Abstract
Monocytes are primary targets for human cytomegalovirus (HCMV) infection and are proposed to be responsible for haematogenous dissemination of the virus. Monocytes acquire different functional traits during polarization to the classical pro-inflammatory M1 macrophage or the alternative anti-inflammatory M2 macrophage. We hypothesized that HCMV induced a pro-inflammatory M1 macrophage following infection in order to promote viral dissemination because, biologically, a pro-inflammatory state provides the necessary tools to drive infected monocytes from the blood into the tissue. To test this hypothesis of monocyte conversion from a normal quiescent phenotype to an inflammatory phenotype, we utilized Affymetrix Human Microarray to acquire a transcriptional profile of infected monocytes at a time point our data emphasized as being a key temporal regulatory point following infection. We found that HCMV significantly upregulated 583 (5.2%) of the total genes and downregulated 621 (5.5%) of the total genes ≥1.5-fold at 4 hours post infection. Further ontology analysis revealed that genes implicated in classical M1 macrophage activation were stimulated by HCMV infection. Specifically, we found that 65% of genes strictly associated with M1 polarization were upregulated, while only 4% of genes solely associated with M2 polarization were upregulated. Analysis of the monocyte chemokinome at the transcriptional level showed that 44% of M1 and 33% of M2 macrophage chemokines were upregulated, respectively. Proteomic analysis using human chemokine antibody arrays confirmed the secretion of these chemotactic proteins from HCMV-infected monocytes. Overall, the results identify that the HCMV-infected monocyte transcriptome displayed a unique M1/M2 polarization signature that was skewed towards the classical M1 activation phenotype.
Introduction
Infection by human cytomegalovirus (HCMV), a member of the Herpesviridae family, leads to morbidity and mortality in immunocompromised individuals including AIDS and organ transplant patients, congenitally infected neonates, and cancer patients undergoing chemotherapy (1-3). HCMV infection causes a wide range of overt organ diseases including retinitis, gastrointestinal disease, hepatitis and interstitial pneumonia due to the broad cellular tropism of the virus in vivo (4). Following initial primary infection of host epithelial cells by contact with HCMV-containing bodily fluids, HCMV replicates and spreads to the peripheral blood where viral dissemination to multiple organ systems occurs (4).
Peripheral blood leukocytes, specifically monocytes, are believed to be responsible for haematogenous spread (4, 5). Monocytes are primary in vivo targets for HCMV (6), are a site of viral latency and persistence (4, 7), are the primary infiltrating cell type found in HCMV-infected organs (8, 9) and their aberrant function following HCMV infection is implicated in atherosclerosis, an inflammatory disease whose development and severity is associated with HCMV infection (10, 11). Furthermore, animal studies indicate that monocyte-associated viremia is a prerequisite for viral pathogenesis (12, 13). In accord with these in vivo observations, we previously provided in vitro evidence suggesting HCMV utilizes monocytes as a vehicle for spread, infiltration into and persistence in host tissue (14-17). Our studies have demonstrated that primary infection of peripheral blood monocytes by HCMV induces a pro-inflammatory state resulting in increased cell motility, firm adhesion to endothelial cells, and transendothelial migration (14-16). These functional attributes are acquired concomitant with the critical process of monocyte-to-macrophage differentiation, where short-lived monocytes (non-permissive for viral replication) differentiate into long-lived macrophages (permissive for viral replication) (15). Although we have shown increased expression of multiple macrophage markers, including human leukocyte-associated antigen-DR (HLA-DR) and CD68 (15), heterogeneity and plasticity are hallmarks of cells belonging to the mononuclear phagocyte system and overall little is known about the phenotypic characteristics of the HCMV activated monocyte/macrophage.
Monocyte and macrophage plasticity is apparent by the distinct morphological and functional responses to particular tissues and to the immunological microenvironment (18-21). Macrophages can be functionally polarized into classically activated M1 macrophages by treatment with IFN-γ alone or in concert with LPS (22) or alternatively activated M2 macrophages by treatment with IL-4 or IL-10 (18, 23). Classically activated macrophages are characterized by an IL-12high, IL-23high, IL-10low phenotype (24) and the production of toxic intermediates (reactive oxygen and nitrogen intermediates) (22) and pro-inflammatory cytokines (IL-1β, and TNF-α) (25). M1 macrophages are potent effector cells efficient at eliminating pathogens and tumour cells. In contrast, alternatively activated macrophages exhibit an IL-12low, IL-23low phenotype (24), produce anti-inflammatory molecules [IL-10 and IL-1 decoy receptor (IL-1Ra)] (21, 26) and express high levels of scavenger, mannose and galactose-type receptors (21, 23). M2 macrophages sublime the inflammatory response, promote angiogenesis and tissue remodelling (21, 26, 27). M1 and M2 macrophages, however, are likely representatives of two extremes along a continuum of possible macrophage biological phenotypes.
‘Classically’ activated monocytes exhibit enhanced antimicrobial activities in a stimulus-dependent [particularly in response to interferon (IFN) γ (28)], but antigen-non-specific manner, through the increased expression of cell surface adhesion receptors and secretion of cytokines and chemokines (29, 30). Elevated levels of cell adhesion molecules promote adhesion to the endothelium, while the increased release of cytokines such as TNF-α and IL-1 can activate the endothelium to further promote transendothelial migration (31, 32). Similar to other pathogenic agents such as gram negative bacteria (via LPS), we previously showed that HCMV activated monocytes exhibit increased expression of cytokines and chemokines, adhesion to endothelial cells, and transendothelial migration (14, 15, 33), suggesting a polarization towards a M1 macrophage phenotype. However, unlike other pathogens, HCMV gains a selective replication advantage from a strong host inflammatory response (15, 34-37). Moreover, LPS and HCMV activated monocytes are morphologically distinct (15); thus, while it appears that monocytes activated by a different directing stimuli may share some functional traits, HCMV must stimulate a distinct M1/M2 macrophage reprogramming to meet its own specific replicative needs. Indeed, analysis of monocyte/macrophage activation by other stimuli such as CCL5 (38), LPS (38), E. coli (39) and Streptococcus pyogenes (40) revealed individually unique expression profiles of macrophage markers characteristic of both the M1 and M2 phenotypes.
To obtain an understanding of the unique changes in monocytes following HCMV infection, particularly in the M1/M2 macrophage reprogramming, we examined the global dysregulation of the infected monocyte transcriptome. Although other studies have examined M1/M2 monocyte/macrophage differentiation induced by various cytokines and bacteria, we for the first time utilized a microarray gene profiling approach to examine the M1/M2 differentiation reprogramming in virally infected monocytes. A cDNA microarray containing 12,626 unique probe sets was used to assess HCMV modulation of genes in peripheral blood monocytes at 4 hours post infection (hpi), which our data has identified as a key temporal point where viral immediate-early proteins are not expressed (15), but a number of critical cellular transcripts associated with early cellular responses are significantly induced (33). HCMV significantly altered the levels of 10.7% (1204 genes) cellular mRNAs in which 5.2% (583 genes) mRNAs were upregulated and 5.5% (621 genes) mRNAs were downregulated. Transcriptional profile comparison analysis revealed a majority of genes strictly associated with the M1 phenotype were induced by HCMV, while a majority of genes associated with the M2 phenotype exhibited no change or a downregulation. The upregulation of monocytic genes implicated in M1 macrophage polarization suggest that HCMV modulates a rapid transition towards a M1 differentiation lineage, supporting our model for haematogenous dissemination of HCMV: HCMV-infection of peripheral blood monocytes forces cells to acquire a M1 pro-inflammatory phenotype to promote infected monocyte infiltration into peripheral tissue, differentiation into long-lived macrophages, and the subsequent establishment of a life-long viral persistence.
Materials and Methods
Virus preparation
HCMV (Towne/E strain; passages 35-45) was cultured as previously described in human embryonic lung (HEL) fibroblasts (33). Virus was purified on a 0.5 M sucrose cushion, resuspended in RPMI 1640 media (Cellgro, Mediatech, Herndon, VA), and used to infect monocytes at a multiplicity of infection (MOI) of 15 for each experiment (14, 15, 33). Monocytes were diluted in RPMI to prevent homotypic aggregation of monocytes and rocked for 4 hrs at 37°C during infection; thus, a high MOI was utilized to ensure that all monocytes would be infected during this short incubation time. We have shown similar HCMV-induced phenotypic changes in monocytes using MOIs of 0.1 to 20 in previous publications (15).
Human peripheral blood monocyte isolation
Blood was drawn by venipuncture and centrifuged through a Ficoll Histopaque 1077 gradient (Sigma, St. Louis, MO). Mononuclear cells were collected and washed with saline (14, 15, 33). Monocytes were then isolated by centrifugation through a Percoll (Pharmacia, Piscataway, NJ) gradient (41). Greater than 95% of isolated peripheral blood mononuclear cells were monocytes as determined by CD14 positive staining (data not shown). Cells were washed and suspended in RPMI 1640 (Cellgro, Mediatech, Herndon, VA) supplemented with 10% human serum (Sigma). University Institutional Review Board and Health Insurance Portability and Accountability Act guidelines were followed for all experimental protocols.
Affymetrix gene array and analysis
Isolated monocytes were HCMV infected or mock infected and incubated nonadherently at 37°C and total RNA harvested 4 hpi with the RNA STAT-60 isolation kit (Tel-Test, Friendswood, TX) according to the manufacturer’s protocol. Affymetrix Human Genome U95Av2 arrays (Affymetrix, Santa Clara, CA), which contain 12,626 characterized sequences, were used to examine the cellular changes in primary human monocytes from 6 different human donors. Total RNA from mock-infected and HCMV-infected monocytes was harvested as described above. A minimum of 5 μg of RNA was used for each array. RNA integrity was assessed by electrophoresis on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA was reverse-transcribed into cDNA and then transcribed in vitro to biotin-labelled cRNA. cRNA from each sample was fragmented and hybridized to GeneChip expression arrays following Affymetrix-recommended protocols. After washing and staining, the microarrays were scanned, and the data quantified. Affymetrix Microarray Suite version 5.0 was used to determine changes in gene expression. Data Mining Tool version 3.0 was used to compile data from each of the replicates and one-way analysis of variance (ANOVA) tests performed to calculate p-values. Spotfire DecisionSite 8.1.1 (Somerville, MA) was used to group genes by ontology, generate scatter plots, and calculate correlation coefficients.
RT-PCR analysis
Isolated monocytes were HCMV infected or mock infected and incubated nonadherently at 37°C on an orbital shaker. At 4 hpi, cells were pelleted, and total RNA isolated using the RNA STAT-60 isolation kit (Tel-Test). 0.5 μg of RNA from each sample was reverse-transcribed by mixing RNA with random hexamers (0.1 μg/μl; Invitrogen, Carlsbad, CA), 1 mM deoxynucleotide triphosphates (Amersham Biosciences, Piscataway, NJ), and ddH20 and incubating at 65°C for 15 minutes. The RNA solution was then chilled for one minute before 1X RT-buffer (Invitrogen), dithiothreitol (DTT; Invitrogen), 80 U of RNasin (Promega, Madison, WI), 400 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen), and ddH20 were added. After one hour of incubation at 37°C, 2 U of RNaseH (Stratagene, La Jolla, CA) was added, and the samples incubated for an additional 30 minutes. PCR was performed using an iCycler (Bio-Rad Model 1708720, Hercules, CA). 2 μg of template DNA was mixed with 1X iTAQ buffer (Bio-Rad), 15 mM MgCl2 (Invitrogen), 50 μM deoxynucleoside triphosphates (Amersham Biosciences), 1.25 U of TAQ polymerase (Bio-Rad), ddH20, and 20 μM forward and reverse primers (Integrated DNA Technologies, Coralville, IA). ICAM-1, integrin β8, integrin α1, IL-1β, TNF-α, CCL2 and glyceraldehydephosphate dehydrogenase (GAPDH) samples were incubated for 5 minutes at 95°C and cycled 35 times for one minute at 95°C, 58°C, and 72°C. Samples were then incubated for 7 minutes at 72°C before storage at 4°C. Controls including samples lacking RT in the reverse transcription reactions and template DNA in the PCR reactions were also performed (data not shown). The following forward and reverse primers were designed with the support of Integrated DNA Technologies: ICAM-1, 5′-AAGCCAAGAGGAAGG AGCAAGACT-3′ (forward) and 5′-TGAACCATGATTGCACCACTGCAC-3′ (reverse); integrin β8, 5′-GCTGATTGATGCGCCACAGACTTT-3′ (forward) and 5′-CAGGCAGACAAATGCAGCGGTAAA-3′ (reverse); integrin α1, 5′-ACGCTCAGTGGAGAACAGATTGGT-3′(forward) and 5′-AATTGTGCTGCCGAGATGACCAGC-3′ (reverse); IL-1β, 5′-AACAGGCTGCTCTGGGATTCTCTT-3′ (forward) and 5′-TGAAGGGAAAGAAGGTGCTCAGGT-3′ (reverse); TNF-α, 5′-ACCCTCAACCTCTTCTGGCTCAAA-3′ (forward) and 5′-AGGCCTAAGGTCCACTTGTGTCAA-3′ (reverse); CCL2, 5′-TCGCTCAGCCAGATGCAATCAATG-3′ (forward) and 5′-AGTTTGGGTTTGCTTGTCCAGGTG-3′ (reverse). To confirm equal cDNA loading, GAPDH RNA was amplified with 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGG GATTTC-3′ (reversed) primers. DNA bands were resolved on an agarose gel and images captured on a GelDoc System using Quantity One software (Bio-Rad).
RNase protection assays
Monocytes were isolated as described above and HCMV infected or mock infected nonadherently at 37°C. At 4 hpi, cells were centrifuged, and total RNA was isolated with the RNA STAT-60 isolation kit (Tel-Test). 2 μg of RNA from each sample was hybridized with an [α32P]uridine 5′-triphosphate (UTP)-labelled human cytokine multiprobe (hCK-2b) template set (BD Biosciences, San Diego, CA) for 12 hours. RNase protection assays were performed with the rabiolabelled RNA using the Multi-Probe RNase Protection Assay System (BD Biosciences, 7th edition) according to the manufacturer’s protocol. Following hybridization, the samples were digested with RNaseA (Promega) and resolved on a denaturing polyacrylamide gel. The gel was then dried, and the images were captured with a phosphorimager (Bio-Rad).
Human chemokine antibody array analysis
Monocytes were isolated as described above and HCMV infected or mock infected nonadherently for 6 hrs at 37°C on an orbital shaker. Following incubation, cells were removed by centrifugation and the supernatant collected and stored at -80°C until their use in the protein microarray assay. Cell-free culture supernatants were assayed for 37 different chemokines by using a RayBio Human Chemokine Antibody Array (RayBiotech Inc., Norcross, GA) according to the manufacturer’s protocol. Data was analyzed by densitometry using Quantity One image analysis software (Bio-Rad).
Phagokinetic track motility assay
Colloidal gold-coated coverslips were prepared as previously described (15). Briefly, glass coverslips were immersed in a 300 Bloom gelatin solution (0.5 g in 300 ml; Sigma), heated at 90°C for 10 min, and dried at 70°C for 45 min. A colloidal gold suspension was prepared by adding 11 ml of tissue culture water (Sigma) and 6 ml Na2CO3 (36.5 mM) to 1.8 ml AuHCl4 (14.5 mM; Fisher Scientific), bringing the solution to a boil, and rapidly adding 1.8 ml of 0.1% formaldehyde (Fisher Scientific). While hot, 2 ml of the colloidal gold suspension was added to each coverslip and incubated at 37°C for 1 h. The coverslips were washed and transferred to 24-well plates.
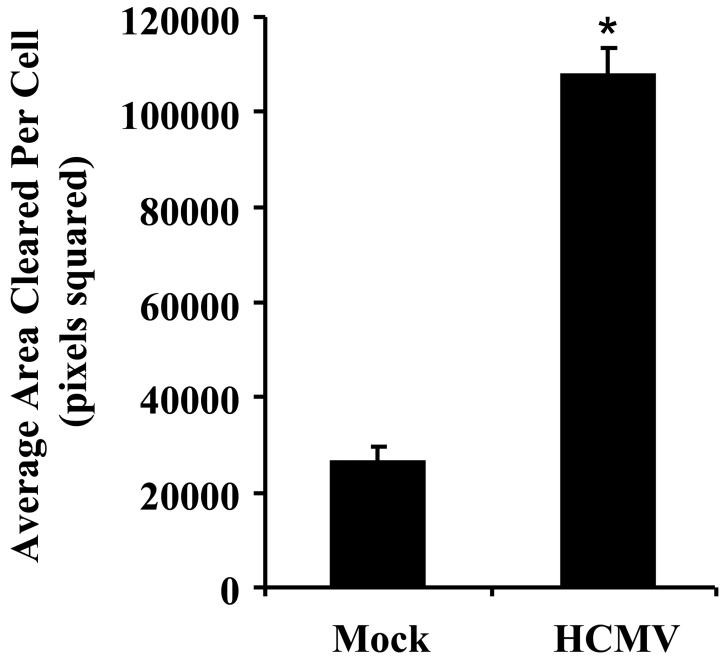
Monocytes were mock infected or HCMV infected and incubated for 45 minutes at 37°C. Cells were then washed extensively with PBS to remove unbound virus and further incubated nonadherently for 6 hours. Supernatants were collected following centrifugation to remove HCMV-infected monocytes. 1 ml of supernatants from mock-infected and HCMV-infected monocytes was added to colloidal gold-covered coverslips in 24 well plates. Next, 500 naïve monocytes from the same donor were added to each well and incubated at 37°C. After 6 hours of incubation, the cells were fixed in 1.5% paraformaldehyde for 15 minutes and mounted onto glass slides with glycerol. Track images of cells were captured using an inverted microscope at 200× original magnification. Average area cleared per cell out of 20 cells per sample was determined by Scion imaging (Scion Corp., Fredrick, MD), and random motility was plotted as mean in arbitrary units (pixels cleared) ± standard error of mean (SEM). The results are representative of 3 independent experiments from different human donors.
Results
Global transcriptome analysis
For compilation of the data from all experiments, the following criteria were used. Genes that had an absent call in more than 2 of the 6 HCMV-infected samples were removed from the pool of genes. The detection algorithm uses probe pair intensities to generate a detection p-value and a predefined Affymetrix threshold was used to assign a present, marginal or absent call. Next, ANOVA tests were performed on the HCMV-infected versus mock-infected samples and p-values were calculated for genes upregulated or downregulated 1.5-fold. A p-value of ≤ 0.05 was used to generate a pool of genes that were statistically significant. Moreover, a fold-change of 1.5 up or down in at least four of six HCMV-infected versus mock-infected samples were considered to be regulated by infection; thus, genes that may otherwise have been eliminated due to anomalous expression by a single donor were accepted. Analysis of 6 independent donors used minimized the number of false positives, thus allowing for a lower fold-change cutoff threshold than other microarray studies (42, 43).
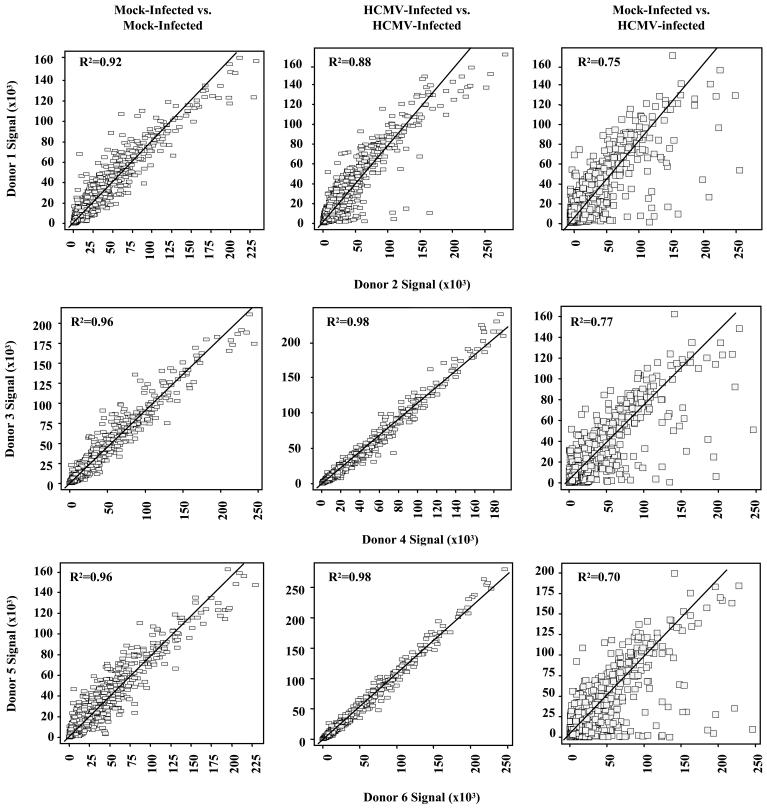
We then tested the validity and reproducibility of hybridization signals from independent preparations of isolated monocyte RNAs from 6 different donors. Scatter plot analysis on the basis of signal was performed on genes from 6 mock-infected and 6 HCMV-infected samples that had a present call in at least 4 of the 6 HCMV-infected samples (Fig. 1). Because of the use of primary cells from different donors, these selection criteria were utilized to account for potential differences in gene expression between donors. Genes from 3 representative pairs of mock-infected vs. mock-infected, HCMV-infected vs. HCMV-infected and mock-infected vs. HCMV-infected sampleswere plotted on the basis of signal and the coefficients of determination ascertained [all possible pairwise analysis were done with similar results obtained (data not shown)]. We found coefficients of determination to be 0.88 or greater between multiple sample comparisons. Conversely, pairwise comparisons of mock-infected and HCMV-infected samples resulted in lower coefficient of determination (∼0.74), verifying that the observed changes in mRNA levels by the HCMV-infected monocyte transcriptome were highly reproducible between donors.
FIGURE 1.
Linear regression analyses of mock-infected and HCMV-infected samples. Monocytes were either mock infected or HCMV infected and incubated nonadherently at 37°C for 4 hours. RNA was harvested and analyzed by Affymetrix gene array. Genes were compiled from six experimental samples and ANOVA tests were performed. P-values were calculated and a p-value cut-off of ≤0.05 was made. To be considered statistically significant, genes had to have a present call in at least 4 of 6 HCMV infected samples and a 1.5-fold or greater change in at least 4 of 6 HCMV samples (see supplemental data S1 for complete list of genes). The significant genes from mock-infected vs. mock-infected, HCMV-infected vs. HCMV-infected and mock-infected vs. HCMV-infected samples were then plotted on the basis of signal. Squares represent individual genes and the “line of best fit” on each graph was used to calculate the coefficients of determination (R2). These scatter plots were generated with Spotfire DecisionSite software. Three representative pairwise comparisons are illustrated although all pairwise comparisons were performed (data not shown).
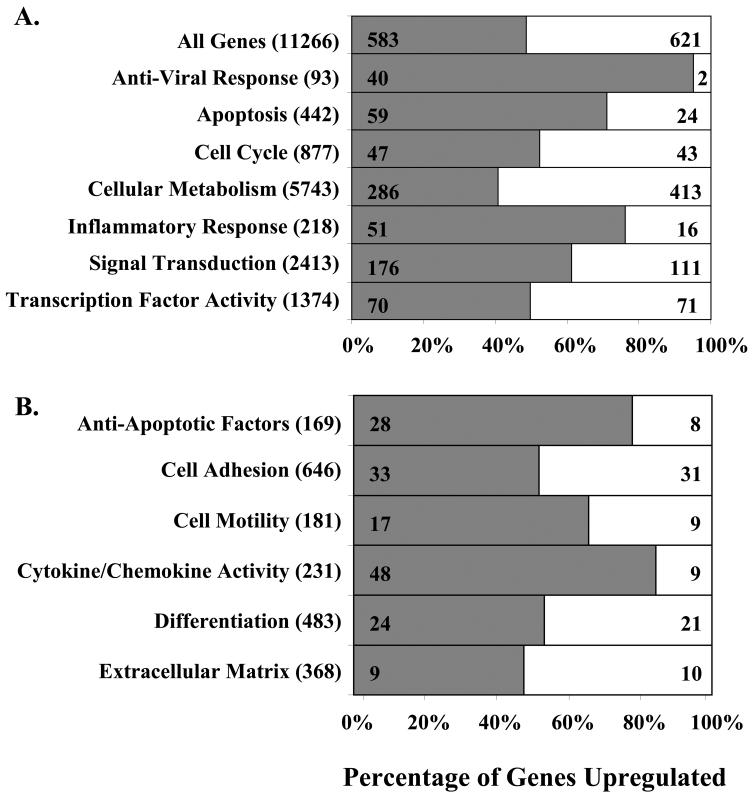
At 4 hpi, HCMV significantly altered the levels of 1204 (10.7% of the total genes examined) cellular mRNAs; where 583 (5.2%) genes were upregulated and 621 (5.5%) genes were downregulated ≥1.5-fold in at least 4 of the 6 replicates (see supplemental data S1 for the complete list of altered transcripts). RT-PCR confirmed that HCMV stimulated ICAM-1, integrin β8, integrin α1, IL-1β, TNF-α, and CCL2 mRNA expression by 4 hpi (Fig. 2). Of the 1204 genes modulated in the monocyte transcriptome following infection, 48.4% were upregulated and 51.6% were downregulated (Fig. 3A). To identify the trends in total cellular gene expression, those genes that were upregulated and downregulated ≥1.5-fold following HCMV infection were grouped by their behavioural patterns. Analysis of specific ontology groups in HCMV-infected monocytes showed altered levels of transcripts involved in the anti-viral response (45% of the total anti-viral response genes examined), apoptosis (18%), cell cycle (10%), cellular metabolism (12%), inflammation (31%), signal transduction (11%), and transcription factor activity (10%) (Fig. 3A). Relative to the total number of genes regulated in the monocyte transcriptome during HCMV binding and entry (10.7%), the ontology analysis indicated genes involved in the anti-viral (45%) and inflammatory responses (31%) to be more substantially modulated. Moreover, within these two ontology clusters, genes were disproportionately upregulated (95% and 77% of genes involved in the anti-viral and inflammatory response, respectively). Because we have previously shown that HCMV induces a pro-inflammatory monocyte to promote the required functional changes in the infected cells necessary for haematogenous dissemination into tissue (14-16), we examined in more detail the gene ontologies known to be involved in inflammation (Fig. 3B, Table I). The results presented in Table I are discussed in more detail below, where we specifically focus on genes implicated in monocyte extravasation into peripheral tissue.
FIGURE 2.
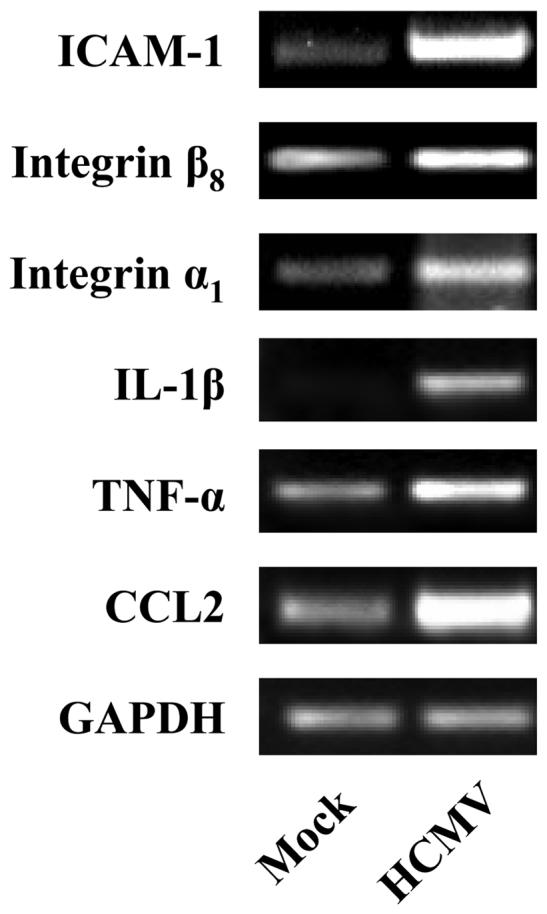
Confirmation of select chemokine and adhesion molecule gene expression by RT-PCR. Monocytes were mock infected or HCMV infected nonadherently for 4 hours at 37°C and RNA was harvested. RT-PCR analysis confirmed that HCMV induced the expression of ICAM-1, integrin β8, integrin α1, IL-1β, TNF-α and CCL2. GAPDH expression is shown as a control,
FIGURE 3.
HCMV alters the monocyte transcriptome at 4 hpi. Percentage of genes upregulated (grey bars) and downregulated (white bars) 1.5-fold or more were grouped into (A) functional categories/ontologies or (B) ontologies involved in inflammation using Spotfire DecisionSite software based on the Gene Ontology Consortium data base. Total genes upregulated and downregulated in each grouping are represented within the bar. The total number of genes analyzed in each ontology category is shown within parenthesis. The percentages represent the percentage of genes that were upregulated following infection.
Table I.
Select statistically significant monocyte mRNAs that increase or decrease ≥1.5-fold 4 hpi with HCMV
| Category and Full Gene Name | Gene Title | Probe Set | Fold Change |
|---|---|---|---|
| Anti-Apoptotic Factors | |||
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 1852_at | 14.6 |
| Secreted phosphoprotein 1 | SPP1 | 2092_s_at | 6.3 |
| Suppressor of cytokine signaling 3 | SOCS3 | 40968_at | 5.7 |
| BCL2-related protein A1 | BCL2A1 | 2002_s_at | 4.9 |
| Interleukin 10 | IL10 | 1548_s_at | 4.6 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 34375_at | 4.1 |
| Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 2 | SERPINB2 | 37185_at | 4.0 |
| CASP8 and FADD-like apoptosis regulator | CFLAR | 1868_g_at | 4.0 |
| Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 9 | SERPINB9 | 34438_at | 3.9 |
| Tumor necrosis factor, alpha-induced protein 8 | TNFAIP8 | 33243_at | 3.6 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | NFKB1 | 1378_g_at | 3.3 |
| Tumor necrosis factor receptor superfamily, member 6 | TNFRSF6 | 1440_s_at | 3.1 |
| Tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 595_at | 2.9 |
| Heat shock 70kDa protein 1A | HSPA1A | 31692_at | 2.5 |
| Baculoviral IAP repeat-containing 3 | BIRC3 | 1717_s_at | 2.5 |
| Immediate early response 3 | IER3 | 1237_at | 2.4 |
| CASP8 and FADD-like apoptosis regulator | CFLAR | 32746_at | 2.3 |
| Heat shock 70kDa protein 9B (mortalin-2) | HSPA9B | 41510_s_at | 1.8 |
| Notch homolog 2 (Drosophila) | NOTCH2 | 38083_at | 1.5 |
| V-rel reticuloendotheliosis viral oncogene homolog A | RELA | 1271_g_at | 1.5 |
| Mucosa associated lymphoid tissue lymphoma translocation gene 1 | MALT1 | 32350_at | -1.6 |
| Transforming growth factor, beta 1 (Camurati-Engelmann disease) | TGFB1 | 1830_s_at | -1.6 |
| Histone deacetylase 1 | HDAC1 | 38771_at | -1.7 |
| RAS p21 protein activator (GTPase activating protein) 1 | RASA1 | 1675_at | -2.0 |
| Annexin A4 | ANXA4 | 37374_at | -2.3 |
| Dimethylarginine dimethylaminohydrolase 2 | DDAH2 | 38621_at | -3.3 |
| Anti-Viral Response | |||
| Chemokine (C-C motif) ligand 4 | CCL4 | 36674_at | 16.2 |
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 1852_at | 14.6 |
| Interferon, alpha 10 | IFNA10 | 1075_f_at | 10.2 |
| Interferon, alpha 2 | IFNA2 | 1791_s_at | 8.0 |
| Chemokine (C-C motif) ligand 8 | CCL8 | 37823_at | 7.4 |
| Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | MX1 | 37014_at | 5.6 |
| Signal transducer and activator of transcription 1, 91kDa | STAT1 | 33338_at | 5.2 |
| Interferon, alpha 1 | IFNA1 | 1666_at | 5.0 |
| Interferon, alpha-inducible protein (clone IFI-15K) | G1P2 | 1107_s_at | 3.9 |
| Interferon-induced protein 35 | IFI35 | 464_s_at | 3.0 |
| 2′,5′-oligoadenylate synthetase 1, 40/46kDa | OAS1 | 38388_at | 3.0 |
| DnaJ (Hsp40) homolog, subfamily C, member 3 | DNAJC3 | 33208_at | 2.8 |
| Tripartite motif-containing 22 | TRIM22 | 36825_at | 2.5 |
| Interferon stimulated gene 20kDa | ISG20 | 33304_at | 2.4 |
| Myxovirus (influenza virus) resistance 2 (mouse) | MX2 | 879_at | 2.3 |
| Interferon, gamma-inducible protein 16 | IFI16 | 1456_s_at | 2.2 |
| Interferon regulatory factor 7 | IRF7 | 36412_s_at | 2.1 |
| Signal transducer and activator of transcription 2, 113kDa | STAT2 | 36770_at | 2.0 |
| Chemokine (C-C motif) ligand 5 | CCL5 | 1404_r_at | 1.8 |
| Interferon (alpha, beta and omega) receptor 2 | IFNAR2 | 1589_s_at | 1.8 |
| Interferon-stimulated transcription factor 3, gamma 48kDa | ISGF3G | 38517_at | 1.6 |
| Protein kinase, interferon-inducible double stranded RNA dependent | PRKR | 1008_f_at | 1.7 |
| Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G | APOBEC3G | 34947_at | 1.7 |
| V-rel reticuloendotheliosis viral oncogene homolog A | RELA | 1271_g_at | 1.5 |
| Protein tyrosine phosphatase, receptor type, C | PTPRC | 40520_g_at | -1.6 |
| Interferon gamma receptor 1 | IFNGR1 | 1038_s_at | -1.6 |
| Cell Adhesion | |||
| Chemokine (C-C motif) ligand 4 | CCL4 | 36674_at | 16.2 |
| Tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 1372_at | 14.7 |
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 1852_at | 14.6 |
| Integrin, beta 8 | ITGB8 | 889_at | 9.3 |
| Fasciculation and elongation protein zeta 1 (zygin I) | FEZ1 | 37743_at | 9.1 |
| Secreted phosphoprotein 1 | SPP1 | 2092_s_at | 6.3 |
| ADAM-like, decysin 1 | ADAMD1 | 34974_at | 6.2 |
| Pleckstrin homology domain containing, family C (with FERM domain) member 1 | PLEKHC1 | 36577_at | 5.2 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 34375_at | 4.1 |
| Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 32640_at | 4.0 |
| Ninjurin 1 | NINJ1 | 41475_at | 3.7 |
| CD44 antigen (homing function and indian blood group system) | CD44 | 1125_s_at | 3.0 |
| Laminin, beta 3 | LAMB3 | 36929_at | 2.9 |
| Scavenger receptor class F, member 1 | SCARF1 | 40034_r_at | 2.7 |
| Integrin, alpha 1 | ITGA1 | 120_at | 2.5 |
| Integrin, alpha 6 | ITGA6 | 41266_at | 2.4 |
| Interleukin 8 | IL8 | 35372_r_at | 2.4 |
| Interleukin 18 (interferon-gamma-inducing factor) | IL18 | 1165_at | 2.0 |
| CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | CD47 | 37890_at | 1.9 |
| Thrombospondin 1 | THBS1 | 867_s_at | 1.9 |
| Heat shock 60kDa protein 1 (chaperonin) | HSPD1 | 37720_at | 1.7 |
| Chemokine (C-C motif) ligand 5 | CCL5 | 1405_i_at | 1.7 |
| Lectin, galactoside-binding, soluble, 1 (galectin 1) | LGALS1 | 33412_at | -1.5 |
| Colony stimulating factor 3 receptor (granulocyte) | CSF3R | 596_s_at | -1.6 |
| Integrin-linked kinase | ILK | 35365_at | -1.7 |
| Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | ITGA5 | 39753_at | -1.7 |
| Rho GDP dissociation inhibitor (GDI) beta | ARHGDIB | 1984_s_at | -1.7 |
| Chemokine (C-C motif) receptor 1 | CCR1 | 1128_s_at | -1.7 |
| Angiogenic factor VG5Q | VG5Q | 35067_at | -1.7 |
| PTK2B protein tyrosine kinase 2 beta | PTK2B | 2009_at | -1.8 |
| Myosin X | MYO10 | 35362_at | -1.8 |
| Integrin, beta 2 (antigen CD18 (p95) | ITGB2 | 37918_at | -1.8 |
| Calcium and integrin binding 1 (calmyrin) | CIB1 | 1020_s_at | -1.9 |
| RAS p21 protein activator (GTPase activating protein) 1 | RASA1 | 1675_at | -2.0 |
| Calsyntenin 1 | CLSTN1 | 41498_at | -2.1 |
| HSPCO34 protein | LOC51668 | 36032_at | -2.1 |
| Collagen, type VII, alpha 1 (epidermolysis bullosa, dystrophic, dominant and recessive) | COL7A1 | 32123_at | -2.1 |
| PTK2B protein tyrosine kinase 2 beta | PTK2B | 33804_at | -2.3 |
| Signal-induced proliferation-associated gene 1 | SIPA1 | 36843_at | -2.3 |
| Pleckstrin homology, Sec7 and coiled-coil domains, binding protein | PSCDBP | 39604_at | -2.4 |
| Zyxin | ZYX | 36958_at | -2.6 |
| CD9 antigen (p24) | CD9 | 39389_at | -2.7 |
| Catenin (cadherin-associated protein), delta 1 | CTNND1 | 40444_s_at | -2.9 |
| Stabilin 1 | STAB1 | 38487_at | -2.9 |
| Endoglin (Osler-Rendu-Weber syndrome 1) | ENG | 32562_at | -3.2 |
| Integrin, alpha M | ITGAM | 38533_s_at | -3.4 |
| Transforming growth factor, beta-induced, 68kDa | TGFBI | 1385_at | -10.1 |
| Chondroitin sulfate proteoglycan 2 (versican) | CSPG2 | 31682_s_at | -14.4 |
| Platelet/endothelial cell adhesion molecule (CD31 antigen) | PECAM1 | 37398_at | -14.7 |
| Cell Motility | |||
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | 431_at | 69.3 |
| Chemokine (C-C motif) ligand 4 | CCL4 | 36674_at | 16.2 |
| Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | PTGS2 | 1069_at | 15.5 |
| Chemokine (C-C motif) ligand 3 | CCL3 | 36103_at | 10.4 |
| Thioredoxin | TXN | 36992_at | 3.7 |
| Myristoylated alanine-rich protein kinase C substrate | MARCKS | 32434_at | 2.8 |
| Plasminogen activator, urokinase receptor | PLAUR | 189_s_at | 2.4 |
| Interleukin 8 | IL8 | 35372_r_at | 2.4 |
| Serine (or cysteine) proteinase inhibitor, clade E, member 2 | SERPINE2 | 41246_at | 2.0 |
| Vanin 2 | VNN2 | 34498_at | 2.0 |
| Jagged 1 (Alagille syndrome) | JAG1 | 35414_s_at | 1.9 |
| Thrombospondin 1 | THBS1 | 867_s_at | 1.9 |
| Chemokine (C-C motif) ligand 5 | CCL5 | 1405_i_at | 1.7 |
| Chloride intracellular channel 4 | CLIC4 | 33891_at | 1.5 |
| Actinin, alpha 4 | ACTN4 | 41753_at | -1.6 |
| Actin related protein 2/3 complex, subunit 1B, 41kDa | ARPC1B | 39043_at | -1.6 |
| Rho GDP dissociation inhibitor (GDI) beta | ARHGDIB | 1984_s_at | -1.7 |
| Mitogen-activated protein kinase 14 | MAPK14 | 37733_at | -2.0 |
| Lymphocyte-specific protein 1 | LSP1 | 36493_at | -2.5 |
| CD9 antigen (p24) | CD9 | 39389_at | -2.7 |
| Coronin, actin binding protein, 1A | CORO1A | 38976_at | -3.0 |
| Formyl peptide receptor-like 2 | FPRL2 | 33092_at | -7.8 |
| Platelet/endothelial cell adhesion molecule (CD31 antigen) | PECAM1 | 37398_at | -14.7 |
| Cytokine/Chemokine | |||
| Interleukin 6 (interferon, beta 2) | IL6 | 38299_at | 280.8 |
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | 431_at | 69.3 |
| Chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | CCL18 | 32128_at | 39.4 |
| Interleukin 12B | IL12B | 563_at | 33.7 |
| Colony stimulating factor 3 | CSF3 | 1334_s_at | 27.4 |
| Chemokine (C-C motif) ligand 20 | CCL20 | 40385_at | 16.5 |
| Chemokine (C-C motif) ligand 4 | CCL4 | 36674_at | 16.2 |
| Chemokine (C-X-C motif) ligand 11 | CXCL11 | 35061_at | 15.2 |
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 1852_at | 14.6 |
| Chemokine (C-C motif) ligand 3 | CCL3 | 36103_at | 10.4 |
| Colony stimulating factor 2 (granulocyte-macrophage) | CSF2 | 1400_at | 10.3 |
| Interferon, alpha 10 | IFNA10 | 1075_f_at | 10.2 |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | INHBA | 40357_at | 9.9 |
| Chemokine (C motif) ligand 1 | XCL1 | 31496_g_at | 8.7 |
| Interferon, alpha 2 | IFNA2 | 1791_s_at | 8.0 |
| Chemokine (C-C motif) ligand 8 | CCL8 | 37823_at | 7.4 |
| Colony stimulating factor 2 (granulocyte-macrophage) | CSF2 | 1401_g_at | 6.8 |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | 408_at | 6.3 |
| Secreted phosphoprotein 1 | SPP1 | 2092_s_at | 6.3 |
| Chemokine (C-X-C motif) ligand 3 | CXCL3 | 34022_at | 6.0 |
| Interleukin 1 receptor antagonist | IL1RN | 37603_at | 5.7 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 1715_at | 5.8 |
| Interleukin 15 | IL15 | 38488_s_at | 5.3 |
| Interferon, alpha 1 | IFNA1 | 1666_at | 5.0 |
| Interleukin 10 | IL10 | 1548_s_at | 4.6 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 34375_at | 4.1 |
| Chemokine (C-C motif) ligand 23 | CCL23 | 36444_s_at | 3.5 |
| Chemokine (C-X-C motif) ligand 5 | CXCL5 | 35025_at | 3.2 |
| Interleukin 1, beta | IL1B | 39402_at | 3.2 |
| Tumor necrosis factor (ligand) superfamily, member 14 | TNFSF14 | 31742_at | 3.1 |
| Chemokine (C-C motif) ligand 7 | CCL7 | 39802_at | 2.5 |
| Interleukin 8 | IL8 | 35372_r_at | 2.4 |
| Interleukin 7 | IL7 | 33966_at | 2.0 |
| Interleukin 18 (interferon-gamma-inducing factor) | IL18 | 1165_at | 2.0 |
| Oncostatin M | OSM | 1579_at | 2.0 |
| Pre-B-cell colony enhancing factor 1 | PBEF1 | 33849_at | 1.9 |
| Lymphotoxin beta (TNF superfamily, member 3) | LTB | 40729_s_at | 1.8 |
| Chemokine (C-C motif) ligand 5 | CCL5 | 1405_i_at | 1.7 |
| CD27-binding (Siva) protein | SIVA | 39020_at | -1.5 |
| IK cytokine, down-regulator of HLA II | IK | 218_at | -1.6 |
| Endothelial cell growth factor 1 (platelet-derived) | ECGF1 | 36879_at | -1.8 |
| Granulin | GRN | 41198_at | -1.9 |
| Tumor necrosis factor (ligand) superfamily, member 8 | TNFSF8 | 33012_at | -2.5 |
| Vascular endothelial growth factor | VEGF | 36101_s_at | -2.5 |
| Differentiation | |||
| Interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) | IL12B | 563_at | 33.7 |
| Colony stimulating factor 3 (granulocyte) | CSF3 | 1334_s_at | 27.4 |
| Aryl-hydrocarbon receptor nuclear translocator 2 | ARNT2 | 35352_at | 14.0 |
| Fms-related tyrosine kinase 1 | FLT1 | 1545_g_at | 13.8 |
| Histone deacetylase 9 | HDAC9 | 37483_at | 10.4 |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | INHBA | 40357_at | 9.9 |
| Fasciculation and elongation protein zeta 1 (zygin I) | FEZ1 | 37743_at | 9.1 |
| Secreted phosphoprotein 1 | SPP1 | 2092_s_at | 6.3 |
| Fms-related tyrosine kinase 1 | FLT1 | 1963_at | 5.0 |
| CD80 antigen (CD28 antigen ligand 1, B7-1 antigen) | CD80 | 35015_at | 4.7 |
| Interleukin 10 | IL10 | 1548_s_at | 4.6 |
| Tumor necrosis factor, alpha-induced protein 2 | TNFAIP2 | 38631_at | 3.5 |
| Growth arrest and DNA-damage-inducible, beta | GADD45B | 39822_s_at | 3.3 |
| Epiregulin | EREG | 34476_r_at | 2.7 |
| Interferon, gamma-inducible protein 16 | IFI16 | 1456_s_at | 2.2 |
| Interleukin 7 | IL7 | 33966_at | 2.0 |
| Serine (or cysteine) proteinase inhibitor, clade E, member 2 | SERPINE2 | 41246_at | 2.0 |
| Agrin | AGRN | 33454_at | 1.8 |
| Metallothionein 3 (growth inhibitory factor (neurotrophic)) | MT3 | 870_f_at | 1.6 |
| Notch homolog 2 (Drosophila) | NOTCH2 | 38083_at | 1.6 |
| Chloride intracellular channel 4 | CLIC4 | 33891_at | 1.5 |
| Purine-rich element binding protein A | PURA | 35221_at | -1.5 |
| Protein tyrosine phosphatase, receptor type, C | PTPRC | 40520_g_at | -1.6 |
| Leukocyte specific transcript 1 | LST1 | 37967_at | -1.6 |
| Endothelial cell growth factor 1 (platelet-derived) | ECGF1 | 36879_at | -1.8 |
| Angiogenic factor VG5Q | VG5Q | 35067_at | -1.8 |
| CCAAT/enhancer binding protein (C/EBP), gamma | CEBPG | 39219_at | -1.8 |
| RAS p21 protein activator (GTPase activating protein) 1 | RASA1 | 1675_at | -2.0 |
| PDZ and LIM domain 7 (enigma) | PDLIM7 | 39530_at | -2.1 |
| V-ski sarcoma viral oncogene homolog (avian) | SKI | 41499_at | -2.2 |
| Alanyl (membrane) aminopeptidase | ANPEP | 39385_at | -2.2 |
| Filamin B, beta (actin binding protein 278) | FLNB | 38078_at | -2.3 |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | YWHAH | 1424_s_at | -2.5 |
| Vascular endothelial growth factor | VEGF | 36101_s_at | -2.5 |
| Bridging integrator 1 | BIN1 | 459_s_at | -2.6 |
| Ubiquitin-conjugating enzyme E2 variant 1 | UBE2V1 | 36959_at | -2.6 |
| CD9 antigen (p24) | CD9 | 39389_at | -2.7 |
| CD86 antigen (CD28 antigen ligand 2, B7-2 antigen) | CD86 | 36270_at | -3.4 |
| Extracellular Matrix | |||
| Matrix metalloproteinase 10 (stromelysin 2) | MMP10 | 1006_at | 12.1 |
| Matrix metalloproteinase 1 (interstitial collagenase) | MMP1 | 38428_at | 9.2 |
| Secreted phosphoprotein 1 | SPP1 | 2092_s_at | 6.3 |
| Matrix metalloproteinase 14 (membrane-inserted) | MMP14 | 34747_at | 5.9 |
| Laminin, beta 3 | LAMB3 | 36929_at | 2.9 |
| Protease inhibitor 3, skin-derived (SKALP) | PI3 | 41469_at | 2.7 |
| Agrin | AGRN | 33454_at | 1.8 |
| Collagen, type VII, alpha 1 (epidermolysis bullosa, dystrophic, dominant and recessive) | COL7A1 | 32123_at | -2.1 |
| Tissue inhibitor of metalloproteinase 2 | TIMP2 | 1375_s_at | -2.1 |
| Vascular endothelial growth factor | VEGF | 36101_s_at | -2.5 |
| Transforming growth factor, beta-induced, 68kDa | TGFBI | 1385_at | -10.0 |
| Chondroitin sulfate proteoglycan 2 (versican) | CSPG2 | 31682_s_at | -14.4 |
HCMV modulates the expression of monocyte transcripts involved in inflammation at 4 hpi
Apoptosis
HCMV encodes a number of immediate-early gene products with anti-apoptotic function (44-46); however, a 3-4 week delay in viral gene expression in HCMV-infected monocytes (15) suggest that HCMV regulates monocyte apoptosis via cellular anti-apoptotic factors. Indeed, detailed examination of transcripts encoding for anti-apoptotic factors revealed 16% were upregulated and only 4% were downregulated at 4 hpi. This data is consistent with previous findings showing that a number of anti-apoptotic related genes such as BCL2A1 (4.9-fold increase) and TNFRSF6 (3.1-fold increase) are specifically upregulated in M1 polarized macrophages (47). Moreover, NF-κB, a transcription factor responsible for the expression of a number of anti-apoptotic and inflammatory genes (48, 49), is upregulated 3.3-fold. These finding suggests that, even in the absence of de novo viral gene expression, HCMV can create a cellular environment skewed towards the inhibition of apoptosis that likely favours monocyte-to-macrophage differentiation and the long term survival of the infected cells.
Cell Adhesion
Adhesion molecules are required for monocyte transendothelial migration during normal and inflammatory processes (50). The tethering, rolling, and firm adhesion of monocytes to the apical surface of vascular endothelial cells is dependent on adhesion molecule expression. Monocytes must first adhere to endothelial cells and then move along the surface of the endothelial cell in search of the endothelial cell junctions (51). Selectins, intercellular adhesion molecules (ICAMs), and integrins are critical mediators of monocyte adhesion to endothelial cells prior to extravasation (52). M1-activated monocytes express significantly higher levels of these receptors; therefore, monocytes with a pro-inflammatory phenotype have a higher propensity to adhere and transmigrate into peripheral tissue (25, 29, 53). We found ICAM-1, which is necessary for the firm adhesion of monocytes to endothelial cells (54), to be upregulated 4-fold at the transcriptional level following HCMV infection (Table I and supplemental data S1). This observation is in agreement with our previous data showing increased ICAM-1 expression on the surface of monocytes following infection and a number of other studies that have demonstrated strong upregulation of surface ICAM-1 expression on endothelial and epithelial cells following infection (55, 56). Additionally, the microarray data indicates that the mRNA of a number of additional cell adhesion molecules such as ninjurin 1, laminin β3, and integrins α1, α6 and β8 were elevated. Conversely, α5, αM, and β2 integrins were downregulated following infection indicating that HCMV can selectively alter integrin message expression. The specific upregulation and downregulation of integrins by HCMV has been previously observed in HUVECs and fibroblasts; however, it remains unclear of the consequences or reasons for such regulation (55, 57, 58). Nonetheless, our previous functional studies showing HCMV-infected monocytes exhibited increased firm adhesion to endothelial cells 6 hpi and increased transendothelial migration 24 hpi demonstrate that the net result of the HCMV mediated modulation of cell adhesion molecules is to increase monocyte adhesion to endothelial cells (14, 15).
Extracellular Matrix Proteins
An important step in monocyte migration to the tissue, following extravasation, is the degradation of the basal lamina. Matrix metalloproteinases (MMPs) are involved in the breakdown of extracellular matrix components. HCMV upregulated the expression of both MMP1 and MMP10 approximately 12-fold. MMP1 dissolves collagen types I, II, III, (59), and MMP10 breaks down proteoglycans and fibronectin (60). HCMV may upregulate the expression of these proteinases in monocytes to promote migration through the extracellular matrix in route to the tissue.
Cytokine Activity
A classical feature of a M1 pro-inflammatory phenotype is the secretion of a milieu of cytokines and chemokines which promote monocyte activation along with cellular recruitment to sites of inflammation (53). Indeed, we find that HCMV infection significantly alters the levels of 25% of cytokine and chemokine mRNAs in monocytes. Consistent with our previous studies, as well as others, pro-inflammatory cytokines IL-1β, IL-6, IL-12 p40, IL-15, inhibin βA and TNF-α, which are associated with the classical activation M1 phenotype, were upregulated following infection (14, 21, 25, 33, 61). HCMV potently induced IL-6, TNF-α and inhibin βA mRNA, reaching a 280-, 14.6- and 9.9-fold increase. HCMV infection also led to a downregulation of certain cytokine genes such as vascular endothelial growth factor (VEGF), which can act as a chemoattractant for T-cells, and tumour necrosis factor ligand superfamily member 8 (CD30), which can stimulate T-cell proliferation. Interestingly, M2-polarization associated anti-inflammatory cytokines, IL-1 receptor antagonist (IL-1Ra) and IL-10 were both upregulated following HCMV infection (21, 25).
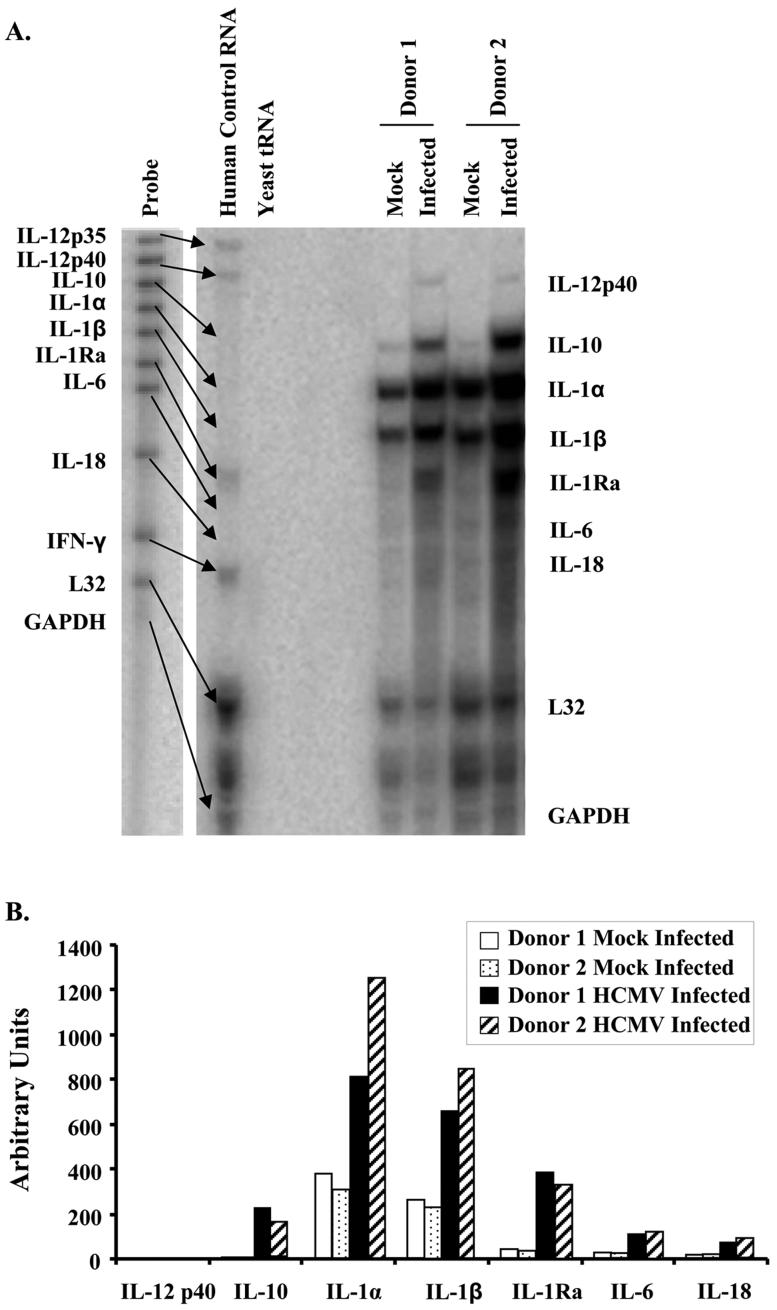
To confirm this unusual M1/M2 cytokine profile, RNase protection assays examining M1 and M2 associated cytokines were performed on RNA harvested from mock-infected and HCMV-infected monocytes 4 hpi isolated from multiple donors. Consistent with our gene profiling analysis, RNase protection assays show the upregulation of cytokine genes implicated in the M1 phenotype (IL-12B, IL-1β, and IL-6) and the M2 phenotype (IL-10, IL-1Ra and IL-18) (Fig. 4A and B). These data highlight a unique reprogramming induced in monocytes following infection with HCMV. We next examine in depth the polarization gene profile of the HCMV-infected monocyte.
FIGURE 4.
HCMV infection induces cytokine expression. Monocytes were mock infected or HCMV infected nonadherently for 4 hours at 37°C and RNA was harvested. (A) RNA from multiple donors was hybidized with a [α32P]UTP-labelled hCM-8 template set and subjected to RNase protection assay using the Multi-Probe RNase protection assay system according to the manufacturer’s protocol. Genes on the left represent the total set of genes probed for with the kit and the genes on the right represent those genes induced following HCMV infection of monocytes. (B) Band intensities from the RNase Protection Assay was determined using by densitometry using Quantity One software (Bio-Rad), normalized to the respective L32 band intensities, and plotted in arbitrary units. Results are representative of 3 independent experiments from 3 donors.
HCMV stimulates monocyte differentiation towards a M1 macrophage phenotype
The rapid production of numerous cytokines is characteristic of a classical M1 monocyte/macrophage activation/differentiation lineage and our transcriptome analysis indicates that HCMV-infected monocytes acquire a pro-inflammatory M1 phenotype. However, the upregulation of transcripts (IL-1Ra and IL-10) implicated in the alternative M2 differentiation lineage suggest a complex and atypical M1/M2 reprogramming of monocytes following infection with HCMV. To further investigate HCMV-induced monocyte-to-macrophage differentiation, we compared the HCMV-infected monocyte transcriptome to the M1/M2 immunophenotypic profile induced by IFN-γ (M1 phenotype) and IL-4 (M2 phenotype) as described by F. Martinez et al. (47). Tables II and III lists genes that were found to be strictly associated with M1 or M2 monocyte polarization, respectively. Transcriptional analysis of monocytes following infection with HCMV revealed that 30 (65%) M1 associated genes were upregulated, 14 (30%) genes showed no change and 2 (5%) genes were downregulated by 4 hpi. Moreover, examination of functional categories indicate that the first M1 associated genes to be modulated during HCMV-induced monocyte polarization are membrane receptors (100%), apoptosis-related genes (100%) and cytokines and chemokines (66%). On the other hand, of the genes associated with the anti-inflammatory M2 phenotype only 2 (4%) were upregulated, 37 (84%) displayed no change and 6 (14%) were downregulated. Taken together these data show a clear transcriptional bias towards a M1 macrophage activation phenotype following infection or monocytes with HCMV, although several transcripts implicated in anti-inflammatory responses typical of alternatively activated macrophages (such as IL-1Ra, IL-10, CCL18 and CCL23) were also upregulated.
Table II.
Comparison of M1 genes expressed in HCMV-infected monocytes/macrophagesa
| Category and Full Gene Name | Gene Title | Probe Set | Fold Change |
|---|---|---|---|
| Membrane Receptors | |||
| Interleukin 15 receptor, alpha | IL15RA | 41677_at | 15.3 |
| Chemokine (C-C motif) receptor 7 | CCR7 | 1097_s_at | 3.0 |
| interleukin 2 receptor, gamma (severe combined immunodeficiency) | IL2RG | 1506_at | 1.9 |
| Interleukin 7 receptor | IL7R | 1370_at | 1.7 |
| Cytokine and Chemokines | |||
| Interleukin 6 (interferon, beta 2) | IL6 | 38299_at | 280.8 |
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | 431_at | 69.3 |
| Interleukin 12B | IL12B | 563_at | 33.7 |
| Chemokine (C-C motif) ligand 20 | CCL20 | 40385_at | 16.5 |
| Chemokine (C-X-C motif) ligand 11 | CXCL11 | 35061_at | 15.2 |
| Tumor necrosis factor (TNF superfamily, member 2) | TNF | 1852_at | 14.6 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 1715_at | 5.8 |
| Interleukin 15 | IL15 | 1036_at | 4.0 |
| Pre-B-cell colony enhancing factor 1 | PBEF1 | 33849_at | 1.9 |
| Chemokine (C-C motif) ligand 5 | CCL5 | 1403_s_at | 1.6 |
| Endothelial cell growth factor 1 (platelet-derived) | ECGF1 | 36879_at | -1.8 |
| Chemokine (C-C motif) ligand 19 | CCL19 | 36067_at | nc |
| Chemokine (C-X-C motif) ligand 9 | CXCL9 | 37219_at | nc |
| Chemokine (C-C motif) ligand 15 | CCL15 | 33789_at | nc |
| Apoptosis-related Genes | |||
| BCL2-related protein A1 | BCL2A1 | 2002_s_at | 4.9 |
| XIAP associated factor-1 | HSXIAPAF1 | 35583_at | 3.4 |
| Tumor necrosis factor receptor superfamily, member 6 | TNFRSF6 | 1440_s_at | 3.1 |
| Baculoviral IAP repeat-containing 3 | BIRC3 | 1717_s_at | 2.5 |
| Growth arrest and DNA-damage-inducible, alpha | GADD45A | 1911_s_at | 2.1 |
| Solute Carriers | |||
| Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | SLC7A5 | 32186_at | 1.6 |
| Solute carrier family 31 (copper transporters), member 2 | SLC31A2 | 34749_at | nc |
| Enzymes | |||
| Adenylate kinase 3 | AK3 | 32331_at | 9.3 |
| Indoleamine-pyrrole 2,3 dioxygenase | INDO | 36804_at | 4.1 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | PFKFB3 | 37111_g_at | 3.6 |
| 2′-5′-oligoadenylate synthetase 2, 69/71kDa | OAS2 | 39263_at | 3.4 |
| Proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | PSME2 | 1184_at | 1.8 |
| Proteasome (prosome, macropain) subunit, beta type, 9 | PSMB9 | 38287_at | 1.7 |
| 2′-5′-oligoadenylate synthetase-like | OASL | 269_at | nc |
| Chitinase 3-like 1 (cartilage glycoprotein-39) | CHI3L1 | 36197_at | nc |
| Hydroxysteroid (11-beta) dehydrogenase 1 | HSD11B1 | 35702_at | nc |
| Phosphofructokinase, platelet | PFKP | 39175_at | nc |
| Proteasome (prosome, macropain) subunit, alpha type, 2 | PSMA2 | 41240_at | nc |
| Extracellular Matrix | |||
| Pentaxin-related gene, rapidly induced by IL-1 beta | PTX3 | 1491_at | 13.5 |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | INHBA | 40357_at | 9.9 |
| Chondroitin sulfate proteoglycan 2 (versican) | CSPG2 | 31682_s_at | -14.4 |
| Insulin-like growth factor binding protein 4 | IGFBP4 | 1737_s_at | nc |
| Apolipoprotein L, 1 | APOL1 | 35099_at | nc |
| Platelet-derived growth factor alpha polypeptide | PDGFA | 1109_s_at | nc |
| DNA-binding Factors | |||
| Interferon regulatory factor 1 | IRF1 | 669_s_at | 3.3 |
| Interferon regulatory factor 7 | IRF7 | 36412_s_at | 2.1 |
| Homeo box (expressed in ES cells) 1 | HESX1 | 35463_at | nc |
| Activating transcription factor 3 | ATF3 | 287_at | nc |
Examined M1 genes are based on the transcriptional profiling work of Martinez et al. (47).
Table III.
Comparison of M2 genes expressed in HCMV-infected monocytes/macrophagesa
| Category and Full Gene Name | Gene Title | Probe Set | Fold Change |
|---|---|---|---|
| Membrane Receptors | |||
| Chemokine (C-X-C motif) receptor 4 | CXCR4 | 649_s_at | -2.5 |
| Transforming growth factor, beta receptor II (70/80kDa) | TGFBR2 | 1814_at | -2.7 |
| Histamine receptor H1 | HRH1 | 35384_at | nc |
| Toll-like receptor 5 | TLR5 | 34473_at | nc |
| Type I transmembrane C-type lectin receptor DCL-1 | DCL-1 | 34760_at | nc |
| Macrophage scavenger receptor 1 | MSR1 | 39981_at | nc |
| G protein-coupled receptor 105 | GPR105 | 33462_at | nc |
| CD209 antigen-like | CD209L | 39270_at | nc |
| CD36 antigen (collagen type I receptor, thrombospondin receptor) | CD36 | 36656_at | nc |
| Mannose receptor, C type 1 | MRC1 | 36908_at | nc |
| Cytokine and Chemokines | |||
| Chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | CCL18 | 32128_at | 39.4 |
| Chemokine (C-C motif) ligand 23 | CCL23 | 36444_s_at | 3.5 |
| Insulin-like growth factor 1 (somatomedin C) | IGF1 | 1501_at | nc |
| Chemokine (C-C motif) ligand 13 | CCL13 | 37454_at | nc |
| Solute Carriers | |||
| Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | SLC4A7 | 34936_at | -1.5 |
| Solute carrier family 38, member 6 | SLC38A6 | 36758_at | nc |
| Enzymes | |||
| Leukotriene A4 hydrolase | LTA4H | 38081_at | -5.5 |
| Cathepsin C | CTSC | 133_at | nc |
| Hexosaminidase B (beta polypeptide) | HEXB | 34888_at | nc |
| Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | LIPA | 38745_at | nc |
| Adenosine kinase | ADK | 168_at | nc |
| Histamine N-methyltransferase | HNMT | 37604_at | nc |
| Tyrosylprotein sulfotransferase 2 | TPST2 | 35172_at | nc |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | HS3ST1 | 41555_at | nc |
| Carbonic anhydrase II | CA2 | 40095_at | nc |
| Arachidonate 15-lipoxygenase | ALOX15 | 34636_at | nc |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | HS3ST1 | 41555_at | nc |
| Extracellular Matrix | |||
| Transforming growth factor, beta 1 (Camurati-Engelmann disease) | TGFB1 | 1830_s_at | -1.6 |
| Fibrinogen-like 2 | FGL2 | 39593_at | -6.7 |
| Selenoprotein P, plasma, 1 | SEPP1 | 34363_at | nc |
| Chimerin (chimaerin) 2 | CHN2 | 33244_at | nc |
| Fibronectin 1 | FN1 | 31719_at | nc |
| DNA-binding Factors | |||
| Growth arrest-specific 7 | GAS7 | 33387_at | nc |
| Early growth response 2 (Krox-20 homolog, Drosophila) | EGR2 | 37863_at | nc |
| V-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | MAF | 41504_s_at | nc |
Examined M2 genes are based on the transcriptional profiling work of Martinez et al. (47).
HCMV induces an atypical M1/M2 macrophage chemokinome
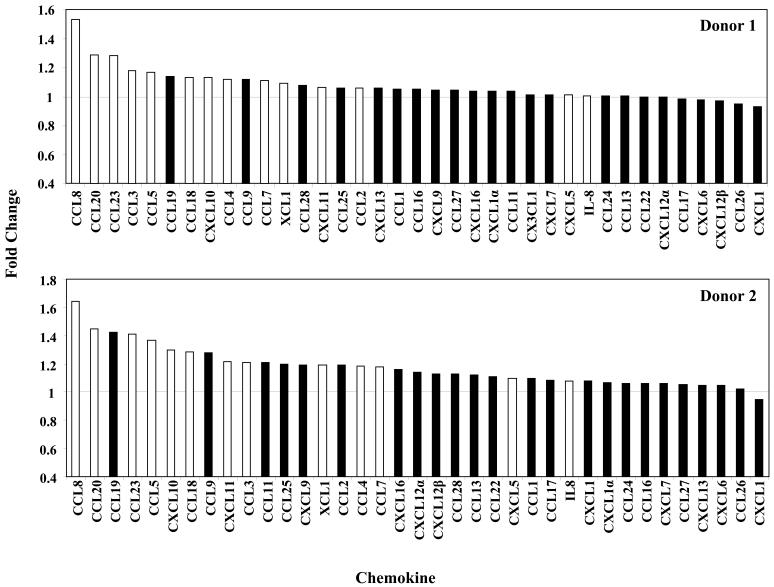
Distinct patterns of chemokines are produced depending on whether a macrophage undergoes a M1 or M2 polarization (47). We found that 16 (44%) chemokine genes were upregulated in monocytes within 4 hpi, while none were downregulated (Table I). Differential analysis showed that 44% and 33% of M1 and M2 associated chemokines to be upregulated, respectively. To confirm that the upregulation of chemokine genes at the level of transcription conferred increased protein production, secretion of chemokines into the supernatant by monocytes 6 hpi was assayed using human chemokine antibody arrays. Examination of 37 chemokines showed that those identified by gene profiling to exhibit elevated gene expression levels also demonstrated the greatest fold increases in protein secretion with the exception of CXCL5 and IL-8, indicating that chemokine protein production strongly correlated with increased gene expression in infected cells (Fig. 5). Contrary, a few chemokines such as CCL19 and CCL9 not identified by the HCMV-infected monocyte transcriptome analysis displayed increased protein secretion, highlighting a complex biological process that likely involves multiple levels of regulation, including transcription, translation and a regulation of the intercellular trafficking. We next examined if the secreted of chemokines from the infected monocyte samples were functional and could induce motility in naïve monocytes. Treatment of uninfected monocytes with supernatant collected from HCMV-infected monocytes significantly stimulated cell motility (Fig. 6). Overall, although HCMV induces an atypical monocyte chemokine signature that does not strictly adhere to characteristics of the M1 or M2 phenotype, we suggest the overwhelming upregulation of chemoattactants promotes viral spread by aiding infected monocyte migration to peripheral tissue and/or naïve monocyte migration to sites of infection to enhance the infected cell pool.
FIGURE 5.
Differential secretion of chemokines by monocytes following infection with HCMV. Monocytes were mock infected or HCMV infected nonadherently for 6 hours at 37°C, and supernatant collected. Cell-free culture supernatants from multiple donors were assayed for 37 different chemokines by using a RayBio Human Chemokine Antibody Array according to the manufacturer’s protocol. Densitometry of the Chemokine Antibody array was performed with Quantity One software (Bio-Rad) and fold change plotted. White bars represent chemokines that were identified by transcriptome profiling analysis to be upregulated following infection with HCMV. Black bars represent chemokines not transcriptionally upregulated at 4 hpi. Results are representative of 4 independent experiments from 3 donors (analysis on one donor was repeated).
FIGURE 6.
Supernatant harvested from HCMV-infected monocytes stimulate naïve monocyte motility. Monocytes were mock infected or HCMV infected and incubated for 45 minutes. Cells were washed extensively to remove unbound virus and incubated nonadherently for 6 hours. Supernatants were harvested from each of the samples following centrifugation and 1 ml added to colloidal gold-covered coverslips. After 500 naïve monocytes were added to each coverslip and at six hours post-addition of naïve monocytes the cells were fixed with 1.5% paraformaldehyde. The average area (arbitrary units2) of colloidal gold cleared by 20 monocytes was determined for each experimental arm from the captured images. Random monocyte motility is plotted as a mean ± S.E.M. of 20 cells per experimental arm. The results represent 3 independent experiments with separate human blood donors. Asterisk denotes a significance of P<0.01.
Discussion
Monocyte extravasation into tissue is a sequential process involving tethering, rolling, adhesion and diapedesis (52). Quiescent monocytes bind to and transmigrate across endothelial cells at very low rates, but the rate of adhesion and transmigration can be significantly increased following classical activation with different pathogenic stimuli including LPS and HCMV (15, 62). However, LPS- and HCMV- activated monocytes are morphologically distinct (15); thus, while it appears that monocytes activated with different stimuli display certain similar phenotypical traits, HCMV must stimulate a distinct M1/M2 monocyte/macrophage polarization, which we advocate serves to promote viral survival and persistence. To examine this possibility, we initiated a study to examine the monocyte transcriptome shortly after infection to obtain a global understanding of the rapid M1/M2 reprogramming of HCMV-infected monocytes.
Global DNA microarray analyses demonstrated that HCMV dramatically altered the transcriptional profile of the monocyte transcriptome following infection. The regulation of gene expression was a highly specific process in which 583 mRNAs were significantly upregulated and 621 mRNAs were significantly downregulated. Ontology analysis revealed a majority of the induced genes are associated with classically activated monocytes/macrophages. Generally, inflammation is utilized by the infected host to help clear infection via the release of antiviral regulatory factors such as IL-1β and TNF-α (63, 64). However, HCMV targets monocytes in vivo (4, 5, 65) and once infected appear to benefit from the induction of a strong host inflammatory response (15, 34-37). Prostaglandin E2, IL-1β, and TNF-α, key mediators in the inflammatory response, promote viral replication via stimulation of the HCMV IE genes (34, 36, 37). Moreover, the induction of M1 associated cytokines and chemokines such as IL-1β, IL-6, and TNF-α can also contribute to the shaping of the classically activated or pro-inflammatory macrophage phenotype, which is important for viral replication (15, 66, 67) and, now as our evidence identifies, for viral dissemination (14, 15).
The absence of viral IE gene expression (15) together with the previous observations that UV-inactivated HCMV and the major viral glycoprotein gB exhibited similar pro-inflammatory modulatory effects on monocyte function (33) suggest the involvement of receptor-ligand interactions. Engagement and activation of cellular surface receptors such as toll-like receptor (TLR) 2, epidermal growth factor (EGFR) and integrins by HCMV (68) followed by the subsequent stimulation of cellular NF-κB and PI(3)K activity (14, 16, 33, 69) is likely the major mechanism for the rapid monocyte gene regulation following infection, although we can not preclude the possible involvement of some tegument proteins which exhibit transactivator activity following viral entry (68). Overall, these studies identify that HCMV binding and/or entry is sufficient to induce a pro-inflammatory monocyte state exhibiting increased cytokine/chemokine secretion, motility, endothelial adhesion, transendothelial migration and monocyte-to-macrophage differentiation.
Although transcriptional profile comparisons of genes differentially expressed in the M1 or M2 phenotype reveal a distinct bias towards the classical activation, a number of genes distinctly associated with alternatively activated monocytes were also upregulated. The anti-inflammatory cytokines, IL-1Ra and IL-10 were both upregulated following HCMV infection. Mice lacking endogenous IL-1Ra were less susceptible to infection by intracellular pathogens supporting the importance of IL-1 in resistance to infection with intracellular organisms (70). HCMV and Epstein Barr virus encode viral homologues to IL-10 (71, 72), which can activate signalling pathways that dampen IFN-γ-triggered microbicidal pathways, inhibit antigen processing and presentation by antigen-presenting cells and inhibit T-cell cytokine production and cytotoxic activity (73). Since the HCMV IL-10 homolog is not synthesized in this early time frame in monocytes, we propose that HCMV utilizes host cytokines instead to inhibit T-cell function. Overall, our data hints that HCMV rapidly and selectively modulates M1 and M2 associated factors resulting in an activated monocyte lying between the ends of the M1/M2 polarization spectrum in order to balance viral spread with immune evasion.
In agreement, examination of the HCMV-infected monocyte chemokine signature showed that the upregulation of chemokines implicated both the classically and alternatively activated phenotype, although again a bias towards the M1 phenotype was observed. Thus, our data indicates that HCMV induces multiple chemoattractants regardless of M1 or M2 association, which we speculate promotes viral spread by driving infected monocyte migration into peripheral tissue and/or the recruitment of naïve monocytes to sites of infection to enhance the infected cell pool. Although beneficial to the virus, chronic secretion of these chemokines could have pathological consequences to the host, such as the development of atherosclerosis where HCMV infection is strongly linked to disease development (10, 11). For example, chemokine (C-C motif) ligand 2 (4.1-fold increase) is believed to be one of the most powerful inducers of monocyte migration into atherosclerotic lesions (74).
Overall, we show that HCMV infection rapidly induces a pro-inflammatory monocyte. Is this activated phenotype a direct consequence of specific M1/M2 reprogramming by the virus or a general response to viruses? HIV studies including microarray analyses indicate pro-inflammatory gene expression resulting in chronically activated HIV-infected monocytes/macrophages occurs in vivo (75-77). Similar to HCMV infected monocytes, release of chemotatic cytokines such as MCP-1 from HIV infected cells enhance dissemination by stimulating recruitment of target CD4 T cells and monocytes/macrophages to sites of infection (78). Furthermore, HIV appears to induce anti-apoptotic and cell cycle associated genes leading to greater cell survival and viral replication, respectively (79-81). Nevertheless, although both HCMV- and HIV-infected monocytes display an activated invasive monocyte phenotype, a comparative analysis of gene expression in monocytes infected with HIV to our results revealed fundamental differences between the two viruses. In HCMV-infected monocytes, expression of inflammatory cytokine genes such as IL-1β, IL-6 and TNF-α were significantly upregulated (presented here) while in HIV-infected monocytes these genes remained unchanged (75). IL-1β and TNF-α have strong anti-viral activity, thus, downregulation by HIV is likely a strategy for immune evasion; however, as mentioned above, HCMV has devised a strategy utilizing the NF-κB activating properties of these cytokines to stimulate its own viral replication (36, 37). The chemokine receptor CCR5 is the major entry coreceptor for HIV and is upregulated in HIV (82), but not in HCMV-infected monocytes. These discussed differences are but a few examples to highlight the transcriptional differences that occur following infection of monocytes with HCMV and HIV. Furthermore, HIV does not appear to force monocyte differentiation into macrophages, as we observed for HCMV (15), but rather as monocytes differentiate they become more susceptible to HIV infection (82). Thus, HCMV- and HIV-infected monocytes are not activated identically in the classical sense and lie at different points along the M1/M2 macrophage polarization continuum. By using microarray technology, we can dissect the unique strategies that these different viruses have evolved to promote dissemination and persistence.
The data presented in this global transcriptional profile study is consistent with our previously proposed model of HCMV dissemination, where HCMV infection classically activates monocytes in order to promote migration into host organ tissue and differentiation into replication permissive macrophages (14-16). Our data also confirms morphological studies suggesting that HCMV infection does not induce all the same characteristics associated with the classically activated M1 phenotype induced by LPS. The HCMV-infected monocyte transcriptome revealed an atypical M1/M2 polarization, with a defined phenotype biased towards the M1 polarization that also incorporated select attributes of the alternatively activated M2 phenotype. Presumably the upregulation of specific genes associated with the anti-inflammatory M2 phenotype, and in particular chemoattractants, would be advantageous for the virus. Overall, our study provides insight into how HCMV reprograms monocytes as a mechanism to promote viral dissemination and demonstrates the complexity of HCMV-induced signaling events, even in the absence of viral gene expression.
Supplementary Material
Acknowledgements
The authors have no financial conflict of interest.
This work was supported by a Malcolm Feist Cardiovascular Research Fellowship and the National Institutes of Health (AI56077 and 1-P20-RR018724).
References
- 1.Masur H, Whitcup SM, Cartwright C, Polis M, Nussenblatt R. Advances in the management of AIDS-related cytomegalovirus retinitis. Ann. Intern. Med. 1996;125:126–136. doi: 10.7326/0003-4819-125-2-199607150-00009. [DOI] [PubMed] [Google Scholar]
- 2.Ho M. Virus infections after transplantation in man. Brief review. Arch. Virol. 1977;55:1–24. doi: 10.1007/BF01314475. [DOI] [PubMed] [Google Scholar]
- 3.Stagno S, Pass RF, Dworsky ME, Henderson RE, Moore EG, Walton PD, Alford CA. Congenital cytomegalovirus infection: The relative importance of primary and recurrent maternal infection. N. Engl. J. Med. 1982;306:945–949. doi: 10.1056/NEJM198204223061601. [DOI] [PubMed] [Google Scholar]
- 4.Sinzger C, Jahn G. Human cytomegalovirus cell tropism and pathogenesis. Intervirology. 1996;39:302–319. doi: 10.1159/000150502. [DOI] [PubMed] [Google Scholar]
- 5.Sinclair J, Sissons P. Latent and persistent infections of monocytes and macrophages. Intervirology. 1996;39:293–301. doi: 10.1159/000150501. [DOI] [PubMed] [Google Scholar]
- 6.Rice GP, Schrier RD, Oldstone MB. Cytomegalovirus infects human lymphocytes and monocytes: virus expression is restricted to immediate-early gene products. Proc. Natl. Acad. Sci. U S A. 1984;81:6134–6138. doi: 10.1073/pnas.81.19.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soderberg-Naucler C, Streblow DN, Fish KN, Allan-Yorke J, Smith PP, Nelson JA. Reactivation of latent human cytomegalovirus in CD14(+) monocytes is differentiation dependent. J. Virol. 2001;75:7543–7554. doi: 10.1128/JVI.75.16.7543-7554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulliam L. Cytomegalovirus preferentially infects a monocyte derived macrophage/microglial cell in human brain cultures: neuropathology differs between strains. J. Neuropathol. Exp. Neurol. 1991;50:432–440. doi: 10.1097/00005072-199107000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Booss J, Dann PR, Griffith BP, Kim JH. Host defense response to cytomegalovirus in the central nervous system. Predominance of the monocyte. Am. J. Pathol. 1989;134:71–78. [PMC free article] [PubMed] [Google Scholar]
- 10.Degre M. Has cytomegalovirus infection any role in the development of atherosclerosis? Clin. Microbiol. Infect. 2002;8:191–195. doi: 10.1046/j.1469-0691.2002.00407.x. [DOI] [PubMed] [Google Scholar]
- 11.Streblow DN, Orloff SL, Nelson JA. Do pathogens accelerate atherosclerosis? J. Nutr. 2001;131:2798S–2804S. doi: 10.1093/jn/131.10.2798S. [DOI] [PubMed] [Google Scholar]
- 12.van der Strate BW, Hillebrands JL, Lycklama a Nijeholt SS, Beljaars L, Bruggeman CA, Van Luyn MJ, Rozing J, The TH, Meijer DK, Molema G, Harmsen MC. Dissemination of rat cytomegalovirus through infected granulocytes and monocytes in vitro and in vivo. J. Virol. 2003;77:11274–11278. doi: 10.1128/JVI.77.20.11274-11278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bale JF, Jr., O’Neil ME. Detection of murine cytomegalovirus DNA in circulating leukocytes harvested during acute infection of mice. J. Virol. 1989;63:2667–2673. doi: 10.1128/jvi.63.6.2667-2673.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MS, Bentz GL, Smith PM, Bivins ER, Yurochko AD. HCMV activates PI(3)K in monocytes and promotes monocyte motility and transendothelial migration in a PI(3)K-dependent manner. J. Leukoc. Biol. 2004;76:65–76. doi: 10.1189/jlb.1203621. [DOI] [PubMed] [Google Scholar]
- 15.Smith MS, Bentz GL, Alexander JS, Yurochko AD. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol. 2004;78:4444–4453. doi: 10.1128/JVI.78.9.4444-4453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MS, Bivins-Smith ER, Tilley AM, Bentz GL, Chan G, Minard J, Yurochko AD. Roles of phosphatidylinositol 3-kinase and NF-κB in human cytomegalovirus-mediated monocyte diapedesis and adhesion: strategy for viral persistence. J. Virol. 2007;81:7683–7694. doi: 10.1128/JVI.02839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentz GL, Jarquin-Pardo M, Chan G, Smith MS, Sinzger C, Yurochko AD. Human cytomegalovirus (HCMV) infection of endothelial cells promotes naive monocyte extravasation and transfer of productive virus to enhance hematogenous dissemination of HCMV. J. Virol. 2006;80:11539–11555. doi: 10.1128/JVI.01016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shnyra A, Brewington R, Alipio A, Amura C, Morrison DC. Reprogramming of lipopolysaccharide-primed macrophages is controlled by a counterbalanced production of IL-10 and IL-12. J. Immunol. 1998;160:3729–3736. [PubMed] [Google Scholar]
- 19.Zhang X, Morrison DC. Lipopolysaccharide-induced selective priming effects on tumor necrosis factor alpha and nitric oxide production in mouse peritoneal macrophages. J. Exp. Med. 1993;177:511–516. doi: 10.1084/jem.177.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redente EF, Orlicky DJ, Bouchard RJ, Malkinson AM. Tumor signaling to the bone marrow changes the phenotype of monocytes and pulmonary macrophages during urethane-induced primary lung tumorigenesis in A/J mice. Am. J. Pathol. 2007;170:693–708. doi: 10.2353/ajpath.2007.060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 22.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, de Waal-Malefyt R, Ottenhoff TH. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc. Natl. Acad. Sci. U S A. 2004;101:4560–4565. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 26.Goerdt S, Orfanos CE. Other functions, other genes: alternative activation of antigen-presenting cells. Immunity. 1999;10:137–142. doi: 10.1016/s1074-7613(00)80014-x. [DOI] [PubMed] [Google Scholar]
- 27.Mosser DM. The many faces of macrophage activation. J. Leukoc. Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 28.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen PF, Wang JE, Almlof M, Thiemermann C, Foster SJ, Solberg R, Aasen AO. Peptidoglycan and lipoteichoic acid modify monocyte phenotype in human whole blood. Clin. Diagn. Lab. Immunol. 2001;8:515–521. doi: 10.1128/CDLI.8.3.515-521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fidler IJ, Nii A, Utsugi T, Brown D, Bakouche O, Kleinerman ES. Differential release of TNF-α, IL 1, and PGE2 by human blood monocytes subsequent to interaction with different bacterial derived agents. Lymphokine Res. 1990;9:449–463. [PubMed] [Google Scholar]
- 31.Kindle L, Rothe L, Kriss M, Osdoby P, Collin-Osdoby P. Human microvascular endothelial cell activation by IL-1 and TNF-α stimulates the adhesion and transendothelial migration of circulating human CD14+ monocytes that develop with RANKL into functional osteoclasts. J. Bone Miner. Res. 2006;21:193–206. doi: 10.1359/JBMR.051027. [DOI] [PubMed] [Google Scholar]
- 32.Hakkert BC, Kuijpers TW, Leeuwenberg JF, van Mourik JA, Roos D. Neutrophil and monocyte adherence to and migration across monolayers of cytokine-activated endothelial cells: the contribution of CD18, ELAM-1, and VLA-4. Blood. 1991;78:2721–2726. [PubMed] [Google Scholar]
- 33.Yurochko AD, Huang ES. Human cytomegalovirus binding to human monocytes induces immunoregulatory gene expression. J. Immunol. 1999;162:4806–4816. [PubMed] [Google Scholar]
- 34.Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc. Natl. Acad. Sci. U S A. 2002;99:3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocarski ES., Jr. Virus self-improvement through inflammation: no pain, no gain. Proc. Natl. Acad. Sci. U S A. 2002;99:3362–3364. doi: 10.1073/pnas.072075899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein J, Volk HD, Liebenthal C, Kruger DH, Prosch S. Tumour necrosis factor alpha stimulates the activity of the human cytomegalovirus major immediate early enhancer/promoter in immature monocytic cells. J. Gen. Virol. 1993;74(Pt 11):2333–2338. doi: 10.1099/0022-1317-74-11-2333. [DOI] [PubMed] [Google Scholar]
- 37.Ritter T, Brandt C, Prosch S, Vergopoulos A, Vogt K, Kolls J, Volk HD. Stimulatory and inhibitory action of cytokines on the regulation of hCMV-IE promoter activity in human endothelial cells. Cytokine. 2000;12:1163–1170. doi: 10.1006/cyto.2000.0689. [DOI] [PubMed] [Google Scholar]
- 38.Locati M, Deuschle U, Massardi ML, Martinez FO, Sironi M, Sozzani S, Bartfai T, Mantovani A. Analysis of the gene expression profile activated by the CC chemokine ligand 5/RANTES and by lipopolysaccharide in human monocytes. J. Immunol. 2002;168:3557–3562. doi: 10.4049/jimmunol.168.7.3557. [DOI] [PubMed] [Google Scholar]
- 39.Mehta A, Brewington R, Chatterji M, Zoubine M, Kinasewitz GT, Peer GT, Chang AC, Taylor FB, Jr., Shnyra A. Infection-induced modulation of m1 and m2 phenotypes in circulating monocytes: role in immune monitoring and early prognosis of sepsis. Shock. 2004;22:423–430. doi: 10.1097/01.shk.0000142184.49976.0c. [DOI] [PubMed] [Google Scholar]
- 40.Goldmann O, von Kockritz-Blickwede M, Holtje C, Chhatwal GS, Geffers R, Medina E. Transcriptome analysis of murine macrophages in response to infection with Streptococcus pyogenes reveals an unusual activation program. Infect. Immun. 2007;75:4148–4157. doi: 10.1128/IAI.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yurochko AD, Liu DY, Eierman D, Haskill S. Integrins as a primary signal transduction molecule regulating monocyte immediate-early gene induction. Proc. Natl. Acad. Sci. U S A. 1992;89:9034–9038. doi: 10.1073/pnas.89.19.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. U S A. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnoult D, Bartle LM, Skaletskaya A, Poncet D, Zamzami N, Park PU, Sharpe J, Youle RJ, Goldmacher VS. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. U S A. 2004;101:7988–7993. doi: 10.1073/pnas.0401897101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U S A. 2001;98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 48.Liu SF, Malik AB. NF-κB activation as a pathological mechanism of septic shock and inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;290:L622–L645. doi: 10.1152/ajplung.00477.2005. [DOI] [PubMed] [Google Scholar]
- 49.Shishodia S, Aggarwal BB. Nuclear factor-κB activation: a question of life or death. J. Biochem. Mol. Biol. 2002;35:28–40. doi: 10.5483/bmbrep.2002.35.1.028. [DOI] [PubMed] [Google Scholar]
- 50.Proost P, Wuyts A, van Damme J. The role of chemokines in inflammation. Int. J. Clin. Lab. Res. 1996;26:211–223. doi: 10.1007/BF02602952. [DOI] [PubMed] [Google Scholar]
- 51.Schenkel AR, Mamdouh Z, Muller WA. Locomotion of monocytes on endothelium is a critical step during extravasation. Nat. Immunol. 2004;5:393–400. doi: 10.1038/ni1051. [DOI] [PubMed] [Google Scholar]
- 52.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 53.Liote F, Boval-Boizard B, Weill D, Kuntz D, Wautier JL. Blood monocyte activation in rheumatoid arthritis: increased monocyte adhesiveness, integrin expression, and cytokine release. Clin. Exp. Immunol. 1996;106:13–19. doi: 10.1046/j.1365-2249.1996.d01-820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mustjoki S, Alitalo R, Elonen E, Carpen O, Gahmberg CG, Vaheri A. Intercellular adhesion molecule-1 in extravasation of normal mononuclear and leukaemia cells. Br. J. Haematol. 2001;113:989–1000. doi: 10.1046/j.1365-2141.2001.02793.x. [DOI] [PubMed] [Google Scholar]
- 55.Shahgasempour S, Woodroffe SB, Garnett HM. Alterations in the expression of ELAM-1, ICAM-1 and VCAM-1 after in vitro infection of endothelial cells with a clinical isolate of human cytomegalovirus. Microbiol. Immunol. 1997;41:121–129. doi: 10.1111/j.1348-0421.1997.tb01177.x. [DOI] [PubMed] [Google Scholar]
- 56.Chan G, Stinski MF, Guilbert LJ. Human cytomegalovirus-induced upregulation of intercellular cell adhesion molecule-1 on villous syncytiotrophoblasts. Biol. Reprod. 2004;71:797–803. doi: 10.1095/biolreprod.104.028118. [DOI] [PubMed] [Google Scholar]
- 57.Warren AP, Owens CN, Borysiewicz LK, Patel K. Down-regulation of integrin α1/β1 expression and association with cell rounding in human cytomegalovirus-infected fibroblasts. J. Gen. Virol. 1994;75(Pt 12):3319–3325. doi: 10.1099/0022-1317-75-12-3319. [DOI] [PubMed] [Google Scholar]
- 58.Ito M, Watanabe M, Ihara T, Kamiya H, Sakurai M. Increased expression of adhesion molecules (CD54, CD29 and CD44) on fibroblasts infected with cytomegalovirus. Microbiol. Immunol. 1995;39:129–133. doi: 10.1111/j.1348-0421.1995.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 59.Pardo A, Selman M. MMP-1: the elder of the family. Int. J. Biochem. Cell Biol. 2005;37:283–288. doi: 10.1016/j.biocel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 60.Gill JH, Kirwan IG, Seargent JM, Martin SW, Tijani S, Anikin VA, Mearns AJ, Bibby MC, Anthoney A, Loadman PM. MMP-10 is overexpressed, proteolytically active, and a potential target for therapeutic intervention in human lung carcinomas. Neoplasia. 2004;6:777–785. doi: 10.1593/neo.04283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 62.Doherty DE, Zagarella L, Henson PM, Worthen GS. Lipopolysaccharide stimulates monocyte adherence by effects on both the monocyte and the endothelial cell. J. Immunol. 1989;143:3673–3679. [PubMed] [Google Scholar]
- 63.Van Damme J, De Ley M, Van Snick J, Dinarello CA, Billiau A. The role of interferon-β1 and the 26-kDa protein (interferon-β2) as mediators of the antiviral effect of interleukin 1 and tumor necrosis factor. J. Immunol. 1987;139:1867–1872. [PubMed] [Google Scholar]
- 64.Hughes TK, Kaspar TA, Coppenhaver DH. Synergy of antiviral actions of TNF and IFN-γ: evidence for a major role of TNF-induced IFN-β. Antiviral Res. 1988;10:1–9. doi: 10.1016/0166-3542(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 65.Taylor-Wiedeman J, Sissons JG, Borysiewicz LK, Sinclair JH. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 1991;72(Pt 9):2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 66.Fish KN, Depto AS, Moses AV, Britt W, Nelson JA. Growth kinetics of human cytomegalovirus are altered in monocyte-derived macrophages. J. Virol. 1995;69:3737–3743. doi: 10.1128/jvi.69.6.3737-3743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ibanez CE, Schrier R, Ghazal P, Wiley C, Nelson JA. Human cytomegalovirus productively infects primary differentiated macrophages. J. Virol. 1991;65:6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yurochko AD. Human cytomegalovirus modulation of signal transduction. In: Stinski MF, Shenk T, editors. Human cytomegaloviruses. Springer-verlag; Berlin Heidelberg: 2008. pp. 205–220. [DOI] [PubMed] [Google Scholar]
- 69.Chan G, Bivins-Smith ER, Smith MS, Yurochko AD. Transcriptome analysis of NF-kappaB- and phosphatidylinositol 3-kinase-regulated genes in human cytomegalovirus-infected monocytes. J. Virol. 2008;82:1040–1046. doi: 10.1128/JVI.00864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirsch E, Irikura VM, Paul SM, Hirsh D. Functions of interleukin 1 receptor antagonist in gene knockout and overproducing mice. Proc. Natl. Acad. Sci. U S A. 1996;93:11008–11013. doi: 10.1073/pnas.93.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salek-Ardakani S, Arrand JR, Mackett M. Epstein-Barr virus encoded interleukin-10 inhibits HLA-class I, ICAM-1, and B7 expression on human monocytes: implications for immune evasion by EBV. Virology. 2002;304:342–351. doi: 10.1006/viro.2002.1716. [DOI] [PubMed] [Google Scholar]
- 72.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc. Natl. Acad. Sci. U S A. 2000;97:1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fiorentino DF, Zlotnik A, Vieira P, Mosmann TR, Howard M, Moore KW, O’Garra A. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J. Immunol. 1991;146:3444–3451. [PubMed] [Google Scholar]
- 74.Charo IF, Taubman MB. Chemokines in the pathogenesis of vascular disease. Circ. Res. 2004;95:858–866. doi: 10.1161/01.RES.0000146672.10582.17. [DOI] [PubMed] [Google Scholar]
- 75.Pulliam L, Sun B, Rempel H. Invasive chronic inflammatory monocyte phenotype in subjects with high HIV-1 viral load. J. Neuroimmunol. 2004;157:93–98. doi: 10.1016/j.jneuroim.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 76.Wen W, Chen S, Cao Y, Zhu Y, Yamamoto Y. HIV-1 infection initiates changes in the expression of a wide array of genes in U937 promonocytes and HUT78 T cells. Virus Res. 2005;113:26–35. doi: 10.1016/j.virusres.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 77.Giri MS, Nebozhyn M, Showe L, Montaner LJ. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J. Leukoc. Biol. 2006;80:1031–1043. doi: 10.1189/jlb.0306157. [DOI] [PubMed] [Google Scholar]
- 78.Izmailova E, Bertley FM, Huang Q, Makori N, Miller CJ, Young RA, Aldovini A. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 2003;9:191–197. doi: 10.1038/nm822. [DOI] [PubMed] [Google Scholar]
- 79.Choi HJ, Smithgall TE. HIV-1 Nef promotes survival of TF-1 macrophages by inducing Bcl-XL expression in an extracellular signal-regulated kinase-dependent manner. J. Biol. Chem. 2004;279:51688–51696. doi: 10.1074/jbc.M410068200. [DOI] [PubMed] [Google Scholar]
- 80.Vazquez N, Greenwell-Wild T, Marinos NJ, Swaim WD, Nares S, Ott DE, Schubert U, Henklein P, Orenstein JM, Sporn MB, Wahl SM. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J. Virol. 2005;79:4479–4491. doi: 10.1128/JVI.79.7.4479-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang M, Li X, Pang X, Ding L, Wood O, Clouse KA, Hewlett I, Dayton AI. Bcl-2 upregulation by HIV-1 Tat during infection of primary human macrophages in culture. J. Biomed. Sci. 2002;9:133–139. doi: 10.1007/BF02256024. [DOI] [PubMed] [Google Scholar]
- 82.Tuttle DL, Harrison JK, Anders C, Sleasman JW, Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J. Virol. 1998;72:4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.