Abstract
Ustilago maydis, a Basidiomycete fungus that infects maize, exhibits two basic morphologies, a yeast-like and a filamentous form. The yeast-like cell is elongated, divides by budding, and the bud grows by tip extension. The filamentous form divides at the apical cell and grows by tip extension. The repertoire of morphologies is increased during interaction with its host, suggesting that plant signals play an important role in generation of additional morphologies. We have used S. cerevisiae and S. pombe genes known to play a role in cell polarity and morphogenesis, and in the cytoskeleton as probes to survey the U. maydis genome. We have found that most of the yeast machinery is conserved in U. maydis, albeit the degree of similarity varies from strong to weak. The U. maydis genome contains the machinery for recognition and interpretation of the budding yeast axial and bipolar landmarks; however, genes coding for some of the landmark proteins are absent. Genes coding for cell polarity establishment, exocytosis, actin and microtubule organization, microtubule plus-end associated proteins, kinesins, and myosins are also present. Genes not present in Saccharomyces cerevisiae and Schizosaccharomyces pombe include a homologue of mammalian Rac, a hybrid myosin-chitin synthase, and several kinesins that exhibit more similarity to their mammalian counterparts. We also used the U. maydis genes identified in this analysis to search other fungal and other eukaryotic genomes to identify the closest homologues. In most cases, not surprisingly, the closest homologue is among filamentous fungi, not the yeasts, and in some cases it is among mammals.
Keywords: polarized growth, cytoskeleton, polarisome, fungal morphogenesis
I. OVERVIEW OF CELL POLARITY AND CELL ORPHOGENESIS
Cell polarity is a fundamental biological process by which asymmetry of cell components (cytoskeleton, cell organelles, and membrane domains) is generated. Polarization of cell components occurs in diverse cells in response to endogenous programming and to external cues. This cellular asymmetry is important in determining cell morphogenesis and the plane of cell division in a variety of eukaryotic cells, and is crucial for development of metazoans. Studies in budding yeast and fission yeast have been instrumental in identification of the machinery involved in cell polarity and morphogenesis. These studies indicate that recognition of a spatial landmark triggers assembly of protein complexes that initiate assembly and polarization of the actin cytoskeleton, which directs vesicle delivery to sites of growth.
Fungal filaments (hyphae) exhibit continuous polarized growth at their tips. New axes of polarized growth (branches) are established along the main axis of polarized growth. Current work in filamentous fungi aims to understand the molecular mechanisms that determine: (a) the axis of polarized growth, (b) new axes of cell polarity, and (c) the site of cytokinesis and septum formation. The availability of the genome sequences of fungi from across the different subdivisions of the kingdom, together with improved tools for generation of knock out strains, regulatable gene expression, and sophisticated imaging techniques promise to reveal new insights about polarized growth and cell morphogenesis in filamentous fungi, and will likely lead in the future to the generation of protein interaction maps for this machinery as has been done in Saccharomyces cerevisiae (Drees et al., 2001).
Here we explore the genome sequence of Ustilago maydis in search of homologues of genes known to play crucial roles in cell morphogenesis and cell polarity in the yeasts. We first provide a synopsis of the morphologies that characterize U. maydis growth in culture and in the plant. We follow with a brief summary of key players of polarized growth in S. cerevisiae, and indicate which components of the machinery are conserved in U. maydis, and where known, describe their role in polarized growth. We conclude from our analysis that most of the yeast machinery is conserved in U. maydis, as it is in other filamentous fungi (Harris, 2006; Philippsen et al., 2005), and in many instances it is used in novel ways.
II. OVERVIEW OF THE MORPHOLOGICAL TRANSITIONS IN THE LIFE CYCLE OF U. maydis
Ustilago maydis is a Basidiomycete fungus that infects maize (Zea mays L) and teosinte (Zea mays spp. mexicana or spp. parviglumis). The fungus exhibits two basic morphologies and is capable of switching from one form to the other (the dimorphic switch) (Fig. 1). This switch is crucial to pathogenicity. The yeast-like form is unicellular, haploid, divides by budding, and is nonpathogenic; the filamentous form is dikaryotic, grows by tip extension, and is pathogenic (reviewed in Banuett, 1995; 2002; Christensen, 1963; Holliday, 1974; Klosterman et al., 2007). The dimorphic switch and other aspects of the life cycle are controlled by two unlinked mating type loci, a and b. The a locus codes for components of a signal transduction pathway (pheromone precursor and receptor genes) and the b locus codes for a combinatorial homeodomain protein. The a locus governs cell fusion of haploid cells and filamentous growth of the dikaryon in vitro but not in planta; the b locus is the major determinant of filamentous growth, in vitro and in planta, and of pathogenicity (reviewed in Banuett, 2002; 2007; Klosterman et al., 2007). In order to complete the life cycle, haploids that fuse must differ at both a and b (for example, a1 b1 + a2 b2). The a locus has two alleles (a1 and a2) and the b locus 25 naturally occurring alleles (b1....b25), and any combination of different b alleles results in an active b protein (reviewed in Banuett 2002; 2007; Klosterman et al., 2007).
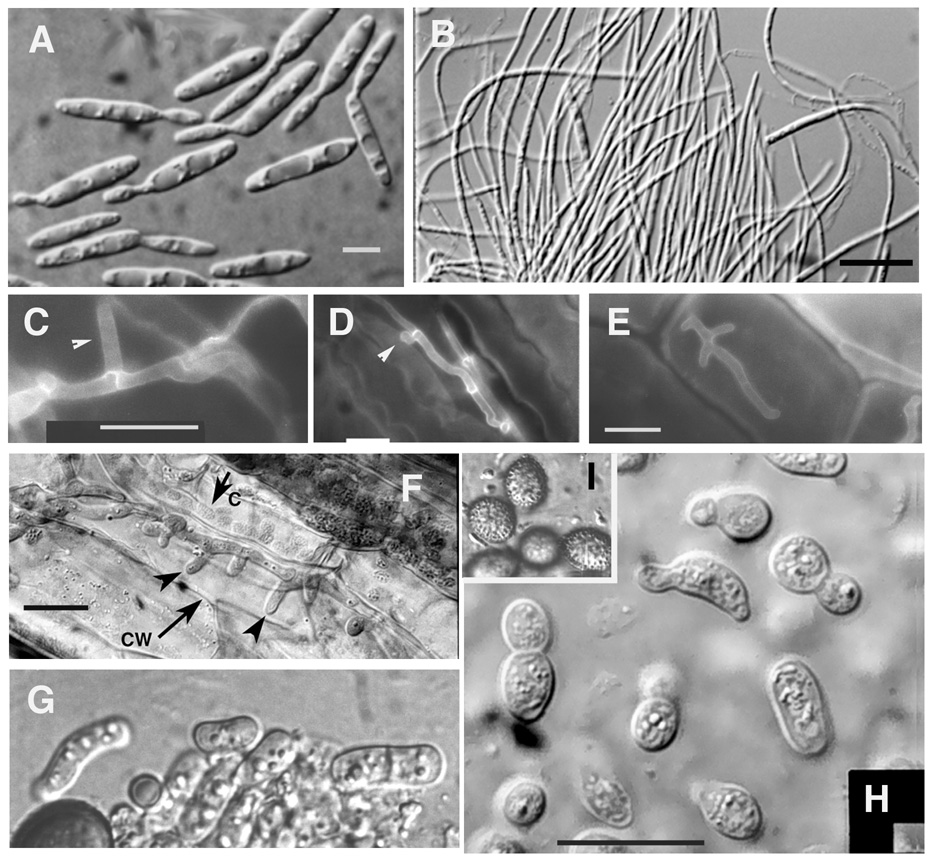
Figure 1. Different morphologies in the life cycle of U. maydis.
U. maydis exhibits two basic morphologies, a yeast-like and a filamentous form. Interaction with the plant increases the repertoire of morphologies observed suggesting that plant signals play a role in reorganization of the machinery for cell polarity, cell morphogenesis, and the cytoskeleton. Panel A. Different stages of budding of haploid yeast-like cells. Panel B. Dikaryotic filaments in culture. No branching is observed. Panels C, D, E, and F. Dikaryotic filaments growing in the plant. Panel C shows a branch (arrowhead). Panel D shows a clamp-like structure (arrowhead) (see Scherer et al., 2006). Panels E and F show multiple branches (arrowheads in Panel F). The cell wall of the plant cell is indicated an arrow. Panel G. Cylindrical cells released upon hyphal fragmentation within the tumors during teliospore formation. Panel H. The cylindrical cells undergo morphological changes during teliospore formation. The cells in Panels G and H are normally embedded in a mucilaginous matrix, which may provide osmotic support during the cell wall remodelling events occurring during these morphological transitions (Banuett and Herskowtiz, 1996). Inset (I). Mature teliospores with echinulate cell wall. The teliospores give rise to haploid yeast-like cells upon meiosis. The scale bar in Panel A is 5 µm, and in Panels B, C, D, E, F, and H, 50 µm.
The repertoire of morphologies is expanded by interaction of U. maydis with its hosts (Fig. 1). This is evidenced by the formation of a specialized structure for penetration, the appressorium, (see for example, Brachmann et al., 2003; Snetselaar and Mims, 1993), by extensive branching on the leaf surface and inside plant cells (Banuett and Herskowitz, 1996), and by formation of clamp-like structures for nuclear distribution (Scherer et al., 2006). Appressoria, branch formation, and clamp-like structures are not observed in culture. Once tumors are formed, dikaryotic hyphae undergo a discrete developmental program characterized by distinct morphologies (cylindrical cells, spherical cells, and other diverse shapes) that arise upon hyphal fragmentation and lead to formation of the teliospore, a round spore with a specialized cell wall (Fig. 1) (Banuett and Herskowitz, 1996). The distinct cell morphologies present in the plant are not observed in vitro. It has thus been proposed that the plant produces signals that trigger fungal differentiation and reorganization of the machinery for polarized growth (Banuett and Herskowitz, 1996). The nature of these signals is not known. Interestingly, the fungus also elicits changes in cell morphology and nuclear position of the host (Banuett and Herskowitz, 1996; Ruiz-Herrera et al., 1999; reviewed in Banuett, 2002). Thus, the interaction of U. maydis with its host appears to be a reciprocal process of signal exchange that results in alterations in cell morphology of both host and fungus.
A. The yeast-like cell morphology
The yeast-like cells are elongated with tapered ends (cigar-shaped). They bud once per cell cycle at one of the cell poles. The mechanism by which one of the cell poles is chosen versus the other is not known. Cells are able to bud at new sites at the cell poles or use a previously chosen site for budding as in apiculate yeasts (Jacobs et al., 1994). In some respects this pattern resembles the bipolar budding pattern of S. cerevisiae. The U. maydis bud grows by tip extension (Banuett and Herskowitz, 2002). There is no phase of isotropic growth, which contrasts with bud growth in S. cerevisiae where there is a short phase of polarized growth early in the cell cycle followed by a switch to isotropic growth in the G2/M phase of the cell cycle (reviewed in Pruyne and Bretscher, 2000a,b). Formation of the U. maydis bud entails several morphological steps that result in the final shape of the bud (Banuett and Herskowitz, 2002).
B. The filamentous form
In culture, the filamentous form grows by tip extension, as occurs in other filamentous fungi, and divides at the apical cell to produce an actively growing tip cell and a subapical cell devoid of cytoplasm, but little is known about division of the apical cell. In planta, most hyphal compartments, not just the tip cell, contain cytoplasm (Banuett and Herskowitz, 1996) and nuclear distribution involves clamp-like structures (Scherer et al., 2006).
C. The Spitzenkörper and filamentous growth
Fungal hyphae exhibit continuous polarized growth; there is no arrest of growth during cytokinesis. Examples of cells that exhibit such highly polarized growth are pollen tubes and root hairs in plants, and neurons in animals.
In filamentous fungi, a fungal-specific phase-contrast opaque body, the Spitzenkörper (apical body), located at or just below the tip of the apical cell, is thought to drive hyphal growth (López-Franco, 1996; Reynaga-Peña et al., 1997). It consists of a heterogeneous population of vesicles surrounding a core that contains polysomes, microtubules, and actin, and is proposed to serve as a supply center for the distribution of vesicles containing materials necessary for tip extension (reviewed in Bartnicki-García, 2002; Harris et al., 2005; Harris, 2006). The Spitzenkörper can be visualized with the amphiphilic styryl dye FM4-64 (Fischer-Parton et al, 2000). Long-range transport of vesicles from other parts of the hypha to the Spitzenkörper is proposed to occur along microtubules, and delivery from the Spitzenkörper to sites of growth is proposed to occur on actin tracks (reviewed in Harris et al., 2005; Harris, 2006). Both microtubules and actin are necessary for hyphal morphogenesis. Disruption of microtubules, kinesin, dynein, and dynactin results in unstable axes of polarized growth but not in absence of polarized growth per se, whereas disruption of actin causes complete loss of cell polarity and results in isotropic growth (Harris et al., 2005; Heath, 2000; Lee et al., 2001; Riquelme et al., 2000; Rupes et al., 1995; Seiler et al., 1999; Torralba et al., 1998a,b; Virag and Griffiths, 2004). Some of these conditions also result in loss or alteration of the position or function of the Spitzenkörper (Crampin et al., 2005; Konzack et al., 2005; Riquelme et al., 2000; Seiler et al., 1999).
There is a dearth of information concerning the molecular composition of the Spitzenkörper. Recent work in different filamentous fungi indicates that formin (a multidomain, actin-nucleating protein), myosin light chain, Sec4 (a Rab GTPase), BemA (homologue of S. cerevisiae Bem1), TeaA and TeaR (homologues of S. pombe end cell markers Tea1 and Mod5, respectively), and two chitin synthases are components of the Spitzenkörper (see below) (Crampin et al., 2005; Leeder and Turner, 2007; Riquelme et al., 2007; Sharpless and Harris, 2000; Takeshita et al., 2008). The presence of formin suggests that the Spitzenkörper may serve as an actin nucleation center. Little is known about the Spitzenkörper in U. maydis.
III. U. maydis HOMOLOGUES OF YEAST GENES INVOLVED IN CELL POLARITY AND CELL MORPHOGENESIS
The yeasts, S. cerevisiae and Schizosaccharomyces pombe have been used extensively in studies of cell polarity and morphogenesis (Chang and Peter, 2003). These studies provide the framework in which to understand these processes in other organisms, in particular the fungi; therefore, we have used these two yeasts as a reference point in our analysis. Below we describe some of the components of the machinery for polarized growth in S. cerevisiae (for S. pombe see supplementary material), and identify components conserved in U. maydis. We describe, where known, their function in U. maydis, and in some filamentous fungi.
A. A hierarchy of GTPase modules controls bud site selection and establishment of polarized growth in S. cerevisiae
1. Recognition of the spatial landmark
S. cerevisiae cells divide by budding and exhibit two budding patterns, axial and bipolar, which are genetically determined by the MAT locus. a or α cells exhibit the axial budding pattern, in which mother and daughter cells bud immediately adjacent to the previous budding site. a/α cells exhibit a bipolar budding pattern, in which daughters bud distally to the site of their birth, whereas the mother can bud distally or proximally to the previous budding site (reviewed in Casamayor and Snyder, 2002; Chant, 1999). A GTPase module consisting of a Ras-like GTPase Rsr1/Bud1, its guanine nucleotide exchange factor (GEF) Bud5, and its GTPase-activating protein (GAP) Bud2 determine the site of budding in response to a spatial landmark for axial or bipolar budding (reviewed in Casamayor and Snyder, 2002; Chant, 1999). The landmark for axial budding consists of septins, Bud3, Bud4, Axl1, Bud10/Axl2, and for bipolar budding of Bud8, Bud9, Bud7, Rax1, and Rax2. The Rsr1/Bud1 GTPase module is not essential for viability; in its absence cells are still capable of polarized growth but in a random manner (reviewed in Casamayor and Snyder, 2002; Chant, 1999; Park and Bi, 2007).
2. Polarity establishment and the Cdc42 GTPase module
The Rsr1/Bud1 GTPase module interprets the spatial landmark (axial or bipolar) and recruits another GTPase module that consists of the Rho GTPase Cdc42, its guanine nucleotide exchange factor (GEF) Cdc24, and its GTPase-activating proteins (GAPs) Bem3 and Rga1/2 (reviewed in Casamayor and Snyder, 2002; Chant, 1999; Park and Bi, 2007). This module in turn recruits the actin cytoskeleton, which polarizes secretion towards the site of growth (reviewed in Pruyne and Bretscher, 2000a,b).
B. U. maydis homologues of landmark proteins and the BUD site selection and polarity establishment GTPase modules
The U. maydis genome lacks coding information for homologues of Bud3 and Bud4 (axial landmark), and Bud8 and Bud9 (bipolar landmark) (Table 1), but contains the information for Bud10/Axl2 (axial landmark), and Rax1 and Rax2 (bipolar landmark). Thus, U. maydis contains a subset of the genes that code for the axial and bipolar landmark proteins. Their role in budding and cell morphogenesis remains to be determined. Interestingly, in Ashbya gossypii, a filamentous fungus closely related to S. cereviasae, AgBud3 appears to act as a landmark for septation (Wendland, 2003). Coding information for components of the Rsr1/Bud1 GTPase module (Rsr1/Bud1, Bud2, and Bud5) and many other recently identified genes required for bipolar budding (BUD13-32; Ni and Snyder, 2001) is present in the U. maydis genome (Table 1). The function of these genes remains to be determined. In A. gossypii, AgRsr1/Bud1 is required for actin organization, for normal hyphal growth and branching, and for position of the polarisome component AgSpa2. Control of the position of a polarisome component by the Rsr1/Bud1 GTPase is a novel function of this GTPase. The Agrsr1 null mutant is characterized by false branch initiation, pausing and reinitiation of growth resulting in bulges along the hypha, and transient appearance and disappearance of AgSpa2 that correlates with phases of pausing and growth (Bauer et al., 2004). In wild type hyphae, AgSpa2 (a homologue of S. cerevisiae Spa2, see below) is present at the hyphal tip continuously. In Candida albicans, another fungus closely related to S. cerevisiae, CaRsr1/Bud1 defects result in random budding in yeast cells and in defects in germ tube emergence (Yaar et al., 1997).
Table 1. U. maydis homologues of the S. cerevisiae machinery for cell polarity, cell morphogenesis, and the cytoskeleton.
S. cerevisiae genes involved in cell polarity, cell morphogenesis, and the cytoskeleton (column 2) were used to probe the U. maydis genome with wuBlast at the MIPS Ustilago database MUMDB (http://mips.gsf.de/genre/proj/ustilago/), and the Ustilago maydis Database at the Broad Institute (http://www.broad.mit.edu/annotation/genome/ustilago_maydis/Home.html). We also used Blast at NCBI (http://www.ncbi.nlm.nih.gov/blast) (Altschul et al., 1997). The genes identified in U. maydis were then used to search for homologues in the Basidiomycota and Ascomycota, and in other eukaryotes using the SIMAP tool at MIPS (see Fig.1S, supplementary material). The source of S. cerevisiae gene products was Drees et al., 2001; Pruyne and Bretscher, 2000; and GO annotation at the Saccharomyces genome database (http://www.yeastgenome.org/). The systematic name for S. cerevisiae genes (column 3) was obtained from (http://www.yeastgenome.org/). U. maydis gene numbers (column 4) were obtained from (http://mips.gsf.de/genre/proj/ustilago/) after BLAST analysis. E values (column 5) indicated are as obtained from wuBlast analysis at MIPS. Other parameters were also used in our Blast analysis, in particular in the comparison across different fungal groups and other eukaryotes (data not shown but available upon request). The last column provides the function of the S. cerevisiae genes (http://www.yeastgenome.org/). The list of genes in this Table is not comprehensive; it does not include many genes involved in cytokinesis, cell separation, secretion, and cell cycle that were used in the analysis shown in Figs 1S (supplementary material).
| CELL POLARITY/CELL MORPHOGENESIS | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 1 | BAG7 | YOR134W | 02902 | 3.3e-48 | RhoGAP; functionally related to Sac7 |
| 2 | BEM1 | YBR200W | 10934 | 3.7e-44 | Establishment of cell polarity. Scaffold for different protein complexes |
| 3 | BEM2 | YER155C | 01161 | 3.4e-42 | RhoGAP |
| 4 | BEM3 | YPL115C | 11852 | 8.9e-24 | GAP for Cdc42 and Rho1 |
| 5 | BEM4 | YPL161C | no hits | Interacts with Cdc42 and Rho1. Bud emergence. | |
| 6 | BNI4 | YNL233W | no hits | Required for localization of CHSIII to bud neck | |
| 7 | BOI1 | YBL085W | 03037 | 1.7e-23 | Bem1-binding protein. Polarized growth |
| 8 | BOI2 | YER114C | 03037 | 1.8e-19 | Bem1-binding protein. Polarized growth |
| 9 | BUD1/RSR1 | YGR152C | 11909 | 7.8e-49 | Bud site selection. RAS superfamily protein |
| 10 | BUD2 | YKLO92C | 01282 | 6.9e-10 | Bud site selection. GAP for Bud1/Rsr1 |
| 11 | BUD3 | YCLO14W | no hits | Bud site selection. Axial landmark | |
| 12 | BUD4 | YJRO92W | no hits | Bud site selection. Axial landmark | |
| 13 | BUD5 | YCRO38C | 11476 | 1.3e-12 | Bud site selection. GEF for Bud1/Rsr1 |
| 14 | BUD6/AIP3 | YLR319C | 11657 | 1.6e-70 | Polarisome component; interacts with formin and actin |
| 15 | BUD7 | YOR299W | 04959 | 5.2e-62 | Export of Chs3 from Golgi to plasma membrane. Bipolar budding. |
| 16 | BUD8 | YLR353W | no hits | Bud site selection. Bipolar landmark | |
| 17 | BUD9 | YGR041W | no hits | Bud site selection. Bipolar landmark | |
| 18 | BUD10/AXL2 | YIL140W | 01289 | 2.8e-24 | Bud site selection |
| 19 | BUD13 | YGL174W | 00191 | 1.1e-10 | Bud site selection. Bipolar budding. |
| 20 | BUD14 | YAR014C | 01012 | 3.4e-14 | Bud site selection. Bipolar budding. |
| 21 | BUD15/EDE1 | YBL047C | 05533 | 4.8e-61 | Bud site selection. Bipolar budding. |
| 22 | BUD16 | YEL029C | 10310 | 1.1e-42 | Bud site selection. Bipolar budding. |
| 23 | BUD17 | YNR027W | 10310 | 1.1e-41 | Bud site selection. Bipolar budding. |
| 24 | BUD20 | YLR074C | 04625 | 2.5e-11 | Bud site selection. Bipolar budding. |
| 25 | BUD21 | YOR078W | no hits | Bud site selection. Bipolar budding. | |
| 26 | BUD22 | YMR014W | 01709 | 4.8e-13 | Bud site selection. Bipolar budding. |
| 27 | BUD23 | YCR047C | 01551 | 8.4e-68 | Bud site selection. Bipolar budding. |
| 28 | BUD25 | YER014C-A | no hits | Bud site selection. Bipolar budding. | |
| 29 | BUD27 | YFL023W | no hits | Bud site selection. Bipolar budding. | |
| 30 | BUD31 | YCR063W | 10057 | 1.0e-46 | Bud site selection. Bipolar budding. |
| 31 | BUD32 | YGR262C | 12089 | 2.3e-20 | Bud site selection. Bipolar budding. |
| 32 | CDC3 | YLR314C | 10503 | 1.1e-117 | Septin |
| 33 | CDC10 | YCR002C | 10644 | 1.6e-96 | Septin |
| 34 | CDC11 | YJR076C | 03449 | 5.0e-77 | Septin |
| 35 | CDC12 | YHR107C | 03599 | 3.0e-95 | Septin |
| 36 | CDC24 | YAL041W | 02422 | 4.2e-75 | GEF for Cdc42. Polarity establishment |
| 37 | CDC42 | YLR229C | 00295 | 1.8e-90 | Small GTP-binding protein of RAS superfamily. Polarity establishment and maintenance. |
| 38 | CHS1 | YNL192W | 10117 | 1.5e-169 | chitin synthase I |
| 39 | CHS2 | YBR038W | 10120 | 3.5e-181 | chitin synthase II |
| 40 | CHS3 | YBR023C | 10277.2 | 1.1e-256 | chitin synthase III |
| 41 | CHS4/SKT5 | YBL061C | 10641 | 1.6e-63 | Activator of chitin synthase III |
| 42 | CHS5 | YLR330W | 02840 | 7.3e-53 | Localization of chitin synthase III |
| 43 | CHS6 | YJL099W | 04959 | 1.2e-32 | Transport of chitin synthase III from Golgi to plasma membrane |
| 44 | CHS7 | YHR142W | 11089 | 7.5e-51 | Export of Chs3 from ER |
| 45 | CKA1 | YIL035C | 01180 | 6.3e-111 | Casein kinase II, alpha subunit |
| 46 | CKA2 | YOR061W | 01180 | 7.6e-106 | Casein kinase II, alpha subunit |
| 47 | CKB1 | YGL019W | 01094 | 9.3e-52 | Casein kinase II, beta subunit |
| 48 | CKB2 | YOR039W | 06107 | 7.8e-59 | Casein kinase II, beta subunit |
| 49 | CLA4 | YNL298W | 10145 | 1.1e-130 | PAK family kinase. Cdc42 effector |
| 50 | CLC1 | YGR167W | 01316 | 1.4e-17 | Clathrin light chain |
| 51 | CMD1 | YBR109C | 03910 | 1.5e-47 | Calmodulin |
| 52 | CYK3 | YDL117W | no hits | Role in cytokinesis | |
| 53 | DFG5 | YMR238W | 06073 | 1.1e-22 | Required for cell elongation, cell polarity |
| 54 | ELM1 | YKL048C | 04755 | 5.4e-26 | Ser/Thr protein kinase in Swe1 pathway |
| 55 | END3 | YNL084C | no hits | Endocytosis and cytoskeletal organization | |
| 56 | ENT1 | YDL161W | 03598 | 2.1e-48 | Endocytosis and cytoskeletal organization |
| 57 | ENT2 | YLR206W | 03598 | 1.6e-66 | Endocytosis and cytoskeletal organization |
| 58 | ENT3 | YJR125C | 05992 | 1.1e-58 | Recruitment of clathrin |
| 59 | ENT4 | YLL038C | 03598 | 2.0e-16 | Endocytosis |
| 60 | FLO8 | YER109C | 01278 | 3.1e-12 | Required for pseudohyphal growth |
| 61 | GIC1 | YHR061C | no hits | Cdc42 effector. Contains CRIB domain | |
| 62 | GIC2 | YDR309C | no hits | Cdc42 effector. Contains CRIB domain | |
| 63 | GIN4 | YDR507C | 03928 | 1.1e-80 | Protein kinase involved in bud growth. Similar to Kcc4 and Hsl1 |
| 64 | HOF1/CYK2 | YMR032W | 00168 | 1.5e-14 | Required for cytokinesis. |
| 65 | HSL1 | YKL101W | 03928 | 1.3e-81 | Nim-related protein kinase; septin ring assembly; Swe1 pathway |
| 66 | HSL7 | YBR133C | 15057 | 1.2e-41 | Involved in Swe1 pathway |
| 67 | HYM1 | YKL189W | 10613 | 3.0e-47 | Cell morphogenesis |
| 68 | IQG1 | YPL242C | 10730 | 2.5e-49 | Cytokinesis. Cell morphology. |
| 69 | KCC4 | YCL024W | 03928 | 3.6e-79 | Nim-related kinase |
| 70 | KEL1 | YHR158C | 15019 | 2.0e-40 | Cell fusion and cell morphology |
| 71 | KEL2 | YGR238C | 15019 | 1.9e-37 | Cell fusion and cell morphology |
| 72 | MSB1 | YOR188W | no hits | Cell morphogenesis. Involved in regulation of Pkc1 and 1,3-beta glucan synthase | |
| 73 | MSB2 | YGR014W | 00480 | 2e-41 | Multicopy suppressor of cdc24 bud emergence defect |
| 74 | MSB3 | YNL293W | 00935 | 3.8e-22 | GTPase activating protein for Sec4; localizes to sites of polarized growth |
| 75 | MSB4 | YOL112W | 00935 | 2.7e-24 | GTPase activating protein for Sec4; localizes to sites of polarized growth |
| 76 | MUB1 | YMR100W | 02587 | 2.0e-50 | Cell separation defect; homologue of A. nidulans SamB |
| 77 | PEA2 | YER149C | no hits | Polarisome component. | |
| 78 | PKC1 | YBL105C | 15023 | 1.4e-230 | Ser/Thr protein kinase; cell wall integrity. Localizes to sites of polarized growth. |
| 79 | PLC1 | YPL268W | 02982 | 2.9e-75 | Phospholipase C. Pseudohyphal growth |
| 80 | PXL1 | YKR090W | 01663 | 2.1e-17 | Maintenance of polarized growth. Similarity to metazoan paxillin |
| 81 | RAS1 | YOR101W | 00986 | 9.9e-65 | GTPase involved in cAMP signalling pathway. |
| 82 | RAS2 | YNL098C | 01643 | 2.3e-49 | Homologue of mammalian Ras proto-oncogene |
| 83 | RAX1 | YOR301W | 00956 | 6.2e-23 | Bipolar budding pattern landmark |
| 84 | RAX2 | YLR084C | 05194 | 2.4e-31 | Bipolar budding pattern landmark |
| 85 | RDI1 | YDL135C | 05693 | 7.8e-33 | Rho GDP dissociation inhibitor for Cdc42 |
| 86 | RGA1 | YOR127W | 03864 | 8.3e-44 | GAP for Cdc42 |
| 87 | RGA2 | YDR379W | 03864 | 2.0e-39 | GAP for Cdc42 |
| 88 | RHO1 | YPR165W | 05734 | 2.7e-78 | Rho subfamily. Regulator of Pkc1 and 1,3-beta glucan synthase |
| 89 | RHO2 | YNL090W | 02494 | 6.4e-54 | Rho subamily protein. |
| 90 | RHO3 | YIL118W | 04070 | 8.7e-52 | Rho subamily protein. |
| 91 | RHO4 | YKR055W | 10663 | 8.9e-48 | Rho subamily protein. |
| 92 | RHO5 | YNL180C | 00295 | 3.5e-52 | Rho subamily protein. |
| 93 | ROM1 | YGR070W | 15070 | 5.9e-104 | GEF for Rho1 |
| 94 | ROM2 | YLR371W | 15070 | 2.0e-137 | GEF for Rho1 |
| 95 | RVS161 | YCR009C | 05283 | 4.1e-66 | Amphiphysin-like lipid raft protein. Regulates polarization of the actin cytoskeleton. |
| 96 | RVS167 | YDR388W | 01748 | 1.4e-51 | Regulation of actin cytoskeleton |
| 97 | SAC7 | YDR389W | 02902 | 1.2e-43 | GAP for Rho1 (Functionally related to Bag7) |
| 98 | SHS1/SEP7 | YDL225W | 03449 | 3.7e-72 | Septin |
| 99 | SPA2 | YLL021W | 04468 | 5.6e-20 | Polarisome component |
| 100 | SPH1 | YLR313C | 04468 | 2.3e-10 | Polarisome component. Homologous to Spa2 |
| 101 | SWE1 | YJL187C | 06337 | 5.3e-40 | Ser/Thr protein kinase; involved in switch from polarized to isotropic growth |
| 102 | WSC1/HCS77 | YOR008C | 11546 | 3.1e-21 | Stretch receptor; cell wall integrity pathway |
| 103 | WSC2 | YNL283C | no hits | Stretch receptor; cell wall integrity pathway | |
| 104 | WSC3 | YOL105C | 15084 | 1.6e-28 | Stretch receptor; cell wall integrity pathway |
| 105 | WHI3 | YNL197C | 04835 | 2.3e-23 | Regulator of cell size. RNA binding protein |
| 106 | YCK1 | YHR135C | 00274 | 3.4e-129 | Casein kinase I isoform. Cell morphogenesis |
| 107 | YCK2 | YNL154C | 00274 | 1.6e-126 | Casein kinase I isoform. Cell morphogenesis. Functionally redundant with Yck1. |
| 108 | ZDS1 | YMR273C | 15061.2 | 4.7e-20 | Regulation of cell polarity |
| 109 | ZDS2 | YML109W | no hits | Regulation of cell polarity. Paralogue of ZDS1 | |
| ACTIN CYTOSKELETON | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 110 | ABP1 | YCR088W | 05340 | 7.9e-23 | Actin-binding protein |
| 111 | ACF4 | YJR083C | Actin cytoskeleton organization | ||
| 112 | ACT1 | YFL039C | 11232 | 2.0e-185 | Actin |
| 113 | AIP1 | YMR092C | 05949 | 3.0e-86 | Actin-interacting protein; actin patch component |
| 114 | APP1 | YNL094W | 04613 | 9.7e-22 | Actin filament organization |
| 115 | ARC15 | YIL062C | 02388 | 2.6e-25 | Subunit of Arp2/3 complex |
| 116 | ARC18 | YLR370C | 10246 | 7.4e-44 | Subunit of Arp2/3 complex |
| 117 | ARC19 | YKL013C | 10173 | 9.8e-58 | Subunit of Arp2/3 complex |
| 118 | ARC35 | YNR035C | 11479 | 1.8e-49 | Subunit of Arp2/3 complex |
| 119 | ARC40 | YBR234C | 05906 | 3.7e-69 | Subunit of Arp2/3 complex |
| 120 | ARK1 | YNL020C | 03081 | 8.7e-61 | Actin regulating Ser/Thr kinase |
| 121 | ARP2 | YDL029W | 05405 | 1.7e-149 | Subunit of Arp2/3 complex |
| 122 | ARP3 | YJR065C | 11265 | 1.2e-139 | Subunit of Arp2/3 complex |
| 123 | BNI1 | YNL271C | 12254 | 1.9e-59 | Formin; actin nucleation |
| 124 | BNR1 | YIL159W | 01141 | 2.3e-31 | Formin; actin nucleation. Functionally redundant with Bni1 |
| 125 | BSP1 | YPR171W | no hits | Involved in actin cytoskeleton organization | |
| 126 | CAP1 | YKL007W | 00423 | 1.1e-24 | F-actin capping protein alpha subunit |
| 127 | CAP2 | YIL034C | 11177 | 2.0e-50 | F-actin capping protein beta subunit |
| 128 | CCT1/TCP1 | YDR212W | 01279 | 3.4e-213 | Chaperonin role for actin and tubulin |
| 129 | CCT2 | YIL142W | 06235 | 5.5e-190 | Chaperonin role for actin and tubulin |
| 130 | CCT3 | YJL014W | 06067 | 1.1e-198 | Chaperonin role for actin and tubulin |
| 131 | CCT4 | YDL143W | 02571 | 9.7e-122 | Chaperonin role for actin and tubulin |
| 132 | CCT5 | YJR064W | 03959 | 4.6e-186 | Chaperonin role for actin and tubulin |
| 133 | CCT6 | YDR188W | 02350 | 6.5e-167 | Chaperonin role for actin and tubulin |
| 134 | CCT7 | YJL111W | 00565 | 8.2e-196 | Chaperonin role for actin and tubulin |
| 135 | CCT8 | YJL008C | 04401 | 2.6e-151 | Chaperonin role for actin and tubulin |
| 136 | CHC1 | YGL206C | 03921 | 0.0 | Clathrin heavy chain |
| 137 | COF1 | YLL050C | 04314 | 3.1e-45 | Actin binding and severing protein |
| 138 | CRN1 | YLR429W | 04417 | 6.5e-107 | Cortical actin cytoskeleton component |
| 139 | LAS1 | YKR063C | 02271 | 6.9e-18 | Bud formation and morphogenesis |
| 140 | LAS17/BEE1 | YOR181W | 03687 | 2.8e-51 | Activator of the Arp2/3 complex. Homologue of human WASP |
| 141 | MLC1 | YGL106W | 11848 | 4.0e-29 | Myosin light chain |
| 142 | MLC2 | YPR188C | 03910 | 5.1e-13 | Myosin light chain |
| 143 | MYO1 | YHR023W | 03286 | 5.4e-183 | Type II myosin |
| 144 | MYO2 | YOR326W | 04555 | 2.4e-276 | Type V myosin |
| 145 | MYO3 | YKL129C | 11115 | 0.0 | Type I myosin |
| 146 | MYO4 | YAL029C | 04555 | 4.5e-258 | Type V myosin |
| 147 | MYO5 | YMR109W | 11115 | 0.0 | Type I myosin |
| 148 | PAN1 | YIR006C | 11804 | 1.3e-40 | Actin patch protein |
| 149 | PFY1 | YOR122C | 10832 | 2.0e-27 | Profilin |
| 150 | PRK1 | YIL095W | 03081 | 9.5e-60 | Ser/Thr protein kinase; regulation of organization and function of actin |
| 151 | SAC6 | YDR129C | 04768 | 2.6e-218 | Fimbrin; actin filament bundling protein |
| 152 | SCP1 | YOR367W | no hits | Component of actin cortical cytoskeleton | |
| 153 | SLA1 | YBL007C | 05337 | 8.3e-57 | Assembly of cortical actin cytoskeleton |
| 154 | SLA2 | YNL243W | 00582 | 1.3e-78 | Assembly of cortical actin cytoskeleton |
| 155 | SRV2 | YNL138W | 10957 | 1.4e-98 | Regulation of actin dynamics and cell morphogenesis |
| 156 | VRP1 | YLR337C | 10566 | 3.7e-43 | Verprolin. Regulation of actin cytoskeleton. Homologue of human WIP |
| EXOCYST | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 157 | SEC3 | YER008C | 01107 | 7.2e-27 | Spatial landmark for secretion |
| 158 | SEC5 | YDR166C | 00710 | 4.9e-22 | Subunit of the exocyst complex |
| 159 | SEC6 | YIL068C | 03632 | 1.6e-67 | Subunit of the exocyst complex |
| 160 | SEC8 | YPR055W | 02554 | 2.6e-35 | Subunit of the exocyst complex |
| 161 | SEC10 | YLR166C | 00329 | 1.2e-61 | Subunit of the exocyst complex |
| 162 | SEC15 | YGL233W | 05494 | 1.1e-29 | Subunit of the exocyst complex |
| 163 | EXO70 | YJL085W | 01845 | 4.3e-14 | Subunit of the exocyst complex |
| 164 | EXO84 | YBR102C | 04147 | 6.5e-31 | Subunit of the exocyst complex |
| OTHER (SECRETION) | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 165 | SEC4 | YFL005W | 03865 | 7.2e-62 | Rab GTPase. Associates with Sec15 |
| 166 | SEC2 | YNL272C | 00553 | 7.3e-23 | GEF for Sec4 |
| MICROTUBULE CYTOSKELETON (SELECTED GENES) DYNEIN/DYNACTIN (per S. cerevisiae)* | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 167 | DYN1/DHC1 | YKR054C | 15045 | 0.0 | Heavy chain of cytoplasmic dynein |
| 04372 | 2.6e-150 | ||||
| 168 | DYN2/SLC1 | YDR424C | 12278 | 9.1e-23 | Dynein light chain |
| 169 | DYN3 | YMR299C | 03459& | no hits | Dynein light intermediate chain |
| 170 | PAC11 | YDR488C | 04598& | no hits | Dynein intermediate chain. Nuclear migration |
| 171 | ACT5/ARP1 | YHR129C | 11692 | 4.2e-105 | Component of the dynactin complex; related to centractin |
| 172 | ARP10 | YDR106W | 05833& | no hits | Component of the dynactin complex |
| 173 | JNM1 | YMR294W | 02603& | no hits | Component of the dynactin complex |
| 174 | NIP100 | YPL174C | 03826 | 6.6e-17 | Component of the dynactin complex. Putative orthologue of mammalian p150Glued |
| KINESINS (per S. cerevisiae)# | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 175 | CIN8 | YELO61C | 10678 | 1.0e-96 | Kinesin. Mitotic spindle assembly; chromosome segregation. Related to Kip 1 |
| 176 | KAR3 | YPR141C | 11986 | 1.6e-80 | Kinesin (- end directed) Related to Um Kin14 |
| 177 | KIP1 | YBL063W | 10678 | 2.5e-107 | Kinesin. Functionally redundant with Cin8 |
| 178 | KIP2 | YPL155C | 00896 | 5.4e-52 | Kinesin related motor protein Related to Um Kin7a and Kin7b |
| 179 | KIP3 | YGL216W | 01560 | 1.7e-118 | Kinesin related motor protein Related to Um Kin8 |
| 180 | SMY1 | YKL079W | 04218 | 3.4e-45 | Interacts with Myo2. N-terminal domain related to motor domain of kinesins. |
| +TIPs | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 181 | BIK1 | YCL029C | 06338 | 2.5e-13 | MT plus end associated protein. Orthologue of mammalian CLIP170 |
| 182 | BIM1/YEB1 | YER016W | 05761 | 7.5e-51 | Together wih Kar9 makes up the MT capture site at the cell cortex |
| 183 | KAR9 | YPL269W | no hits | Nuclear migration | |
| 184 | PAC1 | YOR269W | 03164 | 7.8e-36 | LIS1 /NudF homologue. Nuclear migration |
| TUBULIN | |||||
|---|---|---|---|---|---|
| Sc gene | Systematic name | U. maydis MIPS# | E value | Function in S. cerevisiae | |
| 185 | TUB1 | YML085C | 01221 | 4.4e-181 | α–tubulin |
| 186 | TUB2 | YFL037W | 05828 | 6.7e-185 | β–tubulin |
| 10558 | 2.0e-160 | β–tubulin | |||
| 187 | TUB3 | YML124C | 01221 | 1.5e-180 | α–tubulin |
| 188 | TUB4 | YLR212C | 03803 | 1.1e-90 | γ–tubulin |
Components of the U. maydis dynactin complex shown are per comparison with S. cerevisiae.
Blast analysis indicates that there are no hits for these genes, however, the U. maydis gene numbers provided appears to correspond to the yeast gene.
Kinesins shown are per comparison with S. cerevisiae kinesins. See article by Schuchardt et al., 2005 for details of the kinesin superfamily in U. maydis.
The U. maydis genome contains coding information for homologues of the Rho Cdc42 GTPase module (Cdc42, Cdc24, Bem3, Rga1/2) (Table 1). In addition, U. maydis, like other filamentous fungi, contains a homologue of mammalian Rac, another Rho GTPase, which is absent from the genomes of S. cerevisiae and S. pombe. In mammals, Cdc42 and Rac1 play partially overlapping roles in morphogenesis, cell division, and migration (Jaffe and Hall, 2005). Likewise in filamentous fungi, Cdc42 and Rac appear to have partially overlapping functions in control of cell polarity (see for example, Bassilana and Arkowitz, 2006; Boyce et al., 2001, 2003; Chen and Dickman, 2004; 2006; Rolke and Tudzynski, 2008; Scheffer et al., 2005; Virag et al., 2007).
In U. maydis, Cdc42 is required for cell separation but not for polarized growth, and is not essential for viability (Mahlert et al., 2006; F. Banuett, unpublished results), whereas UmRac1 is required for normal cell morphology of the yeast-like cell and for positioning of the septum (Mahlert et al., 2006). Umcdc42 null mutants form chains of cells that exhibit wild type morphology, separated by a single septum (Mahlert et al., 2006). In wild type cells, there are two septa prior to cell separation, one on the mother side and the other on the bud side. Cell separation occurs in the region between these two septa, the fragmentation zone (Banuett and Herskowitz, 2002; O’Donnell and McLaughlin, 1984; Weinzierl et al., 2002). In S. cerevisiae CDC42 is essential for viability and for polarity establishment: cdc42ts and cdc24 mutants arrest as large unbudded, multinucleate cells (reviewed in Pruyne and Bretscher, 2000a,b). Thus, U. maydis and S. cerevisiae use Cdc42 differently, a theme repeated in other filamentous fungi (see below).
The importance of Rac1 in polarized growth is supported by the following observations: expression of an activated allele of Rac1 results in formation of balloon-like structures at the tips of yeast-like cells; and Umrac1 null mutants exhibit elongated abnormal cells with rounded ends and a septum in the middle; wild type cells, in contrast, have tapered ends (Mahlert et al., 2006). UmRac1 is not required for cell viability, but depletion of UmRac1 in a strain lacking Umcdc42 is lethal: cells arrest as chains of enlarged, aberrant cells, with a single nucleus, and delocalized cell wall material (Mahlert et al., 2006). This observation suggests that Cdc42 and Rac1 share a common target. Rac1 is also necessary for the b-dependent transition from yeast-like to filamentous form: induction of Umrac1 expression in haploid cells (haploids normally lack an active b protein) causes a transition from yeast-like to filamentous form. UmRac1 is thus necessary and sufficient for the dimorphic switch that results in filament formation (Mahlert et al., 2006). The GEF and GAP regulators of the UmRac1 GTPase remain to be identified.
In Aspergillus nidulans Cdc42 is required for hyphal morphogenesis, but it is not required for localization of polarisome components, the Spitzenkörper, or organization of actin, and neither is AnRac1 required for localization of polarisome components or the Spitzenkörper (Virag et al., 2007). In contrast, S. cerevisiae Cdc42 is required for localization of polarisome components and actin organization (reviewed in Chang and Peter, 2003; Pruyne and Bretscher, 2000a, b). AnCdc42 and AnRac likely share a common target because double Anrac1 Ancdc42 mutants are synthetic lethal: the double mutants are unable to polarize and lyse as attached rounded cells separated by septa (Virag et al., 2007). In Penicillium marneffei, Rac is required for normal organization of the actin cytoskeleton. Rac regulates polarized growth of the hypha in concert with Cdc42 but the specific steps controlled by these Rho GTPases differ (Boyce et al., 2001, 2003). The synthetic lethality of cdc42 and rac1 in U. maydis and other filamentous fungi supports the notion that these Rho GTPases play partially overlapping roles in control of cell polarity.
C. U. maydis homologues of effectors of Cdc42
In S. cereviasiae, downstream effectors of Cdc42 include the p21-activated kinases (PAKs) Cla4, Ste20, and Skm1, and two proteins of unknown function, Gic1 and Gic2 (reviewed in Pruyne and Bretscher, 2000a, b). Ste20 and Cla4 are important for the Cdc42-dependent reorganization of the actin cytoskeleton during the cell cycle. The U. maydis genome contains coding information for three Ste20-like kinases (Table 1): Smu1, Cla4, and Don1, but not for Gic1 and Gic2. Smu1 and Cla4 contain the CRIB domain present in p21-activated kinases (PAKs), whereas Don1 lacks this domain, and thus belongs to the germinal center (GCK) subfamily of Ste20-like kinases (Leveleki et al., 2004; Smith et al., 2004). (For further details on Smu1, see the article by García-Pedrajas et al. in this issue). Both UmDon1 and UmCla4 play a role in cytokinesis (Leveleki et al., 2004), just as Cla4 and Ste20 do in S. cerevisiae. Two observations support the idea that UmCla4 is likely to be a downstream effector of UmRac1: both Δcla4 and Δrac1 mutants have a similar phenotype, and they show yeast two-hybrid interactions. Other effectors of Rac1 are likely to exist because Δcla4 does not suppress the lethality observed upon expression of an activated allele of Rac1 (Mahlert et al., 2006).
D. U. maydis homologues of other Rho proteins
The Rho subfamily of GTPases includes Rho, Rac, Cdc42, and others. They act as molecular switches that play important roles in cell polarity, morphogenesis, and cell division (Jaffe and Hall, 2005). S. cerevisiae contains one Cdc42 (described above) and five Rho proteins, Rho1-5. Rho1, an essential gene for viability, is required for polarity establishment. Rho3 and Rho4 are also required for polarized growth. The signal from different Rho proteins (Rho1, Rho3, Rho4, and Cdc42) converges on the formin Bni1, a polarisome component, to control actin organization (see below) (Dong et al., 2003; reviewed in Pruyne and Bretscher, 2000a,b). In addition, Rho1 controls bud morphogenesis via two different outputs: as a catalytic subunit of β-glucan synthase, the enzyme that synthesizes the major component of the cell wall, and as an activator of protein kinase C (Pkc1) that controls the cell wall integrity signal transduction pathway (reviewed in Banuett, 1998; Cabib et al., 1998; Pruyne and Bretscher, 2000a,b). The regulators of Rho1 include two redundant GEFs, Rom1 and Rom2, and two GAPs, Sac7 and Bem3. The U. maydis genome contains coding information for three Rho proteins, Rho1, Rho2, and Rho3, and for homologues of Rho1 regulators (Table 1). Their function has not been reported. Rho proteins in other filamentous fungi are required for polarized growth and it is likely that this function will be conserved in U. maydis. In A. gossypii, rho3 mutants exhibit swelling of hyphal tips indicative of isotropic growth, and rho1 null mutants exhibit lysis of cells in a colony (Wendland and Philippsen, 2001). In A. nidulans, RhoA, a homologue of S. cerevisiae Rho1, is necessary for polarized growth, branching pattern, and cell wall deposition (Guest et al., 2004).
E. U. maydis homologues of polarisome components
In S. cerevisiae, the polarity determining proteins, Spa2, Pea2, Bud6/Aip3, and Bni1, assemble at the bud tip and function as a scaffold for the assembly of other proteins. Mutants lacking either of these proteins exhibit defects in actin organization, apical growth, and have widened necks (reviewed in Pruyne and Bretscher, 2000a,b). The polarisome links Rho GTPases and actin cable assembly through the formin Bni1. Formins are multidomain proteins required for maintenance of cell polarity, cytokinesis, and actin organization, and are central to how inputs from Rho GTPases are transmitted to actin organization (Chang and Peter, 2003; Evangelista et al., 2003; Wallar and Alberts, 2003). Bni1 binds profilin, an actin-monomer binding protein, and has been shown to nucleate actin cables. Bud6 is important for the Bni1 actin-nucleation function (Pruyne et al., 2002; Sagot et al., 2001). Localization of Bni1 to the bud tip requires Spa2 and Pea2. Some polarisome components also localize to the mother-bud neck region during cytokinesis to participate in formation of the actomyosin ring and the septum (reviewed in Pruyne and Bretscher, 2000a,b).
The U. maydis genome contains coding information for homologues of polarisome components Bud6, Spa2, and two formins, UmSepA and UmDia, but not of Pea2 (Table 1). The role of these proteins in cell polarity of the yeast-like and filamentous form in U. maydis remains to be determined. Recent studies in A. gossypii, A. nidulans, and C. albicans (Crampin et al., 2005; Knechtle et al., 2003; Virag and Harris, 2006) are providing insights about the role of the polarisome complex in polarized growth of the hypha. In A. nidulans, homologues of Spa2, Bud6/Aip3, and Bni1 are necessary for maintaining the axis of polarized growth: mutations in either component result in dichotomous branching (branching at the hyphal tip), which is normally not observed in wild type hyphae (Sharpless and Harris, 2002; Virag and Harris, 2006). SpaA::GFP localizes to the hyphal tip, overlapping to some extent the localization of the Spitzenkörper, consistent with a role at the hyphal tip, possibly as a component of the Spitzenkörper (Virag and Harris, 2006). SpaA is not required for localization of the formin SepA (Virag and Harris, 2006), whereas S. cerevisiae Spa2 is required for localization of the formin Bni1 (reviewed in Pruyne and Bretscher, 2000a,b). BudA, the A. nidulans homologue of Bud6/Aip3, localizes to sites of septum formation. The AnbudA mutant phenotype is consistent with a role in maintenance of the axis of polarized growth and determining the area of growth. AnbudA null mutants exhibit reduced hyphal growth rate, increased hyphal diameter and nuclei per hyphal segment, dichotomous branching, and increased branching frequency, and potato-shaped germlings (Virag and Harris, 2006). AnSepA is required for septum formation, for maintenance of the axis of polarized growth, and determining the area of growth, and normal organization of actin at the hyphal tips and septa. SepA null mutants exhibit wider hyphal diameter, dichotomous branching, and lack septa. SepA::GFP localizes to hyphal tips, coincident with localization of the Spitzenkörper, and also to septa (Sharpless and Harris, 2002). This localization led to the hypothesis that the Spitzenkörper acts as an actin-nucleating center (Sharpless and Harris, 2002).
In A. gossypii, the Spa2 homologue, which localizes permanently to the hyphal tip and to sites of branch initiation, appears to determine area of growth, normal growth rate, and branching density (Knechtle et al., 2003). AgBNI1 is necessary for establishment of polarized growth and for actin organization. Disruption of AgBNI1 results in formation of giant potato-shaped cells lacking actin cables and also in the inability to form hyphae (Schmitz et al., 2006). AgBni1 localizes to hyphal tips. A constitutively active AgBni1 results in dichotomous hyphal branching in young hyphae, a type of branching normally restricted to mature hyphae (Schmitz et al., 2006). The localization of these proteins at the hyphal tip supports a role in determining polarized growth.
In C. albicans, CaBni1 and CaMlc1 (myosin light chain) colocalize with the Spitzenkörper, whereas Spa2 and Bud6/Aip3 localize mostly as a cap at the hyphal tip that overlays the Spitzenkörper (Crampin et al., 2005). Thus, in C. albicans it seems that polarized growth of hyphae is determined by two separate entities, the Spitzenkörper and the polarisome, whereas in pseudohyphae and yeast-like cells, the polarisome is responsible for polarized growth; there is no Spitzenkörper in these cells.
F. U. maydis homologues of septins
Septins are GTP binding proteins that are conserved from fungi to mammals and play a role in cytokinesis and other cell processes (Gladfelter et al., 2001; Longtine and Bi, 2003; Pan et al., 2007). In S. cerevisiae, septins form filaments at the mother-bud neck region, arranged longitudinally (along the main cell axis) in an hour-glass shape or as horizontal rings in a cell cycle-dependent manner (Gladfeleter et al., 2001). Assembly of septins requires Cdc42, polarisome components, the PAK kinase, Cla4, and the actin cytoskeleton (Kadota et al., 2004). Septins are proposed to serve a scaffold role and recruit other proteins: bud site selection landmarks, components of the actomyosin contractile ring (CAR), chitin synthases, and other proteins (Gladfelter et al., 2001; Longtine and Bi, 2003). Septins also act as diffusion barriers for integral membrane proteins and have been proposed to serve as organizers of cortical domains of the cell in addition to their scaffold role (Barral et al., 2000; Takizawa et al., 2000).
S. cerevisiae contains 7 septins, Cdc3, Cdc10, Cdc11, Cdc12, Sep7, Spr4, Spr28; the latter two are meiosis-specific and required with other septins during spore maturation. The U. maydis genome contains coding information for homologues of S. cereviase Cdc3, Cdc10, Cdc11, and Cdc12 (Table 1), whereas in other filamentous fungi there appear to be at least five putative septins (see Borkovich et al., 2004). U. maydis Sep3, a homologue of Cdc11, is required for normal cell morphogenesis (Boyce et al., 2005). Haploid Sep3 mutants exhibit aberrant cell morphology: the cells are “lemon drop-shaped”, with a septum in the middle, become multinucleated, and have altered chitin deposition. Sep3 is also required for normal morphology of the promycelium (a short filament produced upon germination and meiosis of the teliospore; Christensen, 1963), and the basidiospores (the haploid cells produced on the promycelium by mitosis of the four primary meiotic products). Genetic analysis indicates that Sep3 acts in the cAMP signaling pathway (Boyce et et al., 2005). The role of other septins in U. maydis remains to be uncovered. In A. nidulans, AspB, a homologue of S. cerevisiae Cdc3, is hypothesized to act as a marker of branch formation. It localizes premitotically as a ring to sites of branching and secondary germ tube emergence. It also localizes postmitotically to sites of septation (Westfall and Momany, 2002).
G. U. maydis homologues of the exocyst complex
In S. cerevisiae, the exocyst is a protein complex required for exocytosis and consists of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84 (TerBush et al., 1996). It is localized to the bud tip during polarized growth and to the neck region during cytokinesis, and is involved in fusion and docking of vesicles transported on actin cables. The Sec3 protein acts as a spatial landmark for the assembly of the exocyst complex (Finger et al., 1998). The assembly of the different protein complexes described above provides a temporal and spatial link between bud site selection with organization of the cytoskeleton, and secretion resulting in cell growth.
The U. maydis genome contains coding information for all the components of the exocyst complex (Table 1) but little is known of their role in polarized growth and cytokinesis. In A. nidulans, SecC, a homologue of S. cerevisiae Sec3, accumulates at the plasma membrane anterior to the Spitzenkörper and may function as a landmark for secretion (Taheri-Talesh et al., 2008).
IV. THE CYTOSKELETON
The cytoskeleton plays an important role in cell morphogenesis and polarized growth in diverse organisms. In U. maydis, both actin and microtubules are required for cell morphogenesis, as in other filamentous fungi.
A. The actin cytoskeleton
The actin cytoskeleton in U. maydis consists of three structures: actin patches, actin cables, and an actin ring as observed in other fungi. The actin cytoskeleton is highly polarized throughout the cell cycle: actin patches concentrate at the presumptive bud site and at the tip of the growing bud, and actin cables polarize towards the actin patches and extend into the mother cell (Banuett and Herskowitz, 2002). During cytokinesis, actin forms a ring in the neck region. Actin patches persist at the growing end during this process. After cell division is complete, both cell ends contain actin patches, with one end having a higher concentration than the other. This distribution of actin is consistent with highly polarized growth at the tip of the cell throughout the cell cycle. There is no evidence for a disorganized actin cytoskeleton at any stage of the cell cycle as occurs during isotropic growth in S. cerevisiae, consistent with observations that there is no isotropic growth during bud morphogenesis in U. maydis (Banuett and Herskowitz, 2002). U. maydis hyphae contain a prominent actin cap at the tip and actin cables polarize towards the tip, as observed in other filamentous fungi (Banuett and Herskowitz, 2002).
1. U. maydis homologues of components of the actin cytoskeleton
In S. cerevisiae, the actin cytoskeleton is involved in polarized secretion. Actin cables serve as tracks on which vesicles move on myosin motors towards sites of active growth and secretion (the bud tip during polarized growth and the neck region during cytokinesis) (Karpova et al., 2000; Pruyne and Bretscher, 2000a,b). All components involved in actin assembly and function in S. cerevisiae appear to be conserved in the U. maydis genome (Table 1). For example, genes coding for the Arp2/3 complex (Winter et al., 1999), responsible for nucleation of branched actin structures, are present in the U. maydis genome, as well as genes coding for homologues of Bee1/Las17, the activator of Arp2/3 (a homologue of the mammalian Wiskott-Aldrich syndrome protein WASP), and Vrp1 (homologue of mammalian Wip, Wiskott-Aldrich syndrome protein interacting-protein). In S. cerevisiae, Arp2/3, Bee1/Las17, and Vrp1 localize to actin patches and form an actin cap during early bud morphogenesis (reviewed in Pruyne and Bretscher, 2000a,b). The S. cerevisae genome contains two tropomyosin isoforms, Tpm1 and Tpm2. These proteins stabilize actin cables (reviewed in Pruyne and Bretscher, 2000a,b). The U. maydis genome contains a single tropomyosin gene, and the putative protein is similar to S. cerevisiae Tpm2. The function of tropomyosin in U. maydis remains to be determined. The U. maydis genome contains coding information for homologues of proteins such as actin binding protein (Abp1), coronin (Crn1), cofilin (Cof1), profilin (Pfy1), fimbrin (Sac6), Pan1, Prk1, Srv2, Sla1 and Sla2, some of which are actin patch components (Table 1). The function of these proteins in U. maydis has not been reported. (See Moseley and Goode, 2006, for a review on the function of these proteins in S. cerevisiae).
2. U. maydis homologues of myosins
As in other filamentous fungi, the U. maydis genome contains coding information for 4 myosins: a class I (Myo1), a class II (Myo2), a class V (Myo5), and a class XVII (myosin-chitin synthase; Csm1) (Table 1) (see Hodge and Cope, 2000, for myosin classification). Myosin-chitin synthase contains an amino-terminal myosin domain and a chitin synthase domain at the carboxy terminus. It is specific to the filamentous fungi; the genomes of S. cerevisaie or S. pombe lack this information. In the yeast-like cell, Ummyo5 is required for cell morphogenesis and cell separation: Ummyo5 null mutants are viable and exhibit slow growth and formation of clusters of shorter and wider cells, which fail to separate (Weber et al., 2003). Young myo5 mutant cells still exhibit polarized growth and polarized chitin at the cell tip, whereas older cells lose polarity and become aberrantly shaped (Weber et al, 2003). In the hypha, Ummyo5 appears to be required for the axis of polarized growth and to determine the area of growth: hyphae deficient in Myo5 are wider in diameter and exhibit an irregular growth pattern (spiral-like), and contain irregular deposition of chitin along the hyphal length (Weber et al., 2003). Myo5::GFP localizes to the incipient bud site, the bud tip in cells with small buds, and becomes more dispersed in large-budded cells; in hyphae, it localizes to the tip, consistent with a role at the cell tip (Weber et al., 2003). The function of Myo1 and Myo2 has not been reported. (See below for role of Csm1).
B. The microtubule cytoskeleton
In many eukaryotes, the microtubule cytoskeleton is required for polarized growth, transport of organelles and vesicles, in addition to its role in spindle assembly and function.
The microtubule cytoskeleton in U. maydis consists of an extensive cytoplasmic array throughout interphase that disassembles in late G2 when a short intranuclear spindle is formed and astral microtubules are assembled from the spindle pole bodies (Banuett and Herskowitz, 2002; Steinberg et al., 2001; Straube et al., 2003). These astral microtubules appear to be involved in nuclear movements to the bud, where the nucleus divides, and then after nuclear division, when one of the nuclei migrates to the mother cell (Banuett and Herskowitz, 2002; Fink et al., 2006; Steinberg et al., 2001). The organization of the microtubule cytoskeleton in U. maydis is more complex than that in S. cerevisiae, and is reminiscent of that observed in S. pombe. In U. maydis yeast-like cells, nucleus-associated spindle pole bodies (SPBs) nucleate astral microtubules, and cytoplasmic microtubule organizing centers (MTOCs), assembled at the base of the bud early during bud morphogenesis, nucleate microtubules towards the bud and mother cell (Straube et al., 2003). The yeast-like cell contains additional motile cytoplasmic MTOCs (Fink and Steinberg, 2006; Straube et al., 2003). In the hypha, the majority of MTs are oriented with their plus ends towards the growing tip; some MTs are oriented with their plus ends towards the basal end of the apical cell (Schuchart et al., 2005).
The microtubule cytoskeleton is required for normal cell morphogenesis in U. maydis (Steinberg et al., 2001), just as it is in S. pombe (Sawin and Nurse, 1998; reviewed in Chang, 2001). In contrast, S. cerevisiae MTs play no role in cell morphogenesis. U. maydis thus resembles S. pombe in the organization of the microtubule cytoskeleton and in the requirement of the microtubule cytoskeleton for cell morphogenesis and for determining the axis of polarized growth. U. maydis may thus combine strategies used by S. cerevisiae and S. pombe in novel ways to establish and maintain polarized growth and to generate cell shape.
1. U. maydis homologues of tubulins
The U. maydis genome contains coding information for an α tubulin (tub1), two β tubulins, and a γ tubulin (tub2) (Table 1). Both α and γ tubulin genes have been characterized and shown to be essential for viability (Steinberg et al., 2001; Straube et al., 2003). γ tubulin is found at the SPBs and also in the cytoplasmic and polar MTOCs and its presence in these structures varies in a cell cycle dependent manner (Straube et al., 2003). α tubulin is required for cell morphology and polarized growth. One β tubulin gene was cloned, but its role in cell polarity and cell morphogenesis was not reported (Gold et al., 1994).
2. U. maydis homologues of microtubule-plus-end-associated proteins (+Tips)
+TIPs are a highly diverse group of proteins that includes MT-dependent motors and nonmotor proteins: CLIP170 (ScBik1), Dynactin (Sc Jnm1, Nip100, Arp1, Arp10), EB1 (Sc Bim1), CLASPS (Sc Stu1), LIS1 (Sc Pac1), Dynein (Sc Dhc1), APC (Sc Kar9), and others (Adkhmanova and Hoogenraad, 2005).
Dynein and dynactin
Cytoplasmic dynein is a multisubunit complex with a MT minus-end directed motor activity. In U. maydis, as in other fungi, there is a single cytoplasmic dynein. The heavy chain of cytoplasmic dynein is encoded by two unlinked genes: dyn1 codes for the putative ATPase region, and dyn2 codes for the microtubule binding region (Straube et al., 2001). Dyn1 and Dyn2 polypeptides interact to form the active protein. Both genes are essential. Analysis of conditional mutations indicates that dynein is required for cell morphogenesis, nuclear migration, microtubule organization, retrograde endosomal transport, and organization of the endoplasmic reticulum (Straube et al., 2001; Wedlich-Soldner et al., 2002). Mutant cells in either dyn1 or dyn2 lose polarity (become rounded), contain multiple nuclei that cluster in the aberrantly shaped cells and microtubules that become disorganized, and develop long projections that exhibit polarized growth (Straube et al., 2001).
Dynactin is a multiprotein complex required for activation of cytoplamic dynein-mediated vesicular transport. It consists of at least ten polypeptides in most eukaryotes; in yeast, four dynactin subunits have been identified (see above). The p150Glued subunit is the largest and interacts with the 74-kDa dynein intermediate chain. Other subunits include the actin-related protein Arp1/centractin (45 kDa); the 50 kDa/dynamitin subunit; dynactin p62, dynactin p27, dynactin, p25, dynactin p24/p22, and CapZ (reviewed in Schroer, 2004; Xiang and Plaman, 2003). In S. cerevisiae the dynactin complex is required for nuclear migration (Lee et al., 2003; Sheeman et al., 2003). In Neurospora crassa, it is also required for nuclear migration, in addition to a role in determining the axis of polarized growth, and position of the Sptizenkörper (Plaman et al., 1994; Riquelme et al., 2000; Seiler et al., 1999). The U. maydis genome contains coding information for homologues of components of the S. cerevisiae dynactin complex (Table 1), and of other dynactin components found in filamentous fungi and other eukaryotes (data not shown), and other +TIPs (Table 1). UmDya1 (the homologue of p150 Glued), UmClip1 (CLIP170 homologue), UmLis1 (LIS1/NudF homologue), and UmPeb1 (homologue of EB1) localize to microtubule plus ends (Lenz et al., 2006; Straube et al., 2003). UmDya1 is required for localization of dynein to microtubule plus ends. UmLis1/NudF is required for activation of dynein and this activation is necessary for retrogade endosomal movement. UmClip1 is not required for retrogade endosomal movement or binding of dynein to microtubule plus ends (Lenz et al., 2006). In S. cerevisiae, Kar9 and Pac1/LIS1 are required for orientation of the mitotic spoindle and nuclear migration (Lee et al., 2003; Miller and Rose, 1998; Sheeman et al., 2003). The U. maydis genome does not contain coding information for a Kar9 homologue.
3. U. maydis homologues of kinesins
In filamentous fungi, as in many other eukaryotes, kinesins are involved in long-distance transport of vesicles. S. cerevisiae contains 6 kinesins whereas filamentous fungi contain 10–12 kinesins (see Borkovich et al., 2004; Xiang and Plamann, 2003). The U. maydis genome contains coding information for 10 kinesins (Kin1, Kin3, Kin4, Kin6, Kin7a, Kin7b, Kin8, Kin9, and Kin14), some of which are more closely related to their metazoan counterparts than to the ascomycetous fungi (Schuchardt et al., 2005). Genetic analysis has shown that all except Kin1 and Kin3 are dispensable for hyphal growth. Null kin1 or kin3 mutants exhibit bipolar growth and short hyphae. Double kin1 kin3 null mutants exhibit the same phenotype as the single mutants, indicating that they likely act in the same pathway (Schuchardt et al., 2005). Interestingly, kin1 and kin3 mutations exacerbate the mutant phenotype conferred by a myo5ts mutation. The residual polar growth in the myo5ts mutant is completely abolished resulting in clusters of misshapen cells that exhibit no polar growth. This synthetic interaction suggests that Kin1 and Kin3 share a common target with Myo5 for polarized growth. Kin1, Kin3, and Myo5 all accumulate at the hyphal apex, the region where the Spitzenkörper localizes; however, localization of the Spitzenkörper in these studies was not reported.
V. U. maydis homologues of chitin synthases
Integrity of the cell wall is crucial for maintenance of cell shape (Latge, 2007; Klis et al., 2006). Chitin and β-glucan are the major components of fungal cell walls (see article by Ruiz-Herrera in this issue). The U. maydis genome contains 7 genes that code for chitin synthases (Chs1, Chs2, Chs3, Chs4, Chs5, Chs6, Chs7) and one gene for a hybrid myosin-chitin synthase protein (Mcs1; see above) (Garcerá-Teruel et al., 2006; Gold and Kronstad, 1994; Weber et al., 2006). Deletion analysis of single genes indicates that Umchs1, Umchs2, Umchs3, Umchs4 are not required for morphology of the yeast-like cell or the hypha, whereas Δchs6 results in cells with wider diameter, and Δmcs1 cells exhibit slightly wider cell diameter and a ballooning of the bud tip, indicative of a role in cell morphogenesis. The Umchs5 or Umchs7 null mutants exhibit a more pronounced phenotype indicative of a role in cell morphology and cell separation, respectively: Δchs5 mutant cells are more elongated than wild type cells, grow irregularly, and have a neck region that is not clearly defined, whereas Δchs7 mutant cells have a cell separation defect but the cells have normal cell morphology, though they are shorter, and in some cases, lose their normal cell shape (Weber et al., 2006). Six of the U. maydis chitin synthases (Chs3-Ch7 and Mcs1) localize to the septal region, and four of them (Chs5, Chs6, Chs7, and Mcs1) also localize to the hyphal tip. Despite the localization of Chs5, 6, 7, and Mcs1 to the hyphal tip, they do not seem to be required for hyphal morphology in culture (Weber et al., 2006; see also Ruiz-Herrera et al., 2006). It is likely that the chitin synthases have partially overlapping functions and are able to substitute for each other when one or two are absent. Deletion of multiple chitin synthases that localize to the hyphal tip will likely be informative (Weber et al., 2006).
VI. FUTURE DIRECTIONS
Our analysis indicates that most of the components of the S. cerevisae machinery for cell polarity and cell morphogenesis, and the cytoskeleton are conserved in U. maydis. Because this analysis was based on S. cerevisiae and S. pombe genes, other components of the cell polarity machinery specific to Ustilago maydis or the Basidiomycetes remain unidentified. Sequence comparisons with other filamentous fungi, where additional genes required for polarized growth have been identified, in combination with genetic screens should prove useful in obtaining a more comprehensive view of the machinery for cell polarity and cell morphogenesis.
The challenge for the future is not only to identify the machinery for cell polarity, but also to understand how the components of the machinery are assembled and rearranged during polarized growth, and how these assemblies generate the diversity of forms that characterize the life cycle of Ustilago maydis.
Supplementary Material
Part A. The U. maydis gene products identified in Table 1, and additional ones (see legend to Table 1) were used as queries to identify highest hit homologues in different fungi and other eukaryotes using SIMAP at MIPS Ustilago database MUMDB (http://mips.gsf.de/genre/proj/ustilago/). The highest hit homologues were found in the Basidiomycota followed by the Ascomycota. The chart represents analysis of 306 U. maydis gene products identified after BLAST analysis with 344 S. cerevisiae gene products, as described in Table 1.
Part B. The chart represents analysis of 276 U. maydis gene products obtained as above and used as queries to identify highest hit homologues in different fungi, exclusively, using SIMAP at MIPS (see above).
Part C. The U. maydis gene products used in this analysis were identified using a subset of S. pombe genes involved in cell polarity, cell morphogenesis, and the cytoskeleton as queries of the U. maydis genome using wuBLAST at MIPS (see above). The identified U. maydis gene products were used as queries to identify highest hit homologues in different fungi and other eukaryotes using SIMAP at MIPS Ustilago database MUMDB (see Part A). The source of S. pombe gene products was La Carbona et al., 2006 and the Schizosaccharomyces pombe Gene DB (http://www.genedb.org/genedb/pombe/). The chart represents analysis of 317 U. maydis gene products obtained after wuBLAST analysis of 340 S. pombe gene products.
Part D. The chart represents analysis of 276 U. maydis gene products obtained as above and used as queries to identify highest hit homologues in different fungi, exclusively, using SIMAP at MIPS (see PartA).
ACKNOWLDEGEMENTS
We apologize to colleagues whose work it was not possible to cite due to space limitations. We thank anonymous reviewers for their useful comments. This work was supported by NIH grants SO6 GM63119 and 2SO6 GM063119 to FB, and, in part, by CONACYT grant 53191 and SNI fellowship to CRP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Banuett F. History of the mating types in Ustilago maydis. In: Heitman J, Kronstad J, Taylor J, Casselton LA, editors. Sex in Fungi: Molecular determination and evolutionary implications. Washington, DC: ASM press; 2007. pp. 351–375. [Google Scholar]
- Banuett F. Pathogenic development in Ustilago maydis: A progression of morphological transitions that results in tumor formation and teliospore production. In: Osiewacz HD, editor. Molecular Biology of Fungal Development. New York, Basel: Marcel Dekker; 2002. pp. 349–398. [Google Scholar]
- Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Molec. Biol. Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genetics. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I. Bud morphogenesis and the actin and microtubule cytoskeletons during budding in the corn smut fungus, Ustilago maydis. Fungal Genet Biol. 2002;37:149–170. doi: 10.1016/s1087-1845(02)00548-0. [DOI] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development. 1996;122:2965–2976. doi: 10.1242/dev.122.10.2965. [DOI] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia S. Hyphal tip growth: Outstanding questions. In: Osiewacz HD, editor. Molecular Biology of Fungal Development. New York, Basel: Marcel Dekker; 2002. pp. 29–58. [Google Scholar]
- Bassilana M, Arkowitz RA. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryot Cell. 2006;5:321–329. doi: 10.1128/EC.5.2.321-329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer Y, Knechtle P, Wendland J, Helfer H, Philippsen P. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Molec. Biol. Cell. 2004;15:4622–4632. doi: 10.1091/mbc.E04-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, Zelter A, Kalkman ER, O'Rourke R, Bowring F, Yeadon J, Ishii C, Suzuki K, Sakai W, Pratt R. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Molec. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, Chang H, D'Souza CA, Kronstad JW. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize. Eukaryot Cell. 2005;4:2044–2056. doi: 10.1128/EC.4.12.2044-2056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J Cell Sci. 2003;1:1249–1260. doi: 10.1242/jcs.00319. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Hynes MJ, Andrianopoulos A. The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. J Bacteriol. 2001;183:3447–3457. doi: 10.1128/JB.183.11.3447-3457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Schirawski J, Müller P, Kahmann R. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 2003;22:2199–2210. doi: 10.1093/emboj/cdg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Drgonova J, Drgon T. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 1998;67:307–333. doi: 10.1146/annurev.biochem.67.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Chang F, Peter M. Yeasts make their mark. Nature Cell Biol. 2003;5:294–299. doi: 10.1038/ncb0403-294. [DOI] [PubMed] [Google Scholar]
- Chang F. Establishment of a cellular axis in fission yeast. Trends Genet. 2001;17:273–278. doi: 10.1016/s0168-9525(01)02279-x. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Chen C, Ha YS, Min JY, Memmott SD, Dickman MB. Cdc42 is required for proper growth and development in the fungal pathogen Colletotrichum trifolii. Eukaryot Cell. 2006;5:155–166. doi: 10.1128/EC.5.1.155-166.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Dickman MB. Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signaling pathways. Mol Microbiol. 2004;51:1493–1507. doi: 10.1111/j.1365-2958.2003.03932.x. [DOI] [PubMed] [Google Scholar]
- Christensen JJ. Corn smut caused by Ustilago maydis. Am. Phytopathol. Soc. Monogr. 1963;No.2:1–41. [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Galman C, Berman J, Sudbery P. Candida albicans hyphae have a Spitzenkörper that is distinct form the polarisome found in yeast and pseudohyphae. J. Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- Dong Y, Pruyne D, Bretscher A. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 2003;161:1081–1092. doi: 10.1083/jcb.200212040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, Kozminski KG, Lau MW, Moskow JJ, Tong A, Schenkman LR, McKenzie A, 3rd, Brennwald P, Longtine M, Bi E, Chan C, Novick P, Boone C, Pringle JR, Davis TN. Fields S, Drubin DG. A protein interaction map for cell polarity development. J Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- Fink G, Steinberg G. Dynein-dependent motility of microtubules and nucleation sites supports polarization of the tubulin array in the fungus Ustilago maydis. Molec. Biol. Cell. 2006;17:3242–3253. doi: 10.1091/mbc.E05-12-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J. 2006;25:4897–4908. doi: 10.1038/sj.emboj.7601354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinson A, Read ND. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 2000;198:246–259. doi: 10.1046/j.1365-2818.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- Garcerá-Teruel A, Xoconostle-Cázares B, Rosas-Quijano R, Ortiz L, León-Ramírez C, Specht CA, Sentandreu R, Ruiz-Herrera J. Loss of virulence in Ustilago maydis by Umchs6 gene disruption. Res Microbiol. 2004;155:87–97. doi: 10.1016/j.resmic.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gold SE, Kronstad JW. Disruption of two genes for chitin synthase in the phytopathogenic fungus Ustilago maydis. Mol Microbiol. 1994;11:897–902. doi: 10.1111/j.1365-2958.1994.tb00368.x. [DOI] [PubMed] [Google Scholar]
- Gold SE, Bakkeren G, Davies JE, Kronstad JW. Three selectable markers for transformation of Ustilago maydis. Gene. 1994;142:225–230. doi: 10.1016/0378-1119(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Guest GM, Lin X, Momany M. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet Biol. 2004;41:13–22. doi: 10.1016/j.fgb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Harris SD, Read ND, Roberson RW, Shaw B, Seiler S, Plamann M, Momany M. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Euk. Cell. 2005;4:225–229. doi: 10.1128/EC.4.2.225-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD. Cell polarity in filamentous fungi: shaping the mold. Intl. Rev. Cytol. 2006;251:41–77. doi: 10.1016/S0074-7696(06)51002-2. [DOI] [PubMed] [Google Scholar]
- Heath IB. Organization and function of actin in hyphal tip growth. In: Steiger C, Baluska F, Volkmann D, Barlow P, editors. Actin: a Dynamic Framework for Mulitple Plant Cell Functions. Dordrecht, Boston & London: Kluwer Academic; 2000. pp. 275–300. [Google Scholar]
- Hodge T, Cope MJ. A myosin family tree. J Cell Sci. 2000;113:3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- Holliday R. Handbook of Genetics. In: King RC, editor. Ustilago maydis. vol 1. New York: Plenum; 1974. pp. 575–595. [Google Scholar]
- Jacobs CW, Mattichak SJ, Knowles JF. Budding patterns during the cell cycle of the maize smut pathogen Ustilago maydis. Can. J. Bot. 1994;72:1675–1680. [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: Biochemistry and Biology. Annu. Rev. Cell Dev. Biol. 2005;21:347–369. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, McNally JG, Moltz SL, Cooper JA. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 1998;142:1501–1517. doi: 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis FM, Boorsma A, De Groot PW. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Perlin MH, García-Pedrajas M, Covert SF, Gold SE. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv. Genet. 2007;57:1–47. doi: 10.1016/S0065-2660(06)57001-4. [DOI] [PubMed] [Google Scholar]
- Knechtle P, Dietrich F, Philippsen P. Maximal growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Molec. Biol. Cell. 2003;14:4140–4154. doi: 10.1091/mbc.E03-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzack S, Rischitor PE, Enke C, Fischer R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol Biol Cell. 2005;16:497–506. doi: 10.1091/mbc.E04-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latge JP. The cell wall: a carbohydrate armour for the fungal cell. Molec. Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- Lee W-L, OBerle JR, Cooper JA. The role of the lissencepahly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Kumar S, Plamann M. Null mutants of the Neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 2001;12:2195–2206. doi: 10.1091/mbc.12.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeder AC, Turner G. Characterization of Aspergillus nidulans polarisome component BemA. Fungal Genet Biol. Epub. 2007 doi: 10.1016/j.fgb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Lenz JH, Schuhardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveleki L, Mahlert M, Sandrock B, Bölker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Lopez-Franco R, Bracker CE. Diversity and dynamics of the Sptizenkörper in growing hyhal tips of higher fungi. Protoplasma. 1996;195:90–111. [Google Scholar]
- Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bölker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2006;59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–930. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KL, McLaughlin DJ. Postmeiotic mitosis, basidiospore development, and septation in Ustilago maydis. Mycologia. 1984;76:486–502. [Google Scholar]
- Pan F, Malmberg RL, Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103–119. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P, Kaufmann A, Schmitz H-P. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 2005;8:1–8. doi: 10.1016/j.mib.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsly JH, Bruno K. Cytoplasmic dynein and centractin are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000a;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. 2000. J. Cell Sci. 2000b;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reynaga-Peña CG, Gierz G, Bartnicki-Garcia S. Analysis of the role of the Spitzenkörper in fungal morphogenesis by computer simulation of apical branching in Aspergillus niger. Proc Natl Acad Sci USA. 1997;94:9096–9101. doi: 10.1073/pnas.94.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M, Bartnicki-García S, González-Prieto JM, Sánchez-León E, Verdín-Ramos JA, Beltrán-Aguilar A, Freitag M. Spitzenkörper localization and intracellular traffic of green fluoresenct protein-labeled CHS-3 and CHS_6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot. Cell. 2007;6:1853–1864. doi: 10.1128/EC.00088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M, Gierz G, Bartnicki-Garcia S. Dynein and dynactin deficiencies affect the formation and function of the Spitzenkörper and distort hyphal morphogenesis in Neurospora crassa. Microbiol. 2000;146:1743–1752. doi: 10.1099/00221287-146-7-1743. [DOI] [PubMed] [Google Scholar]
- Rolke Y, Tudzynski P. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol Microbiol. 2008;68:405–423. doi: 10.1111/j.1365-2958.2008.06159.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J, Xoconostle-Cázares B, Reynaga-Peña CG, León-Ramírez C, Cárabez-Trejo A. Immunolocalization of chitin synthases in the phytopathogenic dimorphic fungus Ustilago maydis. FEMS Yeast Res. 2006;6:999–1009. doi: 10.1111/j.1567-1364.2006.00133.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J, León-Ramírez C, Cabrera-Ponce JL, Martínez-Espinoza AD, Herrera-Estrella L. Completion of the sexual cycle and demonstration of genetic recombination in Ustilago maydis in vitro. Molec. Gen. Genet. 1999;262:468–472. doi: 10.1007/s004380051107. [DOI] [PubMed] [Google Scholar]
- Rupes I, Mao WZ, Astrom H, Raudaskoski M. Effects of nocodazole and brefeldin A on microtubule cytoskeleton and membrane organization in homobasidiomycte Schizophyllum commune. Protoplasma. 1995;185:212–221. [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nature Cell Biol. 2001;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer J, Chen C, Heidrich P, Dickman MB, Tudzynski P. A CDC42 homologue in Claviceps purpurea is involved in vegetative differentiation and is essential for pathogenicity. Eukaryot Cell. 2005;4:1228–1238. doi: 10.1128/EC.4.7.1228-1238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M, Heimel K, Starke V, Kämper J. The Clp1 protein is required for clamp formation and pathogenic development in Ustilago maydis. Plant Cell 2006. 2006;18:2388–2401. doi: 10.1105/tpc.106.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H-P, Kaufmann A, Köhli M, Laissue PF, Philippsen P. From function to shape: a novel role of a formin in fungal morphogenesis of the fungus Ashbya gossypii. Molec. Biol. Cell. 2006;17:130–145. doi: 10.1091/mbc.E05-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchardt I, Assmann D, Thines E, Schuberth C, Steinberg G. Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol Biol Cell. 2005;16:5191–5201. doi: 10.1091/mbc.E05-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Seiler S, Plamann M. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Molec. Biol. Cell. 2003;14:4352–4364. doi: 10.1091/mbc.E02-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S, Plamann M, Schliwa M. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr Biol. 1999;9:779–785. doi: 10.1016/s0960-9822(99)80360-1. [DOI] [PubMed] [Google Scholar]
- Sharpless KE, Harris SD. Functional characterization and localization of the Aspergillus nidulans formin SepA. Molec. Biol. Cell. 2002;13:469–479. doi: 10.1091/mbc.01-07-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Smith DG, García-Pedrajas MD, Hong W, Yu Z, Gold SE, Perlin MH. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot Cell. 2004;3:180–189. doi: 10.1128/EC.3.1.180-189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar KM, Mim CW. Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol. Res. 1994;98:347–355. [Google Scholar]
- Steinberg G, Wedlich-Söldner R, Brill M, Schulz I. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 2001;114:609–622. doi: 10.1242/jcs.114.3.609. [DOI] [PubMed] [Google Scholar]
- Straube A, Brill M, Oakley BR, Horio T, Steinberg G. Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Molec. Biol. Cell. 2003;14:642–657. doi: 10.1091/mbc.E02-08-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Enard W, Berner A, Wedlich-Söldner R, Kahmann R, Steinberg G. A split motor domain in a cytoplasmic dynein. EMBO J. 2001;20:5091–6100. doi: 10.1093/emboj/20.18.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Talesh N, Horio T, Araujo-Bazán L, Dou X, Espeso EA, Peñalva MA, Osmani SA, Oakley BR. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita N, Higashitsuji Y, Konzack S, Fischer R. Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2008;19:339–351. doi: 10.1091/mbc.E07-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Torralba S, Raudaskoski M, Pedregosa AM. Effects of methyl benzimidazole-2-yl carbamate on microtubule and actin cytoskeleton in Aspergillus nidulans. Protoplasma. 1998;202:54–64. [Google Scholar]
- Torralba S, Raudaskoski M, Pedregosa AM, Laborda F. Effect of cytochalasin A on apical growth, actin cytoskeleton organization and enzyme secretion in Aspergillus nidulans. Microbiology. 1998;144:45–53. doi: 10.1099/00221287-144-1-45. [DOI] [PubMed] [Google Scholar]
- Virag A, Lee MP, Si H, Harris SD. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol Microbiol. 2007;66:1579–1596. doi: 10.1111/j.1365-2958.2007.06021.x. [DOI] [PubMed] [Google Scholar]
- Virag A, Harris SD. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Euk. Cell. 2006;5:881–895. doi: 10.1128/EC.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag A, Griffiths AJ. A mutation in the Neurospora crassa actin gene results in multiple defects in tip growth and branching. Fungal Genet Biol. 2004;41:213–225. doi: 10.1016/j.fgb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Weber I, Assmann D, Thines E, Steinberg G. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell. 2006;18:225–242. doi: 10.1105/tpc.105.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I, Gruber C, Steinberg G. A class V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogn Ustilago maydis. Plant Cell. 2003;15:2826–2842. doi: 10.1105/tpc.016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Schulz I, Straube A, Steinberg G. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol Biol Cell. 2002;13:965–977. doi: 10.1091/mbc.01-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl G, Leveleki L, Hassel A, Kost G, Wanner G, Bölker M. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Molec. Microbiol. 2002;45:219–231. doi: 10.1046/j.1365-2958.2002.03010.x. [DOI] [PubMed] [Google Scholar]
- Wendland J. Analysis of the landmark protein Bud3 of Ashbya gossypii reveals a novel role in septum construction. EMBO Rep. 2003;4:200–204. doi: 10.1038/sj.embor.embor727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J, Philippsen P. Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics. 2001;157:601–610. doi: 10.1093/genetics/157.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Momany M. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol Biol Cell. 2002;13:110–118. doi: 10.1091/mbc.01-06-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DC, Choe EY, Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo structural roles of individual subunits. Proc. Natl. Acad. Sci. 1999;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Plamann M. Cytoskeleton and motor proteins in filamentous fungi. Curr Opin Microbiol. 2003;6:628–633. doi: 10.1016/j.mib.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Yaar L, Mevarech M, Koltin Y. A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology. 1997;143:3033–3044. doi: 10.1099/00221287-143-9-3033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Part A. The U. maydis gene products identified in Table 1, and additional ones (see legend to Table 1) were used as queries to identify highest hit homologues in different fungi and other eukaryotes using SIMAP at MIPS Ustilago database MUMDB (http://mips.gsf.de/genre/proj/ustilago/). The highest hit homologues were found in the Basidiomycota followed by the Ascomycota. The chart represents analysis of 306 U. maydis gene products identified after BLAST analysis with 344 S. cerevisiae gene products, as described in Table 1.
Part B. The chart represents analysis of 276 U. maydis gene products obtained as above and used as queries to identify highest hit homologues in different fungi, exclusively, using SIMAP at MIPS (see above).
Part C. The U. maydis gene products used in this analysis were identified using a subset of S. pombe genes involved in cell polarity, cell morphogenesis, and the cytoskeleton as queries of the U. maydis genome using wuBLAST at MIPS (see above). The identified U. maydis gene products were used as queries to identify highest hit homologues in different fungi and other eukaryotes using SIMAP at MIPS Ustilago database MUMDB (see Part A). The source of S. pombe gene products was La Carbona et al., 2006 and the Schizosaccharomyces pombe Gene DB (http://www.genedb.org/genedb/pombe/). The chart represents analysis of 317 U. maydis gene products obtained after wuBLAST analysis of 340 S. pombe gene products.
Part D. The chart represents analysis of 276 U. maydis gene products obtained as above and used as queries to identify highest hit homologues in different fungi, exclusively, using SIMAP at MIPS (see PartA).