Abstract
The disulfide bond between Cys14 and Cys38 of bovine pancreatic trypsin inhibitor (BPTI) lies on the surface of the inhibitor and forms part of the protease binding region. The functional properties of three variants lacking this disulfide, with one or both of the Cys residues replaced with Ser, were examined, and x-ray crystal structures of the complexes with bovine trypsin were determined and refined to the 1.58 Å resolution limit. The crystal structure of the complex formed with the mutant with both Cys residues replaced was nearly identical to that of the complex containing the wild-type protein, with the Ser oxygen atoms positioned to replace the disulfide bond with a hydrogen bond. The two structures of the complexes with single replacements displayed small local perturbations with alternate conformations of the Ser side chains. Despite the absence of the disulfide bond, the crystallographic temperature factors show no evidence of increased flexibility in the complexes with the mutant inhibitors. All three of the variants were cleaved by trypsin more rapidly than the wild-type inhibitor, by as much as 10,000-fold, indicating that the covalent constraint normally imposed by the disulfide contributes to the remarkable resistance to hydrolysis displayed by the wild-type protein. The rates of hydrolysis display an unusual dependence on pH over the range from 3.5 to 8, decreasing at the more alkaline values, as compared to the increased hydrolysis rates for normal substrates under these conditions. These observations can be accounted for by a model for inhibition in which an acyl-enzyme intermediate forms at a significant rate but is rapidly converted back to the enzyme-inhibitor complex by nucleophilic attack by the newly created amino group. The model suggests that a lack of flexibility in the acyl-enzyme intermediate, rather than the enzyme-inhibitor complex, may be a key factor in the ability of BPTI and similar inhibitors to resist hydrolysis.
Keywords: Standard-mechanism protease inhibitors, serine proteases, protein dynamics, disulfide bond
1 Introduction
It is widely believed that internal atomic motions of enzymes play an important role in facilitating catalysis, but it remains difficult to establish clear relationships between specific motions and catalytic mechanisms (1; 2; 3; 4; 5; 6) . The “standard-mechanism”, or “Laskowski-mechanism”, inhibitors of serine proteases may offer a window through which to examine this fundamental question. 1 These proteins, typically of relatively small size, bind to the active sites of their target proteases in a manner similar or identical to that of substrates, but are remarkably resistant to hydrolysis, with turnover times ranging from days to even years (8; 9; 10; 11). Several mechanisms have been proposed to explain the slow rates at which these molecules are cleaved, but the rigidity of the enzyme-inhibitor is generally believed to play an important role (12; 13; 14; 15; 16; 17; 18; 19). By examining variants of either inhibitors or proteases that are associated with higher hydrolysis rates, it may be possible to identify and characterize motions that are necessary for the catalytic reaction.
Rawlings et al. have identified 13 clans of Laskowski-mechanism inhibitors, representing convergent evolution of different three-dimensional structures on a common mechanism.(20) The protease-binding regions of the inhibitors share a very similar conformation, but these segments are supported by three-dimensional scaffolds that vary widely among the clans (9; 10; 21). A common, but not universal, feature of many of these inhibitors is the presence of a disulfide bond linking the protease-binding segment to the structural core (10; 21). Proline residues are also frequently found within the protease-binding segments. These patterns have been taken as evidence that rigidity is an important determinant of inhibitor function.
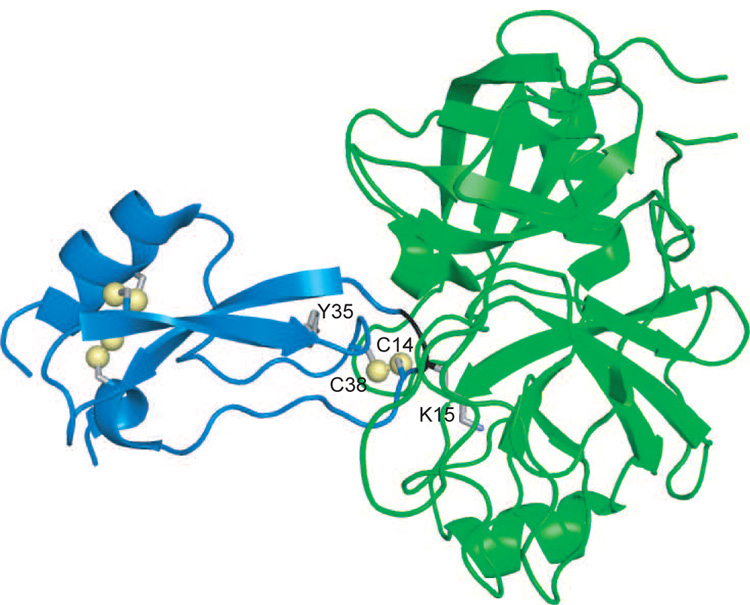
One of the best studied of the Laskowski-mechanism inhibitors is bovine pancreatic trypsin inhibitor (BPTI), a 58 amino-acid residue protein that has served as a model for numerous biochemical and biophysical studies of protein structure, folding and dynamics (22). In this inhibitor, Lys15 binds to the specificity pocket of trypsin, and the Lys15-Ala16 peptide bond is positioned for attack by the catalytic Ser residue (Figure 1). Among the characterized inhibitor-protease complexes, the BPTI-trypsin pair is the most stable thermodynamically, with a dissociation constant (Kd) of approximately 10−13 M, and the slowest to undergo catalysis, with a half-time of several years.(23; 24)
Figure 1.
Ribbon diagram representation of the complex formed between BPTI (blue) and bovine cationic trypsin (green). The sulfur atoms of the six disulfide-bonded Cys residues of BPTI are shown as yellow spheres, and the side-chain atoms of Lys16 and Tyr35 of BPTI are shown as sticks. The backbone ribbon representing Lys15 and Ala16 is colored black to identify the region of the scissile bond. Drawn from the atomic coordinates in entry 2FTL of the Protein Data Bank, using the computer program PyMOL (http://www.pymol.org).
The folded structure of BPTI contains three disulfides, one of which, between Cys residues 14 and 38, lies within the protease-binding site. Because the other two disulfides are buried within the stable core of the inhibitor, the 14–38 disulfide can be selectively reduced, (25; 26) a modification that has been shown to increase the rate of hydrolysis.(27; 28). There are also genetically modified forms of BPTI that are even more sensitive to cleavage after reduction of the disulfide, so that the half-time is only a few minutes. (29; 30) All of these observations are consistent with the idea that the disulfide normally serves to constrain motions that would otherwise allow hydrolysis. However, most of the modified proteins used in the earlier studies contained blocking groups on the sulfur atoms of Cys14 and Cys38, raising the possibility that the increased hydrolysis rates may be due in part to steric perturbations, rather than the loss of the disulfide, per se. In addition, NMR studies have shown that removal of the disulfide has remarkably little effect on the structure or dynamics of the free inhibitor (31; 32).
In this study, we have re-examined the roles of the 14–38 disulfide, using protein variants in which one or both of the Cys residues is replaced with Ser. The abilities of these variants to bind to and inhibit trypsin were measured, and crystal structures of the three enzyme-inhibitor complexes were determined. The structure of the complex containing the mutant with both Cys residues replaced was found to be nearly indistinguishable from that of the wild-type complex, with the two γ-oxygen atoms of the Ser residues forming a hydrogen bond that is geometrically equivalent to the disulfide bond in the normal structure. However, the rate of hydrolysis of the double mutant was approximately 200-fold greater than that of the wild-type protein. The variants in which only one Cys residue was replaced were hydrolyzed more rapidly than the double mutant and displayed local structural perturbations in the complexes with trypsin. These observations suggest that the covalent constraint provided by the disulfide is a major factor in limiting hydrolysis, but that the rate is also quite sensitive to small structural changes in the binding site. Further insights into the inhibition mechanism were obtained from the pH dependence of the hydrolysis rates for the mutant proteins, which suggests the existence of a rapid equilibrium between the enzyme-inhibitor complex and an acyl enzyme intermediate.
2 Results
2.1 Kinetics of hydrolysis
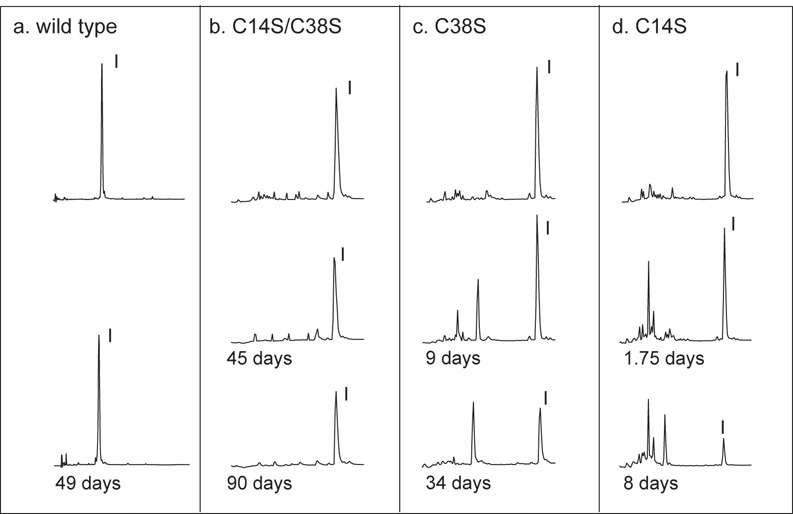
Reversed-phase HPLC was used to monitor the hydrolysis of the BPTI variants by bovine trypsin, as illustrated in Figure 2 and Figure 3. In each experiment, the inhibitor and enzyme were mixed at equal concentrations and incubated at 25°C at pH 7.8 (Figure 2) or 3.4 (Figure 3). At various times, samples were withdrawn, acidified to quench the reactions and fractionated by HPLC. Panel a of Figure 2 shows the HPLC elution profiles for wild-type BPTI immediately after mixing with trypsin and after 49 days at pH 7.8. As expected from previous studies, there was no detectable change in the HPLC profile or decrease in the size of the BPTI peak. When the C14S/C38S variant was incubated with trypsin (panel b), the size of the BPTI peak decreased measurably with time, but was still prominent after 90 days. There were no hydrolysis products detectable in this reaction, presumably because they were rapidly degraded to small peptides that were not retained on the HPLC column. The variants with single replacements of either Cys14 or Cys38 were cleaved more rapidly than the double mutant, as shown in panels c and d of Figure 2. For these proteins, there were several peptide products that accumulated to significant levels during the hydrolysis reaction and eluted prior to the intact inhibitor.
Figure 2.
Hydrolysis of BPTI variants at pH 7.8 monitored by HPLC separation. Each of the indicated BPTI variants was incubated with an equimolar concentration of bovine trypsin at pH 7.8 and 25 °C. Samples of the reaction mixtures were withdrawn at the indicated times, acidified by the addition of HCl and fractionated by reversed-phase HPLC as described in Materials and Methods. The chromatograms were recorded by monitoring UV absorbance at 229 nm, and elution volumes are plotted from left to right. The topmost chromatogram in each panel is of a sample withdrawn from the reaction mixture at time zero. Only the regions of the chromatograms containing BPTI and the hydrolysis products of trypsin and BPTI are shown. Intact trypsin eluted at later times not included in the figure. The peaks containing intact inhibitor are labeled “I”.
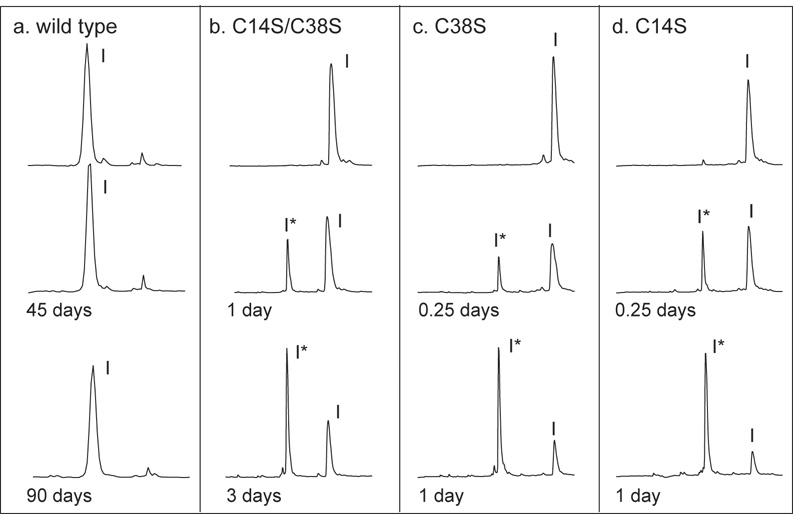
Figure 3.
Hydrolysis of BPTI variants at pH 3.4 monitored by HPLC separation. Hydrolysis reactions and chromatographic separations were carried out as described in the legend to Figure 2, except that the pH of the reaction mixture was 3.4. The peaks containing intact inhibitor and the form cleaved between residues 15 and 16 are labeled as “I” and “I*”, respectively.
When the enzyme-inhibitor complexes were incubated at pH 3.4, quite different HPLC profiles were observed (Figure 3). As at the higher pH, no hydrolysis of the wild-type inhibitor was detectable after 90 days. For the mutants, however, the rate of hydrolysis was much faster at pH 3.4 than at pH 7.8. In addition, the disappearance of the intact inhibitor was associated with the appearance of a new peak in the HPLC profile (marked I* in the figure), which did not disappear significantly with time. For each of the variants, the material in this peak was analyzed by electrospray-ionization mass spectrometry and found to have a mass 18 amu greater than that of the parent molecule, consistent with hydrolysis of a single peptide bond. The disulfides of these products were reduced, resulting in two peptide fragments, which were separated by HPLC and then analyzed by mass spectrometry. For each variant, the peptide masses were consistent with hydrolysis of the peptide bond between Lys15 and Ala16. Although the species identified as I* was not directly observed during the hydrolysis reactions at pH 7.8, it is extremely likely that the initial hydrolysis under these conditions also occurs between Lys15 and Ala16. As shown below, all of the proteins studied here are effective inhibitors at pH 7.8, with dissociation constants less than 10−9 M, and crystal structures show the inhibitors bound to trypsin with the Lys15-Ala16 peptide bond positioned in the active site. In order for the first cleavage to occur at another site, trypsin would have to dissociate from the canonical high-affinity binding site and then bind to an alternative site within the inhibitor, a very unlikely scenario.
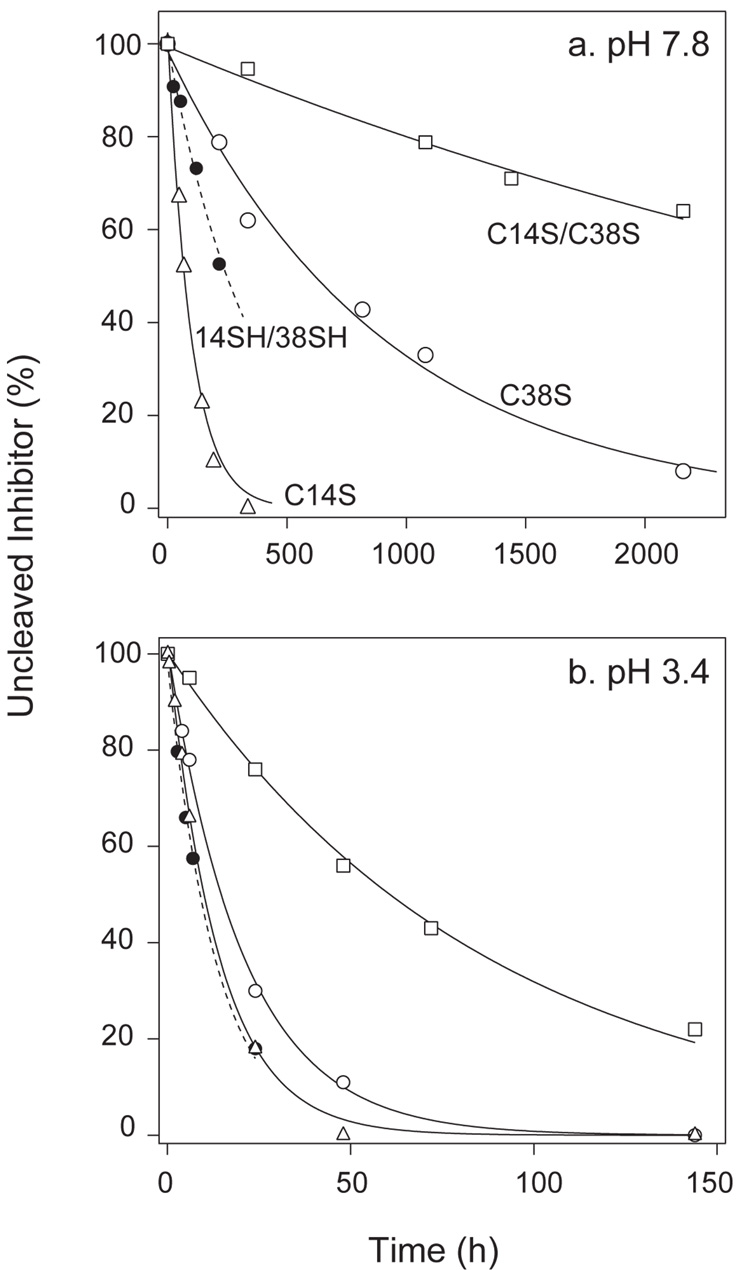
The kinetics of hydrolysis are analyzed quantitatively in Figure 4, where the concentration of uncleaved inhibitor is plotted as a function of time, and the experimental data are fit to a first-order exponential decay function. The figure also includes data for the hydrolysis of the wild-type protein with the 14–38 disulfide selectively reduced (identified here as 14SH/38SH). The rate constants derived from the measurements at pH 7.8 are summarized in Table 1, along with published rates for the wild-type protein and the selectively reduced wild-type protein with the thiols caraboxyamidomethy-lated (14CAM/38CAM). Among the proteins lacking the 14–38 disulfide, the lowest hydrolysis rate was that for the C14S/C38S double mutant, which was cleaved approximately 200-fold more rapidly than the unmodified wild-type protein. The hydrolysis rate for the selectively-reduced wild-type protein is approximately 10-fold greater than for the C14S/C38S mutant, and car-boxyamidomethylation of the reduced Cys residues increases the rate further. (33) The mutants in which only one of the Cys residues was replaced were both hydrolyzed much more rapidly than the double mutant. These results demonstrate that the presence of the disulfide is important for the low hydrolysis rate of the wild-type inhibitor, but that the rate is also quite sensitive to the nature of the replacements or chemical modifications.
Figure 4.
Kinetics of hydrolysis of BPTI variants at (a) pH 7.8 and (b) pH 3.4. Hydrolysis reactions were carried out and monitored by HPLC as illustrated in Figure 2 and Figure 3. The concentrations of uncleaved inhibitor were determined by integration of the chromatogram peaks and are expressed as a percentage of the concentration at time zero. The curves represent fits of the experimental data to a first-order exponential decay function. The data corresponding to the different inhibitor forms are identified directly in panel a, and the same symbols are used in panel b. The estimated rate constants for hydrolysis at pH 7.8 are listed in Table 1. The rates at pH 3.4 were: C14S, 2.0×10−5 s−1; C38S, 1.0×10−5 s−1; C14S/C38S, 3.2×10−6 s−1; 14SH/38SH, 2.1×10−5 s−1.
Table 1.
Kinetic and thermodynamic properties of BPTI variants.
| Variant |
khyd (s−1) |
ΔGd (kcal/mol) |
ΔHd (kcal/mol) |
ΔCp,d (cal/deg·mol) |
|---|---|---|---|---|
| wild-type1 | 3×10−10 | 18 ±0.4 | 14.4 ±0.2 | 410 ±20 |
| 14SH/38SH2 | 8×10−7 | 12 ±0.4 | ||
| 14CAM/38CAM3 | 3×10−6 | 13 ±0.4 | ||
| C14S/C38S | 6×10−8 | >15 | 14 ±0.2 | 450 ±20 |
| C14S | 2×10−6 | 13 | ||
| C38S | 4×10−7 | 13 |
The values of the parameters for wt BPTI are from the following references: khyd (at pH 7.5), ref. [?]; ΔGd, ref. [?], ΔHd and ΔCp,d, ref. [?]
The value for ΔGd for the 14–38 reduced form of wild-type BPTI is from ref. [?]
The value for ΔGd for the 14–38 reduced and carboxyamidomethylated form of wild-type BPTI is from ref. [?] and the rate of hydrolysis of this form was calculated from data in ref. [?].
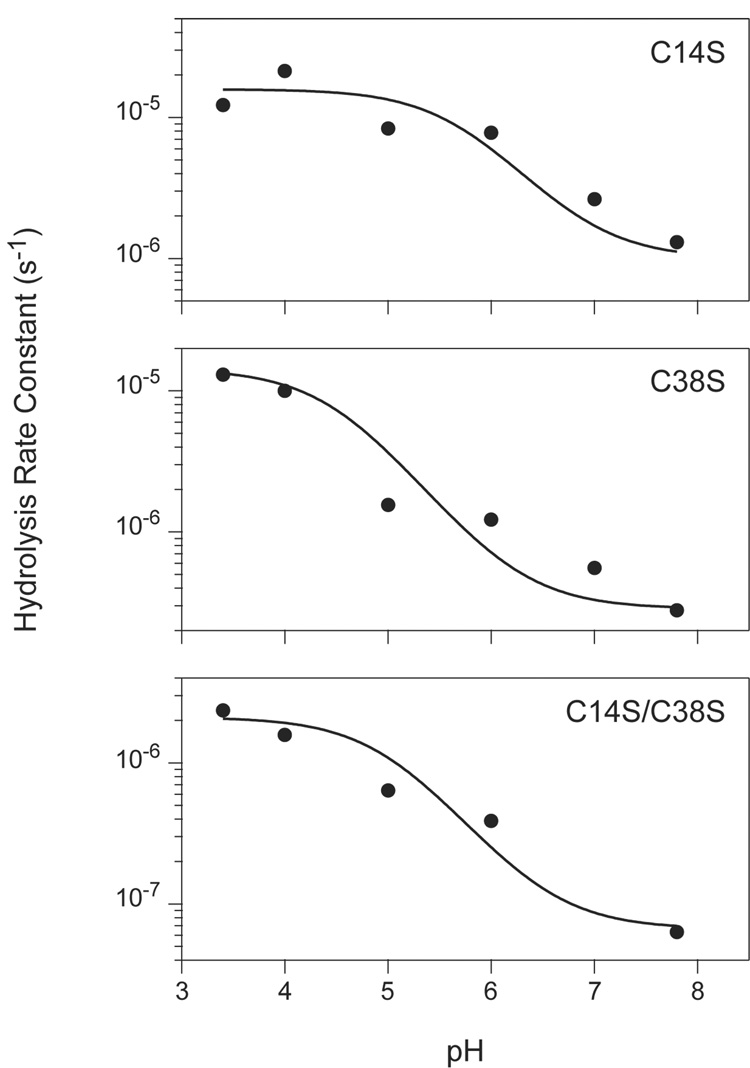
The rates of hydrolysis for the three Cys → Ser variants were also measured at intermediate pH values, and the pH dependence of these rates are plotted in Figure 5. The hydrolysis rate for each of the inhibitors was observed to decrease by approximately 20–50 fold as the pH was increased from 3.4 to 7.8 (Figure 5). This trend contrasts strikingly with the almost universal observation that the turnover rates for serine proteases increase with pH over this range, due to the requirement that the catalytic His residue be in a neutral state in order to promote deprotonation of the catalytic Ser residue (34). The persistence of the cleaved inhibitor (I*) for many hours at pH 3.4 is consistent with the more typical pH dependence. The possible origin and implications of the very different pH dependence observed for the initial hydrolysis reaction is considered in the Discussion.
Figure 5.
Apparent rate constants for hydrolysis of BPTI variants by trypsin versus pH. Hydrolysis reactions were carried out at the indicated pH values and the progress of the reactions were monitored by reversed-phase HPLC, as illustrated in Figure 2 and Figure 3. Rate constants for the reactions were determined by least-squares fitting to a first-order decay function as shown in Figure 4. The curves represent the function described by Equation 7 in the text, with the parameters estimated manually. For all three of the variants, the values of k1 and k2 were both set to 10 s−1. The values used for k−1 for the three mutants were: C14S, 1×108 s−1; C38S, 3.5×108 s−1; C14S/C38S, 1.5×109 s−1. The pKa values used were: C14S, pK1 = 5.7, pK2= 6.9; C38S, pK1 = 5.7, pK2 = 6.9; C14S/C38S, pK1 = 5, pK2 = 6.5.
2.2 Thermodynamics of enzyme-inhibitor dissociation
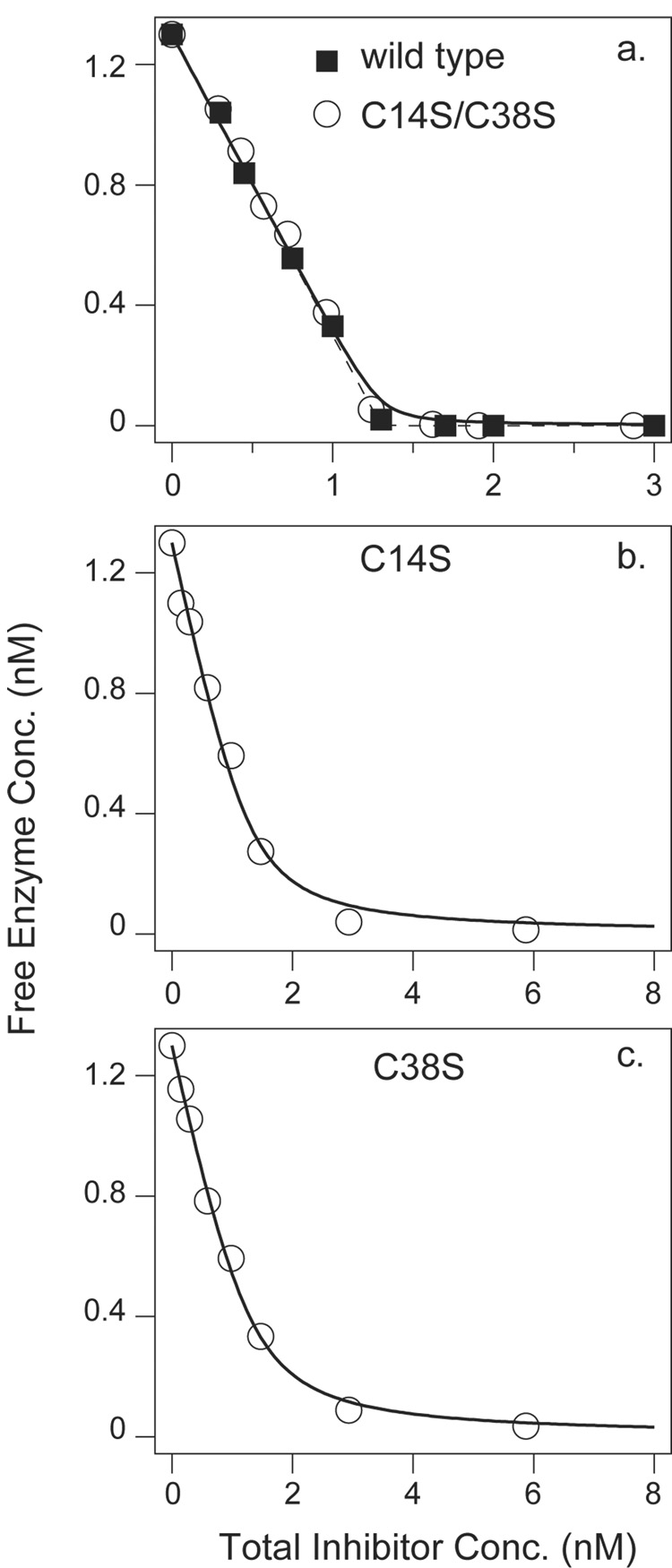
The dissociation constants for the enzyme-inhibitor complexes were determined using a spectrophotometric assay for free trypsin in the equilibrium mixtures. For each inhibitor variant, trypsin at a fixed concentration (1.3 nM) was incubated with various concentrations of inhibitor, and the residual free enzyme was measured, as plotted in Figure 6.
Figure 6.
Equilibrium binding measurements for BPTI variants and bovine trypsin. A fixed concentration of trypsin (1.3 nM) was incubated with the indicated total concentrations of inhibitor at pH 7.8 and 25 °C. The concentration of free enzyme was determined using a spectrophotometric assay with a chromogenic substrate, as described in Materials and Methods. In panel a, the curves are those predicted assuming a single binding site and a dissociation constant of either 6×10−14 M (the value previously determined for the wild-type inhibitor (23), dashed curve) or 6×10−12 M (solid curve). In panels b and c, the curves represent least squares fits of the binding function to the experimental data. The fit values of the dissociation constants were 1.7×10−10 M and 1.4×10−10 M for the C14S and C38S variants, respectively.
For the wild-type inhibitor and the C14S/C38S variant, the concentration of free enzyme decreased linearly with increasing total inhibitor concentration, and there was no detectable activity at the equivalence point. For these inhibitors, the binding is essentially stoichiometric at these concentrations, and it is only possible to place a lower limit on the dissociation constant, Kd < 10−11 M, consistent with previous measurements indicating that the complex with wild-type BPTI has a Kd of approximately 10−13 M (23). For the C14S and C38S inhibitors, however, there was pronounced curvature in the inhibition curves, indicating a significant equilibrium between free and bound inhibitor at nM concentrations. The observed data were fit to a theoretical binding function to estimate the dissociation constants; Kd= 1.7×10−10 M for C14S BPTI and 1.4×10−10 M for the C38S variant. Previous measurements have determined the dissociation constant for the 14–38 reduced form of the wild-type protein to be 1.8×10−9 M (23). Thus, replacing either of the Cys residues in the selectively-reduced inhibitor with Ser increases the affinity of the inhibitor for trypsin by about 10-fold (1.4 kcal/mol), and the combined effect of both substitutions is at least 100-fold (2.7 kcal/mol). Reforming the disulfide of 14SH/38SH, however, increases the stability of the complex by 18,000-fold (6 kcal/mol) (23).
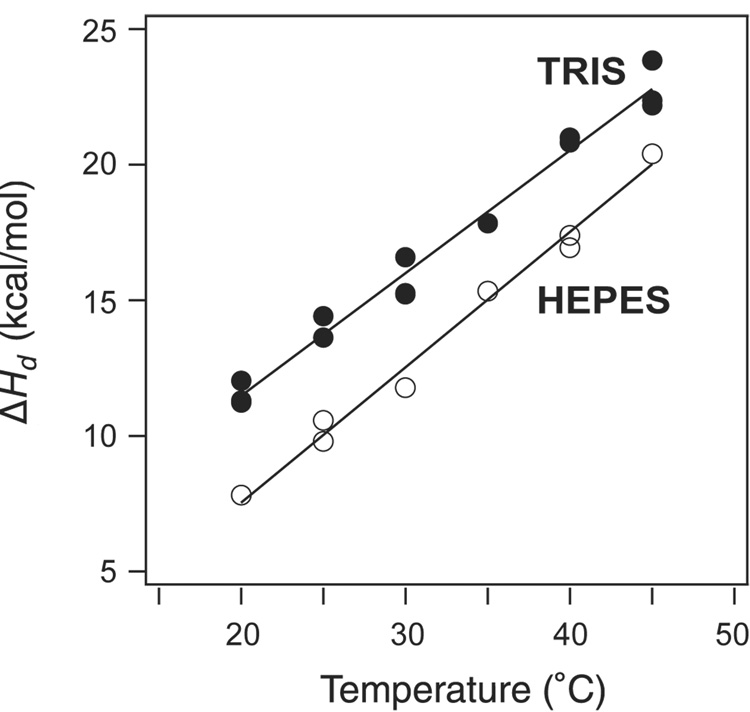
Isothermal titration calorimetry (ITC) was used to further characterize the thermodynamics of the the interaction between trypsin and the C14S/C38S BPTI variant (Figure 7). Under the conditions used for these measurements, the interaction is essentially irreversible, and the observed heat for each injection is a direct measurement of the molar enthalpy for the reaction. The enthalpy of dissociation, ΔHd, was measured at temperatures from 20 to 45°C, and with solutions buffered with either Tris or HEPES. As observed previously for wild-type BPTI and another variant (Y35G),(35) the values of ΔHd observed with Tris buffer were consistently greater than those measured with HEPES, by approximately 3 kcal/mol. The origin of this buffer effect is not known, but it does not appear to be related to any effects of the amino acid replacements. At 25°C, the heat of dissociation for the complex with C14S/C38S BPTI was 14 ± 0.2 kcal/mol(with Tris buffer), very close to the value previously measured for the complex with wild-type BPTI, 14.4 ± 0.2 kcal/mol. (35) The heat capacity change for dissociation, ΔCp,d, was estimated from the slope of the plot of ΔHd versus temperature, and the value for the C14S/C38S mutant, 450± 20 cal/deg·mol, was also similar to that for the wild-type protein, 410 ± 20 cal/deg·mol.(35) These results suggest that the replacement of both Cys14 and Cys38 have very little effect on the direct interactions between the the inhibitor and enzyme.
Figure 7.
Heats of dissociation (ΔHd) for the complex of trypsin with C14S/C38S BPTI. Enthalpy changes were measured by isothermal titration calorimetry at pH 8 in the presence of 20 mM CaCl2 and 50 mM Tris-HCl (filled circles) or 50 mM HEPES (open circles). The lines represent least-squares fits to the experimental data, from which estimates of the heat capacity change, ΔCp,d, and ΔHd at 25 °C were derived. The parameters for the data obtained with Tris-HCl are listed in Table 1. For HEPES, the parameters were: ΔHd = 10.6±0.2 kcal/mol, ΔCp,d = 500±20 cal/deg·mol.
The thermodynamic parameters of the variants studied here are summarized in Table 1, along with values for the native form of the wild-type protein and the form with the 14–38 disulfide reduced and the thiols carboxyamidomethylated (14CAM/38CAM). These values show that the hydrolysis rates and binding affinities vary greatly among the forms lacking the 14–38 disulfide, depending on the nature of the modification, and that increased rates of hydrolysis are loosely correlated with decreased binding affinity.
2.3 Crystal structures of the enzyme-inhibitor complexes
To determine the structural origins of the effects seen upon removing the 14–38 disulfide of BPTI, crystal structures were determined of the complexes formed between bovine trypsin and the C14S, C38S and C14S/C38S BPTI variants. Each of these complexes formed crystals belonging to the same space group as observed previously for complexes of bovine trypsin with wild-type BPTI and other variants (space group I222) (36; 37; 35). X-ray diffraction data with a resolution limit of 1.58 Å were collected using cryopreserved crystals frozen at 100 K. Structures of the three complexes were determined by the method of molecular replacement, using the structure of the complex with wild-type BPTI as an initial model. The final crystallographic residuals, R, were all less than 22%, and Rfree was less than 24% for each structure. Details of the structure calculations are summarized in Table 2.
Table 2.
Crystallographic data collection and refinement statistics
| 2FTL2 (WT) |
2FI3 (C14S/C38S) |
2FI4 (C14S) |
2FI5 (C38S) |
||
|---|---|---|---|---|---|
| A. Diffraction1 | |||||
| Unit cell (Å) a | 74.73 | 74.74 | 74.78 | 74.73 | |
| b | 81.91 | 81.75 | 82.44 | 81.62 | |
| c | 124.26 | 124.27 | 123.96 | 124.07 | |
| Resolution (Å) | 20 - 1.60 | 20 - 1.56 | 20-1.58 | 20-1.58 | |
| Rcrystal (%) | 5.0 | 4.3 | 4.8 | 6.4 | |
| Completeness (%) | 99.9 | 98.6 | 98.5 | 99.8 | |
| B. Refinement | |||||
| Resolution (Å) | 20 - 1.62 | 20 - 1.58 | 20-1.58 | 20-1.58 | |
| Average B-value (Å2) | 17.3 | 14.8 | 16 | 14.8 | |
| R (%) | 21.4 | 20.3 | 21.7 | 20.9 | |
| Rfree (%) | 22.7 | 21.8 | 23.5 | 22.3 | |
| rmsd bonds (Å) | 0.0049 | 0.0050 | 0.0051 | 0.0050 | |
| rmsd angles (°) | 1.30 | 1.33 | 1.34 | 1.33 | |
| Luzatti coordinate error (Å) | 0.21 | 0.19 | 0.21 | 0.20 | |
| C. Data/Parameter | |||||
| Unique reflections | 48,719 | 53,322 | 51,888 | 52,039 | |
| Number of atoms | 2,572 | 2,551 | 2,529 | 2,675 | |
| Data-to-parameter ratio 3 | 4.7 | 5.2 | 5.1 | 4.9 | |
| D. RMS deviation from wild-type structure (Å) | |||||
| Backbone atoms | na | 0.22 | 0.20 | 0.12 | |
| Side-chain atoms | na | 0.42 | 0.39 | 0.31 | |
All diffraction data were collected from crystals cooled to 100 K, and all of the crystals were of the space group I222
from ref. [?]
Data-to-parameter ratio calculated as (Unique reflections) / 4(Number of atoms).
The overall structures of the three complexes examined here were essentially identical to that of the complex with the wild-type inhibitor determined under the same conditions. The RMS deviation between the atomic positions of backbone atoms in the complexes with the mutant and wild-type inhibitors were all less than 0.25 Å, and the RMS deviations for side-chain atoms were all less than 0.45 Å. There were, however, small effects in the region of the substitutions, which are illustrated in Figure 8.
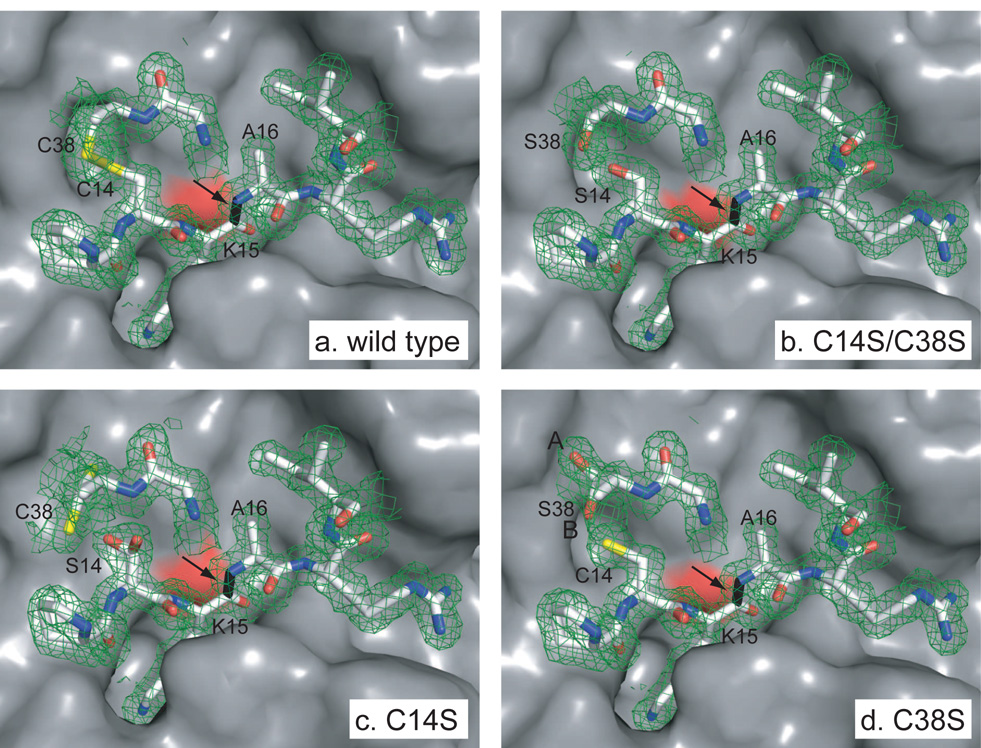
Figure 8.
Structures of complexes of trypsin with BPTI variants containing replacements of Cys14 or Cys38, highlighting the active site region. The orientation shown corresponds approximately to that shown in Figure 1, rotated by 90° about the vertical axis, so that the view is from the inhibitor towards the enzyme active site. The enzyme site is shown in a surface representation, and the primary binding residues of the inhibitors are represented as sticks. Carbon, nitrogen, oxygen and sulfur atoms of the inhibitor are colored white, blue, red and yellow respectively. The scissile peptide bond of the inhibitor is colored black and identified by an arrow. The surface of the side-chain oxygen of the catalytic Ser residue (Ser195) of trypsin is colored red. Electron density maps corresponding to the inhibitor residues are represented as cages, contoured at the level of 1 σ.
The diffraction data for the C14S/C38S mutant were consistent with a single conformation for the two altered residues, which occupied positions nearly identical to those of the Cys sulfur atoms in the wild-type structure. For the mutants with single replacements, C14S and C38S, the calculated electron density maps indicated the presence of alternative conformations for residues 14 and 38. The refined structure of the C14S variant included the alternate conformations shown in the figure, with equal occupancy in each case. In the C38S structure, the two conformations of Cys14 were represented at equal occupancy, but Ser38 was modeled with 75% occupancy of the conformation labeled A and 25% occupancy of conformation B. The alternate conformation of Cys14 in panel d is hidden. The arrow in each panel indicates the scissile peptide bond of the inhibitor, between Lys15 and Ala16. Drawn from the atomic coordinates in PDB entries 2FI3 (C14S/C38S), 2FI4 (C14S) and 2FI5 (C38S).
In the C14S/C38S complex, electron density for the γ-oxygen atoms of the introduced Ser residues is clearly visible and superimposes closely with the positions of the Cys sulfur atoms of the wild-type structure. In the refined structure, the distance between the γ-oxygen atoms of the two Ser residues is 2.63 Å, very close to the expected distance between two atoms of this type linked by a hydrogen bond (2.64 Å)(38). Furthermore, the angles formed between the Cβ-Oγ bond and the Oγ-Oγvector are 118° (Ser14) and 104° (Ser38), indicating that one or the other of the hydroxyl hydrogen atoms would be well positioned to form the hydrogen bond. Thus, the disulfide appears to have been replaced with a hydrogen bond possessing very favorable geometry.
For the two mutants with single Cys → Ser replacements, there were more significant structural perturbations, as shown in panels c and d of Figure 8. In both cases, the electron density for residues 14 and 38 was less well de-fined than for either the wild-type structure or that of the double-replacement mutant, and the final structural models included alternate conformations for both residues.
The pattern of effects seen in the structures of these variants suggests that the differences in the volumes occupied by the Oγor Sγatoms, with van der Waals radii of approximately 1.4 and 1.8 Å respectively, may be an important factor in determining the effects of removing the 14–38 disulfide. In the wild-type structure, the disulfide-bonded sulfur atoms are separated by 2.02 Å, considerably less than the distance between the oxygen atoms of the two Ser residues (2.63 Å) in the C14S/C38S variant. However, the extra distance required for the two oxygen atoms is accommodated with minimal perturbation of the structure. When only one of the two Cys residues is replaced with Ser, however, the minimum distance between the oxygen and sulfur atoms is expected to be 3.23 Å(38). The larger volume required for the thiol sulfur atom appears to cause structural shifts and heterogeneity. With both Cys14 and Cys38 in the thiol form, the minimum interatomic distance is expected to be 3.54 Å, which may cause further disruption of the structure.
In all three of the 14–38 disulfide variants, the geometries of the trypsin active site and the scissile bond of the inhibitor were indistinguishable from those seen in the complex with wild-type BPTI. In particular, there was no evidence of hydrolysis of the scissile bond or deviation from planar geometry.
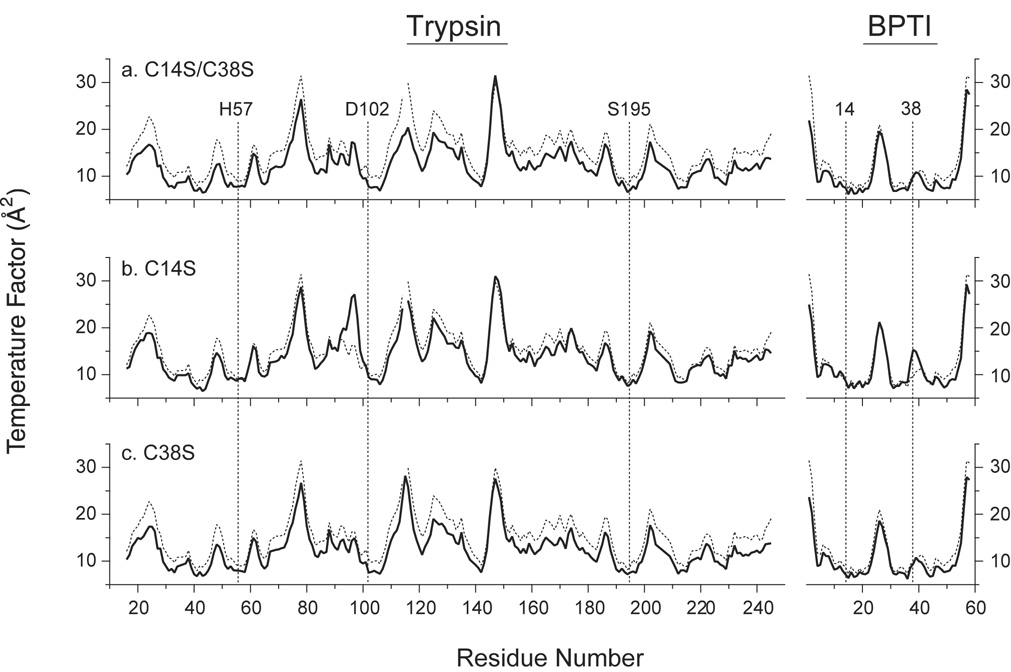
Since the removal of the 14–38 disulfide leads to increased hydrolysis rates for all three of the mutant inhibitors, it might have been expected that the crystal structures would reveal evidence of increased flexibility that could facilitate the catalytic reaction. However, the electron density for the trypsin-binding regions of the inhibitors and the catalytic residues of the enzyme was very well defined. The major exception was the density for Arg39 of the inhibitor, which was poorly defined for all of the structures, including the complex with wild-type inhibitor. Furthermore, the crystallographic temperature factors (B-values) derived from the refinements of the three structures indicate that the Cys → Ser replacements caused minimal changes in the flexibility of the inhibitor or protease in the complex. The B-values for the backbone atoms of both proteins in the three complexes are compared with those for the wild-type complex in Figure 9. For each of the mutant complexes, the average of the B-values (represented with solid curves) was slightly lower than for the wild-type complex (represented with dashed curves), likely reflecting the higher degree of order in these crystals, which displayed slightly improved resolution limits (1.58 Åversus 1.62 Å for the wild-type complex). Within the inhibitor molecules, the only significant difference was an increase in the B-values for Cys38 in the C14S mutant. A lack of order at this site is also indicated by the alternate conformations required to model the electron density of the side chain (Figure 8c). The C14S replacement was also associated with an increase in the B-values for residues 94–99 of the enzyme, which forms a loop on the surface of the enzyme that contacts Cys38 and Arg39 of the inhibitor. Otherwise, however, the substitutions in the inhibitor appeared to cause little or no change in the enzyme, including the catalytic residues Ser195, His57 and Asp102, for which the B-values are among the lowest in each of the complexes.
Figure 9.
Crystallographic temperature factors for backbone atoms of trypsin and BPTI in complexes containing the C14S/C38S, C14S and C38S BPTI variants. Average B-values for the N, Cα, C and O atoms are plotted as a function of residue number. In each panel, the solid curve represents the values for the indicated mutant, and the dashed lines are the values for the complex with wild-type BPTI (PDB entry 2FTL). The vertical dashed lines identify the average B-values for residues 14 and 38 of the inhibitors and the residues of the trypsin catalytic triad.
3 Discussion
The studies described here were designed to help elucidate the factors that determine the remarkably low rates of hydrolysis of BPTI and, by analogy, other Laskowski-mechanism inhibitors by serine proteases. Earlier studies showed that reduction of the 14–38 disulfide in BPTI and modification of the resulting thiols significantly increases the hydrolysis rate, and it has long been thought that this effect is due to some form of enhanced flexibility in the enzyme-inhibitor complex(27; 28). We have extended these observations by studying genetically modified proteins in which the disulfide is removed with minimal additional perturbations and by determining high-resolution crystal structures of the complexes formed by these proteins and trypsin. The results demonstrate that removal of the disulfide increases the hydrolysis rate by 200-fold or more, even when there is no detectable perturbation of the structure and the disulfide is replaced by a hydrogen bond with very favorable geometry (in the C14S/C38S variant). It thus appears that the covalent constraint provided by the disulfide contributes directly to the low hydrolysis rate. On the other hand, removal of the disulfide did not lead to significant increases in the crystallographic temperature factors for atoms in either the enzyme active site or the cleavage site of the inhibitor. Although factors other than the internal dynamics of a molecule can influence B-values,(39) the very similar temperature factors determined for the mutant and wild-type complexes, which were obtained using nearly identical crystals and the same refinement procedures, argue strongly against increased flexibility in the mutant complexes. These observations, and the pH dependence of the hydrolysis rate, offer insights into the mechanism of inhibition, as discussed below.
3.1 Possible inhibition mechanisms
The overall hydrolysis reaction catalyzed by the serine proteases includes two nucleophilic attacks on the carbonyl carbon of the substrate, one by the γ-oxygen of the catalytic Ser residue and the second by a water molecule, as diagrammed in the scheme below (34):
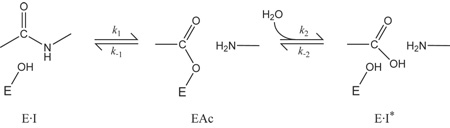 |
(1) |
In this scheme, E · I is the enzyme-inhibitor complex, EAc is the acyl-enzyme intermediate, and E · I* is the complex containing the enzyme and cleaved inhibitor. The very low overall hydrolysis rate for an inhibitor could, in principle, be a consequence of a low rate for either of the forward steps, or rapid reversal of either. Crystal structures of enzyme-inhibitor complexes, such as those described in this paper, invariably show the inhibitor in its uncleaved form, with the scissile bond in a planar configuration (40; 9; 10; 17). The chemical shifts of the carbonyl carbon in 13C-labeled complexes also indicate a planar configuration (41; 42). These observations have generally been taken as evidence that the hydrolysis reaction is blocked at the first step in the scheme shown above, possibly because the rigidity of the complex prevents the atomic motions required to form the tetrahedral transition state (40; 12; 43; 14; 16).
An alternative explanation, described as a “clogged-gutter mechanism” (17), suggests that the acyl intermediate is formed at a significant rate but is rapidly converted back to the intact inhibitor (12; 16; 14; 17). During hydrolysis of a normal peptide substrate, the first product of hydrolysis, containing the new terminal amino group, is free to dissociate from the enzyme after formation of the acyl intermediate. For an inhibitor, however, intramolecular interactions within the inhibitor prevent dissociation, so that the amino group is likely to be well positioned to carry out a nucleophilic attack on the carbonyl group and reform the original peptide bond. In addition, the access of water to the active site may be restricted, reducing the rate of the second step in the reaction. This model is supported by experiments demonstrating that a small fraction (5–10%) of chymotrypsin inhibitor 2 (CI2) molecules in complex with subtilisin form an acyl intermediate, even though the intermediate cannot be detected in crystal structures of the complex(17). A quantum mechanics/molecular mechanics simulation of the BTPI-trypsin complex also suggests that the formation of the acyl intermediate is disfavored both kinetically and thermodynamically (16).
Using the protocol employed by Radisky and Koshland to isolate the acyl-enzyme intermediate for CI2 and subtilisin, we have been unable to detect the formation of the analogous species formed by trypsin and any of the BPTI variants we have studied. This negative result could reflect either very slow formation of the intermediate, or a very low concentration of this species in steady state equilibrium with the EI complex. It is also possible that the conditions used to isolate the intermediate (heating in sodium dodecylsulfate) may promote either reformation of the intact inhibitor or hydrolysis of the intermediate before the proteins dissociate and unfold.
The very similar patterns of B-values observed for complexes with quite different hydrolysis rates suggests that flexibility within the E·I complex is not the determinant of the overall rate, arguing against a model in which the reaction is inhibited at the stage of forming the acyl-enzyme intermediate. On the other hand, modifications of the inhibitor might well enhance the flexibility of the acyl intermediate, thereby facilitating the overall reaction by either decreasing k−1 in the scheme of Equation 1 or increasing k2.
It is also of note that the scissile bonds of the inhibitors all appeared to be intact in the crystal structures, even though the crystals were incubated at room temperature for approximately four weeks, as compared to the half-times for hydrolysis of 4 days and 20 days for the C14S and C38S variants, respectively. A similar result was previously observed for the Y35G BPTI variant, which is cleaved with a half-time of approximately 4 days in solution (35). One possible explanation for the apparent resistance to hydrolysis in the crystals is that the peptide bond is, in fact, hydrolyzed but reforms rapidly because the inhibitor cannot dissociate. However, isotope exchange experiments using 18O-labeled water have not yielded evidence for this process in crystals of the complex with Y35G BPTI (E. Zakharova, W.M. Hansen and D.P. Goldenberg, unpublished results). These observations suggest that crystal packing interactions may inhibit a structural change that is required for one or more steps in the reaction.
3.2 The pH dependence of hydrolysis
The observed pH dependence of the overall hydrolysis rates shown in Figure 5 is particularly striking, since the catalytic rates for serine proteases almost always increase with pH over this range. This trend was first observed by Guttfreund in 1955, who attributed it to the requirement for a His side chain in the unprotonated state (44). This observation has been confirmed many times, for several serine proteases, and the role of the catalytic His residue is well established. (45; 46; 47; 48) However, the opposite trend seen here is not without precedent and has been described previously for the 14CAM/38CAM form of BPTI(33), and for at least two other Laskowski-mechanism inhibitors, soybean trypsin inhibitor (SBTI) (49) and Cucurbita maxima trypsin inhibitor I (CMTI I) (50), both with bovine trypsin. Though these authors did not suggest a specific mechanism to account for the unusual pH dependence, we propose that it is most consistent with a model involving reversible formation of the acyl intermediate and provides strong, if indirect, support for this model.
As shown in the scheme of Equation 1, reformation of the peptide bond of the inhibitor involves a nucleophilic attack by the newly-formed amino group on the carbonyl carbon of the acyl intermediate. To carry out this attack, the amino group must be in its neutral, unprotonated, state. If there is a rapid equilibrium between the enzyme-inhibitor complex (E · I) and the acyl intermediate, low pH will favor higher steady-state concentrations of the latter species and, depending on the balance of rate constants and pKa values, may lead to a higher overall hydrolysis rate.
In order to explore the plausibility of this scenario, a steady-state kinetic model was derived and used to calculate the expected effects of pH on the apparent hydrolysis rate. The model, described in the Materials and Methods section, incorporates ionization equilibria for the His57 side chain, which must be in the deprotonated state for each of the microscopic steps shown in the scheme above, as well as for the newly-formed amino group. In addition, the model assumes that dissociation of the hydrolyzed inhibitor is much faster than reformation of the acyl intermediate, (24) so that the overall hydrolysis rate is determined by the product of the steady state concentration of EAc and the rate constant for the second nucleophilic substitution reaction, k2. The parameters in this model are the pKa values for His57 and the terminal amino group and the rate constants k1, k−1 and k2 (uncorrected for protonation) from the scheme of Equation 1.
With suitable values for the various parameters, the model was able to account for the hydrolysis rates observed for the three Cys variants, as indicated by the curves in Figure 5. The experimental data are not sufficient to define the five parameters in the model, which were adjusted manually to obtain a reasonable fit for each of the mutants, as specified in the legend for Figure 5. Although similar calculated rates can be obtained using a wide range of possible parameter values, the following five arguments were used as constraints:
For several inhibitor-enzyme pairs, it has been shown that inhibitor binding is coupled to reduction of the pKa of His57, from the values typically seen in the free enzyme, ≈ 7, to ≈ 5.(51; 52; 53) The curves shown in Figure 5 were calculated using pKa values of 5 (for C38S and C14S/C38S) or 5.7 (for C14S).
Data from model peptides indicate that the unperturbed pKa for a terminal α-amino group is about 8.(54) Somewhat lower values, (6.5 to 6.7) were required to account for the pH dependence of the hydrolysis rates observed here, but the lower values could reflect the special environment of the new amino group in the complex.
For amide substrates the rate constant for the forward acylation reaction, k1, typically lies in the range of 10–1000 s−1, and the rate constant for hydrolysis of the acyl intermediate, k2, is typically about 50 s−1. (34) A value of 10 s−1 was used for both k1 and k2 for the calculated curves.
In a classic study, Fastrez and Fersht measured the rate of reaction of the amino group of alanine with an acyl-enzyme formed between chymotrypsin and a peptide substrate. (55). The second-order rate constant determined for this intermolecular reaction was 6430 s−1M−1 at pH 9.3. The first-order rate constant for the intramolecular reaction in the enzyme-inhibitor complex (k−1) can be estimated by multiplying the second-order rate constant by an “effective concentration”. Measurements of effective concentrations in enzyme active sites have yielded a wide range of values, from 103 M to 109 M. (56; 57; 58), which would correspond to values as large as 6×1012 s−1 for k−1. The curves shown in Figure 5 were calculated using values for k−1 corresponding to effective concentrations ranging from 1.5×104 to 2.3×105 M.
The failure to detect the acyl-enzyme intermediate either biochemically or crystallographically is consistent with the proposed rate constants, which predict an equilibrium constant that favors the E · I complex relative to the acyl-enzyme by a factor of approximately 107.
These considerations indicate that the observed rates for hydrolysis of the Cys mutants, and the increase of these rates at low pH, can be accounted for by a set of rate constants that do not differ greatly from those observed for normal substates, except for the very high rate of reforming the E · I complex from the acyl intermediate. In addition, the different hydrolysis rates observed for the various mutants can be accounted for by differences in the propensity of the newly-formed amino group to attack the carbonyl of the acyl intermediate (k−1).
Although the rate constants used for the simulations can account for the experimental data, it is important to note that the values of k1 or k2 for the inhibitors could also be significantly smaller than observed for normal substrates, thereby providing an additional mechanism for limiting the overall rate. A low value of k1 in particular, could be due to structural constraints in the complex that inhibit conformational changes necessary for forming the acyl intermediate, as suggested by other authors (59; 60; 61).
The even lower hydrolysis rate for wild-type BPTI could be due to a combination of factors, including a reduced rate for forming the acyl-enzyme intermediate, an increased rate for the reverse reaction or a reduced rate of hydrolysis of the acyl-enzyme. If the values of the forward rate constants are assumed to be comparable to those for a normal substrate (e.g. 10 s−1), then the rate constant for reforming the peptide bond from the acyl intermediate would have to be approximately 1011 s−1, corresponding to an effective concentration of about 107 M. This value approaches the upper limits of what has been observed experimentally (56; 57; 58) or predicted theoretically (62), suggesting that conformational constraints leading to low values of k1 or k2 may be important for limiting the overall rate in this extreme case.
3.3 Conclusions
Taken collectively, the results presented here and in previous studies, particularly those by Radisky and Koshland, suggest that the rigidity of the E · I complex may not be the primary factor inhibiting the hydrolysis of standard-mechanism inhibitors. Instead, the crystallographic and kinetic data appear to be most readily accounted for by a model involving a steady-state equilibrium between the E · I complex and the acyl intermediate. Further support for this model has come from recent studies of human mesotrypsin, which is able to hydrolyze BPTI and other inhibitors much more rapidly than can the major pancreatic enzymes, apparently because an amino acid replacement in the enzyme promotes displacement of the inhibitor leaving group (63; 64; 65). These observations also suggest that further insights into the mechanism of inhibition, and the motions necessary for hydrolysis, may come from studies of complexes representing the later steps in the mechanism, i.e. the acyl intermediate and the complex with the cleaved inhibitor.
4 Materials and Methods
4.1 Protein samples
Bovine cationic trypsin and wild-type BPTI (aprotinin) were obtained from Sigma Chemical Co. and Roche Diagnostics, respectively. Trypsin was further purified by affinity chromatography using immobilized soybean trypsin inhibitor. The C14S, C38S and C14S/C38S BPTI variants were produced by heterologous expression in Escherichia coli and purified as described previously (66; 67; 68). The purified proteins were characterized by electrospray-ionization mass spectrometry, non-denaturing gel electrophoresis and reversed-phase HPLC, all of which indicated that the samples were at least 95% homogeneous. There was no evidence of incorrect disulfide formation in any of the samples.
Wild-type BPTI with the 14–38 disulfide selectively reduced (14SH/38SH) was prepared by treating the native protein with 4 mM dithiothreitol for 1 min at 25 °C, in the presence of 0.1 M Tris-Cl pH 7.8, 0.2 M KCl and 1 mM EDTA. The reaction was quenched by adding HCl to a final concentration of 0.5 M, and the selectively-reduced protein was purified by reversed-phase HPLC. The sample was dried under vacuum in a Speed-Vac centrifugal concentrator and then dissolved in water immediately before being used for kinetic experiments.
4.2 Hydrolysis reactions
To measure the kinetics of hydrolysis of the BPTI variants by bovine trypsin, the inhibitor and enzyme were mixed to yield final concentrations of 8 and 8.5 µM, respectively and incubated at 25 °C. The solutions also contained 20 mM CaCl2 and 150 mM of either Na-citrate (pH 3.4, 4 or 5), MES (pH 6), HEPES (pH 7) or Tris-Cl (pH 7.8). At various times, samples of the reaction mixtures were withdrawn, and the pH was adjusted to 1.5 by the addition of HCl. The samples were then analyzed by reversed-phase HPLC.
The rate of hydrolysis of the 14SH/38SH form of wild-type BPTI was measured under slightly different conditions in order to prevent reformation of the disulfide during the reaction. These reaction mixtures contained 2 mM EDTA and no CaCl2. In addition, the protein solutions were degassed under vacuum before being mixed with the buffer solution, which was degassed by bubbling with N2. The protein concentrations for these experiments were 15.5 µM trypsin and 15 µM 14SH/38SH BPTI.
The primary products of hydrolysis of the C14S, C38S and C14S/C38S variants at pH 3.4 (marked I* in 3) were analyzed by electrospray mass spectrometry using a Micromass Quattro II Triple Quadropole Mass spectrometer. Samples of these species were also subjected to reduction by incubating them with 100 mM dithiothreitol for 30 min at 25 °C in the presence of 6 M guanidinium chloride, 0.1 M Tris-HCl pH 8.7 and 1 mM EDTA. The fragments were then separated by HPLC and analyzed by electrospray mass spectrometry.
4.3 HPLC separations
Reversed-phase HPLC separations were carried out using Vydac C18 columns, which were eluted using gradients of acetonitrile formed by mixing 0.1% TFA (solvent A) with 90% acetonitrile in 0.1% TFA (solvent B). For purification of the 14SH/38SH form of wild-type BPTI, a 1 cm diameter × 25 cm long column was used, with a flow rate of 3 mL/min. For all other separations, a column 0.46 cm diameter × 25 cm long was used, and the flow rate was 1 mL/min.
For separations involving the 14SH/38SH form, the gradient was composed of the following segments: 10% solvent B (0–5 min), 10% to 15% B (5–10 min), 15% to 40 %B (10–40 min). The column temperature was maintained at 37 °C for these separations. All other separations used a column temperature of 45 °C, and the gradient was composed of the following segments: 10% to 20% solvent B (0–5 min), 20% to 29% B (5–15 min), 29% to 55% B (15–110 min).
4.4 Equilibrium binding measurements
Affinity-purified bovine trypsin (1.3 nM) was incubated with the BPTI variants at concentrations ranging from 0.2 to 10 nM. The samples also contained 155 mM Tris-HCl pH 8.0, 4.5 mM CaCl2 and 0.0004% (w/v) triton X-100. After incubations of 1 to 24 h, 800 µL samples were withdrawn and mixed with 20 µL of 10 mM benzoyl-Phe-Val-Arg-4-nitroanilide. The rate of hydrolysis was measured by monitoring absorbance at 405 nm and used to calculate the concentration of free trypsin, relative to that in an uninhibited sample. After incubations of 3 h, no further reduction in the trypsin activity was observed, and the rates measured at 5 and 6 h (for the C14S mutant) or 5, 6 and 24 h (for the C38S and C14S/C38S mutants) were used to estimate the dissociation constant, Kd. The concentration of free trypsin was plotted as a function of total inhibitor (I0) in the mixture, and the data were fit to the following expression:
The total enzyme concentration, E0 was determined using a burst substrate (p-nitrophenyl-p’ -guanido benzoate), (69) and Kd was the only adjustable parameter in the fitting procedure.
4.5 Isothermal titration calorimetry
Protein samples for ITC experiments were extensively dialyzed against buffer solutions containing 10 mM CaCl2 and either 50 mM Tris-HCl or 50 mM HEPES at 4 °C. The buffer solutions were adjusted to pH 8 at the temperature to be used for the calorimetric measurements (20–45 °C). Trypsin and BPTI concentrations were typically about 0.3 mg/mL (0.013 mM) and 1.5 mg/mL (0.23 mM), respectively. Actual concentrations after dialysis were determined by absorbance at 280 nm using extinction coefficients of 3.6×104 cm−1M−1 (trypsin) and 5.4×103 cm−1M−1 (C14S/C38S BPTI). Samples were degassed under vacuum immediately before use.
Heats of binding were measured using a MicroCal VP-ITC titration calorimeter. For each measurement, 1.7 mL of trypsin solution was placed in the calorimeter sample cell, and 20 aliquots of 5 µL each of BPTI were injected at 200 second intervals. Data were analyzed as described in ref. (35).
4.6 X-ray crystallography
Samples for crystallization were prepared by mixing trypsin and the BPTI variant in a molar ratio of 1:1.5, and a total protein concentration of approximately 30 mg/mL. The solutions also contained 20 mM CaCl2 and 10 mM HEPES pH 7.5. Crystals were grown at room temperature using the hanging-drop vapor diffusion technique. Drops contained 1.5 µL of the protein mixture and 1.5 µL of well solution composed of 0.1 M HEPES, 10 mM CaCl2, 0.02% NaN3, and 1.5 – 2.25 M (NH4)2SO4.
Crystals were soaked briefly in reservoir solution plus 20% (v/v) ethylene glycol as cryoprotectant prior to freezing in liquid propane at 100 K. During data collection, crystals were maintained at 100 K in a stream of evaporated nitrogen (Oxford Cryosystems). Data were collected with use of a CCD detector and a rotating anode FR591 generator (Nonius Delft, Netherlands).
Structures of the complexes with C14S, C38S and C14S/C38S BPTI were determined in the I222 space group by molecular replacement using the atomic coordinates for the wild-type BPTI-trypsin complex (PDB entry 2FTL). The structures were refined by iterative cycles of torsion angle simulated annealing, geometry and atom positional minimization, and restrained individual B-factor refinement using maximum likelihood target functions implemented in the computer program CNS (70). Each cycle of refinement was interleaved with manual adjustment and rebuilding using the program O (ref. (71)) while inspecting σA-weighted 2|Fo| − |Fc|, 3|Fo| − 2|Fc| and |Fo| − |Fc| difference maps. As phases improved, peaks in the |Fo| − |Fc| difference maps with greater than 4.8σ intensity and with appropriate geometry and disposition of H-bonding donors or acceptors were assigned to solvent molecules including water molecules, sulfate ions and, in a few instances, ethylene glycol molecules. Where indicated by residual positive and negative electron density peaks, certain residues were modeled with two or three alternate conformations.
As observed previously for the wild-type and Y35G complexes, (35) the initial electron density maps for the complexes with C14S and C38S BPTI showed evidence for a non-standard amino acid residue at the position expected for Asn115 of trypsin. For the C14S complex, the diffraction data were fit best by a model incorporating isoaspartate, the expected product of deamidation and peptide isomerization, (72) at this site. For the complex with the C14S/C38S variant, the data were most consistent with a mixture of both Asn (Asp) and isoaspartate at position 115. No evidence of isomerization was observed for the C38S variant.
4.7 Kinetic model
To consider the possible origins of the pH dependence of the hydrolysis reactions, the model described by Equation 1 was used. In the following, we assume that dissociation of E·I can be ignored and that dissociation of the E·I* complex is much faster than reformation of the acyl intermediate, as shown previously for bovine trypsin and wild-type BPTI. (24) We also assume that reformation of E · I is much more rapid than hydrolysis of EAc to produce E · I*, and that there is a steady-state equilibrium between E · I and EAc.
Each of the steps in scheme of Equation 1 is expected to be influenced by one or more ionization equilibria. For the conversion of E · I to EAc, His57 of the catalytic triad must be in its neutral form. If the microscopic rate constant for the unprotonated form is k1, then the apparent rate constant, is given by:
| (2) |
where pK1 is the pKa for the His side chain. For the reverse of this reaction, His57 must be neutral, and the newly-generated terminal amino group must be uncharged. If the two ionization reactions are assumed to be independent, the apparent rate constant for this reaction depends on pH according to:
| (3) |
where pK2 is the pKa for the new terminal amino group. For the second catalytic step, hydrolysis of the acyl intermediate, the His57 side chain must again be neutral. In this step, however, the solution pH does not significantly affect the concentration of the reactive nucleophile, a neutral water molecule. The pH dependence of the apparent rate constant is given by:
| (4) |
If it is assumed that E · I and EAc are in a steady state equilibrium, the apparent first-order rate constant for hydrolysis of the enzyme-bound inhibitor is given by the product of and the fraction of the total complex that is in the acyl intermediate form:
| (5) |
The fraction of complex in the EAc form is given by:
| (6) |
Substitution gives the following expression for the apparent first-order rate constant:
| (7) |
4.8 Protein Data Bank accession numbers
The atomic coordinates for the three structures described in this paper have been deposited in the Protein Data Bank, with accession codes 2FI3 (C14S/C38S), 2FI4 (C14S) and 2FI5 (C38S).
Acknowledgments
This research was supported by grants GM42494 (to DPG) and GM067994 (to MPH) from the U.S. National Institutes of Health. Mass spectrometry services were supported by U.S. National Science Foundation grant CHE-9708413 and the University of Utah Research Instrumentation Fund.
Abbreviations used
- BPTI
bovine pancreatic trypsin inhibitor
- 14SH/38SH
wild-type BPTI with the 14-38 disulfide selectively reduced
- 14CAM/38CAM
wild-type BPTI with the 14-38 disulfide selectively reduced and carboxyamidomethylated. Amino acid replacements are indicated by the wild-type residue type (using the one letter code for the 20 standard amino acid residues), followed by the residue number and the mutant residue type; Elsewhere, amino acid residue types are indicated by the standard 3-letter code
- HEPES
N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- MES
2-(N-morpholino)ethane sulfonic acid
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The “standard mechanism” was initially defined and extensively studied by the late Professor Michael Laskowski Jr., and the authors of the MEROPS database(7) of peptidases and inhibitors have suggested that the term “Laskowski mechanism” be adopted as a fitting tribute to his seminal contributions to this field.
References
- 1.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 2.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci., USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsson MHM, Parson WW, Warshel A. Dynamical contributions to enzyme catalysis: critical tests of a popular hypothesis. Chem. Rev. 2006;106:1737–1756. doi: 10.1021/cr040427e. [DOI] [PubMed] [Google Scholar]
- 4.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem. Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 5.Callender RH, Dyer RB. Advances in time-resolved approaches to characterize the dynamical nature of enzymatic catalysis. Chem. Rev. 2006;106:3031–3042. doi: 10.1021/cr050284b. [DOI] [PubMed] [Google Scholar]
- 6.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 7.Rawlings ND, Morton FR, Kok C, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. http://merops.sanger.ac.uk/ [DOI] [PMC free article] [PubMed]
- 8.Laskowski M, Jr, Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 9.Bode W, Huber R. Natural proteinase inhibitors and their interactions with proteinases. Eur. J. Biochem. 1992;204:433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- 10.Laskowski M, Jr, Qasim MA. What can the structures of enzyme-inhibitor complexes tell us about the structures of enzyme substrate complexes? Biochim. Biophys. Acta. 2000;1477:324–337. doi: 10.1016/s0167-4838(99)00284-8. [DOI] [PubMed] [Google Scholar]
- 11.Otlewski J, Jelen F, Zakrzewska M, Oleksy A. The many faces of protease-protein inhibitor interaction. EMBO J. 2005;24:1303–1310. doi: 10.1038/sj.emboj.7600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longstaff C, Campbell AF, Fersht AR. Recombinant chymotrypsin inhibitor 2: Expression, kinetic analysis of inhibition with α-chymotrypsin and wild-type and mutant subtilisin BPN’ and protein engineering to investigate inhibitory specificity and mechanism. Biochemistry. 1990;29:7339–7347. doi: 10.1021/bi00483a025. [DOI] [PubMed] [Google Scholar]
- 13.Beeser SA, Goldenberg DP, Oas TG. Enhanced protein flexibility caused by a destabilizing amino acid replacement in BPTI. J. Mol. Biol. 1997;269:154–164. doi: 10.1006/jmbi.1997.1031. [DOI] [PubMed] [Google Scholar]
- 14.Lu W-Y, Starovansnik MA, Dwyer JJ, Kossiakoff AA, Kent SBH, Lu W. Deciphering the role of the electrostatic inter-actions involving Gly70 in Elgin C by total chemical protein synthesis. Biochemistry. 2000;39:3575–3584. doi: 10.1021/bi992292q. [DOI] [PubMed] [Google Scholar]
- 15.Cai M, Gong Y-X, Wen L, Krishnamoorthi R. Correlation of binding-loop internal dynamics with stability and function in potato I inhibitor family: Relative contributions of Arg50 and Arg52 in Cucurbita maxima trypsin inhibitor-V as studied by site-directed mutagenesis and NMR spectroscopy. Biochemistry. 2002;41:9572–9579. doi: 10.1021/bi0258952. [DOI] [PubMed] [Google Scholar]
- 16.Peräkylä M, Kollman PA. Why does trypsin cleave BPTI so slowly? J. Am. Chem. Soc. 2000;122:3436–3444. [Google Scholar]
- 17.Radisky ES, Koshland DE., Jr A clogged gutter mechanism for protease inhibitors. Proc. Natl. Acad Sci., U S A. 2002;99:10316–10321. doi: 10.1073/pnas.112332899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radisky ES, King DS, Kwan G, Koshland DE., Jr The role of the protein core in the inhibitory power of the classic serine protease inhibitor, chymotrypsin inhibitor 2. Biochemistry. 2003;42:6484–6492. doi: 10.1021/bi034275d. [DOI] [PubMed] [Google Scholar]
- 19.Radisky ES, Lu C-JK, Kwan G, Koshland DE., Jr Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2. Biochemistry. 2005;44:6823–6830. doi: 10.1021/bi047301w. [DOI] [PubMed] [Google Scholar]
- 20.Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem. J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krowarsch D, Clerplckl T, Jelen F, Otlewski J. Canonical protein inhibitors of serine proteases. Cell. Mol. Life Sci. 2003;60:2427–2444. doi: 10.1007/s00018-003-3120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, Menegatti E. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): A milestone protein. Curr. Protein Pept. Sci. 2003;4:231–251. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- 23.Vincent J-P, Lazdunski M. Trypsin-pancreatic trypsin inhibitor association. Dynamics of the interaction and role of disulfide bridges. Biochemistry. 1972;11:2967–2977. doi: 10.1021/bi00766a007. [DOI] [PubMed] [Google Scholar]
- 24.Quast U, Engel J, Steffen E, Tschesche H, Kupfer S. Stopped-flow kinetics of the resynthesis of the reactive site peptide bond in kallikrein inhibitor (Kunitz) by β-trypsin. Biochemistry. 1978;17:1675–1682. doi: 10.1021/bi00602a015. [DOI] [PubMed] [Google Scholar]
- 25.Kress LF, Laskowski M., Sr The basic trypsin inhibitor of bovine pancreas VII. Reduction with borohydride of disulfide bond linking half-cysteine residues 14 and 38. J. Biol. Chem. 1967;242:4925–4929. [PubMed] [Google Scholar]
- 26.Creighton TE. Interactions between cysteine residues as probes of protein conformation: The disulphide bond between Cys-14 and Cys-38 of the pancreatic trypsin inhibitor. J. Mol. Biol. 1975;96:767–776. doi: 10.1016/0022-2836(75)90151-5. [DOI] [PubMed] [Google Scholar]
- 27.Kress LF, Wilson KA, Laskowski M., Sr The basic trypsin inhibitor of bovine pancreas. VIII. Changes in activity following substitutions of reduced half-cystine residues 14 and 38 with sulfhydryl reagents. J. Biol. Chem. 1968;243:1758–1762. [PubMed] [Google Scholar]
- 28.Jering H, Tschesche H. Preparation and characterization of the active derivative of bovine trypsin-allikrein inhibitor (Kunitz) with the reactive site lysine-15 - alanine -16 hydrolyzed. Eur. J. Biochem. 1976;61:443–452. doi: 10.1111/j.1432-1033.1976.tb10038.x. [DOI] [PubMed] [Google Scholar]
- 29.Goldenberg DP, Frieden RW, Haack JA, Morrison TB. Mutational analysis of a protein folding pathway. Nature. 1989;338:127–132. doi: 10.1038/338127a0. [DOI] [PubMed] [Google Scholar]
- 30.Goldenberg DP, Berger JM, Laheru DA, Wooden S, Zhang J-X. Genetic dissection of pancreatic trypsin inhibitor. Proc. Natl. Acad. Sci., USA. 1992;89:5083–5087. doi: 10.1073/pnas.89.11.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naderi HM, Thomason JF, Borgias BA, Anderson S, James TL, Kuntz ID. 1H NMR assignments and three-dimensional structure of Ala14/Ala38 bovine pancreatic trypsin inhibitor based on two-dimensional NMR and distance geometry. In: Nall B, Dill K, editors. Conformations and Forces in Protein Folding. Washington, D.C: American Association for the Advancement of Science; 1991. pp. 86–114. [Google Scholar]
- 32.Beeser SA, Oas TG, Goldenberg DP. Determinants of backbone dynamics in native BPTI: Cooperative influence of the 14–38 disulfide and the Tyr 35 side chain. J. Mol. Biol. 1998;284:1581–1596. doi: 10.1006/jmbi.1998.2240. [DOI] [PubMed] [Google Scholar]
- 33.Wilson KA, Laskowski M., Sr The basic trypin inhibitor of bovine pancreas. XI. Sequence of tryptic cleavages in an artificially prepared temporary inhibitor. J. Biol. Chem. 1971;246:3555–3561. [PubMed] [Google Scholar]
- 34.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 35.Hanson WM, Domek GJ, Horvath MP, Goldenberg DP. Rigidification of a flexible protease inhibitor variant upon binding to trypsin. J. Mol. Biol. 2007;366:230–243. doi: 10.1016/j.jmb.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber R, Kukla D, Bode W, Schwager P, Bartels K, Deisenhofer J, Steigmann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 Å resolution. J. Mol. Biol. 1974;89:73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- 37.Helland R, Otlewski J, Sundheim O, Dadlez M, Smalås AO. The crystal structures of the complexes between bovine β-trypsin and ten P1 variants of BPTI. J. Mol. Biol. 1999;287:923–942. doi: 10.1006/jmbi.1999.2654. [DOI] [PubMed] [Google Scholar]
- 38.Tsai J, Taylor R, Chothia C, Gerstein M. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 39.Ringe D, Petsko GA. Study of protein dynamics by x-ray diffraction. Methods Enzymol. 1986;131:389–433. doi: 10.1016/0076-6879(86)31050-4. [DOI] [PubMed] [Google Scholar]
- 40.Read RJ, James MNG. Introduction to the protein inhibitors. In: Barrett A, Salvesen G, editors. Proteinase Inhibitors. Amsterdam: Elsevier; 1986. pp. 301–336. [Google Scholar]
- 41.Baillargeon MW, Laskowski M, Jr, Neves DE, Porubcan MA, Santini RE, Markley JL. Soybean trypsin inhibitor (Ku-nitz) and its complex with trypsin. Carbon-13 nuclear magnetic resonance studies of the reactive site arginine. Biochemistry. 1980;19:5703–5710. doi: 10.1021/bi00566a006. [DOI] [PubMed] [Google Scholar]
- 42.Richarz R, Tschesche H, Wüthrich K. Carbon-13 nuclear magnetic resonance studies of the selectively isotope-labeled reactive site peptide bond of the basic pancreatic trypsin inhibitor in the complexes with trypsin, trypsinogen and anhydrotrypsin. Biochemistry. 1980;19:5711–5715. doi: 10.1021/bi00566a007. [DOI] [PubMed] [Google Scholar]
- 43.Kojima S, Kumaga I, Miura K-I. Requirement for a disulfide bridge near the reactive site of protease inhibitor SSI (Streptomyces subtilisin inhibitor) for its inhibitory action. J. Mol. Biol. 1993;230:395–399. doi: 10.1006/jmbi.1993.1157. [DOI] [PubMed] [Google Scholar]
- 44.Gutfreund H. The characterization of the catalytic site of trypsin. Trans. Faraday Soc. 1955;51:441–446. [Google Scholar]
- 45.Robillard G, Shulman RG. High resolution nuclear magnetic resonance study of the histidine-Aspartate hydrogen bond in chymotrypsin and chymotrypsinogen. J. Mol. Biol. 1972;71:507–511. doi: 10.1016/0022-2836(72)90366-x. [DOI] [PubMed] [Google Scholar]
- 46.Bachovchin WW, Roberts JD. Nitrogen-15 nuclear magnetic reesonance spectroscopy. The state of histidine in the catalytic triad of α-lytic protease. Implications for the charge relay mechanism of peptide-bond cleavage by serine proteases. J. Am. Chem. Soc. 1978;100:8041–8047. [Google Scholar]
- 47.Kossiakoff AA, Spencer SA. Neutron diffraction identifies His 57 as the catalytic base in trypsin. Nature. 1980;288:414–416. doi: 10.1038/288414a0. [DOI] [PubMed] [Google Scholar]
- 48.Steitz TA, Shulman RG. Crystallographic and NMR studies of the serine proteases. Annu. Rev. Biophys. Bioeng. 1982;11:419–444. doi: 10.1146/annurev.bb.11.060182.002223. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski M, Jr, Finkenstadt WR, Mattis JA, McKee RE. Thermodynamics and kinetics of the interaction of trypsin with trypsin inhibitors. In: Peeters H, editor. Protides of biological fluids. Vol. 23. Oxford, UK: Pergamon; 1975. pp. 85–91. [Google Scholar]
- 50.Otlewski J, Zbyryt T. Single peptide bond hydrolysis/ resynthesis in squash inhibitor so serine proteinases. 1. Kinetics and thermodynamics of the interaction between squash inhibitors and bovine β-trypsin. Biochemistry. 1994;33:200–207. doi: 10.1021/bi00167a026. [DOI] [PubMed] [Google Scholar]
- 51.Vincent J-P, Lazdunski M. The interaction between α-chymotrypsin and pancreatic trypsin inhibitor (Kunitz inhibitor): kinetic ant thermodynamic properties. Eur. J. Biochem. 1973;38:365–372. doi: 10.1111/j.1432-1033.1973.tb03069.x. [DOI] [PubMed] [Google Scholar]
- 52.Ascenzi P, Aduci P, Amiconi G, Ballio A, Guaragna A, Menegatti E, Schnebil HP, Bolognesi M. Binding of the recombinant proteinase inhibitor elgin c from leach Hirudo medicinalis to serine (Pro)enzymes: A comparative thermodynamic study. J. Mol. Recognit. 1991;4:113–119. doi: 10.1002/jmr.300040402. [DOI] [PubMed] [Google Scholar]
- 53.Baker BM, Murphy KP. Dissecting the energetics of a protein-protein interaction: The binding of ovomucoid third domain to elastase. J. Mol. Biol. 1997;268:557–569. doi: 10.1006/jmbi.1997.0977. [DOI] [PubMed] [Google Scholar]
- 54.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups in proteins. Protein Sci. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fastrez J, Fersht AR. Demonstration of the acyl-enzyme mechanism for the hydrolysis of peptides and anilides by chymotrypsin. Biochemistry. 1973;12:2025–2034. doi: 10.1021/bi00735a001. [DOI] [PubMed] [Google Scholar]
- 56.Jencks WP. On the attribution and additivity of binding energies. Proc. Natl. Acad Sci., U S A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sievers A, Wolfenden R. The effective molarity of the substrate phosphoryl group in the transition state for yeast OMP decarboxylase. Bioorganic Chem. 2005;33:45–53. doi: 10.1016/j.bioorg.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Miller BG, Wolfenden R. Catalyic proficiency: The unusual cae of OMP decarboxylase. Annu. Rev. Biochem. 2002;71:847–885. doi: 10.1146/annurev.biochem.71.110601.135446. [DOI] [PubMed] [Google Scholar]
- 59.Coombs GS, Rao MS, Olson AJ, Dawson PE, Madison EL. Revisiting catalysis of chymotrypsin family serine proteases using peptide substrates and inhibitors with unnatural main chains. J. Biol. Chem. 1999;274:24074–24079. doi: 10.1074/jbc.274.34.24074. [DOI] [PubMed] [Google Scholar]
- 60.Fodor K, Harmat V, Neutze R, Szilágyi L, Gráf L, Katona G. Enzyme:substrate hydrogen bond shortening during the acylation phase of serine protease catalysis. Biochemistry. 2006;45:2114–2121. doi: 10.1021/bi0517133. [DOI] [PubMed] [Google Scholar]
- 61.Liu B, Schofield CJ, Wilmouth RC. Structural analysis on intermediates in serine protease catalysis. J. Biol. Chem. 2006;281:24024–24035. doi: 10.1074/jbc.M600495200. [DOI] [PubMed] [Google Scholar]
- 62.Page MI, Jencks WP. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc. Natl. Acad. Sci., U.S.A. 1971;68:1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katona G, Berglund GI, Hajdu J, Gráf L, Szilágyi L. Crystal structure reveals basis for the inhibitor resistance of human brain trypsin. J. Mol. Biol. 2002;315:1209–1218. doi: 10.1006/jmbi.2001.5305. [DOI] [PubMed] [Google Scholar]
- 64.Szmola R, Kukor Z, Sahin-Tóth M. Human mesotrypsin is a unique digestive protease specialized for the degradation of trypsin inhibitors. J. Biol. Chem. 2003;278:48580–48589. doi: 10.1074/jbc.M310301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salameh MA, Soares AS, Hockla A, Radisky ES. Structural basis for accelerated cleavage of bovine pancreatic trypsin inhibitor (BPTI by human mesotrypsin. J. Biol. Chem. 2008;283:4115–4123. doi: 10.1074/jbc.M708268200. [DOI] [PubMed] [Google Scholar]
- 66.Goldenberg DP. Kinetic analysis of the folding and unfolding of a mutant form of bovine pancreatic trypsin inhibitor lacking the cysteine-14 and −38 thiols. Biochemistry. 1988;27:2481–2489. doi: 10.1021/bi00407a034. [DOI] [PubMed] [Google Scholar]
- 67.Bulaj G, Goldenberg DP. Early events in the disulfide-coupled folding of BPTI. Protein Sci. 1999;8:1825–1842. doi: 10.1110/ps.8.9.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J-X, Goldenberg DP. Mutational analysis of the BPTI folding pathway: I. Effects of aromatic → Leu substitutions on the distribution of folding intermediates. Protein Sci. 1997;6:1549–1562. doi: 10.1002/pro.5560060719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chase T, Jr, Shaw E. p-Nitrophenyl-p’-guanidobenzoate HCl: A new active site titrant for trypsin. Biochem. Biophys. Res. Comm. 1967;29:508–514. doi: 10.1016/0006-291x(67)90513-x. [DOI] [PubMed] [Google Scholar]
- 70.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewsk J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 71.Jones TA, Zou J-Y, Cowan SW. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 72.Reisnner KJ, Aswad DW. Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals. Cell. Mol. Life Sci. 2003;60:1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]