Abstract
The non-random positioning of chromosome territories in eukaryotic cells is largely correlated with gene density and is conserved throughout evolution. Gene rich chromosomes are predominantly central while gene poor chromosomes are peripherally localized in interphase nuclei. We previously demonstrated that artificially introduced human chromosomes assume a position equivalent to their endogenous homologues in the diploid colon cancer cell line DLD-1. These chromosomal aneuploidies result in a significant increase in transcript levels, suggesting a relationship between genomic copy number, gene expression and chromosome position. We previously proposed that each chromosome is marked by a “zip-code” that determines its non-random position in the nucleus. Here we investigated (1) whether mouse nuclei recognize such determinants of nuclear position on human chromosomes to facilitate their distinct partitioning and (2) if chromosome positioning and transcriptional activity remain coupled under these trans-species conditions. Using 3D-FISH, confocal microscopy and gene expression profiling, we show (i) that gene poor and gene rich human chromosomes maintain their divergent but conserved positions in mouse-human hybrid nuclei and (ii) that a foreign human chromosome is actively transcribed in mouse nuclei. Our results suggest a species-independent conserved mechanism for the non-random positioning of chromosomes in the 3-dimensional interphase nucleus.
Keywords: Mouse, MMCT, Transcription, Chromosome territories, 3D-FISH, DLD-1, Aneuploidy
Introduction
It is now well established that chromosomes are non-randomly positioned in the interphase nuclei of many eukaryotes such as humans, mice, chicken and plants (Croft, et al. 1999; Habermann, et al. 2001; Mayr, et al. 2003; Mayer, et al. 2005). In most cell types across species, gene dense chromosomes are positioned predominantly towards the nuclear center while chromosomes with lower gene densities are positioned towards the nuclear periphery (Croft, et al. 1999; Cremer, et al. 2001; Habermann, et al. 2001; Mayer, et al. 2005; Lanctot, et al. 2007). Despite extensive genomic rearrangement throughout evolution, such a non-random arrangement has been conserved in higher primates over a span of 30 million years, suggesting a strong functional significance of this higher order nuclear architecture (Tanabe, et al. 2002; Tanabe, et al. 2005; Mora, et al. 2006; Neusser, et al. 2007). Despite the apparent importance of this biological phenomenon, little is known about the underlying mechanism. Parameters such as size, gene density and transcriptional activity of chromosomes have been proposed as factors responsible for determining chromosome positioning.
A tissue specific and non-random arrangement of chromosomes has also been demonstrated in a variety of mouse primary cells (Parada et al 2004). Perhaps because the variation in gene density and chromosome size is not as large in mice as it is in humans (Δdensity 2.5 vs. 4.6; Δsize 1.7 vs. 3, respectively), chromosome positioning is not as polarized and has been shown to correlate with both gene density and chromosome size (Mayer, et al. 2005). Flat human fibroblasts during the G0 stage of the cell cycle have been shown to have a greater correlation of non-random position with chromosome size than gene density (Bolzer, et al. 2005). A similar result was obtained in a comparative study of chromosomes 6, 12, 13 and 17 in humans and the territories of their orthologous chromosomes in new world monkeys (Mora, et al. 2006).
There is, however, more evidence supporting the role of gene density. In the chicken, for instance, early replicating gene dense microchromosomes are more centrally positioned than late replicating gene poor macrochromosomes (Habermann, et al. 2001). The homogeneously sized chromosomes of the primate species Wolf’s Guenon are distinctly positioned with gene dense chromosomes predominantly in the nuclear center (Neusser, et al. 2007). Both 2D and 3D-FISH analyses of cells containing translocation chromosomes show that the more gene dense partner is predominantly more centrally positioned than the gene poor partner (Croft et al 1999, Cremer et al 2003). Flat human fibroblasts showed a higher density of Alu rich sequences, typically found in the gene dense R-bands of chromosomes, in the center of the nucleus, suggesting that while size may have had a greater influence on territory positioning, gene density based correlations are relevant and might be crucial in establishing non-random chromosome positioning patterns (Bolzer, et al. 2005). This has been observed for individual genes within their respective territories. The gene rich sub-domains of human chromosomes (HSA) 11, 12, 18 and 19 were shown to be oriented more towards the nuclear interior than the gene poor sub-domains (Kupper, et al. 2007).
We previously proposed that the non-random 3D nuclear positioning of chromosome territories is established through a chromosome-associated “zip-code”. Hereafter we use the term “determinants” rather than “zip-code” in order to distinguish it from zip-code binding proteins involved in RNA localization (Deshler, et al. 1998). In the present study we examined the position of human chromosomes in mouse-human hybrid cell lines in order to assess whether the “determinants” of chromosome positioning could be interpreted in different species and if the positioning mechanism worked to the same extent. Using this trans-species system we were also hoping to observe an uncoupling of transcription and nuclear position.
Materials and Methods
Cell culture
A9 mouse-human monochromosomal hybrid fibroblast cells were grown in DMEM-F12 media, 10% FBS, with Penicillin (50 units/ml), Streptomycin (50 μg/ml), G418 (800 μg/ml) in the presence of 5% CO2 at 37ºC (Tanabe, et al. 2000). The cells were grown on chamber slides for 3D-FISH experiments.
Cell fixation and permeabilization
Mouse-human hybrid cell lines A9+7, A9+18, and A9+19 were fixed independently and processed as previously described to preserve the nuclear morphology (Sengupta, et al. 2007). Briefly, cells were permeabilized in CSK buffer for 5 minutes on ice, fixed in 4% PFA (5 minutes), re-permeabilized with 0.5% TX-100 (10 minutes), liquid nitrogen freeze-thawed (3 times), and denatured in 0.1N HCl for 10 minutes and stored in 50%FA/2x SSC (pH = 7.4) over night at 4ºC. Care was taken to ensure that the slides did not dry at any point.
FISH
3D FISH Briefly, flow sorted human chromosomes 7, 18 and 19 (purchased from M.A Ferguson-Smith and Patricia O’Brien, Univ. of Cambridge, U.K.) were individually DOP-PCR labeled with spectrum orange (Vysis). The chromosome painting probes (1.25 μg) were precipitated with human Cot-1 DNA (12.5 μg) (Invitrogen). Hybridization and detection were performed as described (Sengupta, et al. 2007). 2D FISH FISH was performed on metaphase spreads from A9+19 cells using arm specific paints for chromosomes 19p (Rhodamine110) and 19q (Cy3) (Padilla-Nash, et al. 2001). FISH was also performed for chromosome 7 and 18 labeled with spectrum orange on metaphase preparations from A9+7 and A9+18 cell lines (data not shown). (http://www.riedlab.nci.nih.gov/protocols.asp).
Confocal Imaging
Hybridized and DAPI stained A9 hybrid nuclei were imaged on a Zeiss LSM 510 NLO Meta system (Carl Zeiss, Inc., Thornwood, NY, USA) mounted on an Axiovert 200M microscope using a Plan-Apochromat 100× 1.4 oil DIC objective. The imaging was performed sequentially in a multi track, 2-channel mode. The Z-stacks were acquired using a frame size of 512 × 512 with a pixel depth of 8 bit. All nuclei were imaged at a constant voxel size of 0.087 μm × 0.087 μm × 0.3 μm, scan zoom and line averaging in order to facilitate radial distance measurement comparisons with DLD-1 nuclei as previously described (Sengupta et al 2007).
3D reconstructions, volume and distance measurements
Nuclei were individually cropped from a given field, surface rendered and subjected to 3D measurements using 3D-constructor and Image-Pro Plus (v 6.1) software packages (Media Cybernetics, Inc., Bethesda, MD, USA). The appropriate thresholds were visually adjusted to best-fit the raw image data for the red and blue channels. Surface rendering was performed as described (Sengupta, et al. 2007) using the above mentioned software packages. The count option in the 3D-constructor menu segments the iso-surface and gives the raw volume (μm3) of each chromosome territory, without implementing iso-surface simplification or sub-sampling of the data sets. All 3D radial-distance measurements were performed on 3D-reconstructions of nuclei from confocal images stacks. The geometric centers of the DAPI stained nucleus and chromosome territories were determined. The location of each chromosome territory was calculated as a percent of its distance from the center of the nucleus to the nuclear periphery as described (Tanabe, et al. 2002; Sengupta, et al. 2007). Minimum of 118 chromosome territories were analyzed for each cell line to obtain distance measurements of HSA7, HSA18 and HSA19.
Statistical analyses
Mann-Whitney-Wilcoxon sum rank test was used to examine if there were significant differences in median values of the radial distance measurements and Kolmogorov-Smirnov (K-S) two-tailed test was independently applied to assess if there were significant (P < 0.05) differences in the shapes of the distributions of 3D-radial distance profiles of chromosome territories. Student’s t-test was used to determine if a significant difference existed between the mean volumes of the human chromosome territories in A9 and DLD-1 derived cells. GraphPad InStat (v 3.06), Stata/SE (v 9.0) and Sigma Plot (v 9.0) were used for statistical analyses and graphical representations.
Gene expression analyses
RNA was isolated from actively growing A9, A9+7, A9+18 and A9+19 cell cultures using Trizol (Ambion) with minor modifications to the manufacturer’s instructions (http://www.riedlab.nci.nih.gov/protocols.asp). Cell line RNA and human reference RNA (Stratagene) were labeled with Cy3 and Cy5 (Perkin-Elmer), respectively, following the instructions in the Agilent labeling kit and a dual color hybridization was performed on an Agilent 4 × 44K whole human expression array in order to assess the transcriptional activity of the human chromosome in the hybrid cell lines. Results were analyzed and plotted along the length of the human chromosome ideograms using CGH Analytics (Agilent) gene expression software. The expression level of all genes mapping to a chromosome arm relative to the human reference RNA were averaged to generate an arm expression average.
Array Comparative Genomic Hybridization (aCGH)
Oligonucleotide-based Human Genome Microarray CGH was performed according to the protocol provided by the manufacturer (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis, protocol version 4.0, June 2006, Agilent Technologies, Santa Clara, CA), with minor modifications. Briefly, 1 μg of genomic DNA from the A9 mouse cell line and from A9+19 hybrid cell line were digested for 2 hours with AluI and RsaI (Promega, Madison, WI) at 37°C. Test DNA from A9+19 cells was labeled with Cy3-dUTP, and A9 DNA was used as reference and labeled with Cy5-dUTP (PerkinElmer, Waltham, MA). The labeling reaction was performed at 37°C for 2 hours using the Bioprime Array CGH Genomic Labeling Module (Invitrogen). Unincorporated nucleotides were eliminated using Microcon YM-30 columns (Millipore, Bedford, MA). Cy3 and Cy5-labeled samples were combined in equal amounts according to the incorporation of labeled nucleotides as measured using a Nanodrop. A 185K oligonucleotide-based Human Genome Microarray G4411A (Agilent Technologies) was subjected to hybridization for 40h at 65°C, washed using the manufacturers’ recommended conditions, and scanned using a laser scanner (G2565BA, Agilent Technologies). Agilent Feature Extraction™ software (version 9.1, Agilent Technologies) was applied for image analysis. To visualize the aCGH data we used Agilent CGH Analytics 3.4 software (Agilent Technologies).
Results
3D-FISH with chromosome-specific painting probes for Homo sapiens HSA7, HSA18 and HSA19 was performed on A9 mouse-human monochromosomal hybrid fibroblasts cell lines. Imaging of these nuclei shows intense DAPI staining chromocenters typical of mouse nuclei, which are known to cluster in a cell type specific manner (Mayer, et al. 2005; Weidtkamp-Peters, et al. 2006) (Fig. 1 a, c, e). Confocal microscopy confirmed that HSA7 was confined to discrete chromosome territories in both mouse A9+7 and human DLD-1+7 nuclei as illustrated in the merged confocal image stacks represented in Figures 1a and 1b. Similar results were obtained for HSA18 in A9+18 and DLD-1+18 nuclei (Fig. 1c, d) as well as HSA19 in A9+19 and DLD-1+19 nuclei (Fig. 1e, f).
Fig. 1.
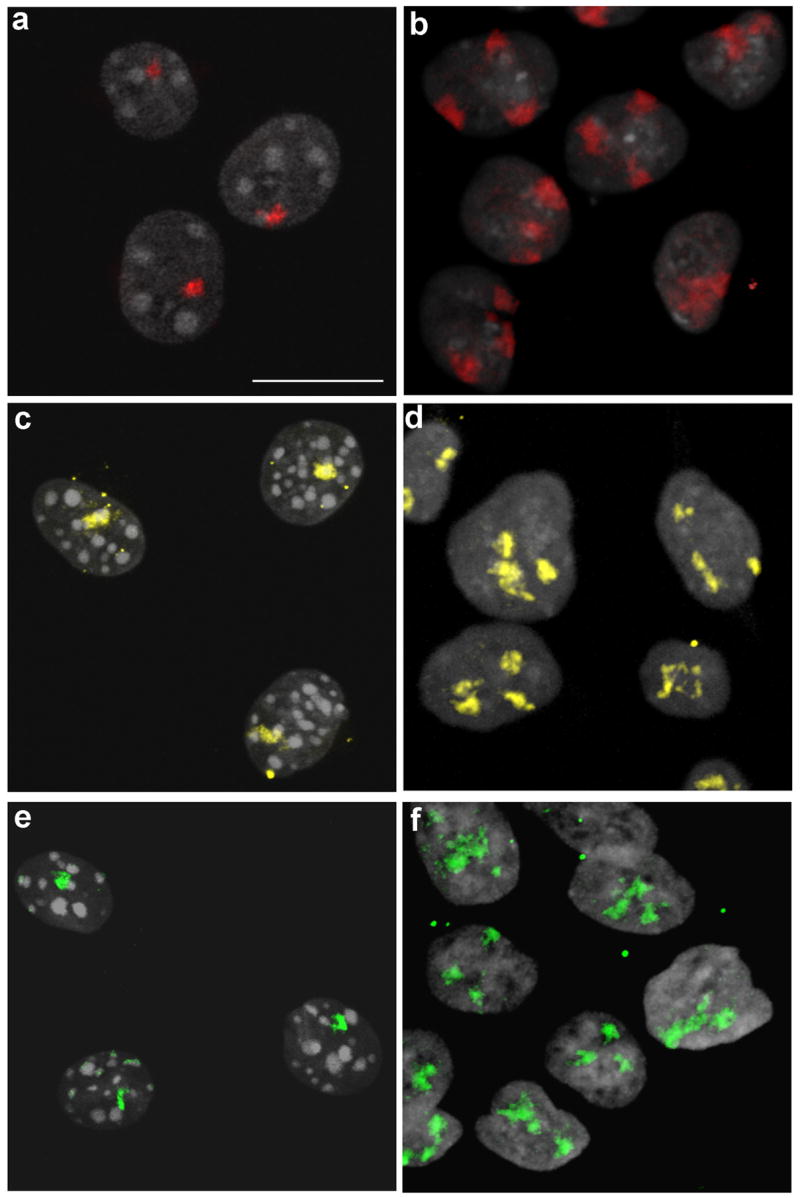
Human chromosomes occupy discrete territories in both mouse and human nuclei. Structurally preserved nuclei were hybridized with painting probes specific for HSA7 (red), HSA18 (yellow) and HSA19 (green). Confocal image stacks were merged and maximum intensity projections were generated from each of the A9 mouse-human monochromosomal fibroblast nuclei (a, c and e) as well as from DLD-1 derivatives (b, d and f), Scale bar: 10 μm. (a and b) HSA7 territories visualized in A9+7 (a) and DLD-1+7 (b). (c and d) HSA18 territories visualized in A9+18 (c) and DLD-1+18 (d). (e and f) HSA19 territories visualized in A9+19 (e) and DLD-1+19 (f)
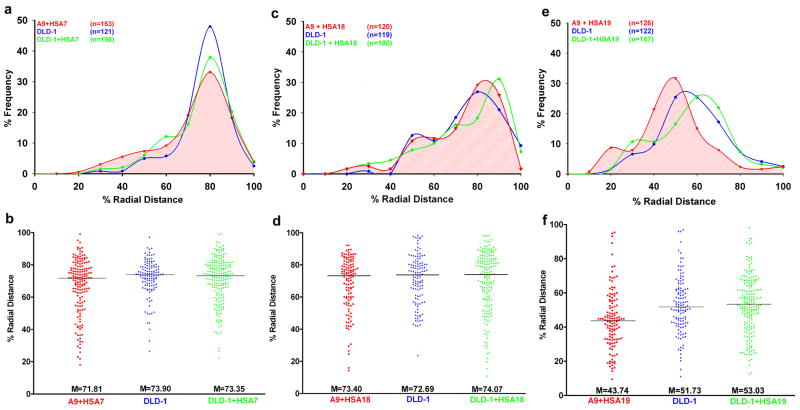
3D reconstructions, distance measurements and statistical analyses were performed for each series of nuclei. The radial distance measurements showed that the gene poor HSA7 territory was predominantly located towards the nuclear periphery (Fig. 2a), with a median value of 71.81 (Fig. 2b). This positioning was comparable to that of HSA7 in both DLD-1 (M = 73.90) and DLD-1+7 (M = 73.35) nuclei (Fig. 2a, b). Application of the Mann-Whitney-Wilcoxon (MW) sum-rank test, used to assess differences in the median values (ΔM), revealed that the slightly more internal positioning of HSA7 in mouse A9+7 nuclei had a marginally significant difference compared to DLD-1 (ΔM = −2.09, PMW = 0.03, ΔM > 0: peripheral, ΔM < 0: internal), but not DLD-1+7 (ΔM = −1.54, PMW = 0.11). As reported previously, no significant difference was identified in HSA7 position between DLD-1 and DLD-1+7 (ΔM = −0.55, PMW = 0.51) (Fig. 2b, Table 1). Comparable results were obtained using the Kolmogorov-Smirnov (K-S) two-tailed test which calculates the significance based on the shape of the distribution curves.
Fig. 2. Human chromosomes assume a conserved radial position in both mouse and human nuclei that correlates with their gene densities.
(a, c and e) 3D radial distance profiles of chromosome territories HSA7 (a), HSA18 (c) and HSA19 (e). X-axis: % Radial Distance, 0% - nuclear center, 100% - nuclear periphery and Y-axis: % Frequency of chromosome territories.
(b, d and f) Raw distributions of 3D radial distance measurements. X-axis: Cell line, Y-axis: % Radial Distance.
Table 1.
Statistical analyses of radial distance measurements using Kolmogorov-Smirnov (KS) and Mann-Whitney-Wilcoxon (MW) tests
| DLD-1+/DLD-1 | A9+/DLD-1 | A9+/DLD-1+ | |
|---|---|---|---|
| HSA7 | PKS = 0.15 | PKS = 0.02 | PKS = 0.36 |
| PMW = 0.51 | PMW = 0.03 | PMW = 0.11 | |
| ΔM = −0.55 | ΔM = −2.09 | ΔM = −1.54 | |
| HSA18 | PKS = 0.34 | PKS = 0.76 | PKS = 0.17 |
| PMW = 0.94 | PMW = 0.33 | PMW = 0.25 | |
| ΔM = +1.38 | ΔM = +0.71 | ΔM = −0.67 | |
| HSA19 | PKS = 0.73 | PKS = 0.000 | PKS = 0.000 |
| PMW = 0.73 | PMW = 0.0001 | PMW = 0.0001 | |
| ΔM = +3.29 | ΔM = −8.02 | ΔM = −9.55 |
HSA: Homo sapiens
DLD-1+: DLD-1 derived cell lines, DLD-1: DLD-1 diploid cell line
A9+: A9 derived cell lines
PKS: P-value from Kolmogorov-Smirnov test (P < 0.05)
PMW: P-value from Mann-Whitney Wilcoxon test (P < 0.05)
M: Difference in median values, ΔM > 0: shift away from nuclear center,
M < 0: shift towards nuclear center
Radial distance profiles demonstrated the peripheral position of HSA18 (~70–80%) in A9+18 mouse nuclei, consistent with previous results from DLD-1 and DLD-1+18 nuclei (Fig. 2c) (Cremer, et al. 2003; Sengupta, et al. 2007). This was confirmed through a comparison of the median radial distances of HSA18 in A9+18 (M = 73.40), DLD-1 (M = 72.69) and DLD-1+18 (M = 74.07) (Fig. 2d). Mann-Whitney-Wilcoxon test did not identify a significant difference in the position of HSA18 between A9+18 and DLD-1 (ΔM = +0.71, PMW = 0.33) or A9+18 and DLD-1+18 nuclei (ΔM = −0.67, PMW = 0.25) (Fig. 2c, d and Table 1). This was again substantiated by the K-S test (Table 1).
Our subsequent analysis of A9+19 nuclei recapitulated the predominant positioning of HSA19 towards the nuclear center (Fig. 2e). Unlike the statistical analyses of HSA7 and HSA18, the radial distance of HSA19 (M = 43.74) in A9+19 nuclei showed a very significant shift of HSA19 to a more internal position relative to both DLD-1 (M = 51.73, ΔM = −8.02, PMW = 0.0001) and DLD-1+19 nuclei (M = 53.03, ΔM = −9.55, PMW = 0.0001) (Fig. 2f and Table 1). This was again consistent with the K-S test. Taken together, our results demonstrate that the polarized partitioning of gene poor (HSA7 and HSA18) and gene rich (HSA19) human chromosomes is conserved and is effectively recapitulated in the mouse nucleus. The measurements indicated that the volumes of chromosome territories were not significantly different for chromosomes 18 and 19 when human (DLD-1 derived) and mouse nuclei (A9 derived) were compared. However, we noted a slightly increased volume of HSA7 in the mouse nuclei (P = 0.02) as summarized in Table 2.
Table 2.
Comparison of chromosome territory volumes in A9+ and DLD-1+ derived nuclei
| A9+ | DLD-1+ | A9+/DLD1+ | |
|---|---|---|---|
| HSA7 | 10.01 ± 1.18(n = 161) | 8.17 ± 0.95 (n = 147) | P = 0.02 |
| HSA18 | 6.32 ± 0.73 (n = 113) | 5.87 ± 0.66 (n = 162) | P = 0.37 |
| HSA19 | 5.18 ± 0.52 (n = 118) | 5.04 ± 0.52 (n = 166) | P = 0.68 |
Volumes are in μm3 ± 2xSEM (Standard Error of Means)
HSA: Homo sapiens
A9+: A9 derived cell lines
DLD-1+: DLD-1 derived cell lines
P: P-value from students t-test (P < 0.05)
n: number of chromosome territories
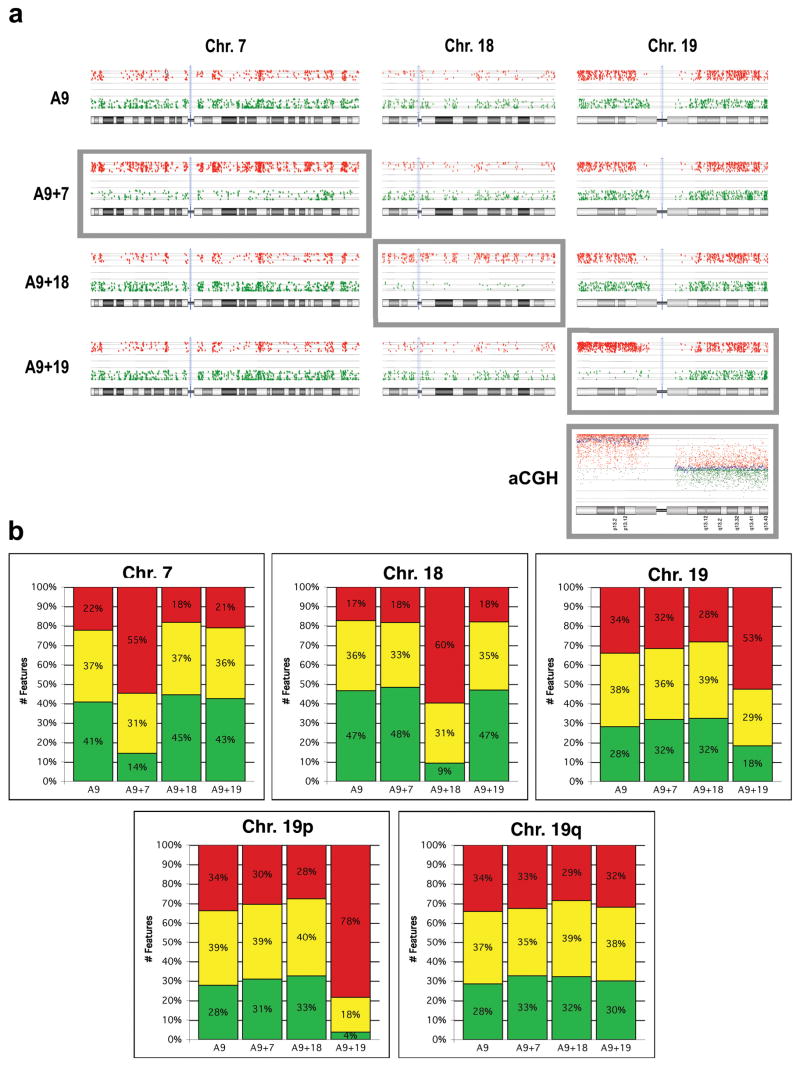
We next wanted to assess gene transcriptional activity from the artificially introduced human chromosome and determine if there was concordance with a conserved chromosome position in the mouse nuclei. RNA isolated from A9 and each of the A9 monochromosomal hybrid mouse cell lines was labeled with Cy3, combined with Cy5 labeled human reference RNA and hybridized onto whole human genome oligonucleotide arrays. Any hybridization of the A9 RNA onto these human arrays would be the result of extensive sequence conservation between the mouse mRNA and the array oligonucleotide corresponding to the homologous human gene. The expression ratio plots for array features (some genes are represented by more than one feature) mapping to human chromosomes 7, 18 & 19 clearly showed that hybridization signal can be detected for a considerable number of features. In fact, the normalized intensity values for many features were more than 2.8-fold (1.5-fold log2) different from those observed in the human reference RNA (Fig. 3a, red dots in top row labeled A9). On the other hand, it was perhaps not surprising that even more features had a normalized intensity much lower than in the human reference (Fig. 3a, green dots in top row labeled A9). Thus, the intensity ratios observed in the A9 cells for genes mapping to HSA7, HSA18 and HSA19 represent the normalized background “expression noise”. Any alteration in that distribution of ratios in the A9 hybrid cell lines would then be attributable to the detection of mRNA from the human chromosome.
Fig. 3. Human chromosomes are transcriptionally active in mouse A9 cells.
(a) Gene expression profiles of HSA7, 18 and 19 in A9 and A9 hybrid cells. Red dots: cell line/human reference ratio >1.5 log2, Green dots: cell line/human reference ratio < -1.5 log2. Grey boxes indicate the human chromosome in each cell line. aCGH: Genomic profile of A9+19 relative to the A9 mouse cell line on a human oligonucleotide-based CGH microarray. Increased copy number for chromosome arm 19p is clearly indicated, while no change in 19q is observed.
(b) Top Panel: Quantification of Fig. 3a displaying the percentage of genes showing greater than a 1.5 log2 increase (red), more than a 1.5 log2 decrease (green) or no significant change (yellow) in gene expression levels for each of the human chromosomes in the A9 monochromosomal hybrid cell lines. Bottom Panel: HSA19 was further divided into p and q arms to demonstrate the difference in detected expression of these two chromosome arms.
The resulting gene expression profile from the three A9 hybrid cell lines, each containing a unique human chromosome, are presented in the lower three panels of Fig. 3a. What was observed in all three instances was a general shift in the signal intensity ratios of genes mapping to the introduced chromosomes (grey boxes). This was manifested as an increase in the number of red features and a corresponding reduction in the number of green features for that chromosome relative to A9 and the A9 hybrids lacking that particular chromosome. This was not observed for genes mapping to the non-introduced chromosomes, such as HSA18 and HSA19 in A9+7 for example. In other words, transcriptional activity of the introduced chromosome resulted in the generation of mRNA encoding human genes. Addition of this human-specific hybridization signal to the background signal caused an increase in the intensity ratios. The quantitative results of these ratio plots are graphically represented in Fig. 3b.
While this shift was uniform across the entire length of chromosomes 7 and 18, it was only observed for the short arm of chromosome 19. This was more apparent when the percentage of features with an increased ratio was calculated separately for the p and q arms (Fig. 3b, bottom panel). Clearly human-specific transcripts only from the genes mapping to 19p was detected. Having observed this discrepancy, we examined the status of human chromosome 19 in A9+19 cells using microarray comparative genomic hybridization (aCGH). The results revealed the presence of 19p, and not 19q, in this cell line. Unfortunately the absence of features on the array corresponding to the highly repetitive pericentromeric region of chromosome 19 only enabled us to determine that the breakpoint occurred somewhere between position 24,168,233 and 32,545,047 bp (Fig. 3a, aCGH). We therefore performed FISH analyses on metaphase spreads using differentially labeled arm specific paint probes. These clearly demonstrated the presence of 19p (green) and a small region of 19q (red) (Fig. 4). Combining the aCGH and FISH results, we can state that the break occurred in the gene poor pericentromeric heterochromatic region of 19q somewhere between the centromere and position 32,545,047 bp, thereby explaining the absence of transcriptional activity for genes mapping to the remainder of the long arm of chromosome 19 (Fig. 3a).
Fig. 4.
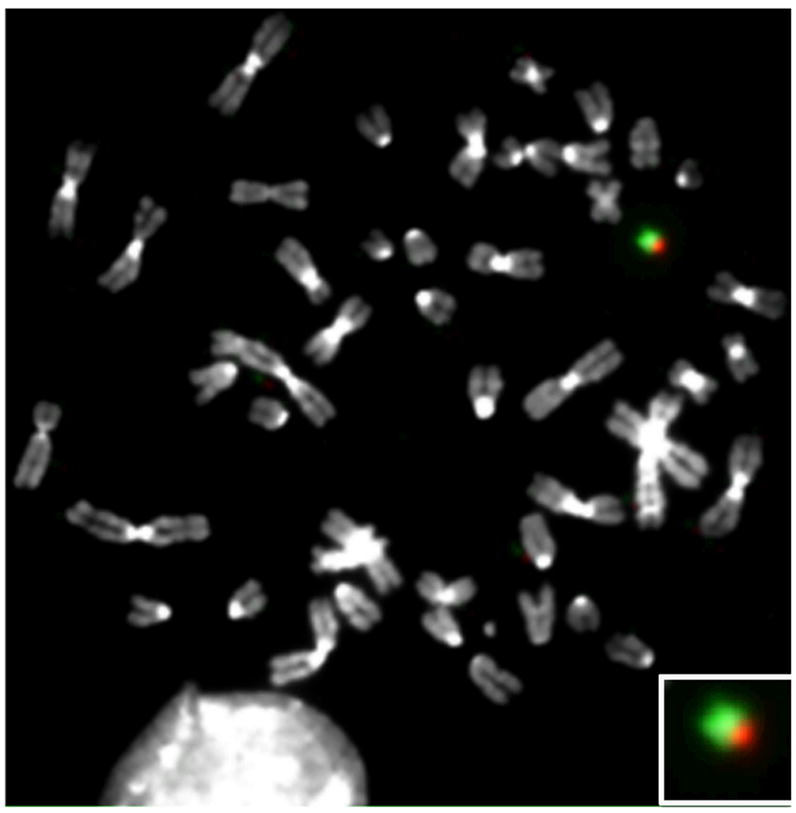
Human chromosome 19 is partially deleted in A9+19 mouse-human hybrid cells. FISH analyses of A9+19 metaphase reveals deleted HSA19q. Green: 19p and Red: 19q Inset: enlarged image of HSA19.
Discussion
We previously showed that both artificially introduced gene poor or gene rich human chromosomes assume a position equivalent to their endogenous homologues in the 3D interphase nucleus of human DLD-1 colorectal cancer cells (Sengupta, et al. 2007). Artificial introduction of the gene poor chromosome 7 into DLD-1 cells resulted in a significant increase in average transcript levels of genes mapping to chromosome 7 (DLD-1+7) (Upender, et al. 2004). Taken together these studies revealed for the first time that conservation in chromosome positioning also extended to artificially introduced chromosomes and showed a concordance with transcriptional activity (Upender, et al. 2004; Sengupta, et al. 2007). What determines the conservation of chromosome position in the nucleus is not known. In the present study, we examined whether the positioning mechanism in a mouse nucleus was capable of recognizing such a “determinant” on a human chromosome. The conserved placement of the gene-poor chromosomes 7 and 18 towards the nuclear periphery and the gene-dense chromosome 19 near the nuclear interior provides evidence for conservation of both the “determinant” and the positioning mechanism that recognizes it.
We also wanted to take advantage of this mixed species experimental design to determine if the nuclear positioning was coupled with transcriptional activity of genes along the entire length of the human chromosome. Our gene expression profiling results confirm that the artificially introduced human chromosomes are transcriptionally active in the mouse genome. Thus, it remains a possibility that gene-density and/or transcriptional activity of a chromosome serves as the “determinant” by which the 3D positions of chromosome territories are determined in mammalian nuclei. These results and their significance in the context of available literature are detailed in the following sections.
Conservation of chromosome positioning in human and mouse nuclei
3D FISH studies on a variety of cell types have shown a chromosome positioning pattern correlated with gene density. In spherically shaped human lymphoblastoid nuclei, HSA18 and HSA19 were distributed radially at ~70–80% and ~40–50% from the nuclear center, respectively (Croft, et al. 1999). A distinct correlation of chromosome positioning was observed with both gene density and chromosome size in a variety of mouse cell types such as lymphocytes, fibroblasts, myotubes, myoblasts and macrophages (Mayer, et al. 2005). In this context it is important to emphasize that mouse chromosomes do not vary extensively in terms of gene density. The most gene dense Mus musculus chromosome (MMU7; ~19 genes/Mbp) has slightly more than twice the density of the most gene sparse chromosome (MMU18; ~9.04 genes/Mbp), not taking into consideration MMUY with 3.3 genes/Mbp. 3D FISH studies and quantitative estimates of radial distances of chromosomes in spherically shaped mouse lymphoblast nuclei showed a maximum difference of ~15% in Peak Radial Distance (PRD) between gene rich MMU11 (15.68 genes/Mbp, (PRD = ~60%)) and gene poor MMUX (9.36 genes/Mbp, PRD = ~75%) (Mayer, et al. 2005). While in other mouse fibroblasts, ES cells and macrophages the difference in PRD was only ~5–8% (Mayer, et al. 2005). Our 3D FISH studies on normal primary kidney epithelial cells (day 2) cultured from mice reiterate the correlation between gene density and non-random radial position of MMU7 (gene rich) and MMU18 (gene poor) in these nuclei as well as the small PRD (Sengupta & Padilla-Nash, unpublished data). Therefore, the relatively small positional differences in nuclei of murine epithelial or flat fibroblast cells are likely due to the relative uniformity of gene density.
In contrast, HSA19 is ~4.6 fold more gene dense than HSA18 and they have conspicuously divergent nuclear positions as evidenced by their PRD of ~22% in DLD-1. In A9 hybrids, a 28–30% difference in PRD was recorded between the most gene poor/peripheral (HSA7 and HSA18) and gene dense/central (HSA19) chromosomes, comparable to that of human DLD-1 and DLD-1 derived nuclei (Fig. 2a–f). This is perhaps the largest difference in peak radial distance shown so far in mouse nuclei. Our results demonstrate for the first time that mouse cells have the ability to (1) correctly place human chromosomes in the same conserved 3D position they would assume inside a human nucleus and (2) partition gene poor (HSA7, HSA18) and gene rich (HSA19) chromosomes with greater divergence, given a large enough difference in their gene density.
A significant difference in HSA19 position was recorded in A9+19 nuclei compared with both DLD-1 and DLD-1+19 nuclei (Fig. 2e, f). The fact that HSA19 is 25% more gene dense than the most dense mouse chromosome may account for this greater internalization. Another plausible explanation is that a large portion of HSA19 is absent in the A9+19 cells, thereby making it physically easier to move this smaller gene rich chromosome (19pter-19q11; 28.5 Mbp, 27.6 genes/Mbp) further internally. It is not simply a matter of size, however, as HSA7 (159 Mbp) is twice the size of HSA18 (76 Mbp) but they have a similar nuclear distribution towards the nuclear periphery. Thus chromosome size does not seem to serve as a general “determinant” of chromosome positioning in either human or mouse nuclei.
Mouse A9 fibroblast nuclei in our 3D FISH preparations are predominantly spherical with a volume nearly twice that of DLD-1 nuclei, which are flat-ellipsoidal. Notwithstanding these differences in nuclear shape, the relative positioning of the human chromosomes is maintained in both the mouse and human cells. This further suggests that chromosome positioning is independent of nuclear shape. It remains unclear how non-random chromosome positioning coordinates with transcriptional activity, and whether the transcriptome of a given cell type determines chromosome positioning or vice versa. In support of the latter, it has been demonstrated that disruption of Lamin function in mouse fibroblast cells (Lmnb1−/− and its endoproteolysis gene Rce1−/−) resulted in a relocalization of chromosome 18 away from the nuclear periphery. This had widespread effects on gene expression patterns, including the upregulation of a 4 Mbp region on chromosome MMU18 (Malhas, et al. 2007). Likewise, in cells derived from patients with laminopathies, a similar striking alteration in chromosome position was observed, supporting a role for lamins in controlling chromosome positioning and gene expression (Meaburn, et al. 2007).
Transcriptional activity of human chromosomes in mouse-human hybrid cells
Having previously found that introduction of an extra chromosome resulted in a significant increase in gene expression levels in DLD-1 derived cells, we were curious as to what extent a human chromosome would be transcriptionally active in the mouse nucleus. Various assays have previously demonstrated transcripts of human origin in mouse cells. For instance, expression of human specific isozymes was detected in A9 monochromosomal hybrid cells (Koi, et al. 1989). Mouse-human hybrid cells in which the entire mouse chromosomes were retained but the human chromosomes were selectively lost expressed human antigens on the cell surface (Weiss, et al. 1967). An in vitro assay system using MMCT was used to identify the imprinting status of various genes on human chromosomes and A9 monochromosomal hybrids appropriately maintained the parental expression pattern and methylation status of the human imprinted genes (Kugoh, et al. 1999; Inoue, et al. 2001). Mouse ES cells containing fragments of human chromosomes 2, 14 and 22 showed tissue specific expression of the human genes in chimeric mice, suggesting a whole chromosome might also exhibit expression in the mouse (Tomizuka, et al. 1997). MMCT that was used to generate a mouse model of Down syndrome recapitulated the phenotype, providing further evidence for transcriptional activity of human chromosomes in nuclei from different species (O’Doherty, et al. 2005).
In the present study, gene expression analyses of A9 hybrid cells unambiguously detected transcriptional activity originating from the human chromosomes. The “background” signal observed in the A9 cells in the absence of any human chromosomes (Fig. 3a) is the result of both the normalization process as well as cross hybridization of mouse mRNA to the oligonucleotides on the human expression array. This is perhaps not surprising given the high degree of sequence identity in coding regions between the two species. Against the backdrop of interspecies hybridization, we clearly determined that a significant fraction of genes were expressed from HSA7, HSA18 and HSA19. The specificity of these analyses was further substantiated by analyses of A9+19 cells in which only transcripts corresponding to genes residing on HSA19p were detected, consistent with subsequent FISH and aCGH analyses demonstrating that HSA19q was mostly deleted (Fig. 3 and Fig. 4). Thus, our gene expression analyses demonstrate for the first time that genes along the entire length of a human chromosome are transcriptionally active in the nucleus of another species (Fig. 3a). It was infeasible to establish whether the extent of transcription from each of the human chromosomes was similar in the mouse and human cells. As a result, it remains impossible to elucidate whether the correct positioning of each territory resulted in physiologic expression levels of the encoded genes. Thus, we could not assess the extent to which the level of transcription was related to chromosome positioning.
Species independent mechanism of non-random chromosome positioning
Various lines of evidence from studies on nuclei from humans and other primate species strongly suggest that the nuclear center is a particularly favorable milieu for placing gene dense chromosomes, while the nuclear periphery with its associated heterochromatin favors gene poor chromosomes (Craig, et al. 1997; Bolzer, et al. 2005; Neusser, et al. 2007). Heterochromatin cannot be the only determining factor, however, since even chromosomes found in the nuclear interior contain heterochromatic regions that are largely transcriptionally inactive. There is also a large body of evidence showing the relocalization of individual gene loci or entire domains to the periphery of their chromosome territory upon transcriptional activation (Lanctot, et al. 2007). Although these regions also move to a more internal nuclear position, a concomitant shift of the entire chromosome territory to a more central localization within the nucleus has not been described. One could speculate that the nuclear interior is a more favorable location for transcription, however there is currently no evidence that the transcription machinery is localized in a gradient with higher concentrations in the interior of the nucleus. Rather transcription factories appear to be randomly distributed throughout the nuclear volume (Spector 2003). The extent to which chromosome positioning and chromosome transcriptional activity are mechanistically coupled with one another remains elusive.
In total, the current evidence suggests that gene density is perhaps one of the most decisive factors in determining the non-random positioning of chromosomes within the nucleus. It is tempting to speculate that nuclei of eukaryotes are somehow endowed with the ability to quantify the gene density of each chromosome in order to distribute them non-randomly in the 3D nucleus. Our studies for the first time demonstrate the presence of a species independent mechanism responsible for arranging chromosomes at strikingly contrasting positions on the basis of gene density.
Acknowledgments
We thank Stephen Wincovitch and Amalia Dutra (NHGRI/NIH) for use of the confocal microscope and for valuable discussions. We thank Media Cybernetics for help with 3D measurements. Buddy Chen, Tom Ellerman & Joseph Cheng for IT support. We are grateful to Sudhir Varma, NCI, for critical comments and useful discussions on the manuscript. Hesed Padilla-Nash kindly provided the chromosome arm-specific painting probes. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Boyle S, Perry P, Bickmore WA. Scaffold attachments within the human genome. J Cell Sci. 1997;110(Pt 21):2673–2682. doi: 10.1242/jcs.110.21.2673. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I. Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res. 2001;9:569–584. doi: 10.1023/a:1012447318535. [DOI] [PubMed] [Google Scholar]
- Inoue J, Mitsuya K, Maegawa S, Kugoh H, Kadota M, Okamura D, Shinohara T, Nishihara S, Takehara S, Yamauchi K, Schulz TC, Oshimura M. Construction of 700 human/mouse A9 monochromosomal hybrids and analysis of imprinted genes on human chromosome 6. J Hum Genet. 2001;46:137–145. doi: 10.1007/s100380170101. [DOI] [PubMed] [Google Scholar]
- Koi M, Shimizu M, Morita H, Yamada H, Oshimura M. Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn J Cancer Res. 1989;80:413–418. doi: 10.1111/j.1349-7006.1989.tb02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugoh H, Mitsuya K, Meguro M, Shigenami K, Schulz TC, Oshimura M. Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res. 1999;6:165–172. doi: 10.1093/dnares/6.3.165. [DOI] [PubMed] [Google Scholar]
- Kupper K, Kolbl A, Biener D, Dittrich S, von Hase J, Thormeyer T, Fiegler H, Carter NP, Speicher MR, Cremer T, Cremer M. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol. 2007;176:593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005;6:44. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Jasencakova Z, Meister A, Schubert I, Zink D. Comparative analysis of the functional genome architecture of animal and plant cell nuclei. Chromosome Res. 2003;11:471–484. doi: 10.1023/a:1024978711705. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, Kill IR, Bridger JM. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Mora L, Sanchez I, Garcia M, Ponsa M. Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma. 2006;115:367–375. doi: 10.1007/s00412-006-0064-6. [DOI] [PubMed] [Google Scholar]
- Neusser M, Schubel V, Koch A, Cremer T, Muller S. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma. 2007;116:307–320. doi: 10.1007/s00412-007-0099-3. [DOI] [PubMed] [Google Scholar]
- O’Doherty A, Ruf S, Mulligan C, Hildreth V, Errington ML, Cooke S, Sesay A, Modino S, Vanes L, Hernandez D, Linehan JM, Sharpe PT, Brandner S, Bliss TV, Henderson DJ, Nizetic D, Tybulewicz VL, Fisher EM. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Nash HM, Heselmeyer-Haddad K, Wangsa D, Zhang H, Ghadimi BM, Macville M, Augustus M, Schrock E, Hilgenfeld E, Ried T. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. [DOI] [PubMed] [Google Scholar]
- Sengupta K, Upender MB, Barenboim-Stapleton L, Nguyen QT, Wincovitch SM, Sr, Garfield SH, Difilippantonio MJ, Ried T. Artificially introduced aneuploid chromosomes assume a conserved position in colon cancer cells. PLoS ONE. 2007;2:e199. doi: 10.1371/journal.pone.0000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- Tanabe H, Kupper K, Ishida T, Neusser M, Mizusawa H. Inter- and intra-specific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys. Cytogenet Genome Res. 2005;108:255–261. doi: 10.1159/000080824. [DOI] [PubMed] [Google Scholar]
- Tanabe H, Muller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci U S A. 2002;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Nakagawa Y, Minegishi D, Hashimoto K, Tanaka N, Oshimura M, Sofuni T, Mizusawa H. Human monochromosome hybrid cell panel characterized by FISH in the JCRB/HSRRB. Chromosome Res. 2000;8:319–334. doi: 10.1023/a:1009283529392. [DOI] [PubMed] [Google Scholar]
- Tomizuka K, Yoshida H, Uejima H, Kugoh H, Sato K, Ohguma A, Hayasaka M, Hanaoka K, Oshimura M, Ishida I. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat Genet. 1997;16:133–143. doi: 10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Rahn HP, Cardoso MC, Hemmerich P. Replication of centromeric heterochromatin in mouse fibroblasts takes place in early, middle, and late S phase. Histochem Cell Biol. 2006;125:91–102. doi: 10.1007/s00418-005-0063-3. [DOI] [PubMed] [Google Scholar]
- Weiss MC, Green H. Human-mouse hybrid cell lines containing partial complements of human chromosomes and functioning human genes. Proc Natl Acad Sci U S A. 1967;58:1104–1111. doi: 10.1073/pnas.58.3.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]