer mixture was added per well. During the five minutes prior to and throughout the entire 30 minute light exposure, the slides were placed in a plastic bag with argon flow to reduce oxygen in the atmosphere. The light source was an Acticure (Exfo) high pressure mercury lamp with an in-house internal bandpass filter (350 nm –650 nm) and an external 490 nm longpass filter (Edmund Optics) positioned at the end of a light guide and a collimating lens. The light intensity was measured as 260 mW/cm2 using an International Light radiometer with no filter on the radiometer. After polymerization, unreacted monomer was removed with three five minute water rinses, followed by air drying. Each formulation investigated was polymerized from at least three eosin arrays in three separate polymerization sessions.
2.3.2. Real time IR (RTIR) monitoring of polymerizations
A Nicolet Magna-IR 760 E.S.P. spectrometer adapted with a horizontal sample holder was used to monitor polymerizations in real time, tracking the peak centered at approximately 6175 cm−1. C=C conversion was calculated by the change in peak area. Sample chambers were prepared using binder clips to clamp two glass slides on either side of a 1 mm silicone gasket cut to have a 2 cm × 1 cm hole for monomer. Argon (or pure oxygen where indicated) was bubbled through monomer for 20 minutes, then monomer was introduced to the sample chamber with a syringe and needle. The same light setup was used as described above, except the intensity was 540 mW/cm2. Each condition was tested in triplicate. In the case of surface-mediated polymerization, the bottom slide of the sample chamber comprised an eosin-coated slide. In solution-initiated polymerizations, eosin-isothiocyanate instead of eosin was used to employ a dye as similar as possible to that present when the streptavidin-eosin conjugate is used on the surface.
2.4. Characterization of Microarray films
The thickness of the fully dehydrated polymer films grown from microarray spots was measured using a Dektak 6M surface profilometer with 12.5 µM diameter tip and a stylus force of 1mg. Plots of film thickness versus eosin surface density were made and the data were fit to the equation [thickness = k*(b-density)a] using a least squares parameter estimation. This equation is proposed because it incorporates the threshold behavior observed and because it relates thickness to initiator surface density raised to some power, which is an expected relationship since it is generally observed that the polymerization rate scales with the initiation rate to some power [31], that power classically being 0.5 under ideal conditions. The purpose of the curve fit is primarily to guide the eyes, as the calculated parameters do not reveal any obvious trends.
3. Results and discussion
3.1. Variations to a PEGDA, MDEA, and vinyl pyrrolidone aqueous formulation
3.1.1 Microarray results
Aqueous monomer formulations are used to form hydrogels for a wide variety of applications including cell encapsulation [5,6], biodetection [10,11,13,14], and electrophoresis [32]. Surface modification of materials by photografted or polymerized hydrogels is also frequently used. [1,4,10,11] The standard formulation used here contains 22% PEGDA (423mM), 210 mM methyldiethanolamine (MDEA), and 35 mM 1-vinyl-2-pyrrolidone (VP) in water, resembling similar formulations published by Cruise and by Kizilel [5,6,25–27], except for the utilization of triethanolamine instead of MDEA, and a pH of 8, instead of pH = 9. Cruise and Kizilel varied PEGDA, VP, and amine coinitiator concentrations by two to four-fold and reported that film thicknesses are affected by a factor of about two or less, though the model Kizilel presents suggests that differences on the order of 10-fold may be achieved [5,6,27]. Our initial trials revealed that two-fold variations of any of the components had little effect (data not shown), so larger variations in concentrations were evaluated. The PEGDA concentration was increased two and four-fold from the initial value, MDEA concentration was varied by a factor of ten, and VP concentration was varied up by ten-fold or alternatively no VP was used. All formulations were adjusted to pH = 9 using 1N HCl or 1N NaOH.
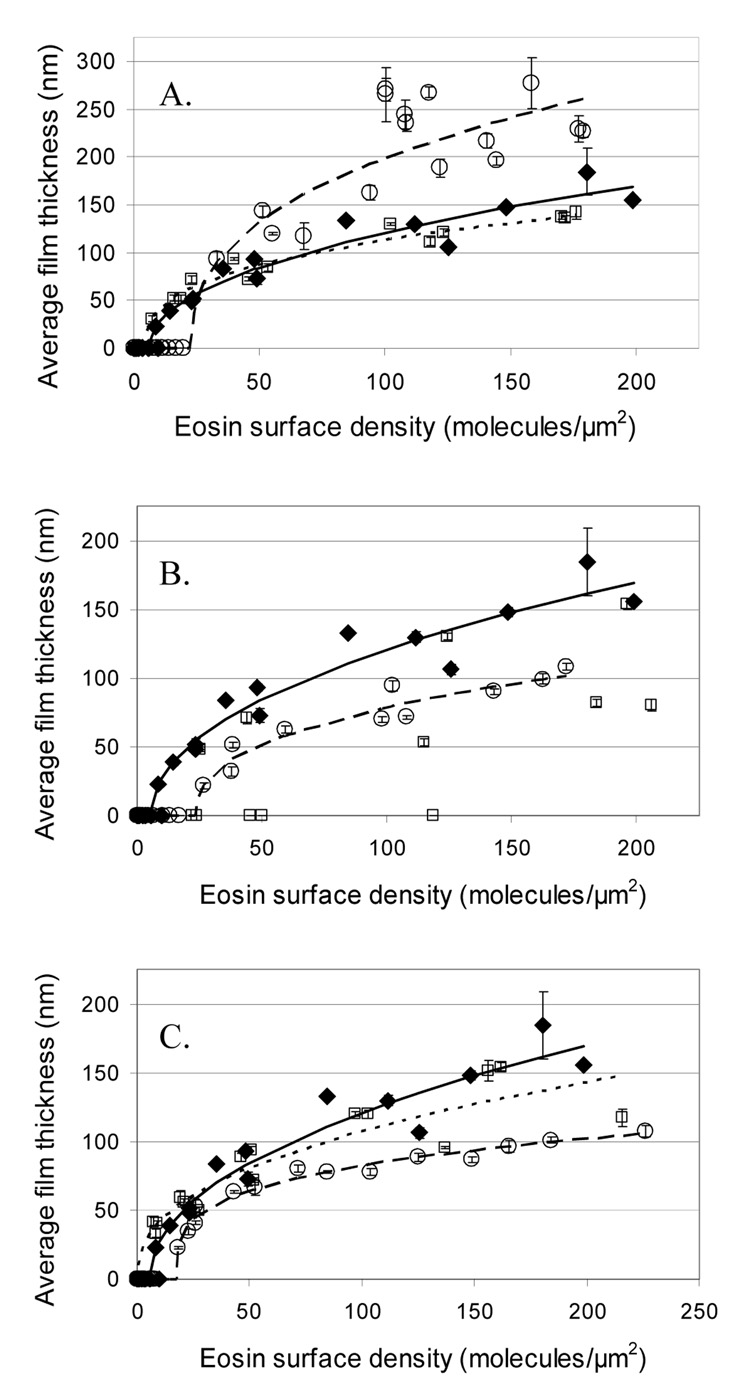
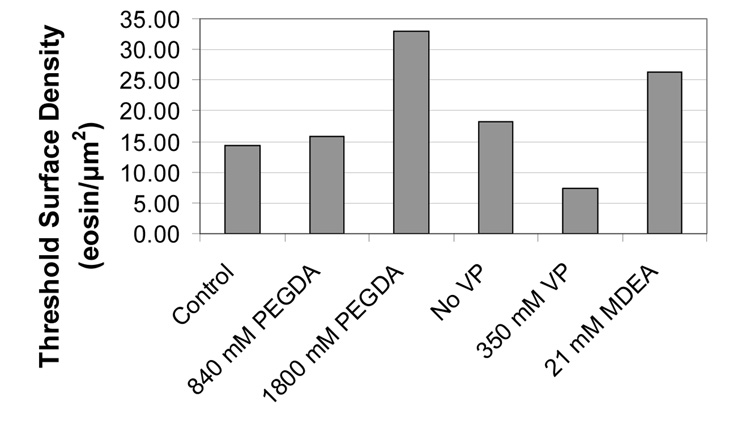
Surprisingly, this polymerization system is incredibly amenable and robust in its response to drastic changes in the concentrations of these components (Fig. 1). All formulations grew polymer films at high eosin surface densities. The thickest films were seen with the 1800 mM PEGDA formulation (a nonaqueous formulation) which had films approaching 300 nm, compared to less than 200 nm for the other concentrations. Unfortunately, 1800 mM PEGDA resulted in reduced sensitivity, with a threshold eosin surface density of 33 eosins per µm2, while the standard formulation could generate visible films from as few as 14 eosins per µm2 (Fig.2). 420 mM and 840 mM PEGDA formulations had very similar film thicknesses across all of the eosin surface densities evaluated. Reducing MDEA by ten-fold resulted in thinner films and a higher threshold eosin surface density, while increasing MDEA ten-fold was the least favorable condition of all, lacking consistent film growth until more than 180 eosins per µm2 were on the surface. While omitting VP resulted in thinner films, increasing VP ten fold resulted in only slightly thinner films, yet had the best sensitivity of all the formulations, with a threshold eosin surface density of only 7 eosins per µm2.
Fig. 1.
Effect on film thickness of varying the concentration of PEGDA, MDEA, and VP. Curves are fit to the data using the equation thickness = k*(b-density)a. A) Vary PEGDA concentration: 420 mM (◆ and ——), 480 mM (□ and ■■■), and 1800 mM (○ and — —); B) Vary MDEA concentration: 21 mM (○ and — —), 210 mM (◆ and ——), and 2100 mM (□); C) Vary VP concentration: no VP (○ and — —), 35 mM VP (◆ and — —), and 350 mM VP (□ and ■■■). Unless otherwise indicated, PEGDA is 420 mM, MDEA is 210 mM and VP is 35 mM, which is considered the standard formulation, represented in all cases by ◆ and — —. Error bars represent the standard error from five replicate spots on the array. No curve was fit to the 2100 mM MDEA formulation because there were too few non-zero thicknesses.
Fig. 2.
Minimum (threshold) surface density of eosin that consistently resulted in visible polymer films. In all cases, the light intensity was 260 mW/cm2 light and the exposure time was 30 minutes.
In comparing our results with those published previously by Cruise and Kizilel [5,6,27], a few important considerations should be noted. Firstly, our gel thicknesses are measured after the hydrogels have completely dried (this approach is necessary since gels of 500 µm diameter dry very quickly when removed from water). Conversely, Cruise measured swollen hydrogel thicknesses and Kizilel does not specify whether the gels are hydrated. This difference helps to explain why others report films on the order of tens to hundreds of microns, while the films we measure are on the order of tens to hundreds of nanometers. Also, the fact that we measure dry film thicknesses may explain why tenfold adjustments in MDEA and VP concentrations were required to see differences in film thickness. It is possible that differences in film thicknesses may be more pronounced in swollen gels, which are affected both by crosslink density (which is a strong function of polymerization conditions) and the amount of polymer present [33]. Another important consideration is that these other results utilize only up to three eosin surface densities whose values are not reported. In the experiments presented here, a wide range of well-characterized eosin surface densities are investigated. Since the observed trends change with eosin surface density, surfaces prepared by different methods, and thus at different densities, are likely to exhibit varying trends.
Taking these considerations into account, comparisons can be made between the data presented here and the results presented by Cruise and Kizilel. Both Cruise and Kizilel find that film thickness is increased by using a higher monomer concentration, which is in agreement with our finding that 1800 mM PEGDA grows thicker films than 420 mM and 840 mM PEGDA at high eosin surface densities. Cruise finds that decreased VP results in thinner films, while Kizilel predicts that increasing VP will decrease thickness. We find that both increasing and decreasing VP may reduce film thickness at high eosin surface densities, but that at low initiator densities, the high VP condition yields polymer films when the lower VP conditions do not. Finally, both Cruise and Kizilel find or predict that increasing amine coinitiator concentration reduces film thickness, while we report that both increasing and decreasing MDEA can limit film thickness, depending on the remainder of the formulation and initiation conditions.
3.1.2 RTIR surface-mediated polymerization results
To understand better the effect of PEGDA, MDEA, and VP concentrations on surface-mediated polymerization with eosin, we used real-time IR spectroscopy (RTIR) to monitor these polymerizations. Since film thickness is dependent on the material mass bound to the surface, we hypothesize that faster polymerization rates and larger conversions observed by RTIR will correlate to thicker films and lower threshold eosin surface densities in the microarray format. The same formulations evaluated above were injected into sample chambers comprised of streptavidin-eosin bound to a biotin-coated glass slide, with a gasket spacer and an unmodified glass slide placed on top to form a 1 cm × 2 cm × 1 mm space within the gasket material for monomer. The sample chamber was irradiated to initiate polymerization, while serial IR measurements were taken. The slides prepared for these experiments had 570 (+/−20) eosins per µm2. The hydrogels formed in these chambers spanned the entire 1 mm width of the chamber (except in the case of 1800 mM PEGDA). When the polymerization time is reduced, the hydrated film thickness can be significantly less than the maximum possible 1 mm. Control experiments were performed to confirm that no polymerization occurs in the absence of surface-bound eosin on these time scales.
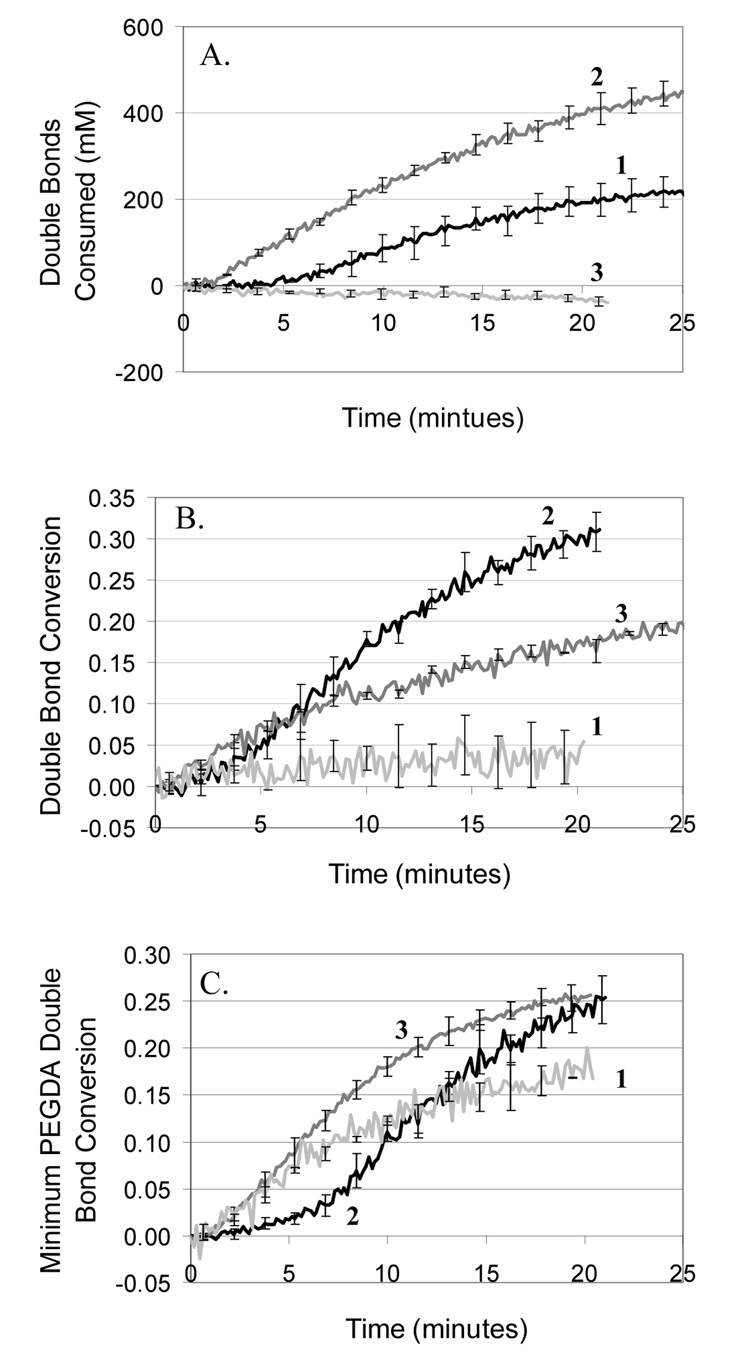
Because polymerization rate scales with monomer concentration under ideal conditions, it is expected that doubling the monomer concentration will approximately double the polymerization rate, which agrees with the observations for 420 mM and 840 mM PEGDA (Figure 3A). Surprisingly, with 1800 mM PEGDA (non-aqueous) no polymerization is detected by IR. When these samples were evaluated by profilometry, very thin films on the order of 100nm were found, indicating that polymerization did occur, but could not be detected by IR because the polymer films were too thin. It is likely that growth of a thick film from the 1800 mM PEGDA formulation is prevented by diffusion limitations. As the polymer film becomes thicker, it becomes especially critical for the initiating MDEA radical to be able to diffuse away from the surface to react with free monomer. Due to the high viscosity of PEGDA compared to water, the nonaqueous gel would greatly retard the diffusion of MDEA radicals compared to a hydrogel environment. An additional plausible explanation is that exposure of streptavidin to a nonaqueous environment may cause the protein to rearrange such that its hydrophilic regions where eosin modification has occurred are now buried within the protein’s interior, sterically hindering interaction with the coinitiator and ultimately reducing the initiation efficiency.
Fig. 3.
Effect of PEGDA, MDEA, and VP concentration on surface-mediated polymerization rate. A) Compare PEGDA concentrations: 420 mM (1), 840 mM (2), 1800 mM (3). B) Compare MDEA concentrations: 21 mM (1), 210 mM (2), 2100 mM (3). C) Compare VP concentrations: 0 mM (1), 35 mM (2), 350 mM (3). Approximately 570 eosins per µm2 were present on the surface. Since PEGDA and VP double bonds have different molar absoptivities, a minimum PEGDA double bond conversion is calculated assuming that PEGDA and VP undergo only alternating polymerization until all the VP is consumed.
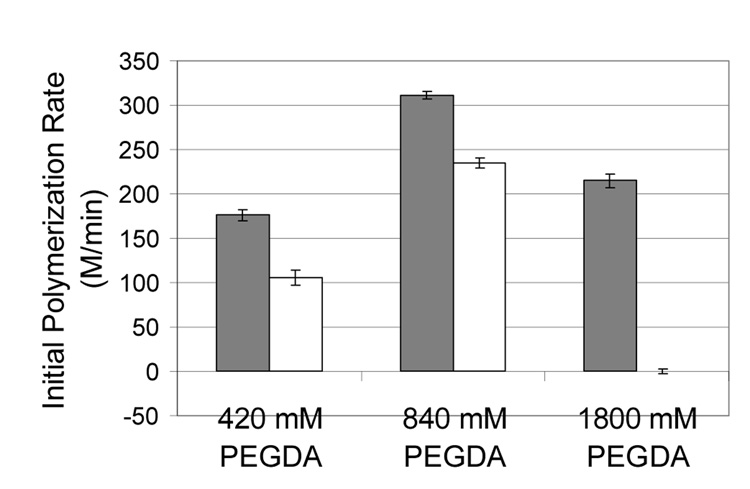
These hypotheses were further examined by RTIR monitoring of polymerization from eosin versus streptavidin-eosin in solution (not surface-mediated) (Figure 4). As expected, polymerization with unconjugated eosin results in an approximately doubled initial polymerization rate as PEGDA is increased from 420 mM to 840 mM. However, 1800 mM PEGDA has a much lower than expected initial polymerization rate (about 600 M/min is expected, but we see only 200 M/min). This observation may be explained by diffusion limitations of MDEA and eosin in the more viscous monomer formulation. When looking at the effect of conjugating streptavidin to eosin, we see a slight decrease in initial rates for the 420 and 840 mM conditions, and a complete inhibition of polymerization in the 1800 mM condition. The more drastic inhibitory effect seen with 1800 mM PEGDA is may be due to protein rearrangement in the nonaqueous environment that causes eosin to become sterically inaccessible. Consequently, the results from both the surface-mediated and the solution-based polymerizations are consistent with the hypotheses of diffusion limitation and protein structure disruption caused by the nonaqueous monomer formulation.
Fig. 4.
Effect of PEGDA concentration and eosin conjugation to streptavidin on the initial polymerization rate when the initiator is in solution (not surface-mediated polymerization). Dark bars represent eosin and light bars represent eosin-streptavidin conjugate. In all cases, the eosin concentration is 0.3 µM, the monomer formulation contains 210 mM MDEA and 35 mM VP, and the light intensity is 260 mW/cm2.
These RTIR results help to explain the high threshold initiator densities required for microarray format polymerization with 1800 mM PEGDA. This nonaqueous formulation is likely causing the surface-bound streptavidin to rearrange, resulting in fewer eosin moieties actually being available to initiate polymerization. The RTIR and microarray results for varying PEGDA concentrations reveal an incongruous result in that the microarray films from the 420 mM and 840 mM PEGDA formulations have similar thicknesses, while RTIR indicates that the 840 mM condition polymerizes twice as quickly as the 420 mM condition. A possible explanation is that the 840 mM sample may indeed bind more material to the slide, yet form a higher density gel upon drying.
Consistent with the microarray results, both too much and too little MDEA may result in decreased polymerization rates (Fig. 3B), depending on the other initiation conditions. Since MDEA is a component of the initiation mechanism, too little MDEA results in decreased initiation rates. Additionally, MDEA can act as a chain transfer agent, which, when present in an appropriate concentration, may help extend the kinetic chain development further into the region of free monomer, enhancing film growth by helping to overcome the transport limitations of long kinetic chains and the more highly crosslinked polymer. Having too little MDEA may hinder this phenomenon. At the highest MDEA concentration the polymerization may be hindered by excessive chain transfer to the point that network formation is slowed and termination becomes more facile. Also, the excess MDEA radicals that would initiate polymerization may actually start terminating kinetic chains as the concentration of monomer decreases, a phenomenon referred to as primary radical termination.
To analyze RTIR results for varying the VP concentration, a minimum PEGDA concentration was calculated (Fig. 3C). The molar absorptivity of the VP double bond at the peak area of interest is approximately twice that of PEGDA, making it impossible to distinguish which part of the peak area decrease is caused by PEGDA versus VP, which becomes a significant concern when the concentration of VP approaches that of PEGDA, as in the 350 mM VP condition. It was found that under the polymerization conditions we use, VP does not readily homopolymerize. Therefore, a minimum PEGDA C=C conversion was calculated assuming that the polymerization proceeds entirely in an alternating fashion until all VP is consumed. This approach is additionally supported by the fact that is has been proposed that VP accelerates acrylate polymerization by favorable heteropolymerization kinetics [34]. Further, the molecular weight of VP is only 111, as compared to 575 for PEGDA, indicating that the largest weight fraction of the polymer is PEGDA, implying that this monomer contributes more to the polymer film thickness than does VP. RTIR results indicate that the highest VP (350 mM) condition yields the greatest minimum PEGDA conversions, and that the 35 mM VP condition also out performs the no VP condition at long reaction times (Fig. 3C), which is consistent with the assertion that VP acts as an accelerant. The microarray and RTIR data for VP are fairly consistent, with the no VP condition performing poorly in both and the high VP yielding conversion improvements in the RTIR format and better sensitivity in the microarray format.
3.2 Comparison of PEGDA vs. Acrylamide
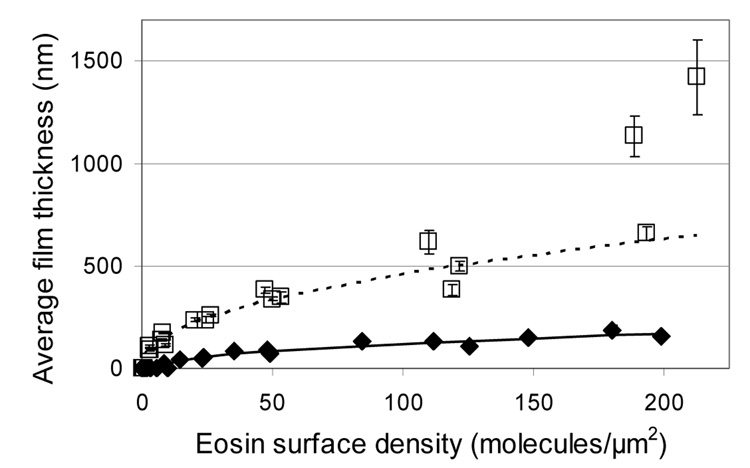
None of the variations to the standard PEGDA formulation were able to yield both thicker films and a lower threshold eosin surface density for visible film growth. Therefore, acrylamide was investigated as an alternative aqueous monomer system that has a history of use with eosin initiation systems predominantly in the formation of holographic recording media [28,29]. Eosin microarrays were reacted with a 40wt% acrylamide formulation (5.2 M acrylamide and 130 mM bisacrylamide in water with 210 mM MDEA and 35 mM VP) and compared to the standard 22% PEGDA formulation (420 mM PEGDA, in water with 210 mM MDEA and 35 mM VP). Figure 5 demonstrates that the acrylamide formulation results in thicker polymer films at all eosin surface densities tested, and that the improvement in thickness ranges from four to eight-fold (maximum polymer thickness increased from 180 nm to 1420 nm). In addition, higher sensitivity is achieved using acrylamide, with a threshold eosin surface density of only 2.8 eosins per µm2, compared to 14 eosins per µm2 for the PEGDA formulation and 7.3 eosins per µm2 for the 350 mM VP condition which had the best sensitivity of all the PEGDA formulations. Notably, two of the three acrylamide data points near 200 eosin/µm2 do not follow the same trend as the rest of the microarray data which was fit to the equation [thickness = k*(b-density)a]. These two data points represent experimental sets in which films greater than 1 micron were achieved—approximately twice the thickness predicted by the equation used for fitting. This break from the trend could indicate that at very high eosin concentrations there is a fundamental difference in how the polymerization proceeds.
Fig. 5.
Comparison of film thicknesses achieved using the standard PEGDA formulation versus the acrylamide formulation. Curves are fit to the data using the equation thickness = k*(b-density)a. ◆ and —— represent the standard PEGDA formulation (420 mM PEGDA, 210 mM MDEA, and 35 mM VP). □ and ■■■ represent the acrylamide formulation (5.2 M acrylamide, 130 mM bisacrylamide, 210 mM MEA, 35 mM VP). The three acrylamide data points near 200 eosin/µm2 were not included in the curve fit, because the equation appears to be inappropriate for that region of the data.
RTIR was used to compare surface polymerization rates between the acrylamide and PEGDA formulations. Streptavidin-eosin was coated onto epoxy-functionalized glass (epoxy forming covalent linkages with amines and thiols on the protein surface), then reaction chambers were constructed as described above. Epoxy slides were used here instead of biotin slides to achieve a lower eosin surface density (160 +/− 10 fluors per µm2) so that a more stringent comparison could be made. Figure 6 demonstrates that acrylamide polymerization has a smaller inhibition time before onset of polymerization and that it polymerizes faster, reaching a higher conversion. Although acrylamide has a lower molecular weight than PEGDA (71 vs. 575), the acrylamide formulation is 40 wt% in water, while PEGDA is only 22% in water. This difference implies that for equal monomer conversion, we would actually expect more mass reacted to the surface in the acrylamide formulation. Thus, the acrylamide formulation is clearly preferable for growing thick films from surface-bound eosin. The low molecular weight of acrylamide and the lower crosslinking content of this monomer formulation likely favor its faster surface-mediated polymerization. A less highly crosslinked gel may allow initiating MDEA radicals to diffuse more readily throughout the growing film to regions of higher monomer concentration.
Fig. 6.
RTIR monitoring of surface-mediated polymerization. Comparison of the standard PEGDA formulation (420 mM PEGDA, 210 mM MDEA, and 35 mM VP) (1) and the acrylamide formulation (5.2 M acrylamide, 130 mM bisacrylamide, 210 mM MDEA, 35 mM VP) (2). Approximately 160 eosins per µm2 were present on the surface.
3.3 Use of Ascorbic acid as a coinitiator
Articles have been published concerning polymerization with acrylamide using eosin and ascorbic acid instead of an amine coinitiator. This polymerization system has been reported to be either insensitive to the presence of oxygen [1], or to actually require oxygen to proceed [30]. For a biodetection application, it would be desirable to reduce oxygen inhibition so that the assay may be performed in the absence of a purge gas. Therefore, we investigated using ascorbic acid as a coinitiator in place of MDEA. Polymerization was attempted from streptavidin-eosin microarray spots using 5.2 M acrylamide, 130 mM bisacrylamide, 210 mM ascorbic acid, and 35 mM VP (pH = 4) in either an argon or an oxygen environment, but no visible polymer films were generated, even from 280 eosins/µm2 spots. Additionally, polymerization from biotin glass slides with streptavidin-eosin attached (870 +/− 40 eosin/µm2) were used in the RTIR polymerization format with either argon or oxygen bubbled through the monomer, yet no polymerization was detected by IR and no film was apparent.
To investigate further the lack of polymerization with this formulation, RTIR was used to monitor polymerization using high concentrations of eosin in solution (not surface initiated). Ten-fold differences in eosin concentration were tested to determine the eosin concentration for which at least 5% or higher conversion could be achieved by 20 minutes. The highest conversion achieved at a concentration below the nominal cut-off was 2% which is difficult to distinguish from instrument noise. Since some reports have found that oxygen is required for polymerization in the ascorbic acid system, we investigated bubbling oxygen versus argon through the monomer formulations prior to polymerization. Threshold eosin solution concentrations were also determined for each formulation without any coinitiator added, since at very high eosin concentrations, polymerization can be achieved in the absence of any coinitiator as has been published previously [35].
Acrylamide with ascorbic acid and argon could not reach 5% conversion even with 30 µM eosin, which was the highest concentration investigated (Table 1). However, with oxygen bubbled through the monomer instead of argon, 30 µM eosin yielded 20% conversion at 20 minutes, thus reproducing the previous report that oxygen helps to facilitate this polymerization system. Interestingly, in the absence of coinitiator, acrylamide and argon at pH 4 can yield a 24% conversion with 30 µM eosin, but adding ascorbic acid under these anaerobic conditions inhibits polymerization. Therefore, we observe that ascorbic acid requires oxygen to initiate polymerization and that in anaerobic conditions ascorbic acid actually inhibits the polymerization. The lowest threshold eosin solution concentration (0.03 µM) was achieved using MDEA and argon with PEGDA or acrylamide (pH 9). The second best was PEGDA or acrylamide without any coinitiator at pH 9 and with argon, which was 100 fold less sensitive than their MDEA counterparts, yet still 10-fold more sensitive than acrylamide with ascorbic acid and oxygen. The fact that acrylamide with ascorbic acid requires 1000 times more eosin to polymerize than PEGDA or acrylamide with MDEA explains why no surface polymerization was observed in the microarray format. Perhaps with higher eosin surface density or longer reaction times, acrylamide with ascorbic acid may be able to generate a surface-initiated film.
Table 1.
Comparison of threshold eosin solution concentrations for 5% conversion by 20 minutes with various monomer formulations and reaction conditions. All formulations are in water and contain 35 mM VP. PEGDA, when used, is 420 mM. Acrylamide, when used, is 5.2 M acrylamide and 130 mM bisacrylamide. MDEA and Ascorbic acid, when used, are 210 mM.
| Coinitiator | Monomer formulation | pH | Argon | Oxygen | Threshold Eosin Solution Concentration (µM) | Double Bond Conversion at 20 min. |
|---|---|---|---|---|---|---|
| MDEA | PEGDA | 9 | + | 0.03 | 0.22 +/− 0.03 | |
| MDEA | Acrylamide | 9 | + | 0.03 | 0.07 +/− 0.01 | |
| Ascorbic Acid | Acrylamide | 4 | + | >30 | 0.02 +/− 0.003 | |
| Ascorbic Acid | Acrylamide | 4 | + | 30 | 0.21 +/− 0.07 | |
| none | PEGDA | 9 | + | 3 | 0.05 +/− 0.005 | |
| none | Acrylamide | 9 | + | 3 | 0.06 +/− 0.003 | |
| none | Acrylamide | 4 | + | 30 | 0.25 +/− 0.03 | |
| none | Acrylamide | 4 | + | >30 | 0.00 +/− 0.001 |
4. Summary and conclusions
40 wt% acrylamide with MDEA was found to generate four to eight-fold thicker films and form visible films from five-fold lower eosin surface densities compared to a similar 22wt% PEGDA formulation. A reasonable dynamic response in film thickness from changes in eosin surface density was observed. Variations to the concentrations of PEGDA, VP and MDEA yielded more modest improvements, with 1800 mM PEGDA producing a maximum polymer thickness of 280nm compared to only 180 nm for 420 mM PEGDA. It was also seen that increasing VP from 35 mM to 350 mM decreased the threshold eosin surface density for polymerization down to 7.3 eosins/µm2 from 14 eosins/µm2. Polymerization of acrylamide by eosin in solution with ascorbic acid as a coinitiator was found to proceed only in the presence of oxygen, yet failed to initiate polymerization from the eosin-coated surfaces, an outcome that reflects the much higher threshold eosin concentration required for polymerization with this formulation
In situations where using a low initiator concentration is desirable (either in surface-mediated or non surface-mediated polymerizations), a rapid comparison of various monomer formulations may be made using a dilution series of initiator surface densities prepared in a microarray format. This method rapidly revealed that polymerization of acrylamide with ascorbic acid requires a much higher eosin concentration than the other formulations investigated. RTIR monitoring of surface-mediated polymerization revealed interesting trends that helped explain the microarray results; however, RTIR does not appear to be a reliable method to predict which formulations will generate thicker films and have lower threshold initiator densities in the microarray format.
Acknowledgements
This material is based upon work supported under a National Science Foundation Graduate Research Fellowship. This work has also been supported by NIH R21 CA 127884.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Satoh M, Shirai K, Saitoh H, Yamauchi T, Tsubokawa N. J Polym Sci Part A Polym Chem. 2005;43:600–606. [Google Scholar]
- 2.Raghuraman GK, Rühe J, Dhamodharan R. J Nanopart Res. 2008;10:415–427. [Google Scholar]
- 3.Chen Y, Deng Q, Xiao J, Nie H, Wu L, Zhou W, et al. Polymer. 2007;48:7604–7613. [Google Scholar]
- 4.Fan X, Lin L, Messersmith PB. Biomacromolecules. 2006;7:2443–2448. doi: 10.1021/bm060276k. [DOI] [PubMed] [Google Scholar]
- 5.Cruise GM, Hegre OD, Scharp DS, Hubbell JA. Biotechnol Bioeng. 1998;57:655–665. doi: 10.1002/(sici)1097-0290(19980320)57:6<655::aid-bit3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 6.Cruise GM, Scharp DS, Hubbell JA. Biomaterials. 1998;19:1287–1294. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 7.Shi Z, Zhou Y, Yan D. Polymer. 2006;47:8073–8079. [Google Scholar]
- 8.Sebra RP, Ansenth KS, Bowman CN. J Polym Sci Part A Polym Chem. 2006;44:1404–1413. [Google Scholar]
- 9.Sikes HD, Hansen RR, Johnson LM, Jenison R, Birks JW, Rowlen KL, et al. Nat Mater. 2008;7:52–56. doi: 10.1038/nmat2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen RR, Sikes HD, Bowman CN. Biomacromolecules. 2008;9:355–362. doi: 10.1021/bm700672z. [DOI] [PubMed] [Google Scholar]
- 11.Hansen RR, Avens HJ, Shenoy R, Bowman CN. Anal Bioanal Chem. 2008 doi: 10.1007/s00216-008-2259-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebra RP, Masters KS, Cheung CY, Bowman CN, Anseth KS. Anal Chem. 2006;78:3144–3151. doi: 10.1021/ac052246y. [DOI] [PubMed] [Google Scholar]
- 13.Lou X, Lewis MS, Gorman CB, He L. Anal Chem. 2005;77:4698–4705. doi: 10.1021/ac050706h. [DOI] [PubMed] [Google Scholar]
- 14.He P, Zhen W, Tucker EZ, Gorman CB, He L. Anal Chem. 2008;80:3633–3639. doi: 10.1021/ac702608k. [DOI] [PubMed] [Google Scholar]
- 15.Khire VS, Harant AW, Watkins AW, Anseth KS, Bowman CN. Macromolecules. 2006;39:5081–5086. [Google Scholar]
- 16.Khire VS, Benoit DSW, Anseth KS, Bowman CN. J Polym Sci Part A Polym Chem. 2006;44:7027–7039. [Google Scholar]
- 17.Sebra RP, Masters KS, Bowman CN, Anseth KS. Langmuir. 2005;21:10907–10911. doi: 10.1021/la052101m. [DOI] [PubMed] [Google Scholar]
- 18.Reddy SK, Sebra RP, Anseth KS, Bowman CN. J Polym Sci Part A Polym Chem. 2005;43:2134–2144. [Google Scholar]
- 19.Schmelmer U, Jordan R, Geyer W, Eck W, Gölzhäuser A, Grunze M, et al. Angew Chem Int Ed. 2003;42:559–563. doi: 10.1002/anie.200390161. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Huang W, Miller MD, Baker GL, Bruening ML. J Polym Sci Part A Polym Chem. 2003;41:386–394. [Google Scholar]
- 21.Tsujii Y, Ejaz M, Sato K, Goto A, Fukuda T. Macromolecules. 2001;34:8872–8878. [Google Scholar]
- 22.Jordan R, Ulman A, Kang JF, Rafailovich MH, Sokolov J. J Am Chem Soc. 1999;121:1016–1022. [Google Scholar]
- 23.Jordan R, West N, Ulman A, Chou Y, Nuyken O. Macromolecules. 2001;34:1606–1611. [Google Scholar]
- 24.An Y, Hubbell JA. J Controlled Release. 2000;64:205–215. doi: 10.1016/s0168-3659(99)00143-1. [DOI] [PubMed] [Google Scholar]
- 25.Kizilel S, Pérez-Luna VH, Teymour F. Langmuir. 2004;20:8652–8658. doi: 10.1021/la0496744. [DOI] [PubMed] [Google Scholar]
- 26.Kizilel S, Sawardecker E, Teymour F, Pérez-Luna VH. Biomaterials. 2006;27:1209–1215. doi: 10.1016/j.biomaterials.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 27.Kizilel S, Pérez-Luna VH, Teymour F. Macromol Theory Simul. 2006;15:686–700. [Google Scholar]
- 28.Blaya S, Murciano A, Acebal P, Carretero L, Ulibarrena M, Fimia A. Appl Phys Lett. 2004;84:4765–4767. [Google Scholar]
- 29.Ortuño M, Gallego S, García C, Pascual I, Neipp C, Beléndez A. Phys Scr. 2005;T118:66–68. [Google Scholar]
- 30.Tigulla P, Vuruputuri U. J Chem Sci. 2004;116:115–118. [Google Scholar]
- 31.Odian G. Principles of Polymerization. 4th ed. New Jersey: John Wiley and Sons; 2004. Chapter 3. [Google Scholar]
- 32.Panyim S, Chalkley R. ArchBiochem and Biophys. 1969;130:337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- 33.Elliott JE, Macdonald M, Nie J, Bowman CN. Polymer. 2004;45:1503–1510. [Google Scholar]
- 34.White TJ, Liechty WB, Guymon CA. J Polym Sci Part A Polym Chem. 2007;45:4062–4073. [Google Scholar]
- 35.Neckers DC, Raghuveer KS, Valdes-Aguilera O. Polym Mater Sci Eng. 1989;60:15–16. [Google Scholar]