Abstract
The role of short-range hydrogen bond interactions at the interface between electron transfer proteins cytochrome c2 (cyt) and reaction center (RC) from Rb. sphaeroides was studied by mutation (to Ala) of RC residues Asn M187, Asn M188 and Gln L258 which form inter-protein hydrogen bonds to cyt in the cyt:RC complex. The largest decrease in binding constant KA (8-fold) for single mutation was observed for Asn M187, which forms an intra-protein hydrogen bond to the key residue Tyr L162 in the center of the contact region having low solvent accessibility. Interaction between Asn M187 and Tyr L162 was also implicated in binding by double mutation of the two residues. The hydrogen bond mutations did not significantly change the second order rate constant, k2 , indicating the mutations did not change the association rate for forming the cyt:RC complex, but increased the dissociation rate. The first order electron transfer rate, ke, for the cyt:RC complex was reduced by up to a factor of 4 (for Asn M187). The changes in ke were correlated with the changes in binding affinity but were not accompanied by increases in activation energy. We conclude that short-range hydrogen bond interactions contribute to the close packing of residues in the central contact region between the cyt and RC near Asn M187 and Tyr L162. The close packing contributes to fast electron transfer by increasing the electronic coupling and contributes to the binding energy holding the cyt in position for times long enough for electron transfer to occur.
Inter-protein electron transfer plays an important role in biological energy conversion in photosynthesis and respiration where mobile electron transfer proteins carry electrons between fixed membrane-bound protein complexes. (1) The reaction between electron transfer partners requires binding followed by electron transfer processes that are governed by protein-protein interactions at the contact interface. These interactions can influence both the binding energy and pathway for electron transfer. The role of long-range electrostatic interactions has been extensively documented. However, short-range van der Waals and hydrogen bond interactions between proteins have not been as well studied. In this work, we examine the role of short-range interactions due to hydrogen bonds on the binding and electron transfer between cytochrome c2 (cyt) and the reaction center (RC) from Rhodobacter sphaeroides.
The RC is a membrane bound protein responsible for the initial light induced electron transfer in photosynthesis (2). Light absorbed by the RC induces an electron transfer from a bacteriochlorphyll dimer, the primary electron donor D, through a series of intermediate acceptors to a quinone acceptor Q. Electrons from Q are passed through a cyclic electron transfer circuit involving cytochrome bc1 and return to the RC carried by a water soluble electron carrier, cytochrome c2. (3).
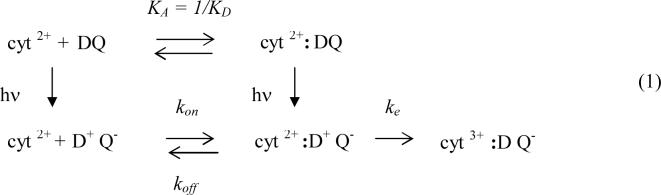
The reaction between cyt and RC has been extensively studied by pulsed laser kinetics experiments (4) (5-7) which allow the determination of microscopic rate constants. The simplest scheme for the light induced electron transfer reactions for RCs from Rb. sphaeroides is shown below.
Prior to illumination, the reduced cyt and RC are in equilibrium between a bound and free state as shown in the top line (association constant KA =1/KD∼106 M−1). Light absorption leads to electron transfer forming the oxidized donor/ reduced quinone acceptor D+Q− (bottom line). In the fraction of RCs having a bound cyt the first order electron transfer reaction occurs in the time scale of microseconds (ke ∼ 106 s−1) (4). This reaction is rate limited by electron tunneling as shown by its dependence on driving force. (8) For the fraction of RCs lacking bound cyt, the reaction occurs via a second order reaction k2. For the reaction scheme shown in eq (1) the value of k2 is governed by the equation
| (2) |
where kon and koff are the association and dissociation rates (1). At low ionic strength ke>>koff and k2 is in the diffusion limit k2 =kon ∼109 s−1M−1 (4). In this regime electron transfer is limited by the rate of protein association and independent of the electron transfer rate ke, consistent with the finding that k2 is independent of the driving force for electron transfer. (8) The two key functional features of the reaction is that the diffusion limited second order rate constant is fast (k2∼109 s−1M−1) and the dissociation rate of the cyt ( koff∼103 s−1) is fast enough not to be the rate limiting step in turnover. These features are well optimized for cyclic electron transfer.
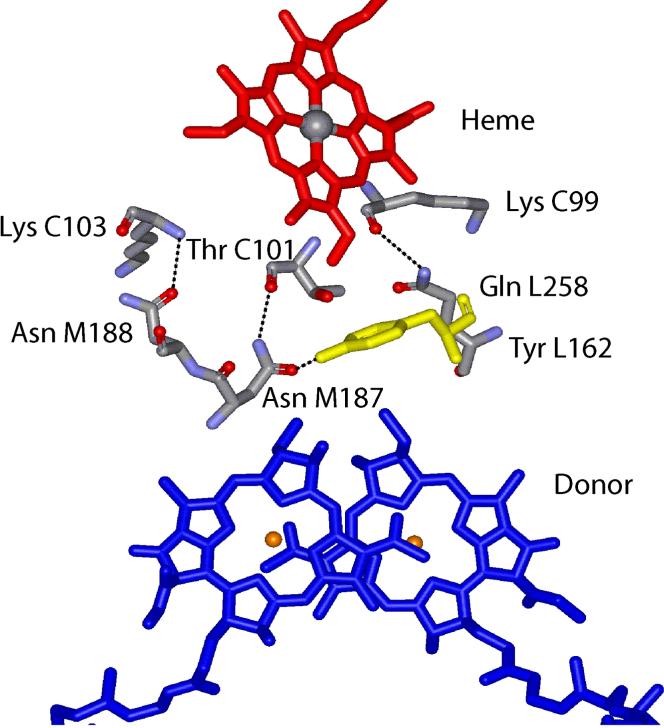
The molecular basis for these rates was revealed by the structure of the complex between cyt c2 and RC from Rb. sphaeroides determined by x-ray crystallography (9). Measurement of the electron transfer rate in the co-crystal gave the same value as in solution, ke =1.0×106 s−1, indicating that the cyt:RC complex in the crystal is in the active configuration. In the crystal structure, the cyt c2 is bound on the surface of the RC with the redox active cofactors in close proximity to each other - heme edge close to BChl2 (Figure 1). The interface region between the two proteins has a central short-range interaction domain with residues from the two proteins making van der Waals, cation-pi and hydrogen bond contacts. At the center of this domain is Tyr L162 on the RC which separates the two electron transfer cofactors (heme and BChl2) and is a hot spot for binding and electron transfer in Rb. sphaeroides (10) (11). This region is surrounded by an electrostatic domain of solvent separated charged residues which exhibit long-range electrostatic interactions.
Figure 1.
Structure of the cyt c2:RC complex (PDB ID 1L9J) showing the inter-protein hydrogen bonds (dotted lines) from three amide residues on the RC to backbone atoms on the bound cyt c2. The redox active heme cofactor (red) on the cyt and the BChl2 donor (blue) on the RC, buried in their respective proteins, are in close contact with the key RC residue Tyr L162 (yellow) in the docked structure. (9)
The role of charged residues in the interface has been established by the dependence of the electron transfer reaction rate k2 on ionic strength (12), chemical modification of charged residues (13), cross-linking (14, 15) and site directed mutagenesis of charged residues (7, 16, 17). Mutation of charged residues in the interface region changed KA and k2 but had little effect on ke. The effects on KA and k2 were explained quantitatively by electrostatic interactions important for docking the positively charged heme exposed surface of the cyt onto the negatively charged periplasmic side of the RC. (18) (19). The lack of affect on ke shows that the electron tunneling interactions in the bound state were not changed by modifying charged residues in the peripheral electrostatic domain of the contact interface.
The role of short range interactions was studied by mutation of hydrophobic residues (10) (11) and residues involved in the cation pi interaction (20). Mutation of hydrophobic residues changed both the KA and ke. The changes in ke show the importance of short range tunneling contacts in the central region for electron transfer, particularly those involving Tyr L162 consistent with theoretical studies of electron transfer (21-23). The effect of hydrophobic mutations on k2 was a function of the binding affinity of cyt c2 to the mutant RC (11). For small changes in binding, k2 was unchanged. However, if the binding affinity was reduced further, the value of k2 decreased greatly. The pattern of changes to k2 indicate that hydrophobic interactions do not alter kon but have their major effect on koff. The decrease in k2 results when koff > ke and dissociation occurs before electron transfer. The role of water molecules in the interface region in binding (24) (25) and electron transfer (23) has been studied.
What is the role of hydrogen bonding to the inter-protein electron transfer? An additional structural feature of the cyt:RC interface is the presence of inter-protein hydrogen bonds between three amide residues (Asn M187, Asn M188 and Gln L258) on the RC and backbone atoms on the cyt c2. (Figure 1). In addition to inter-protein hydrogen bonds an intra-protein hydrogen bond is formed from Asn M187 to the critical Tyr L162 residue. In order to determine the role of these hydrogen bond interactions in the inter-protein electron transfer process, the amide residues on the RC were mutated to Ala. In order to assess the role of the hydrogen bonding to Tyr L162 a double mutant having changes Tyr L162 -> Phe, and Asn M187 -> Ala was constructed to compare with the single mutant RCs. The effect of these mutations on binding and electron transfer was studied using pulsed laser kinetics to study the role of hydrogen bonding on the dynamics and energy landscape for inter-protein electron transfer. A preliminary report of this work has been presented (26).
MATERIALS AND METHODS
Site Directed Mutagenesis
The site directed mutations on the RC were constructed as previously described using the QuickChange Mutagenesis kit (Stratagene) and a Perkin Elmer PCR System (27). The mutation was confirmed by DNA sequencing
Protein Isolation and Purification
The bacteria harboring the modified RC gene was grown in the dark as described (27). RCs from Rb. sphaeroides carotenoidless strain R26 (native) and mutant strains were isolated in 15mM Tris-HCl pH 8, 0.025% lauryl dimethylamine-N-oxide (LDAO), 0.1mM EDTA following published procedures (27).. The final ratio of absorbance, A280/A800, was ≤ 1.5. The RC samples were then dialyzed for two days against HM (10mM Hepes pH 7.5, 0.04% dodecyl-β-D-maltoside (Anatrace)).Native and mutant cyt c2 proteins were isolated and purified as previously reported (28). Samples were purified to an absorbance ration A280/A412 ≤ 0.3.
Quinone/Quinol Preparations
Quinone (Q0) (2,3-dimethoxy-5-methylbenzoquinone) was obtained from Aldrich with ≥ 99% purity. Quinol (Q0H2) was synthesized by reducing quinone with hydrogen gas in the presence of platinum black also obtained from Aldrich.
Electron Transfer Measurements
Electron transfer kinetics between the RC and cyt c2 were measured by flash absorption spectroscopy monitoring changes at 865 nm as described (11). For measurements of ke in the microsecond range absorbance changes were also monitored at 600 nm (to minimize the flash artifact) and a pulsed zenon flash was used as the source for the measuring beam to improve the signal/noise ratio. In the latter case, measurements were corrected for the small decay of the flash intensity during the measurement period. Most measurements were conducted at 23° C in a buffer of 10 mM Hepes and 0.04% β-maltoside at pH 7.5. Samples included 50 μM Q0 and Q0H2 as a redox buffer to ensure that the cyt c2 was reduced prior to each laser flash. For measurements of the binding constant and second order rate constant, a low RC concentration (< 0.3 μM) was used to achieve a reasonably high fraction of unbound cyt and to avoid problems due to aggregation. For measurements of ke, higher RC concentrations (∼ 3μM) were used to increase the signal/noise ratio necessary to measure the fast electron transfer rates.
The binding affinity and second-order rate constants were determined by monitoring the absorbance changes due to oxidation of D to D+ at 865 nm. The association constant KA was determined by monitoring the absorbance changes of the fast and slow phases ΔAf and ΔAs fit to the relation
| (3) |
where ke is the fast rate constant associated with electron transfer from the bound cyt in the complex and ks is the rate constant for the slow phase of electron transfer due to the association of the free cyt with the RC. The values of KA were obtained from the fraction of bound cyt determined from the dependence of the amplitudes of the fast and slow phases on the concentration of free cyt.
| (4) |
The change in free energy ΔΔG due to mutation of a residue is calculated by
| (5) |
where kB is Boltzmann's constant and T is the absolute temperature. The second-order rate constant, k2, was determined from the observed rate of the slow phase of the electron transfer, kS, under conditions where the concentration of free cyt c2, was large compared to the unbound RC concentration, so that the reaction is pseudo first-order. The rate constant, ks
| (6) |
was plotted versus free cyt c2 concentration to obtain the second-order rate constant, k2.
Solvent Accessibility
The solvent accessible surface area (SASA) was determined for mutated residues in the structure of the cyt:RC complex using the GETAREA 1.1 program (29) with a rolling ball of radius 1.4Å.
RESULTS
Flash kinetic experiment
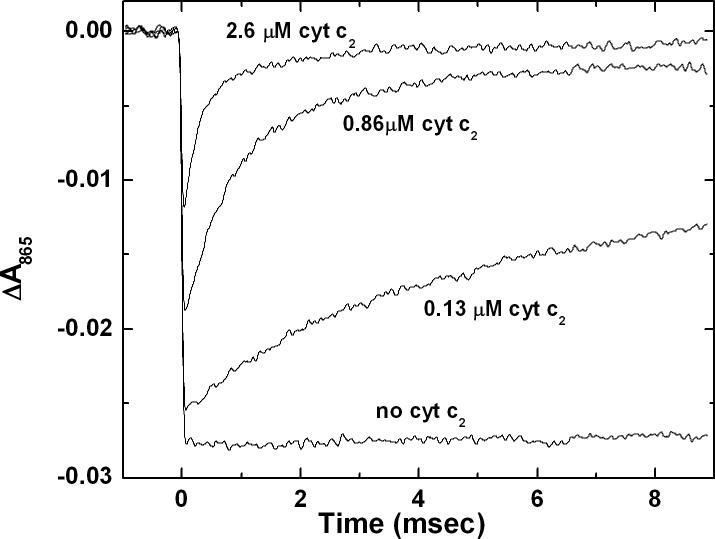
The cytochrome binding and second order rates of electron transfer were determined by laser kinetic experiments done at low RC concentrations (<0.3 μM) which reduced effects of aggregation. A typical measurement is shown in Figure 2 for the NA (M187) mutant. The electron transfer from the RC to cyt was monitored by the recovery of the flash induced absorbance at 865 nm. The reactions were biphasic. The fast phase is due to electron transfer from the fraction of RCs having a bound cyt (not resolved in Figure 2). The slow phase is due to the second order reaction of the fraction of RCs lacking a bound cyt. The relative proportion of fast and slow phases was dependent on the cyt concentration and was used to determine the binding constant, KA.
Figure 2.
Absorbance changes due to electron transfer from cyt c2 to NAM187 RC. In RC samples with no cyt c2, oxidation of D (monitored at 865 nm) is observed after a laser flash at t=0 with no recovery on this time scale. In the presence of cyt, two phases of recovery are observed: a fast phase near t=0, unresolved in these traces due to rapid electron transfer ( τ ∼ 4 μs) from bound cyt (See Fig. 4) and a slow phase due to second order electron transfer from unbound cyt. The fraction of fast component and the rate of the slow phase increase with increasing cyt concentration and were used to determine KA and k2. The RC concentration was 0.3 μM and the values for the free cyt are as indicated. (10 mM Hepes and 0.04% β-maltoside at pH 7.5, T=23 C)
Effect of mutations on the binding constant, KA
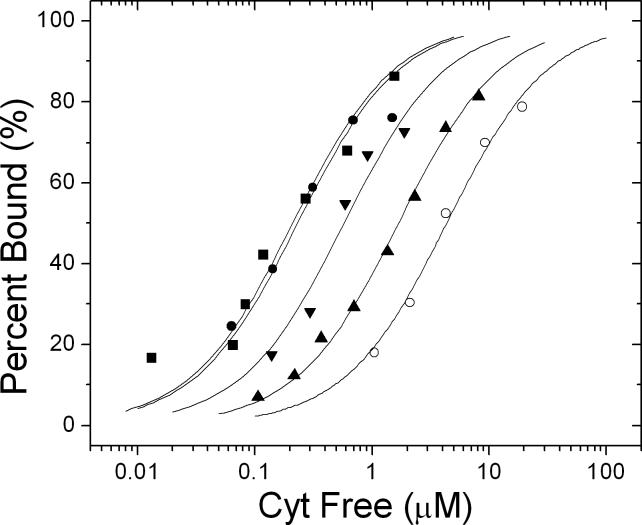
The effects of mutating each of the hydrogen bonding residues (Asn M188, Gln L258 and Asn M187) on the RC surface to Ala resulted in different changes in binding constants. (see Table 1) . The largest change for single mutations was observed for mutation of Asn M187 (factor of 8) (Figure 3). The triple H-bond mutant (NA(M187)/NA(M188)/QA(L258) having all three residues converted to Ala showed a larger decrease in KA (factor of 18). The sum of the changes in free energies ΔΔG for the three single mutants (69 meV) is within uncertainty the same as the change in free energy of the triple mutant (74 meV) indicating the additive effect of the individual mutations showing that these mutations act as independent contributions to the binding energy. The effects of the intra-protein hydrogen bond between Asn M187 and Tyr L162 was assessed by comparing the change in the binding energy of the double mutant YF(L162)/ NA(M187) to that of the sum changes for single mutants. The double mutant had a change in free energy (64 meV) which was much smaller than the sum of the changes in free energy for the single mutants (126 meV). This indicates that the two residues do not bind independently to the cyt and suggest a role for an intra-protein hydrogen bond between Tyr L162 and Asn M187 in the binding process.
Table 1.
Binding Constants, Free Energy Changes, Electron Transfer Rates and Activation Energies for Native and Mutant RCs.a and Solvent Accessible Surface Area of the Mutated Residueb.
| RC strain | KA | ΔΔGB | ke | Ea | k2 | SASAb |
|---|---|---|---|---|---|---|
| (μM−1) | (meV) | (106 s−1) | (meV) | (109 s−1M−1) | A2 | |
| Native | 4.0 | 0 | 1.00 | 65 | 1.7 | - |
| NA(M188) | 5.0 | −6 | 0.93 | 21 | 2.0 | 23 |
| QA(L258) | 1.7 | 22 | 0.82 | 5 | 1.4 | 14 |
| NA(M187) | 0.5 | 53 | 0.26 | 21 | 2.0 | 9 |
| Triple H-bond | 0.22 | 74 | 0.10 | 35 | 1.5 | - |
| YF(L162) | 0.23 | 73 | 0.25 | - | 0.8 | - |
| YF(L162)/NA(M187) | 0.33 | 64 | 0.50 | - | 1.2 | - |
KA is the binding constant, ke the first order electron transfer rate constant, k2 the second order electron transfer rate constant, ΔΔGB the change in the binding free energy with respect to the native RC, Ea the activation energy for the first order electron transfer rate
SASA is the solvent accessible surface area of the mutated amino acid side chain in the Native cyt:RC complex. Statistical uncertainties: measure rate constants are ± 10%, KA ± 10%, Ea ± 20meV, ΔΔGB ± 4 meV,.
Figure 3.
Plot of the fraction of RCs with bound cyt c2, versus the free cyt c2 concentration for native and mutant RCs. The symbols for different RCs are: native (●), NA(M188) (■), QA(L258) (▼), NA(M187) (▲) and the triple mutant (o). The curves are the fits of equation 3 to the measured data for each RC. The values determined for KA are shown in Table 1. The triple mutant has the largest decrease binding affinity. (Conditions: 0.2 μM RC in 10 mM Hepes and 0.04% β-maltoside at pH 7.5.)
Effect of mutations on the second order rate constant, k2
The rate of the slow phase of the electron transfer reaction shown in Figure 2 is proportional to the free cyt concentration and was used to determine the second order rate constant. The second order electron transfer rates were found to be relatively unchanged due to mutation of the hydrogen bonding residues. (see Table 1). This is similar to the effect of mutating hydrophobic residues and can be explained if the short range interactions are not yet formed in the transition state for rate limiting protein association. (11).
Effect of hydrogen bond mutations on the electron transfer rate in the bound state, ke
The fast kinetic data for Native RC and NA M187 RC at high cyt concentration (100 μM) monitored at 600 nm are shown in Figures 4a and 4b. The NA (M187) RCs exhibited a significantly slower electron transfer rate than Native RC. The decays for both traces could be well fit by two exponential rate constants. For NA(M187) RC the fast decaying major component (∼80%) was assigned to the electron transfer rate in the bound state, ke = 2.5 ×105 s−1, a reduction of 4-fold compared to native. In addition, a small amount of slow component (∼20%) was required to fit the data, ks = 5×104 s−1. The fast rates were confirmed by taking data on multiple samples at both 600nm and 865nm. The triple H-bond mutant showed the greatest reduction in ke (10 fold), and the NAM188 and QAL258 exhibited a fast rate very similar to Native RCs. The results are shown in Table 1.
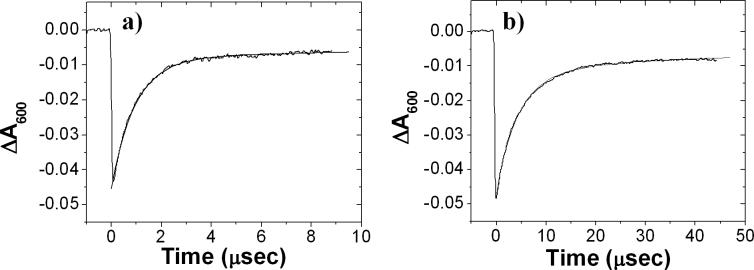
Figure 4.
Absorbance changes due to electron transfer monitored at 600nm for a) native and b) NA(M187) RCs in the presence of high concentration of cyt c2. The electron transfer rate in the mutant RCs were slower than in Native RCs. (Note the difference in time scales). The native RC = 3.5 μM and cyt = 70μM. The NAM187 RC = 4μM and cyt = 100μM. The data were fit with equation 3. For native RCs, ke = 106 s−1(75%), ks = 3×104 s−1 (25%) and for NA(M187) RCs, ke = 0.26×106 s−1 (80%), ks =5×104 s−1(20%). (10 mM Hepes and 0.04% β-maltoside at pH 7.5, T=26 C)
The slow phase could be due to a small amount of a second order phase or to heterogeneity. To test these ideas, the electron transfer rates were studied as a function of cyt concentration. The rate constant for the fast phase was independent of the cyt concentration confirming the assignment to ke. (see supplementary Figure S1) The rate constant for the slow phase was concentration dependent and gave a kobs ∼3×104 s−1 at a cyt concentration of 100 μM (see supplementary Figure S1). This value is in the range of the observed values of ks = 2−5×104 s−1 seen in Figure 4. These results suggest that the slow phase at high cyt concentration may be due to a small amount of second order reaction.
The lack of saturation of the binding site even at high cyt concentration may be due to aggregation of RC at the higher concentration of RCs necessary for fast electron transfer measurements. This aggregation may also account for the observation that full recovery of the signal to the baseline is not observed in some samples. (e.g. Figure 4b). Evidence for aggregation comes from the finding that the amount of cyt needed to achieve half saturation was greater at the high RC concentration needed to measure the fast kinetics than at low concentrations used in the binding measurements (Supplementary Figure S2) This concentration dependence varied for different preparations of RC and is attributed to an aggregation of RCs that alters the cyt binding. Similar reports of aggregation of RCs have been described by Tiede. (30)
Temperature dependence of ke
The activation energy, Ea, of ke observed in native and hydrogen bond mutant RCs were determined by measuring the temperature dependence of the fast rate constant. As the temperature was lowered, the kinetic traces showed an increased amount of a slow phase. (Figure 5). This result is due to a reduced binding of the cyt at lower temperature as shown by measurements made at lower RC and cyt concentration (data not shown). The result indicate that the enthalpy for binding is positive (ΔH = +54 ± 5 meV for native RC, [RC] =0.3 μM ) and the binding is driven by entropy.
Figure 5.
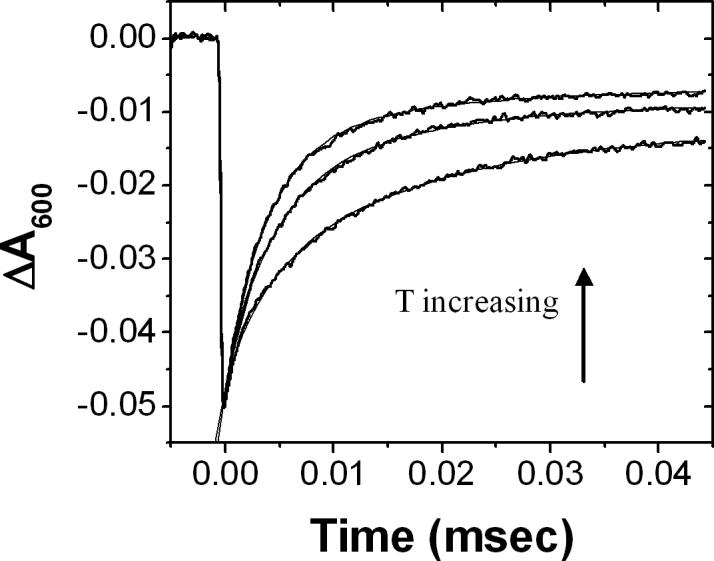
Temperature dependence of fast electron transfer between cyt and NA M187 RC. The data is shown for the electron transfer at T= 1, 18, and 36 °C (increasing from bottom to top). The decrease in rate at low temperature is due mainly to an decrease in the fraction of RCs with bound cyt. The rate of the fast component was relatively temperature independent. (Figure 6) Other conditions same as for Figure 4.
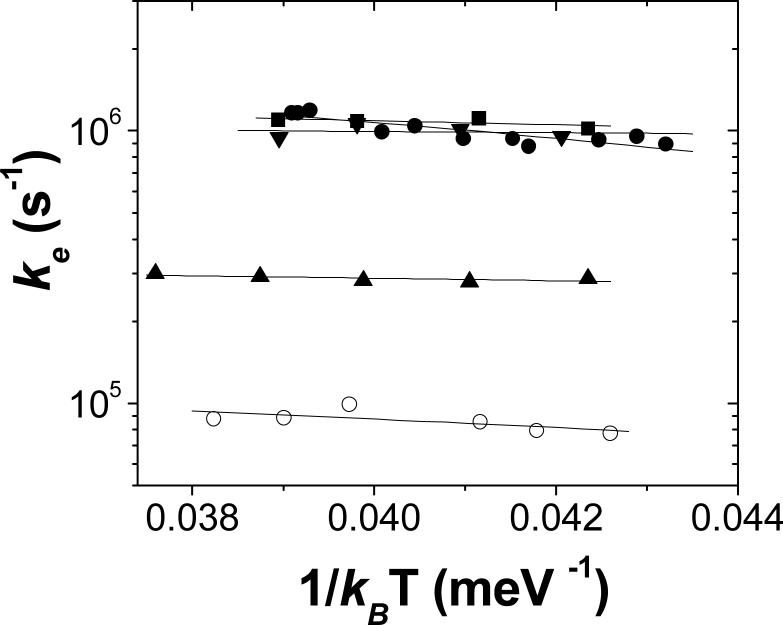
After correcting for the contribution due to the slow component, the values for ke at different temperatures were found to be relatively temperature independent. The activation energies, Ea, were calculated using the Arrhenius equation. ke = koexp-(Ea/kBT) (see Figure 6). The values for the activation energies for ke were all relatively low and were not increased by mutation of hydrogen bonds. (Table 1). The activation energy for Native RCs found in the present study (Ea = 65±12 meV ) is in good agreement with the activation energy Ea = 58 meV computed using the Marcus equation (31)
| (7) |
using the values of λ= 500 meV and ΔGe = −160 meV (8). The activation energy measured here is lower than that obtained previously (32) under different conditions, 60% glycerol, where a larger reorganization energy was measured (λ = 1000 meV) (33). A preliminary report (34) indicating a larger activation energies for mutant RCs was in error, due to contributions from the second order phase to the fast electron transfer reaction.
Figure 6.
Plot of ke versus 1/kBT for native and mutant RCs. The symbols for different RCs are: native (●), NA(M188) (■), QA(L258) (▼), NA(M187) (▲) and the triple mutant (o). The slope of the line fit to the data were used to obtain the activation energy Ea for electron transfer (Table 1). (Conditions: 3 μM RC in 10 mM Hepes and 0.04% β-maltoside at pH 7.5.)
DISCUSSION
In this study three residues on the RC surface that make inter-protein H-bonds to cyt in the cyt:RC complex were mutated to Ala. The mutations were found to make appreciable changes in the binding affinity KA indicating a role of hydrogen bonds in binding. Although the mutations did not appreciably change the second order rate constant k2, the rate of electron transfer between the two proteins in the bound state, ke, decreased in parallel with decreasing binding affinity, indicating that the pathway for electron transfer has been altered by removing hydrogen bonds.
Effects on Binding. KA
The effects of mutating hydrogen bonded residues resulted in a range of changes to the binding free energy (Table 1). The residue whose mutation produces the largest change in binding energy is Asn M187. This residue is unique is several respects. It is located near the center of the small cluster of residues that make hydrophobic contacts with the cyt (9). As a result, Asn M187 has a low solvent accessibility (SASA = 9 Å2 ) in the cyt:RC complex (Table 1). In contrast, the other two residues are at the outer edge of the short-range contact domain and are much more solvent exposed (SASA =14 and 23 Å2 ). The changes in binding energy ΔΔG are correlated with solvent accessibility (Table 1). This is consistent with the proposal that mutations of residues in solvent accessible regions may be compensated by interactions with solvent molecules with relatively little cost in energy while mutation of deeply buried residues necessarily entails a larger energy cost. (35) (36). In addition, mutation of Asn M187 results in the loss of the intra-protein hydrogen bond to Tyr L162 (Figure 1) which may allow the phenolic ring to assume alternate configurations which could hinder cytochrome binding. The mobility of the aromatic residue may also account for the decreased binding of the YF(L162) mutant in which Phe replaces Tyr. The importance of the intra-protein hydrogen bond between Asn M187 and Tyr L162 is indicated by the finding that the change in binding energy for the double mutant YF(L162)/NA(M187) was less than the sum of energies for the single mutants (Table 1) This indicates that part of the effect of mutating Asn M187 is due to the steric interference with binding due to increased mobility of the aromatic group.
In summary, the explanation for the large effect on binding due to mutation of Asn M187 is the energetic consequence of disruption of both intra and inter-protein close packed contacts in the short-range interaction domain. The high specificity of the H-bond both in bond distance and angle strongly restricts the structures capable of H-bond formation. The finding that H-bonds are involved in binding in the cyt:RC complex argues for the importance of a specific bound complex optimized for electron transfer.
Effects on the second order rate constant, k2
The second order rate constant governs the maximum rate of turnover and can thus be viewed as controlling electron transfer in the biological system. The mutations of residues involved in H-bonding interactions do not appreciably change the second order rate constant, k2 (Table 1) indicating that these mutation have little effect on kon. This is consistent with the model for the reaction in which the association rate kon is the rate limiting step and the transition state is one in which the cyt c2 is located relatively far (∼10Å) from the position in the bound state. Thus in the transition state, the short range interactions such as H-bonds have not yet been formed (19).
The main effect of short range interactions such as hydrogen bonds is to decrease the dissociation rate, koff. For small changes in koff , the observed value of k2 remains insensitive to changes in koff as long as koff < ke. However, when koff > ke , as has been found for some of the more disruptive hydrophobic mutants (11), the second order rate goes to a fast exchange regime in which dissociation occurs before electron transfer and the observed value of k2 depends on the fraction of RC having a bound cyt in steady state equilibrium, i.e. k2 = keKA. Thus, short-range interactions such as H-bonds and hydrophobic interactions play an important role in electron transfer by retaining the cyt in the bound active configuration such that electron transfer occurs faster than dissociation. These features provide a measure of robustness to the inter-protein electron transfer process.
Effects on the first order electron transfer rate, ke
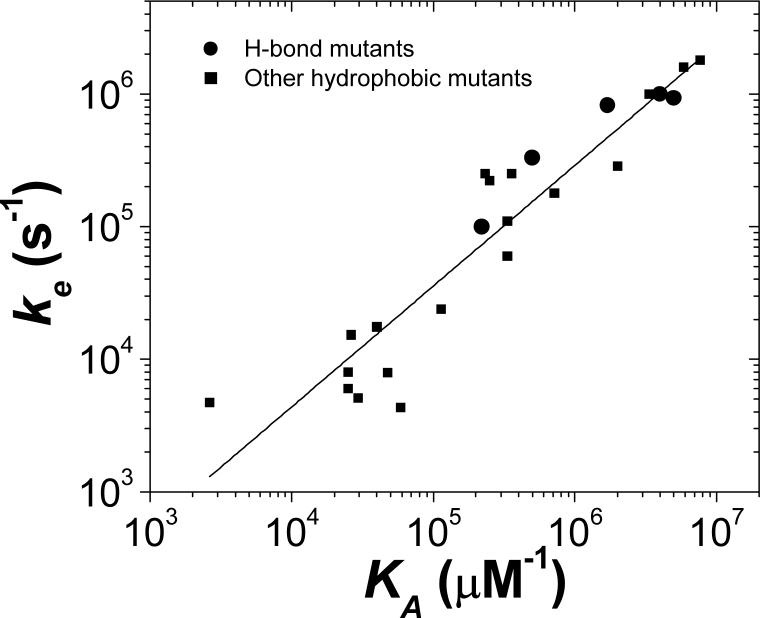
The changes in ke for different mutant RCs were roughly proportional to changes in KA (see Table 1). These parallel changes in ke and KA due to mutation of hydrogen bonding residues are similar to the changes previously observed due to mutation of hydrophobic residues involved binding at the cyt:RC interface (11) The values of ke and KA resulting from mutation of hydrogen bonding residues from the present study are shown in Figure 7 together with the values resulting from mutation of hydrophobic residues.
Figure 7.
The cyt c2 :RC first order electron transfer rate constant ke versus the binding constant KA for native and mutant RCs. The rates for H-bond mutants determined in this study (●) are plotted with rates previously determined (■) for hydrophobic mutant RCs (11). The H-bond mutants follow the same general trend in which changes in ke are roughly proportional to changes in KA
The results indicate that the changes in ke and KA due to mutation of residues involved in short range interactions are strongly correlated. How can this close correlation be explained? For conventional long distance electron tunneling, the rate is proportional to the electronic coupling V and Franck-Condon factor (FC).
| (8) |
V decreases exponentially with distance (37) (38) while the main contribution to FC is exp-(Ea/kBT) where Ea varies with the free energy change upon electron transfer ΔGe and the reorganization energy λ as given in equation (7). (31). For changes to short-range interactions, i.e. hydrogen bonds and hydrophobic interactions, in the interface region of the cyt RC complex it is likely that the donor-acceptor distances would be altered. On the other hand since the surface residues are not in direct contact with the cofactors, the values of ΔGe and λ (and thus FC) are not expected to change. A simple model to explain the slower rates of electron transfer is that the donor-acceptor distance changes due to mutation. Such a change in distance would also affect the binding constant. However, there is no a priori reason to expect such a strong correlation electron transfer rate and binding constant over a wide range of mutations over an ∼20-fold change in the rate of electron transfer.
An alternative model to consider is a dynamic docking model in which the cyt binds in several positions on the RC surface each with different activity for electron transfer which are in dynamic equilibrium on the time scale of electron transfer as has been proposed for docking of a cytochrome (b5) and myoglobin (39). One interesting possibility to explain the lower rate of electron transfer is a dynamic docking model in which the conformation active in electron transfer has a higher energy than the conformation inactive in electron transfer. In this case a lower rate of electron transfer would be associated with a larger activation energy. The results (Table 1) for the hydrogen bonding mutants indicate that this is not the case and in fact for all mutant RCs the activation energies are similar to and even slightly lower than that of the native RC. We thus conclude that the lower rate of electron transfer in the hydrogen bond mutants occurs from the lowest energy conformation of the cyt:RC complex. The low activation energies also rule out the possibility that the slower rates are due to changes in driving force or reorganization energy. Thus, the slower rates in the mutant RCs are due to smaller electronic coupling interaction V between heme and BChl2 in the cyt:RC complex.
The correlation between the electron transfer rate and binding changes remains to be explained quantitatively at the molecular level. Possible approaches to gain insight include detailed pathway calculations using static or dynamic models. The binding energy should include contributions from short range interaction, electrostatic interactions and desolvation (18). The rate may be influenced by electron transfer through solvent water at the interface (23) (40) or the dynamics of conformational inter-conversion in a reaction-diffusion model (41). Further studies of this problem may provide insights on the structure and dynamics of short range protein-protein interactions.
CONCLUSIONS
Mutation of hydrogen bonded residues, particularly Asn M187 which is closely associated with Tyr L162, in the interface region in the cyt:RC complex shows that these residues play a role in binding of the cyt to the RC in a position for fast electron transfer. Hydrogen bonding facilitates the close contact and tight packing at the interface leading to strong electronic coupling resulting in fast electron transfer. The hydrogen bonds do not greatly affect the rate of association of the cyt on to the RC but play a significant role in decreasing the dissociation rate and thus allow the cyt to be bound long enough for electron transfer in the complex to occur. These results show the importance of short range interactions in the inter-protein electron transfer process.
Supplementary Material
Supplementary Figure S1 Biphasic electron transfer rates vs cyt c2 concentration for NA(M187) (black) and triple mutant (magenta) RCs. The fast rates (triangles) were independent of free cyt concentration and are assigned to the first order rate, ke . Average values of ke = 8.6×104 s(−1) and 2.3×105 s(−1) were found for NA(M187) and triple mutant respectively. The slow rates (circles) are proportional to cyt concentration and represent a second order reaction. The RC concentration was ∼3 μM. (10 mM Hepes and 0.04% β-maltoside at pH 7.5)
Supplementary Figure S2. KD as function of RC concentration. The value of the cyt concentration needed for half saturation KD increases as a function of the RC concentration. This result varied for different preparations of RCs and is attributed to an aggregation effect. (10 mM Hepes and 0.04% β-maltoside at pH 7.5)
ACKNOWLEDGMENTS
DNA sequencing was performed by the DNA Sequencing Shared Resource, UCSD Cancer Center, which is funded in part by NCI Cancer Center Support Grant # 2 P30 CA23100−23.
Abbreviations
- cyt c2
cytochrome c2
- RC
reaction center
- Rb
Rhodobacter
- Q0
2,3-dimethoxy-5-methylbenzoquinone
- D
primary electron donor (bacteriochlorophyll dimer)
- D+
photo-oxidized primary electron donor
- ke
first order electron transfer rate
- k2
second order rate constant
- kon
association rate
- koff
dissociation rate
- KA
binding constant
- KD
dissociation constant
- Ea
activation energy
- ΔΔG
change in binding free energy
- SASA
solvent accessible surface area
Footnotes
This work was supported by NIH grant GM 41637.
SUPPORTING INFORMATION AVAILABLE
Figure S1 Biphasic electron transfer rates vs cyt c2 concentration for NA(M187) and triple mutant RCs.
Figure S2. KD as function of RC concentration. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Bendall D. In: Protein electron transfer. Bendall D, editor. Bios Scientific Publishers Ltd; Oxford, UK: 1996. pp. 43–68. [Google Scholar]
- 2.Blankenship RE. Molecular mechanisms of photosynthesis. Blackwell Science Inc; London: 2002. [Google Scholar]
- 3.Dutton PL, Prince RC. In: The photosynthetic bacteria. Clayton RC, W. R. S., editors. Plenum Press; New York: 1978. pp. 525–570. [Google Scholar]
- 4.Tiede D, Dutton P. In: The photosynthetic reaction center. Deisenhofer J, Norris J, editors. Academic Press; San Diego: 1993. pp. 258–288. [Google Scholar]
- 5.Overfield RE, Wraight CA, Devault DC. Microsecond photooxidation kinetics of cytochrome c2 from rhodopseudomonas sphaeroides: In vivo and solution studies. FEBS Lett. 1979;105:137–142. doi: 10.1016/0014-5793(79)80903-5. [DOI] [PubMed] [Google Scholar]
- 6.Moser C, Dutton PL. Cytochrome c and c2 binding dynamics and electron transfer with photosynthetic reaction center protein and other integral membrane redox proteins. Biochemistry. 1988;27:2450–2461. doi: 10.1021/bi00407a031. [DOI] [PubMed] [Google Scholar]
- 7.Tetreault M, Rongey SH, Feher G, Okamura M. Interaction between cytochrome c2 and the photosynthetic reaction center from rhodobacter sphaeroides: Effects of charge-modifying mutations on binding and electron transfer. Biochemistry. 2001;40:8452–8462. doi: 10.1021/bi010222p. [DOI] [PubMed] [Google Scholar]
- 8.Lin X, Williams JC, Allen J, Mathis P. Relationship between rate and free energy difference for electron transfer from cytochrome c2 to the reaction center in rhodobacter sphaeroides. Biochemistry. 1994;33:13517–23. doi: 10.1021/bi00250a002. [DOI] [PubMed] [Google Scholar]
- 9.Axelrod HL, Abresch EC, Okamura MY, Yeh AP, Rees DC, Feher G. X-ray structure determination of the cytochrome c2:Reaction center electron transfer complex from rhodobacter sphaeroides. J.Mol. Biol. 2002;319:501–515. doi: 10.1016/S0022-2836(02)00168-7. [DOI] [PubMed] [Google Scholar]
- 10.Farchaus J, Wachtveitl J, Mathis P, Oesterhelt D. Tyrosine 162 of the photosynthetic reaction center l-subunit plays a critical role in the cytochrome c2 mediated rereduction of the photooxidized bacteriochlorophyll dimer in rhodobacter sphaeroides. 1. Site-directed mutagenesis and initial characterization. Biochemistry. 1993;32:10885–93. doi: 10.1021/bi00091a044. [DOI] [PubMed] [Google Scholar]
- 11.Gong X, Paddock M, Okamura M. Interactions between cytochrome c2 and photosynthetic reaction center from rhodobacter sphaeroides: Changes in binding affinity and electron transfer rate due to mutation of interfacial hydrophobic residues are strongly correlated. Biochemistry. 2003;42:14492–500. doi: 10.1021/bi035603c. [DOI] [PubMed] [Google Scholar]
- 12.Prince RC, Cogdell RJ, Crofts AR. The photo-oxidation of horse heart cytochrome c and native cytochrome c2 by reaction centres from rhodopseudomonas spheroides r-26. Biochim. Biophys. Acta. 1974;347:1–13. doi: 10.1016/0005-2728(74)90194-7. [DOI] [PubMed] [Google Scholar]
- 13.Long J, Durham B, Okamura M, Millett F. Role of specific lysine residues in binding cytochrome c2 to the rhodobacter sphaeroides reaction center in optimal orientation for rapid electron transfer. Biochemistry. 1989;28:6970–6974. doi: 10.1021/bi00443a029. [DOI] [PubMed] [Google Scholar]
- 14.Drepper F, Dorlet P, Mathis P. Cross-linked electron transfer complex between cytochrome c2 and the photosynthetic reaction center of rhodobacter sphaeroides. Biochemistry. 1997;36:1418–27. doi: 10.1021/bi961350u. [DOI] [PubMed] [Google Scholar]
- 15.Rosen D, Okamura MY, Abresch EC, Valkirs GE, Feher G. Interaction of cytochrome c with reaction centers of rhodopseudomonas sphaeroides r-26: Localization of the binding site by chemical cross-linking and immunochemical studies. Biochemistry. 1983;22:335. doi: 10.1021/bi00271a016. [DOI] [PubMed] [Google Scholar]
- 16.Caffrey MS, Bartsch RG, Cusanovich MA. Study of the cytochrome c2-reaction center interaction by site-directed mutagenesis. J. Biol. Chem. 1992;267:6317–6321. [PubMed] [Google Scholar]
- 17.Tetreault M, Cusanovich M, Meyer T, Axelrod H, Okamura M. Double mutant studies identify electrostatic interactions that are important for docking cytochrome c2 onto the bacterial reaction center. Biochemistry. 2002;41:5807–15. doi: 10.1021/bi012053e. [DOI] [PubMed] [Google Scholar]
- 18.Miyashita O, Onuchic JN, Okamura MY. Continuum electrostatic model for the binding of cytochrome c2 to the photosynthetic reaction center from rhodobacter sphaeroides. Biochemistry. 2003;42:11651–11660. doi: 10.1021/bi0350250. [DOI] [PubMed] [Google Scholar]
- 19.Miyashita O, Onuchic J, Okamura M. Transition state and encounter complex for fast association of cytochrome c2 with bacterial reaction center. Proc Natl Acad Sci U S A. 2004;101:16174–9. doi: 10.1073/pnas.0405745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paddock M, Weber K, Chang C, Okamura M. Interactions between cytochrome c2 and the photosynthetic reaction center from rhodobacter sphaeroides: The cation-pi interaction. Biochemistry. 2005;44:9619–25. doi: 10.1021/bi050651d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aquino A, Beroza P, Beretan D, Onuchic J. Docking and electron transfer between cytochrome c2 and the photosynthetic reaction center. Chemical Physics. 1995;197:277–288. [Google Scholar]
- 22.Miyashita O, Okamura MY, Onuchic JN. Theoretical understanding of the interprotein electron transfer between cytochrome c2 and the photosynthetic reaction center. Journal of Physical Chemistry B. 2003;107:1230–1241. [Google Scholar]
- 23.Miyashita O, Okamura M, Onuchic J. Interprotein electron transfer from cytochrome c(2) to photosynthetic reaction center: Tunneling across an aqueous interface. Proc Natl Acad Sci USA. 2005;102:3558–3563. doi: 10.1073/pnas.0409600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogorelov T, Autenrieth F, Roberts E, Luthey-Schulten Z. Cytochrome c(2) exit strategy: Dissociation studies and evolutionary implications. J. Phys. Chem. B. 2007;111:618–34. doi: 10.1021/jp064973i. [DOI] [PubMed] [Google Scholar]
- 25.Autenrieth F, Tajkhorshid E, Schulten K, Luthey-Schulten Z. Role of water in transient cytochrome c2 docking. J. Phys. Chem. B. 2004;108:20376–87. [Google Scholar]
- 26.Abresch E, Villalobos M, Paddock M, Chang C, Okamura M. The importance of buried h-bonds on binding and electron transfer in the cytochrome c2:Reaction center complex. 2006 Biophysical Society Annual Meeting Abstract, Biophysical J. 2006;90 770-Plat. [Google Scholar]
- 27.Paddock ML, Adelroth P, Chang C, Abresch EC, Feher G, Okamura MY. Identification of the proton pathway in bacterial reaction centers: Cooperation between asp-m17 and asp-l210 facilitates proton transfer to the secondary quinone (q(b)) Biochemistry. 2001;40:6893–6902. doi: 10.1021/bi010280a. [DOI] [PubMed] [Google Scholar]
- 28.Bartsch R. In: The photosynthetic bacteria. Clayton R, Sistrom W, editors. Plenum Press; New York: 1978. pp. 249–279. [Google Scholar]
- 29.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 1998;19:319–333. [Google Scholar]
- 30.Tiede DM, Littrell K, Marone PA, Zhang R, Thiyagarajan P. Solution structure of a biological bimolecular electron transfer complex: Characterization of the photosynthetic reaction center-cytochrome c2 protein complex by small angle neutron scattering. J. Appl. Cryst. 2000;33:560–564. [Google Scholar]
- 31.Marcus RA, Sutin N. Electron transfer in chemistry and biology. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- 32.Venturoli G, Mallardi A, Mathis P. Electron transfer from cytochrome c2 to the primary donor of rhodobacter sphaeroides reaction centers. A temperature dependence study. Biochemistry. 1993;32:13245–53. doi: 10.1021/bi00211a037. [DOI] [PubMed] [Google Scholar]
- 33.Venturoli G, Drepper F, Williams J, Allen J, Lin X, Mathis P. Effects of temperature and deltago on electron transfer from cytochrome c2 to the photosynthetic reaction center of the purple bacterium rhodobacter sphaeroides. Biophysical Journal. 1998;74:3226–40. doi: 10.1016/s0006-3495(98)78029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abresch E, Paddock M, Okamura MY. Inter-protein electron transfer from cytochrome c2 to reaction center: Electron transfer in the bound complex reflects dynamic conformational fluctuations. 2007 Biophysical Society Meeting Abstracts,Biophys J. 2007:503a. [Google Scholar]
- 35.Bogan A, Thorn K. Anatomy of hot spots in protein interfaces. Journal of Molecular Biology. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 36.Lo Conte L, Chothia C, Janin J. The atomic structures of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 37.Moser CC, Keske JM, Warnke K, Farid RS, Dutton PL. The nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 38.Beratan D, Betts J, Onuchic J. Protein electron transfer rates set by the bridging secondary and tertiary structure. Science. 1991;252:1285–1288. doi: 10.1126/science.1656523. [DOI] [PubMed] [Google Scholar]
- 39.Liang ZX, Kurnikov IV, Nocek JM, Mauk AG, Beratan DN, Hoffman BM. Dynamic docking and electron-transfer between cytochrome b5 and a suite of myoglobin surface-charge mutants. Introduction of a functional-docking algorithm for protein-protein complexes. J Am Chem Soc. 2004;126:2785–98. doi: 10.1021/ja038163l. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Balabin I, Beratan D. The nature of aqueous tunneling pathways between electron-transfer proteins. Science. 2005;310:1311–3. doi: 10.1126/science.1118316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Lin S, Allen J, Williams J, Blankert S, Laser C, Woodbury N. Protein dynamics control the kinetics of initial electron transfer in photosynthesis. Science. 2007;316:747–50. doi: 10.1126/science.1140030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Biphasic electron transfer rates vs cyt c2 concentration for NA(M187) (black) and triple mutant (magenta) RCs. The fast rates (triangles) were independent of free cyt concentration and are assigned to the first order rate, ke . Average values of ke = 8.6×104 s(−1) and 2.3×105 s(−1) were found for NA(M187) and triple mutant respectively. The slow rates (circles) are proportional to cyt concentration and represent a second order reaction. The RC concentration was ∼3 μM. (10 mM Hepes and 0.04% β-maltoside at pH 7.5)
Supplementary Figure S2. KD as function of RC concentration. The value of the cyt concentration needed for half saturation KD increases as a function of the RC concentration. This result varied for different preparations of RCs and is attributed to an aggregation effect. (10 mM Hepes and 0.04% β-maltoside at pH 7.5)