Abstract
Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative diseases that include Creutzfeldt-Jakob disease, bovine spongiform encephalopathy and sheep scrapie. Although one of the earliest events during TSE infection is the cellular uptake of protease resistant prion protein (PrP-res), this process is poorly understood due to the difficulty of clearly distinguishing input PrP-res from either PrP-res or protease-sensitive PrP (PrP-sen) made by the cell. Using PrP-res tagged with a unique antibody epitope, we examined PrP-res uptake in neuronal and fibroblast cells exposed to three different mouse scrapie strains. PrP-res uptake was rapid and independent of scrapie strain, cell type, or cellular PrP expression, but occurred in only a subset of cells and was influenced by PrP-res preparation and aggregate size. Our results suggest that PrP-res aggregate size, the PrP-res microenvironment, and/or host cell-specific factors can all influence whether or not a cell takes up PrP-res following exposure to TSE infectivity.
Keywords: protein aggregation, PrP, PrP-res, PrP-sen, prion, scrapie, transmissible spongiform encephalopathy, TSE
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative diseases that include kuru, Creutzfeldt-Jakob disease, and Gerstmann-Sträussler-Scheinker syndrome in humans as well as bovine spongiform encephalopathy, sheep scrapie, and chronic wasting disease (CWD) in deer and elk (Priola & Vorberg, 2006). Partially protease resistant prion protein (PrP-res) associates closely with infectivity and is considered to be a marker of TSE infection. PrP-res is derived from the normal protease sensitive form of PrP, PrP-sen. The post-translational, conformationally driven conversion of PrP-sen to PrP-res is a critical, though poorly understood, event during TSE pathogenesis.
Certain cell types when exposed to scrapie infected brain homogenate will produce PrP-res over multiple serial passes, and when injected into experimental animals, cause disease. These cells are considered to be persistently infected. A number of studies have shown that both neuronal and non-neuronal cell lines can become persistently infected with certain strains of mouse scrapie, sheep scrapie or CWD (Baron, Magalhaes et al., 2006), (Butler, Scott et al., 1988), (Race, Fadness et al., 1987), (Raymond, Olsen et al., 2006), (Rubenstein, Carp et al., 1984), (Schatzl, Laszlo et al., 1997), (Vorberg, Raines et al., 2004a), (Vorberg, Raines et al., 2004b). The vast majority of what is understood about the process of cell infection and PrP-res formation has been the result of studies on such persistently infected cells. These studies have shown that PrP-res formation in infected cells occurs at the cell surface and/or along the endocytic pathway (Beranger, Mange et al., 2002), (Borchelt, Taraboulos et al., 1992), (Caughey & Raymond, 1991) and that cell associated PrP-res has a long half life (Borchelt, Scott et al., 1990), (Caughey & Raymond, 1991) and can accumulate in endolysosomal compartments (Caughey, Raymond et al., 1991), (McKinley, Taraboulos et al., 1991), (Pimpinelli, Lehmann et al., 2005), (Taraboulos, Scott et al., 1994). Although cells persistently infected with scrapie agent have provided important insights into the cellular biology of PrP-res formation during infection, very little is known about how PrP-res is initially taken up by cells.
Several recent studies have looked at the earlier stages of TSE infection with some data suggesting that PrP-res is taken up rapidly (i.e. within 24 hrs) (Hijazi, Kariv- Inbal et al., 2005), (Horonchik, Tzaban et al., 2005), (Mohan, Hopkins et al., 2005), (Paquet, Daude et al., 2007) and other data indicating that uptake may occur more slowly (i.e. over days) (Bergstrom, Jensen et al., 2006),(Magalhaes, Baron et al., 2005). Most of these earlier studies were limited by the difficulty of specifically distinguishing PrP-res in the inoculum from PrP-res newly made in the cells which may help to explain the conflicting results. Even in instances where partially purified PrP-res was uniquely labeled, it was not always possible to absolutely distinguish the labeled PrP-res from other labeled, but non-relevant, protein contaminants (Magalhaes, Baron et al., 2005). Additionally, few studies have examined more then one strain of TSE agent meaning that any potential differences in strain-specific PrP-res uptake early during infection remain largely unresolved.
Using uniquely epitope tagged PrP-sen molecules we have previously shown that, regardless of scrapie strain, both mouse neuroblastoma (MNB) cells as well as mouse fibroblast cells form de novo PrP-res after only 4 hrs of scrapie exposure (Vorberg, Raines et al., 2004a). This finding strongly suggests that the association of PrP-res with a cell occurs rapidly. We have now used PrP-res uniquely tagged with an epitope to the mouse monoclonal antibody 3F4 to examine the interactions between PrP-res from different strains of mouse scrapie and different cell types during the acute stage of scrapie infection (0 – 72 hrs). Our results show that PrP-res uptake is indeed very rapid and independent of cell type, scrapie strain, and host cell PrP-sen expression. However, PrP-res uptake was influenced by PrP-res aggregate size and only a certain subpopulation of cells detectably internalized PrP-res. Furthermore, PrP-res in brain homogenate was taken up more efficiently by cells then either partially purified PrP-res or PrP-res in brain microsomes. Our results suggest that, while PrP-res uptake during acute infection appears to be similar for different scrapie strains and cell types, internalization of PrP-res can be influenced by PrP-res aggregate size, PrP-res microenvironment, and/or an as yet unidentified host factor or factors.
RESULTS
PrP-res3F4 uptake by mouse neural cells
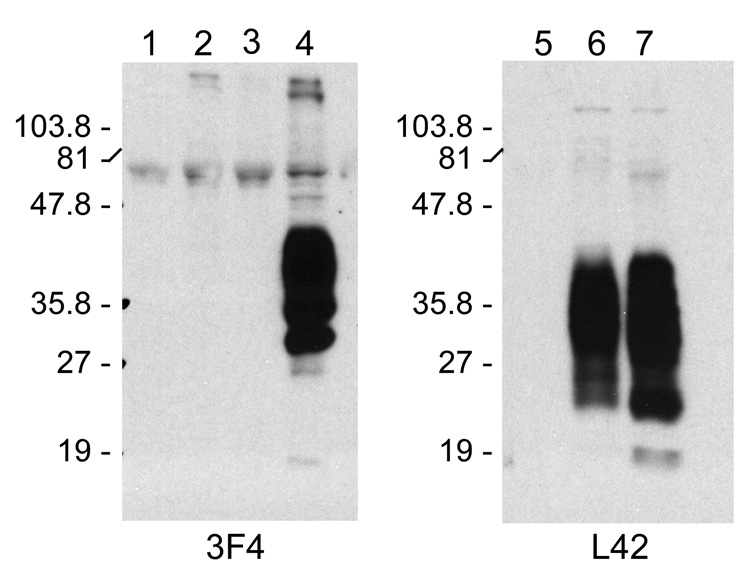
To determine whether or not our mouse neural cells were able to take up PrP-res3F4, MoL42-CFD5 cells were exposed to 22L(3F4), ME7(3F4), Obi(3F4) or uninfected Mock(3F4) brain homogenates. MoL42-CFD5 cells are mouse neuronal PrP0/0 cells that have been modified to express mouse PrP-sen that is recognized by the mouse monoclonal antibody L42 allowing us to distinguish it, if necessary, from mouse PrP in the brain homogenate. MoL42 PrP-sen is not recognized by the 3F4 mouse monoclonal antibody (Fig. 1).
Figure 1. Cell line expression of PrP.
Western blot detection of PrP-sen with either mouse monoclonal antibody 3F4 (left panel) or mouse monoclonal antibody L42 (right panel) for cell lines CF10 (lanes 1, 5), MoL42-CFD5 (lanes 2, 6), and MoL42-Ψ2A2 (lanes 3, 7). As a positive control for the specificity of the 3F4 antibody, Lane 4 shows CF10 cells expressing Mo3F4.
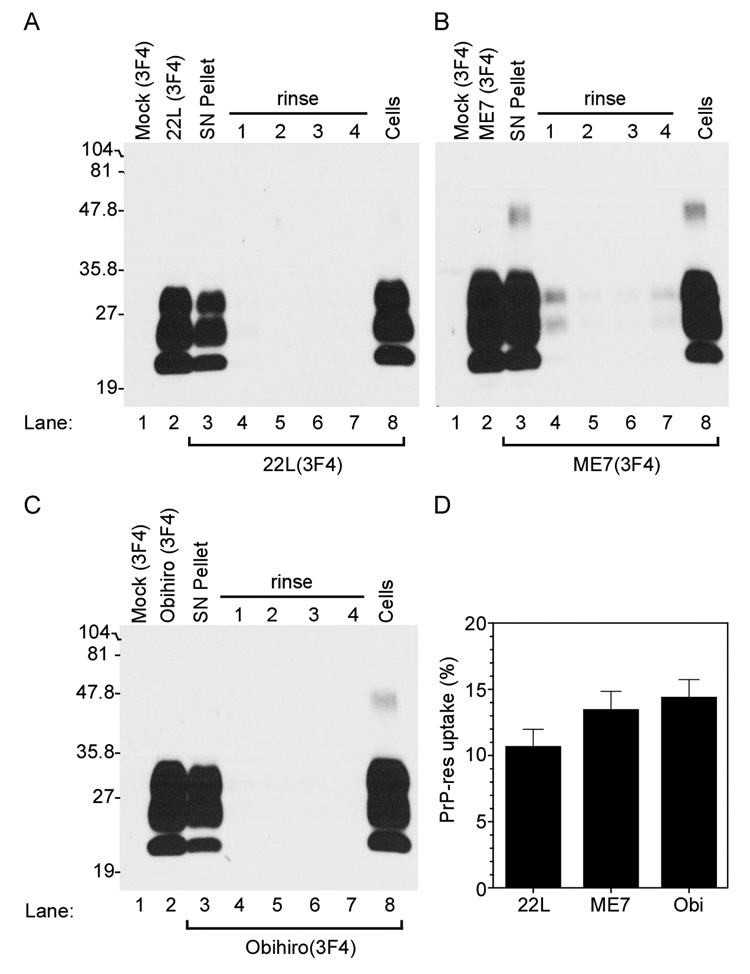
After 24 hrs incubation time, cells were rinsed to remove any unbound PrP-res3F4, lysed and assayed for PrP-res3F4 by Western blot as shown in Fig. 2. Unbound, excess PrP-res3F4 was effectively washed from the cells by four rinses of fresh media (Fig. 2A–C) and no PrP-res3F4 could be detected in cells exposed to Mock(3F4) brain homogenate (Fig. 2A–C, lane 1). Thus, residual, unbound PrP3F4 could be efficiently removed from the cell monolayer. PrP-res3F4 associated with detached cells, cell debris, and/or unbound brain homogenate was observed in the pellet from a low speed spin of the cellular supernatant (Fig. 2 A–C, SN pellet) but significant amounts remained cell-associated (Fig. 2A–C, Cells). By 24 hours, the amount of PrP-res3F4 that was cell associated was 10–15% of the total PrP-res added to the cells (Fig. 2D), a percentage that did not significantly change at later timepoints (Fig. 4 and data not shown). Thus, unbound PrP-res3F4 (i.e. PrP-res3F4 in both the SN pellet and the clarified supernatant) remained in excess throughout the course of the experiment. These data suggest that the majority of PrP-res3F4 in the brain homogenate did not interact with the cells.
Figure 2. Mouse neural cells take up PrP-res3F4.
Western blot analysis of PrP-res3F4 uptake into MoL42-CFD5 cells. Brain homogenate from 22L, ME7 or Obihiro scrapie-infected Tg(WT-E1) mice were PK treated and used as positive controls (Panels A–C, lane 2) while PK treated brain homogenate from mock-infected Tg(WT-E1) mice was used as negative controls (Panel A–C, lane 1). For both the positive and negative controls, 1/20th of the total homogenate used was loaded onto the gel. Brain homogenates (200µl of a 1% brain homogenate) from 22L(3F4) (panel A), ME7(3F4) (panel B), or Obi(3F4) (panel C) were added to MoL42-CFD5 cells for 24 hours and then removed from the cell monolayer. Cell debris and dead cells were spun out of the supernatant and the supernatant pellet was PK treated, methanol precipitated and the entire sample loaded onto the gel (SN Pellet). Cells were then rinsed with PBS four times (rinse 1–4), lysed, and both the cell lysate and rinses were PK treated, methanol precipitated and the entire sample loaded onto the gel. PrP-res3F4 was detected in the lysed and PK treated cells (Cells). All blots were analyzed using the mouse monoclonal antibody 3F4 and developed using ECL (Amersham). The percentage of total PrP-res3F4 taken up by the cells after 24 hours is shown in Panel D. PrP-res3F4 was quantified as detailed in the materials and methods. There was no significant difference between the three strains in the amount of PrP-res taken up by the MoL42-CFD5 cells (p>0.05 using 1-way Anova with Bonferroni’s multiple comparison test). Data are shown as mean ± S.D. for N=12.
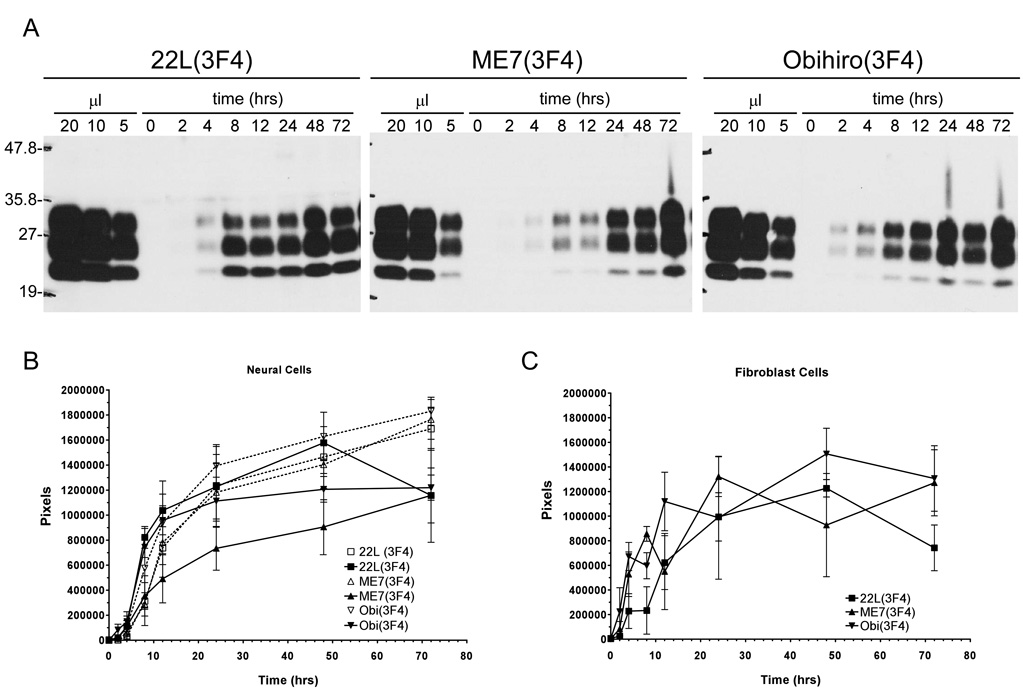
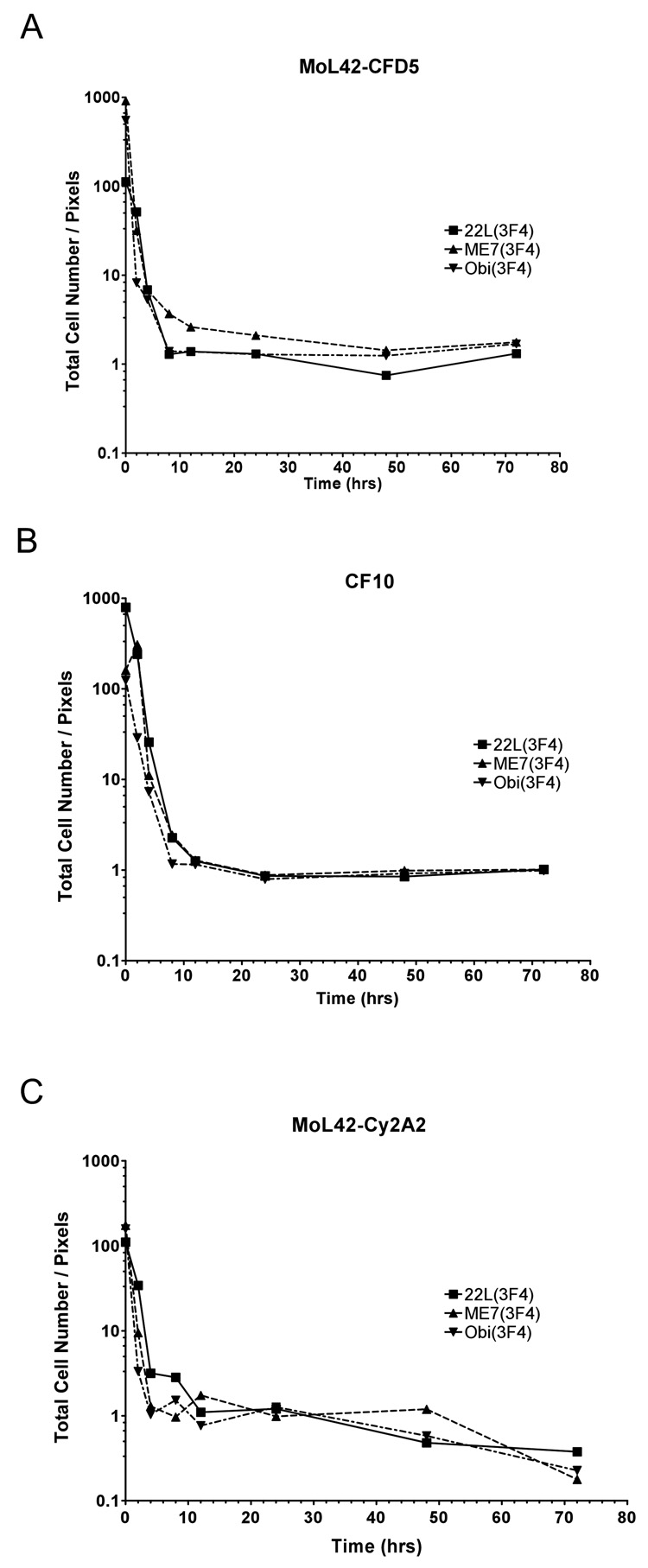
Figure 4. Kinetics of PrP-res3F4 uptake into cells.
A) Representative Western blot analysis of PrP-res3F4 uptake into MoL42-CFD5 cells from 0 – 72 hrs. Scrapie-infected brain homogenates from 22L(3F4), ME7(3F4) or Obi(3F4) were analyzed. The first three lanes of each gel show PK-treated scrapie-infected brain homogenate for the appropriate strain loaded in 20 ul, 10 ul, and 5 ul volumes. These lanes were used as internal standards to establish a constant film exposure time from experiment to experiment (4min) in order to quantify the data within the linear range of the film. The amount of PrP-res internalized by the cells increased over time. B) Graphical representation of total cell-associated 22L(3F4), ME7(3F4), or Obi(3F4) taken up by either MoL42-CFD5 cells (solid lines N=5 or 6) or CF10 cells (dotted lines N=3) from 0–72 hrs. The error bars represent SEM. No statistically significant differences were found between the scrapie strains (p>0.05, Two-way ANOVA) or between the different cell types (p>0.05, Mann Whitney test) for each scrapie strain. C) Graphical representation of total cell-associated 22L, ME7 or Obihiro PrP-res3F4 taken up by MoL42-ψ2A2 fibroblast cells (N=3) from 0 – 72 hrs where the error bars represent SEM. No statistically significant differences were found between the scrapie strains (p>0.05, Two-way ANOVA) or between the fibroblast and neuronal cell types (p>0.05, Mann Whitney test). All blots were analyzed using the mouse monoclonal antibody 3F4. For all data, the amount of PrP-res is expressed in pixels and was quantified by analysis of ECL developed western blots using the UN-SCAN-IT software as detailed in the Methods.
PrP-res is internalized by cells
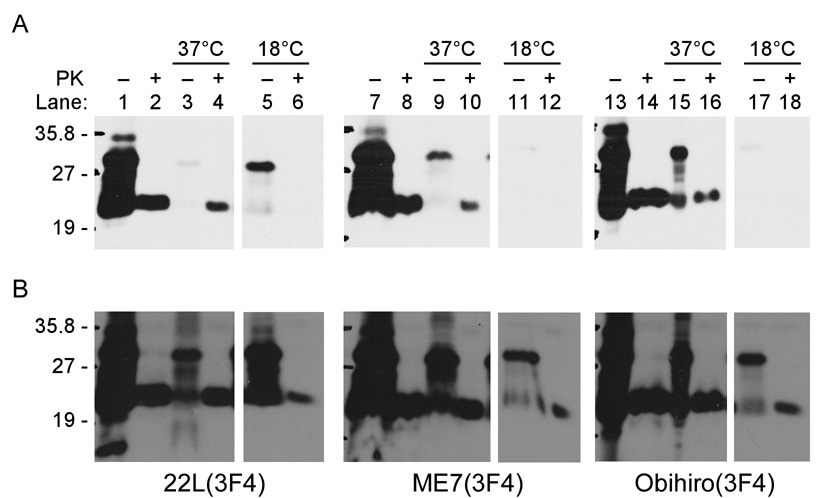
To determine if cell-associated PrP-res3F4 was bound to the cells non-specifically or internalized via an active cellular process such as endocytosis, MoL42-CFD5 cells were exposed to scrapie-infected brain homogenates and incubated either at 18°C, which blocks endocytosis, or at 37°C. Without PK digestion, some PrP3F4 was bound and/or internalized in the cells at both 37 and 18°C (Fig. 3A), although in the case of 22L(3F4) a longer gel exposure was required to detect it (Fig. 3B). This material was also observed when uninfected brain homogenate was overlayed onto the cells (data not shown) suggesting that at least some fraction of the PrP-sen3F4 in the brain homogenate was aggregated and interacting with the cells. As shown in Fig. 3A, cells exposed to scrapie-infected brain homogenates at 37°C were positive for PrP-res3F4 while cells which were exposed to scrapie-infected brain homogenates at 18°C appeared to be negative for PrP-res3F4. However, upon longer exposure of the gel, it was clear that a small amount of PrP-res3F4 was also cell associated at 18°C (Fig. 3B). Given the fact that very long gel exposures were required to reliably detect PrP-res3F4 associated with cells at 18°C, our data suggest that the majority of the cell-associated PrP3F4 is likely being actively internalized by the cells and not simply attaching to the cell surface.
Figure 3. PrP-res3F4 uptake into neural cells is significantly decreased at 18°C.
MoL42-CFD5 cells were exposed to infected brain homogenate from 22L(3F4), ME7(3F4) or Obi(3F4) scrapie for 24 hours at either 37°C or 18°C. Samples were PNGaseF treated to remove complex glycans. Non-PK digested infected brain homogenates were run to illustrate total PrP levels (lanes 1,7,13) while PK-digested brain homogenates were run as a positive control for PrP-res (lanes 2,8,14). Some PrP-sen3F4 was taken up by cells at 37°C (−PK lanes 3, 9, 15) and PrP-res3F4 was detected in cells which had been exposed to scrapie brain homogenate and incubated at 37°C (+PK lanes 4, 10, 16). Exposure time for Panel A was 8 minutes. After overnight exposures of the gels, some PrP-sen3F4 (−PK lanes 5, 11, 17) and a low level of PrP-res3F4 (+PK lanes 6, 12, 18) were detected in cells which had been exposed to scrapie brain homogenate and incubated at 18°C (Panel B). All blots were analyzed using the mouse monoclonal antibody 3F4 and developed using ECL (Amersham).
Rapid PrP-res3F4 uptake into cells is independent of scrapie strain, cell type and host cell PrP-sen expression
In order to determine the kinetics of PrP-res3F4 uptake into cells during exposure to TSE infectivity, MoL42-CFD5 cells were exposed to PrP-res3F4 in 22L, ME7, or Obihiro scrapie infected brain homogenates for 0–72 hrs. For each strain, cell-associated PrP-res3F4 was detected by 2 hours post-exposure and the amount of PrP-res3F4 internalized by the cell increased over time (Fig. 4A). Quantification of total PrP-res3F4 uptake into the cells for each given time point showed that there were no significant differences in PrP-res3F4 uptake amongst the different scrapie strains. For each strain, approximately half of the total PrP-res3F4 eventually taken up by the cells was cell-associated by 10–15 hrs post-inoculation and the majority was taken up by 24hrs. Although ME7 appeared to be taken up more slowly by MoL42-CFD5 cells (Fig. 4B, ME7 solid line), this difference was not statistically significant when compared to the other strains (p>.05 by 2-Way ANOVA).
Previous studies have suggested that cells exposed to scrapie associate with PrP-res independently of host cell PrP expression (Hijazi, Kariv-Inbal et al., 2005),(Paquet, Daude et al., 2007). However, given that both the rabbit epithelial cells expressing doxycyline inducible sheep PrP-sen (Rov cells) (Paquet, Daude et al., 2007) and the Chinese hamster ovary (CHO) cells (Hijazi, Kariv-Inbal et al., 2005) used in these studies may still express low levels of PrP-sen (Hijazi, Kariv-Inbal et al., 2005), a role for PrP-sen in PrP-res uptake could not be completely ruled out. To determine whether or not host cell PrP-sen expression was needed for the internalization of PrP-res, we utilized a cell line (CF10) derived from PrP0/0 mice. CF10 cells are neural cells and are the parent cell line of MoL42-CFD5 cells. However, CF10 cells do not express PrP-sen (Fig. 1). PrP-res3F4 was taken up by the CF10 cells similarly to MoL42-CFD5 cells (Fig. 4B, dashed lines). Thus, the uptake of PrP-res derived from different strains of mouse scrapie is independent of PrP-sen expression by the host cell.
We next determined if cell type could influence PrP-res uptake into cells. To test this, non-neuronal MoL42-ψ2A2 cells (a fibroblast cell line) were analyzed using the same kinetics assay employed on both the MoL42-CFD5 and CF10 cell lines. Results from this assay showed that, for all strains tested, the MoL42-ψ2A2 cells took up PrP-res3F4 from scrapie brain homogenate similarly to either MoL42-CFD5 or CF10 cells (Fig. 4C). Thus, the kinetics of PrP-res uptake were similar for both neuronal and non-neuronal cell types, at least with respect to fibroblast cells.
Increased PrP-res uptake correlates with increased cell number
The observed increase in cell-associated PrP-res3F4 over 72 hours could be due either to an increase in the number of cells harboring PrP-res3F4 or to an increase in the amount of PrP-res3F4 per cell. To determine if increased PrP-res3F4 uptake correlated with cell number, the number of cells at each time point was divided by the amount of PrP-res3F4 taken up by the cells as quantified by Western blot. This ratio of cell number to cell-associated PrP-res3F4 was then plotted against the time cell cultures were exposed to scrapie brain homogenate. For all strains and cell lines tested, the ratio of cells to total PrP-res3F4 was high 2 hours after exposure of the cells to the infected homogenate. However, over time that ratio decreased and, as indicated by the linear portion of the curve, after 8 hours was essentially 1:1 (Fig. 5A–C). These data suggest that the system had reached a saturation point and that the population of cells within the culture that could take up PrP-res was limited (Fig. 5A–C).
Figure 5. Increased levels of cellular PrP-res3F4 correlate with increased cell number.
Ratio of total cell number to total cell-associated PrP-res3F4 for scrapie strains 22L(3F4), ME7(3F4) or Obi(3F4). PrP-res3F4 was quantified using ECL developed western blots analyzed with the UN-SCAN-IT software. A) Ratio of the number of MoL42-CFD5 cells to total PrP-res3F4 (N=5). B) Ratio of the number of CF10 cells to total PrP-res3F4 (N=6). C) Ratio of the number of MoL42-ψ2A2 cells to total PrP-res3F4 (N=6). The variability of the data at later time points was likely due to increased cell death over time (data not shown) rather than any cell-type differences.
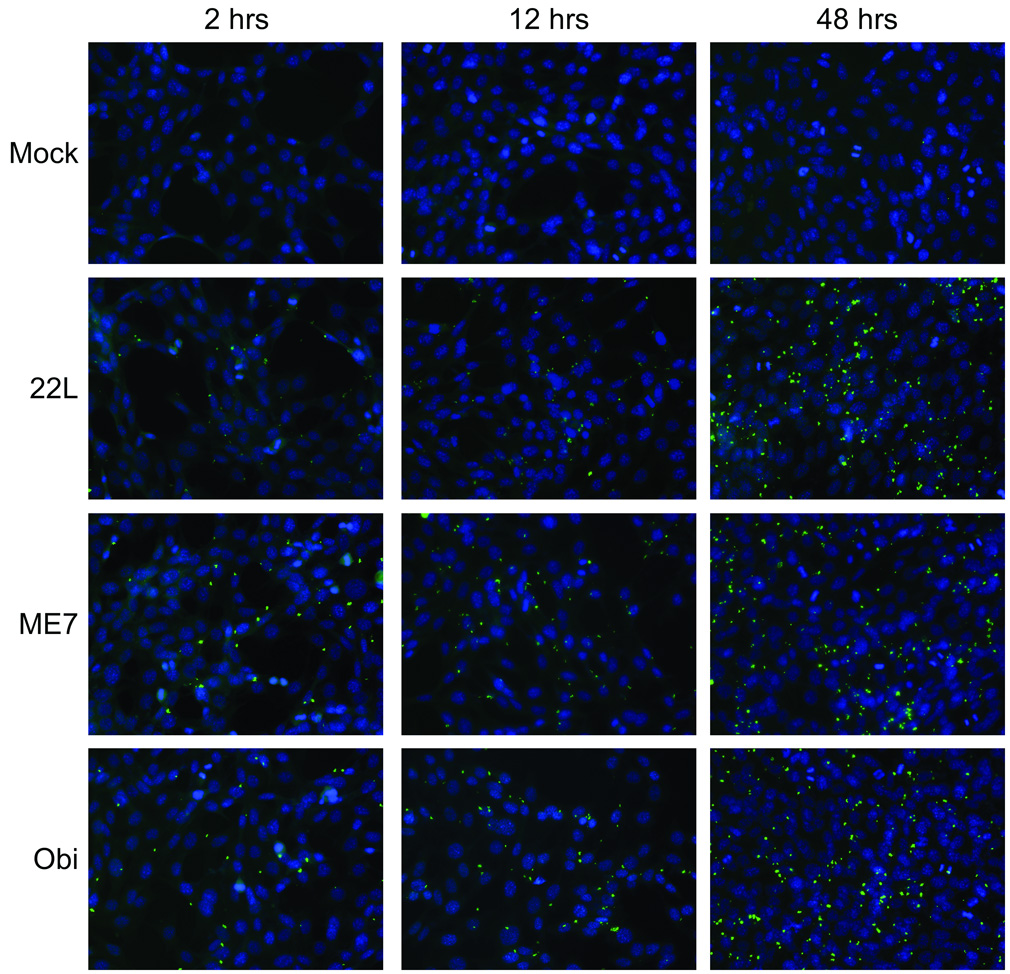
In order to determine if the number of PrP-res3F4 positive cells increased over time, the uptake of partially purified PrP-res3F4 into MoL42-CFD5 cells was assessed by immunofluorescent microscopy for 22L(3F4), ME7(3F4) and Obi(3F4) PrP-res3F4. Similar to the western blot data in Fig. 4, cell-associated PrP-res3F4 was detected as early as 2 hours. PrP-res3F4 was observed as small, punctate aggregates generally localized to both cytoplasmic and perinuclear regions (Fig. 6). Consistent with the data in Figure 5, as cell numbers increased so too did the number of cells positive for PrP-res3F4. However, for all time points examined, only 30%–40% of the cells were positive for PrP-res3F4. Our results suggest that there is a select subpopulation of cells able to take up PrP-res which, over time, divide and increase in number leading to the observed increase in the amount of cell-associated PrP-res.
Figure 6. PrP-res3F4 uptake into neural cells using immunofluorescence microscopy.
PrP-res3F4 partially purified from 22L(3F4), ME7(3F4), Obi(3F4) or mock brain homogenates was added to MoL42-CFD5 cells for 2 – 48 hrs. Cells were rinsed, fixed and immunolabeled with the mouse monoclonal antibody 3F4. Anti-mouse FITC labeled antibody (green) was used to detect PrP-res3F4 while DAPI stain (blue) denotes cell nuclei. All images were taken with a 40X objective.
PrP-res aggregate size influences PrP-res binding and uptake
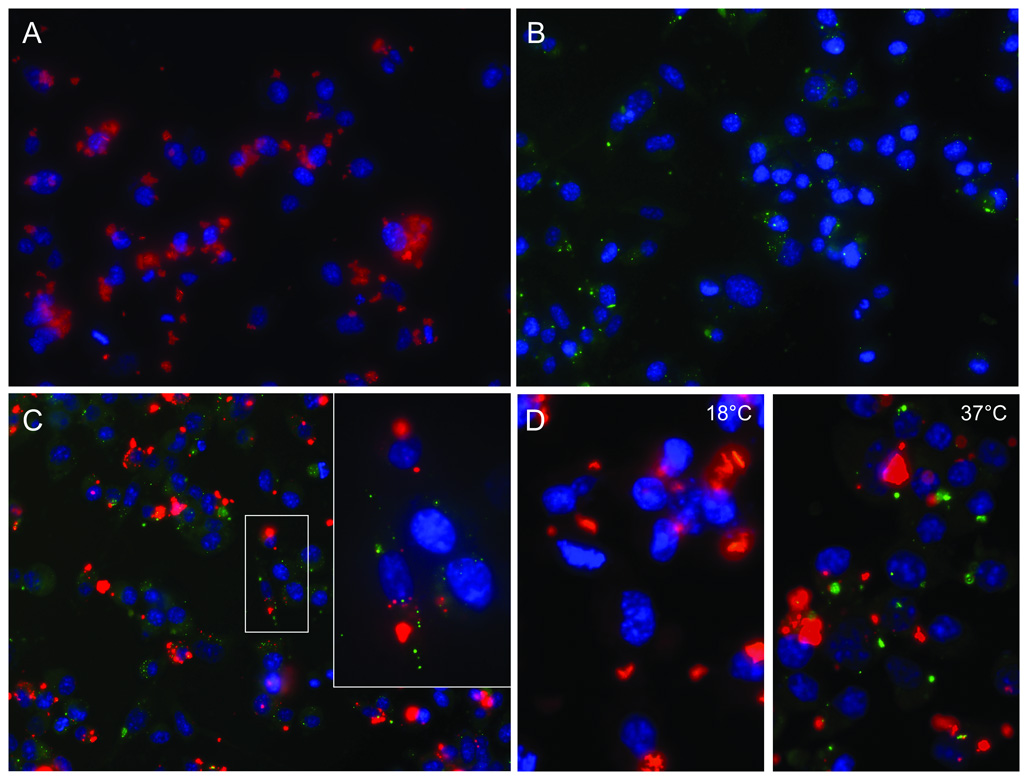
Uptake of PrP-res3F4 in cells in our assay was quite different from that of previous studies where large, fluorescent PrP-res aggregates were not fully internalized by SN56 neuronal cells until several days post PrP-res exposure (Baron, Magalhaes et al., 2006), (Magalhaes, Baron et al., 2005). To test whether differences in cell type or PrP-res preparation could account for these discrepancies, we exposed differentiated SN56 neuronal cells for 24 hrs to either 22L PrP-res3F4 or 22L PrP-res3F4 conjugated to an Alexa-Fluor-596 fluorescent tag (Fig. 7). SN56 cells exposed to Alexa-Fluor labeled 22L PrP-res3F4 showed very large PrP-res3F4 aggregates (Fig. 7A) that were indistinguishable from those described previously (Baron, Magalhaes et al., 2006),(Magalhaes, Baron et al., 2005). By contrast, SN56 cells exposed to non-Alexa-Fluor labeled 22L PrP-res3F4 (Fig. 7B) showed the same small, punctate aggregates of PrP-res3F4 observed in MoL42-CFD5 cells (Fig. 6). When both Alexa-Fluor labeled PrP-res from 22L scrapie-infected wild type mice and non-Alexa- Fluor labeled 22L PrP-res3F4 were added to the same monolayer of SN56 cells, the difference in PrP-res aggregate size was even more apparent (Fig. 7C). The difference in aggregate size between 22L PrP-res and Alexa-Fluor labeled 22L PrP-res strongly suggests that the larger PrP-res aggregates were primarily a consequence of the Alexa- Fluor labeling process.
Figure 7. Large PrP-res aggregates bind to the cell surface and are not rapidly internalized.
Differentiated SN56 cells were exposed to Alexa-Fluor-596 labeled (red) or unlabeled (green) 22L PrP-res3F4 for 24 hours. Cells were then rinsed, fixed and when necessary, immunolabeled with the mouse monoclonal antibody 3F4 (Panels B–D). Anti-mouse FITC labeled antibody (green) was used to detect 22L PrP-res3F4, while DAPI stain (blue) denotes cell nuclei. Alexa-Fluor-596 labeled proteins are red. All images were taken with a 40X objective. Cells were exposed to A) Alexa-Fluor labeled partially purified 22L PrP-res3F4 (red), B) partially purified 22L PrP-res3F4 (green), C) Alexa- Fluor labeled partially purified PrP-res from a 22L scrapie-infected wild type mouse (red) and partially purified 22L PrP-res3F4 (green) (inset taken with a 60X objective). In panel D, cells were exposed for 24 hours at either 18°C or 37°C to Alexa-Fluor labeled partially purified PrP-res from a 22L scrapie-infected wild type mouse (red) and 22L PrP-res3F4 (green). The larger Alexa-Fluor labeled PrP-res aggregates do not colocalize with the smaller, more punctate PrP-res3F4 aggregates.
When both Alexa-Fluor labeled 22L PrP-res from scrapie-infected wild type mice and non-Alexa-Fluor labeled 22L PrP-res3F4 were added to the same monolayer of SN56 cells, there was almost no co-localization between the PrP-res molecules in the two preparations (Fig. 7C). To help resolve this difference in co-localization, we conducted a temperature shift analysis of SN56 cells exposed to both Alexa-Fluor labeled 22L PrP-res from wild type mice and non-Alexa-Fluor labeled PrP-res3F4 (Fig. 7D). Cells incubated at 37°C (Fig. 7D) showed localization patterns similar to Figure 7C. However, cell-associated PrP-res3F4 was not detected at 18°C suggesting that the amount of PrP-res3F4 bound or internalized by the cells at was very low (Fig. 7D). By contrast, significant amounts of Alexa-Fluor labeled PrP-res were cell-associated at 18°C (Fig. 7D) suggesting that it was bound to the cell surface via either PrP-res3F4 or the Alexa-Flour dye moiety. The fact that Alexa-Fluor labeled PrP-res was largely out of the plane of focus of the cell when examined by immunofluorescence (note the faint halo effect around the red Alexa-Fluor labeled PrP-res aggregates in Fig. 7) also support the conclusion that these aggregates were primarily localized at the cell surface. Our data suggest that during the first 24 hrs of infection, large PrP-res aggregates are localized primarily at the cell surface while smaller aggregates are rapidly internalized.
Infectious brain homogenate PrP-res is taken up most efficiently by cells
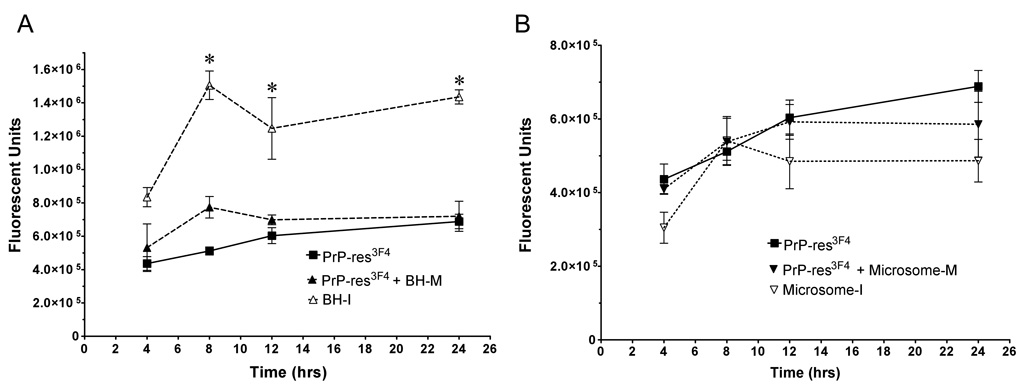
A number of PrP-res preparation methods have been employed to infect cells in vitro (Baron, Magalhaes et al., 2006), (Baron, Wehrly et al., 2002), (Bendheim, Barry et al., 1984), (Vorberg & Priola, 2002). To test if different PrP-res preparation methods would alter PrP-res uptake into cells, equal amounts of Obihiro PrP-res3F4 derived either from partially purified PrP-res or infectious crude brain homogenate was added to MoL42- CFD5 cells and the uptake of PrP-res into the cells was assayed by Western blot. By 8 hours post infection, infectious crude brain homogenate appeared to be taken up significantly more efficiently then partially purified PrP-res (Fig. 8A, open triangles). To determine whether or not this discrepancy in PrP-res uptake was due to a difference in total protein, mock infected brain homogenate was added to the partially purified PrP-res in order to match the total protein content found in the infectious brain homogenate. Protein adjusted partially purified PrP-res was taken up with the same efficiency as partially purified PrP-res alone (Fig. 8A). Microsome PrP-res was also taken up by cells with the same efficiency as either partially purified PrP-res or total protein adjusted partially purified PrP-res (total protein adjusted with mock infected microsome preparation) (Fig. 8B). Taken together, our results suggest that there is an increased efficiency in the uptake of PrP-res when it is associated with an infectious brain homogenate.
Figure 8. Infectious brain homogenate PrP-res3F4 is taken up more efficiently then either microsome or partially purified PrP-res3F4.
Graphical representation of PrP-res3F4 uptake from 0 – 24 hrs into MoL42-CFD5 cells (N = 6) where error bars represent SEM. A) Kinetics of PrP-res3F4 uptake using either infectious brain homogenate PrP-res3F4 (BH-I), partially purified PrP-res3F4 (PrP-res3F4) or partially purified PrP-res3F4 with mock infected brain homogenate (PrP-res3F4 + BH-M) added to match the total protein content of BH-I (* p<0.05, t = 8–24 hrs, Bonferroni test). B) Kinetics of PrP-res3F4 uptake using either infectious microsome PrP-res3F4 (Microsome-I), purified PrP-res3F4 (PrP-res3F4) or partially purified PrP-res3F4 with mock infected microsomes (PrP-res3F4 + Microsome-M) added to match total protein content of infectious microsomes (p>0.05 for all time points, Bonferroni test). All data was obtained with IR-dye 800CW developed western blots where the PrP-res3F4 level was quantified as fluorescent units using the Li-Cor Odyssey imaging system and associated software.
DISCUSSION
The use of PrP-res tagged with a unique antibody epitope has allowed us to examine for the first time the cellular uptake of PrP-res present in an infectious inoculum in the absence of any confounding background from host cell derived PrP-res or PrP-sen. Our data show that PrP-res uptake is cell type and scrapie strain independent and are consistent with previous work where the acute conversion of cellular PrP-sen to PrP-res was also found to be cell type and scrapie strain independent (Vorberg, Raines et al., 2004a). Furthermore, the current study demonstrates that during the first three days post-scrapie exposure cells take up PrP-res from different strains at a similar rate (Fig. 4, Fig. 6) and that this process absolutely does not require host cell expression of PrP-sen (Fig. 4). Thus, although PrP-sen is necessary for persistent PrP-res formation and scrapie infection, its absence in cells does not inhibit acute uptake of PrP-res.
Regardless of strain, PrP-res uptake into cells was detectable by 2 hrs (Fig. 4, Fig. 6) and, after 8 hrs, was apparently restricted by total cell number (Fig. 5). This change in the kinetics curve may be related to the fact that most cells are still rapidly dividing and in log phase during the first 8 hours of exposure to PrP-res. Over time, the cells become more confluent and PrP-res uptake may be reduced as cellular division slows. This interpretation is consistent with the recent observation that cell division can also influence PrP-res levels within mouse neuroblastoma cells persistently infected with scrapie (Ghaemmaghami, Phuan et al., 2007).
For all mouse strains tested, our results show that 10–15% of PrP-res3F4 in the brain homogenate was taken up by the cells (Fig. 2D). This is in stark contrast to a recent study showing that the amount of PrP-res taken up by the cell during acute TSE infection was strain dependent and could exceed 80% (Paquet, Daude et al., 2007). One possible explanation for this discrepancy is that the population of cells susceptible to TSE infection is higher in the epithelial cells used in the previous study than in the cells used here. Another possibility is that strain-specific differences in the size of the PrP-res aggregate may influence how much PrP-res the cells take up. The recent observation that PrP-res particle size can influence scrapie infectivity (Silveira, Raymond et al., 2005), the demonstration that large Alexa-Fluor PrP-res aggregates are broken down over several days into smaller aggregates which are then internalized by the cell (Magalhaes, Baron et al., 2005), as well as our data demonstrating that large AlexFluor-labeled PrP-res aggregates are not rapidly internalized (Fig. 7A), are all consistent with this possibility.
Interestingly, at any given timepoint only 30–40% of the cells were able to take up detectable levels of PrP-res (Fig. 6) despite the fact that it was always supplied in excess. The most likely explanation for this result is that there is a limited population of cells which can detectably take up PrP-res. Over time, these cells divide and increase in number leading to the observed increase in the amount of cell-associated PrP-res, a process that may also be important once persistent infection has been established (Ghaemmaghami, Phuan et al., 2007), (Weissmann, 2004). Alternatively, it may be that only certain cells have the necessary cell surface ligands to endocytose PrP-res. A third possibility may relate to the observation that the pathway of cellular internalization of particles such as viruses and bacteria is influenced by both particle size and the composition of plasma membrane microdomains (Cheng, Singh et al., 2006) Perhaps only a certain subpopulation of cells within the culture have the plasma membrane microdomains needed to efficiently interact with and internalize PrP-res. Whatever the explanation, our data are consistent with the hypothesis that PrP-res aggregate size can influence not only how PrP-res initially interacts with the cell but also its uptake.
Mouse PrP-res from all strains tested formed small, punctate aggregates that appeared to localize primarily in the perinuclear and cytoplasmic regions of the cell (Fig. 6). This localization is consistent with earlier studies of PrP-res in persistently infected cells (Caughey, Raymond et al., 1991), (McKinley, Taraboulos et al., 1991), (Pimpinelli, Lehmann et al., 2005), (Taraboulos, Scott et al., 1994) and these punctate aggregates likely act as PrP-res seeds that help propagate new PrP-res formation (Vorberg, Raines et al., 2004a). Despite the evidence that cell-type can influence establishment of a persistent infection (Vorberg, Raines et al., 2004a), for all cell types and strains tested we were unable to detect any differences in the cellular localization of PrP-res. For example, both ME7 and 22L PrP-res appeared to be in similar cytoplasmic and perinculear locations (Fig. 6), despite the fact that the 22L scrapie strain infects fibroblast cells while ME7 does not (Vorberg, Raines et al., 2004a). This suggests that the initial cellular location of the inoculum PrP-res may not be a determining factor in whether or not a cell becomes persistently infected. However, given the fact that our current results do not allow us to clearly distinguish different cellular compartments, further studies looking at the co-localization of input PrP-res3F4 with different cellular markers will be needed to determine if this is the case.
Previous studies using the hamster scrapie strain 263K and CHO cells found that hamster PrP-res remained cell associated at 18°C and likely bound to cells via cell surface heparan sulfate (Hijazi, Kariv-Inbal et al., 2005). It was therefore somewhat surprising that we found that only a small amount of mouse PrP-res3F4 was cell-associated at 18°C (Fig. 3 and Fig. 7D). Our data comparing AlexaFluor labeled PrP-res to unlabeled PrP-res demonstrate that PrP-res aggregate size can influence whether or not PrP-res can detectably bind to the cell surface. Thus, one explanation for the difference between the mouse and hamster results could be that hamster PrP-res aggregates are larger than mouse PrP-res aggregates. Alternatively, it is possible that at 18°C the receptor for internalization of mouse PrP-res is largely absent from the cell surface but still present for hamster PrP-res. However, the low level of mouse PrP-res cell surface binding and uptake at 18°C (Fig. 3, Fig. 7D) suggests that mouse PrP-res is taken up into cells through an active cellular process such as endocytosis that may not necessarily be mediated by a specific cell surface ligand.
PrP-res within an infectious brain homogenate was taken up more efficiently then either partially purified PrP-res or PrP-res in microsome preparations (Fig. 8). This suggests that there may be a co-factor or some type of PrP-res associated microenvironment that is specific to infectious brain homogenate which is removed or disrupted during either of the alternative PrP-res preparations. Consistent with the idea that PrP-res may have to be in a specific microenvironment to efficiently infect cells (Baron, Magalhaes et al., 2006), the simple addition of brain homogenate protein to partially purified PrP-res preparations did not enhance the uptake of partially purified PrP-res (Fig. 8A). It is also unlikely that microsomes provide the appropriate microenvironment given that microsome PrP-res was not taken up as efficiently as PrP-res in brain homogenate (Fig. 8B). Our results do not negate the possibility that microsomes may be more infectious (Baron, Magalhaes et al., 2006), but suggest that the apparent increased efficiency with which microsomes infect cells is not due to more efficient uptake of PrP-res during the early stages of infection as previously proposed (Baron, Magalhaes et al., 2006).
In combination with earlier studies (Vorberg, Raines et al., 2004a), our data suggest that there may be at least two blocks to the persistent infection of a cell with scrapie. The first occurs during the acute stage of infection (0–72hrs) when factors such as PrP-res aggregate size, the microenvironment of the PrP-res inoculum, and/or the presence of a specific population of cells help to determine whether or not PrP-res is bound to the cell and internalized. This block would not be strictly dependent upon scrapie strain or cell type but rather would depend upon heterogeneity in both the PrP-res and cell populations. Once a cell culture becomes persistently infected, cellular heterogeneity may also influence both the level and stability of infectivity over long term passage (Bosque & Prusiner, 2000), (Enari, Flechsig et al., 2001), (Weissmann, 2004).
We have previously shown that the mouse Ψ2 fibroblast cells used in the present study can be infected with the 22L, but not the ME7, strain of mouse scrapie (Vorberg, Raines et al., 2004a). The fact that ME7 acts similarly to 22L during acute infection (Fig. 4–Fig. 5 and (Vorberg, Raines et al., 2004a)) suggests that the second block to infection is scrapie-strain dependent and occurs 1) after uptake and localization of the inoculum PrP-res and, 2) after an acute burst of new PrP-res formation in the cell (Vorberg, Raines et al., 2004a).
MATERIAL AND METHODS
PrP0/0 Neural Cell Line
Pregnant mice in which the PRNP gene has been knocked out (PrP0/0) (Manson, Clarke et al., 1994) were anesthetized with isofluorane at gestation day 15. Embryos were isolated, decapitated and the heads were transferred to cold dissecting solution (phosphate buffered saline (PBS), Ca2+/Mg2+ free, 5% glucose). Brains were removed and freed of meninges and blood vessels, cut into 5 mm2 pieces, and disassociated by aspiration in Dulbecco’s phosphate buffered saline (DPBS) with 10% FBS (Gibco Inc.), 0.5 mM glutamine, and B27. The cell suspension was centrifuged at 160 × g, 4°C, 5 min and cells were resuspended in DMEM with 5% FBS, and F12. Cells were adjusted such that 40,000 cells/well were plated on poly-L-lysine coated 24 well plates. After 4 hrs incubation at 37°C, 5% CO2, medium was replaced with 1 ml/well of DMEM with growth supplements FBS (3%), B27, and N2. Transfection of cells for immortalization was done 48 hrs after plating, using the plasmid vector pSV3-neo (ATCC, #37150) (Southern & Berg, 1982) and lipofectamine (Invitrogen) according to the manufacturer’s instructions. Two days after transfection, G418 was added to the medium to select for transfected, immortalized cells. Immortalized clones were pooled and single cell clones were established. A single cell clone (CF10) was selected and maintained in Opti-MEM supplemented with 10% FBS and 1% penicillin streptomycin (PS). CF10 cells are strongly positive for nestin (data not shown) which is predominantly found in stem cells of the CNS. Therefore, it is likely that they are neural stem cells.
MoL42-CFD5 and Mo3F4-CF10 cell lines were derived by limiting dilution cloning from CF10 cells that had been transduced with a retrovirus encoding the mutant PrP molecule of interest (Mann, Mulligan et al., 1983). The Mo3F4-CF10 cells express mouse PrP with the epitope to the mouse monoclonal antibody 3F4 (Mo3F4), while the MoL42-CFD5 cells express mouse PrP with the epitope to the mouse monoclonal antibody L42 (MoL42) (Vorberg, Buschmann et al., 1999). The L42 epitope can be used to distinguish mouse PrP in the cells from mouse PrP in brain homogenate.
Fibroblast cells (ψ2) are susceptible to infection with the mouse scrapie strain 22L but not with the ME7 strain (Vorberg, Raines et al., 2004a). MoL42-ψ2A2 is a cell line derived by limiting dilution cloning from ψ2 fibroblast cells (Mann, Mulligan et al., 1983;Miller & Buttimore, 1986) that had been transduced by the same retrovirus used to make the MoL42-CFD5 cells. MoL42-ψ2A2 cells express both endogenous mouse PrP as well as mouse PrP with the L42 epitope. MoL42-ψ2A2 cells were maintained in RPMI with 10% FBS supplemented with 1% penicillin/streptomycin (final concentration of 100 units/ml penicillin G and 100 ug/ml streptomycin) (Invitrogen). SN56 cells are neuronal derived cells from mouse septum neurons (Hammond, Lee et al., 1990) and were used with the kind permission of Dr. Bruce Wainer (Department of Pathology, Emory University of Medicine, Atlanta, GA).
PrP-res preparation
Scrapie strains 22L and ME7 were the kind gift of Dr. James Hope (Vitechnologies, Boston, Ma) while the Obihiro scrapie strain (Shinagawa, Takahashi et al., 1985) was the kind gift of Dr. Motohiro Horiuchi University of Obihiro, Hokkaido, Japan). Tg(WT-E1) mice were from the laboratory of Dr. David Harris (Washington University, St. Louis, Mo). These mice express high levels of PrP-sen with an epitope tag (Bolton, Seligman et al., 1991) for the mouse monoclonal antibody 3F4 (PrP-sen3F4).
The mouse-adapted scrapie strains 22L, ME7, and Obihiro (Obi) in DPBS were passaged once in Tg(WT-E1) mice (Chiesa, Piccardo et al., 1998) to generate the brain homogenates 22L(3F4), ME7(3F4), and Obi(3F4). Tg(Wt-E1) mice inoculated with uninfected C57Bl/10 mouse brains were designated as mock controls (Mock(3F4)). Brains from terminally ill Tg(WT-E1) mice or mock infected, age matched controls were harvested and stored at −70°C. To study the uptake of PrP-res from crude brain homogenate, brains were dounce homogenized in DPBS (10% w/v), sonicated, and then stored at −70°C as described previously (Vorberg, Raines et al., 2004a). Crude brain microsome fractions were prepared as described in detail by Baron and colleagues (Baron, Wehrly et al., 2002), while PrP-res was partially purified as previously described (Raymond & Chabry, 2004) but without proteinase K (PK) digestion. Partially purified PrP-res was quantified by western blot using a standard protein concentration curve derived from recombinant hamster PrP-sen. For experiments where partially purified PrP-res preparations were compared to less purified microsome or brain homogenate preparations, total protein content was normalized using microsomes or brain homogenate from mock-infected Tg(WT-E1) mice. Alexa-Fluor-596 labeled PrP-res preparations were made as previously described by Magalhāes and collegues (Magalhaes, Baron et al., 2005).
Cellular uptake of PrP-res
To analyze PrP-res uptake over time, experiments were performed in 24 well microtiter plates as previously described (Vorberg, Raines et al., 2004a). Briefly, different cell numbers were initially plated in order to ensure similar cell numbers at time of harvest. The number of cells plated were: MoL42-CFD5 cells (0–24 hrs, 3 × 105 cells/ml; 48hrs 2 × 105 cells/ml; and 72 hrs 1.5 × 105 cells/ml), CF10 and MoL42-ψ2A2 cells (0–24 hrs 4 × 105 cells/ml; 48hrs 2 × 105 cells/ml; and 72 hrs 1.5 × 105 cells/ml). After incubation at 37°C for 24 hrs, the medium was removed and replaced with 200 µl of brain homogenate, microsomes, or partially purified PrP-res diluted in Opti-MEM. The cells were exposed to equivalent amounts of PrP-res for each strain (10 ng for each preparation). Mock preparations were diluted 1:10 in Opti-MEM. Samples were then incubated at 37°C for 2–4 hrs followed by the addition of 400 µl/well of fresh media. At each time point, cells were washed 4 times with 300 µl of fresh medium and either removed from the plate using trypsin-EDTA (Invitrogen) and counted with a hemacytometer or lysed directly in 3X lysis buffer (3 mM Tris-HCl, pH 7.4, 420 mM NaCl, 15 mM EDTA, 1.5% sodium deoxycholate, and 1.5% Triton x–100). Lysates were treated with benzonase (0.167 U/ul) (Novagen) for 30 min at 37°C, then PK (60 µg/ml) (Roche) for 1 hr at 37°C followed by the addition of 1.2 µg of 4-(2-aminoethyl)benzenesulfonylfluoride (PEFABLOC) (Roche). PK digested lysates were precipitated in four volumes of cold methanol for 2 hrs at −20°C followed by centrifugation at 20,800 × g for 30 min. Pellets were sonicated into sample buffer (2.5% SDS, 3 mM EDTA, 2% β-mercaptoethanol, 5% glycerol, 0.02% bromphenol blue, and 63 mM Tris-HCL, pH 6.8), boiled for 3 min, optionally PNGaseF treated for 12 hours according to the manufacturer’s instructions (New England Biolabs), and loaded on 16% Tris-Glycine precast gels (Invitrogen). PrP was detected by western blot analysis using the mouse monoclonal antibody 3F4 (1:3,000) followed by secondary ECL-anti-mouse IgG (1:5,000) (Amersham) or anti-mouse IR-dye 800CW (1:10,000) (Li-Cor).
Quantitative ECL data was generated using the UN-SCAN-IT software (Silk Scientific Corp.) according to the manufacturer’s instructions. In order to quantify the data within the linear range of the film, the first three lanes of each gel were used as internal standards to establish a constant film exposure time which was then used for every experiment. PrP-res levels were quantified from gels exposed to film for a set time (4 min) using a fixed parameter box (i.e. the box was the same volume for every experiment) and the UN-SCAN-IT software. Pixel count totals within the fixed parameter box were summed and presented as pixels. When the secondary antibody anti-mouse IR-dye 800CW was used to develop the western blot, quantitative data were obtained using the Li-Cor Odyssey IR scanner and the software provided with the system (Li-Cor).
To quantify the percentage of the input brain homogenate PrP-res3F4 that was taken up by the cells, MoL42-CFD5 cells were plated into 24 well plates as detailed above and scrapie-infected brain homogenate containing 8ng of PrP-res3F4 was added to the cells for 24 or 48 hrs. Following removal of the supernatant and four rinses of the cells with PBBS, the cells were lysed and cell associated PrP-res3F4 was isolated as described above. For each strain, a standard curve consisting of serial two-fold dilutions of the input PrP-res3F4 brain homogenate was used to quantify the samples. These serial two-fold dilutions (8ng-1ng) were loaded on every gel and used to establish a standard curve to quantify only the samples run on the same gel. Two experiments were run for a total of 12 samples for each strain and timepoint. Quantitative data were obtained using the Li-Cor Odyssey IR scanner and the software provided with the system (Li-Cor).
Analysis of PrP-res uptake by fluorescent microscopy
Cells were plated in Lab-Tek II chamber slides (8-well) (Nalge Nunc Inc.) at the densities listed above. After 24 hrs at 37°C, the media was removed and immediately replaced with 200 µl of Opti-MEM containing 3 ng of partially purified PrP-res from 22L(3F4), ME7(3F4), or Obi(3F4) or volume matched partially purified PrP-res Mock(3F4) preparations. Following incubation from 2 to 48 hrs, the media was aspirated and the cells were washed 4 times with fresh medium. 200ul of 10% formaldehyde was added to each well for 30 min, followed by 200 µl of 0.4% triton-X-100 for 10 min. The wells were then rinsed with 500 µl of PBS (2X), and select chambers were treated with 200 µl of PK (10ug/ml) for 9 min at 37°C. The PK was then removed and excess PK blocked with 200 µl of PEFABLOC (10mM). Wells were washed again with PBS and treated with guanidine thiocyanate (4M, 200 µl/chamber) for 30 min at 25°C. After rinsing in PBS, 200 µl of the mouse 3F4 monoclonal antibody (1:200 dilution) was added for 30 min followed by PBS washes (3X) and the addition of 200 µl of goat anti-mouse FITC conjugated antibody (1:400 dilution) for 30 min. Following the antibody incubations, the slides were rinsed in PBS and coverslip mounted with ProLong Gold antifade reagent with DAPI (Invitrogen). Slides were viewed on an Olympus BX51 fluorescent microscope with both 20X and 40X air immersion objectives. All images were taken with an Olympus DP71 camera and were processed and analyzed with Microsuite Software (Olympus).
For comparison of PrP-res to Alexa-Fluor-596 conjugated PrP-res, SN56 cells were plated in Lab-Tek II chamber slides (8-well) (Nalge Nunc Inc.) at a 1:20 dilution from a confluent 25 cm2 flask and differentiated with cAMP (1mm) in serum free Opti-MEM for 24 hrs. PrP-res or Alexa-Fluor-596 conjugated PrP-res (8 ng) was added to the cells and the cells were incubated for 24 hrs at 37°C. Cells were then fixed, labeled and observed by florescent microscopy as described above.
ACKNOWLEDGMENTS
We wish to thank Anita Mora and Gary Hettrick for technical support and Dr. Byron Caughey, Dr. Sonja Best, and Dr. Kristin McNally for critical reading of the manuscript. This research was supported in part by the University of Montana department of Biomedical and Pharmaceutical Sciences Neuroscience program and the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (Project #1-Z01-AI000752-12). All animals were treated in accordance with the regulations and guidelines of the Animal Care and Use Committee of the Rocky Mountain Laboratories and the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Baron GS, Magalhaes AC, Prado MA, Caughey B. Mouse-adapted scrapie infection of SN56 cells: greater efficiency with microsome-associated versus purified PrP-res. J. Virol. 2006;80:2106–2117. doi: 10.1128/JVI.80.5.2106-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, Wehrly K, Dorward DW, Chesebro B, Caughey B. Conversion of raft associated prion protein to the protease-resistant state requires insertion of PrP-res (PrP(Sc)) into contiguous membranes. EMBO J. 2002;21:1031–1040. doi: 10.1093/emboj/21.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendheim PE, Barry RA, DeArmond SJ, Stites DP, Prusiner SB. Antibodies to a scrapie prion protein. Nature. 1984;310:418–421. doi: 10.1038/310418a0. [DOI] [PubMed] [Google Scholar]
- Beranger F, Mange A, Goud B, Lehmann S. Stimulation of PrP(C) retrograde transport toward the endoplasmic reticulum increases accumulation of PrP(Sc) in prion-infected cells. J.Biol.Chem. 2002;277:38972–38977. doi: 10.1074/jbc.M205110200. [DOI] [PubMed] [Google Scholar]
- Bergstrom AL, Jensen TK, Heegaard PM, Cordes H, Hansen VB, Laursen H, Lind P. Short-term study of the uptake of PrP(Sc) by the Peyer's patches in hamsters after oral exposure to scrapie. J.Comp Pathol. 2006;134:126–133. doi: 10.1016/j.jcpa.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Bolton DC, Seligman SJ, Bablanian G, Windsor D, Scala LJ, Kim KS, Chen CM, Kascsak RJ, Bendheim PE. Molecular location of a species-specific epitope on the hamster scrapie agent protein. J. Virol. 1991;65:3667–3675. doi: 10.1128/jvi.65.7.3667-3675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Scott M, Taraboulos A, Stahl N, Prusiner SB. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Taraboulos A, Prusiner SB. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J.Biol.Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- Bosque PJ, Prusiner SB. Cultured cell sublines highly susceptible to prion infection. J. Virol. 2000;74:4377–4386. doi: 10.1128/jvi.74.9.4377-4386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler DA, Scott MR, Bockman JM, Borchelt DR, Taraboulos A, Hsiao KK, Kingsbury DT, Prusiner SB. Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Raymond GJ. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J.Biol.Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- Caughey B, Raymond GJ, Ernst D, Race RE. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Singh RD, Marks DL, Pagano RE. Membrane microdomains, caveolae, and caveolar endocytosis of sphingolipids. Mol.Membr.Biol. 2006;23:101–110. doi: 10.1080/09687860500460041. [DOI] [PubMed] [Google Scholar]
- Chiesa R, Piccardo P, Ghetti B, Harris DA. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- Enari M, Flechsig E, Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc.Natl.Acad.Sci.U.S.A. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Phuan PW, Perkins B, Ullman J, May BC, Cohen FE, Prusiner SB. Cell division modulates prion accumulation in cultured cells. Proc.Natl.Acad.Sci.U.S.A. 2007;104:17971–17976. doi: 10.1073/pnas.0708372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond DN, Lee HJ, Tonsgard JH, Wainer BH. Development and characterization of clonal cell lines derived from septal cholinergic neurons. Brain Res. 1990;512:190–200. doi: 10.1016/0006-8993(90)90626-M. [DOI] [PubMed] [Google Scholar]
- Hijazi N, Kariv-Inbal Z, Gasset M, Gabizon R. PrPSc incorporation to cells requires endogenous glycosaminoglycan expression. J.Biol.Chem. 2005;280:17057–17061. doi: 10.1074/jbc.M411314200. [DOI] [PubMed] [Google Scholar]
- Horonchik L, Tzaban S, Ben-Zaken O, Yedidia Y, Rouvinski A, Papy-Garcia D, Barritault D, Vlodavsky I, Taraboulos A. Heparan sulfate is a cellular receptor for purified infectious prions. J.Biol.Chem. 2005;280:17062–17067. doi: 10.1074/jbc.M500122200. [DOI] [PubMed] [Google Scholar]
- Magalhaes AC, Baron GS, Lee KS, Steele-Mortimer O, Dorward D, Prado MA, Caughey B. Uptake and neuritic transport of scrapie prion protein coincident with infection of neuronal cells. J.Neurosci. 2005;25:5207–5216. doi: 10.1523/JNEUROSCI.0653-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R, Mulligan RC, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, Hope J. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol.Neurobiol. 1994;8:121–127. doi: 10.1007/BF02780662. [DOI] [PubMed] [Google Scholar]
- McKinley MP, Taraboulos A, Kenaga L, Serban D, Stieber A, DeArmond SJ, Prusiner SB, Gonatas N. Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab Invest. 1991;65:622–630. [PubMed] [Google Scholar]
- Miller AD, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol.Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan J, Hopkins J, Mabbott NA. Skin-derived dendritic cells acquire and degrade the scrapie agent following in vitro exposure. Immunology. 2005;116:122–133. doi: 10.1111/j.1365-2567.2005.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet S, Daude N, Courageot MP, Chapuis J, Laude H, Vilette D. PrPc does not mediate internalization of PrPSc but is required at an early stage for de novo prion infection of Rov cells. J. Virol. 2007 doi: 10.1128/JVI.01137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli F, Lehmann S, Maridonneau-Parini I. The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur.J.Neurosci. 2005;21:2063–2072. doi: 10.1111/j.1460-9568.2005.04049.x. [DOI] [PubMed] [Google Scholar]
- Priola SA, Vorberg I. Molecular aspects of disease pathogenesis in the transmissible spongiform encephalopathies. Mol.Biotechnol. 2006;33:71–88. doi: 10.1385/MB:33:1:71. [DOI] [PubMed] [Google Scholar]
- Race RE, Fadness LH, Chesebro B. Characterization of scrapie infection in mouse neuroblastoma cells. J.Gen.Virol. 1987;68(Pt 5):1391–1399. doi: 10.1099/0022-1317-68-5-1391. [DOI] [PubMed] [Google Scholar]
- Raymond GJ, Chabry J. Methods and tools in biosciences and medicine. Basel, Switzerland: Birkhäuser Verlag; 2004. Purification of the pathological isoform of prion protein (PrPsc or PrPres) from transmissible spongiform encephalopathy-affected brain tissue; pp. 16–26. [Google Scholar]
- Raymond GJ, Olsen EA, Lee KS, Raymond LD, Bryant PK, III, Baron GS, Caughey WS, Kocisko DA, McHolland LE, Favara C, Langeveld JP, van Zijderveld FG, Mayer RT, Miller MW, Williams ES, Caughey B. Inhibition of protease-resistant prion protein formation in a transformed deer cell line infected with chronic wasting disease. J. Virol. 2006;80:596–604. doi: 10.1128/JVI.80.2.596-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein R, Carp RI, Callahan SM. In vitro replication of scrapie agent in a neuronal model: infection of PC12 cells. J.Gen.Virol. 1984;65(Pt 12):2191–2198. doi: 10.1099/0022-1317-65-12-2191. [DOI] [PubMed] [Google Scholar]
- Schatzl HM, Laszlo L, Holtzman DM, Tatzelt J, DeArmond SJ, Weiner RI, Mobley WC, Prusiner SB. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol. 1997;71:8821–8831. doi: 10.1128/jvi.71.11.8821-8831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa M, Takahashi K, Sasaki S, Doi S, Goto H, Sato G. Characterization of scrapie agent isolated from sheep in Japan. Microbiol.Immunol. 1985;29:543–551. doi: 10.1111/j.1348-0421.1985.tb00856.x. [DOI] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern PJ, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J.Mol.Appl.Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- Taraboulos A, Scott M, Semenov A, Avrahami D, Prusiner SB. Biosynthesis of the prion proteins in scrapie-infected cells in culture. Braz.J.Med.Biol.Res. 1994;27:303–307. [PubMed] [Google Scholar]
- Vorberg I, Buschmann A, Harmeyer S, Saalmuller A, Pfaff E, Groschup MH. A novel epitope for the specific detection of exogenous prion proteins in transgenic mice and transfected murine cell lines. Virology. 1999;255:26–31. doi: 10.1006/viro.1998.9561. [DOI] [PubMed] [Google Scholar]
- Vorberg I, Priola SA. Molecular basis of scrapie strain glycoform variation. J.Biol.Chem. 2002;277:36775–36781. doi: 10.1074/jbc.M206865200. [DOI] [PubMed] [Google Scholar]
- Vorberg I, Raines A, Priola SA. Acute formation of protease-resistant prion protein does not always lead to persistent scrapie infection in vitro. J.Biol.Chem. 2004a;279:29218–29225. doi: 10.1074/jbc.M402576200. [DOI] [PubMed] [Google Scholar]
- Vorberg I, Raines A, Story B, Priola SA. Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J.Infect.Dis. 2004b;189:431–439. doi: 10.1086/381166. [DOI] [PubMed] [Google Scholar]
- Weissmann C. The state of the prion. Nat.Rev.Microbiol. 2004;2:861–871. doi: 10.1038/nrmicro1025. [DOI] [PubMed] [Google Scholar]