Abstract
We report the crystal structure of the non-selective cation channel NaK from b. cereus at a resolution of 1.6 Å. The structure reveals the intracellular gate in an open state compared to the closed form reported previously, making NaK the only channel for which the three-dimensional structures of both conformations are known. Channel opening follows a conserved mechanism of inner helix bending utilizing a flexible glycine residue, the gating hinge, seen in MthK and most other tetrameric cation channels. Additionally, distinct inter and intra-subunit rearrangements involved in channel gating are seen and characterized for the first time along with inner helix twisting motions. Furthermore, we identify a residue deeper within the cavity of the channel pore, Phe92, which likely forms a constriction point within the open pore, restricting ion flux through the channel. Mutating this residue to Ala causes a subsequent increase in ion conduction rates as measured by 86Rb flux assays. The structures of both the open and closed conformations of the NaK channel correlate very well with those of equivalent K+ channel conformations, namely MthK and KcsA, respectively.
The opening and closing of ion channel pores in response to external stimuli such as voltage or ligand binding is a process of extreme importance to ion channel physiology[1]. Up till now, our knowledge of the open and closed conformations of tetrameric cation channel pores comes predominantly from the crystal structures of various K+ channels, among which the structures of KcsA and MthK have been generally accepted as reasonable models for the closed and open conformations of tetrameric cation channel pores, respectively [2-9]. The closed conformation is characterized by near straight inner helices and the subsequent bundle crossing formed by interactions between their c-terminal residues. In the open conformation seen in the MthK structure, the inner helices appear to bend at a conserved glycine residue, the gating hinge also seen in most other cation channels, resulting in the disruption of the bundle crossing. However, structural information on both states from the same channel has so far been lacking.
In the process of studying the ion selectivity properties of the NaK channel which shares overall sequence and structural similarities with KcsA except in the selectivity filter region and conducts both Na+ and K+ [10, 11], we crystallized a truncated construct (NaKNΔ19) lacking its N-terminal M0 helix and determined its structure at a resolution of 1.6 Å. To our surprise, the NaKNΔ19 structure reveals the intracellular gate in an open conformation. Combined with the closed conformation structure of NaK, the new structure provides a unique opportunity to analyze in detail the structural features underlying pore opening and closing within the same channel. We observe both inner helix twisting and bending at the glycine gating hinge upon pore opening. Our data clearly demonstrate the conservation of central sequence and structural motifs involved in channel opening, while also offering insight into the intricate rearrangements of interactions within and between channel subunits upon undergoing gating transitions. In addition, the high resolution structures also allow us to carry out an analysis of the molecular details of ion binding in the NaK selectivity filter, which we report in our companion paper.
Results
Overall structure of the open NaK channel
NaKNΔ19 was crystallized in the presence of various monovalent cations (NaCl, KCl, or RbCl) (Methods). Numerous structures of NaKNΔ19 in different salts were determined by molecular replacement and refined to resolutions between 1.6 Å and 2.0 Å. Only the 1.6 Å structure is discussed here whereas all other structures are discussed in the companion paper focusing on ion selectivity. All crystals were of space group I4 and contained two subunits per asymmetric unit. Interestingly, each subunit uses three crystallographic 4-fold related counterparts to form a functional channel tetramer. In other words, the molecular 4-fold coincides with the crystallographic tetrad and the crystal contains two sets of channel tetramers with virtually identical structures except for minor differences in B-factor distribution.
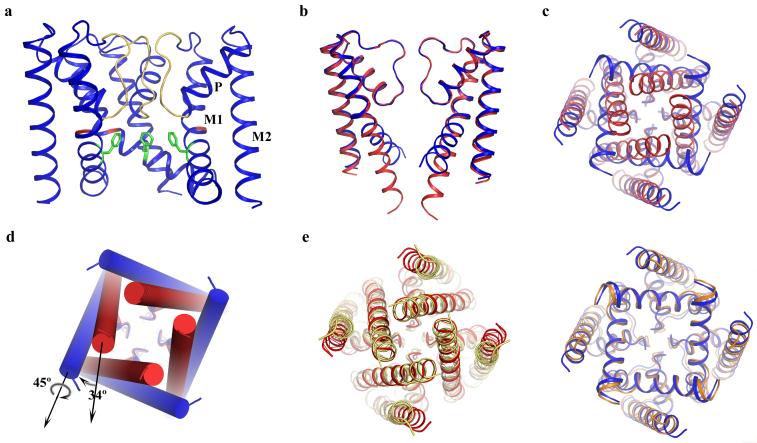
In comparison to NaK, the NaKNΔ19 structure apparently adopts an open conformation at its intracellular gate (Fig. 1a). Superimposition of the structures of NaKNΔ19 and NaK with its M0 helix removed, viewed along the plane of the membrane with proximal and distal subunits removed (Fig. 1b) and from the intracellular side (Fig. 1c), reveal conformational changes of the NaK intracellular gate upon opening and closing. The regions surrounding the selectivity filter in both structures (residues 35 to 85) are almost identical with a main chain root-mean-square deviation (RMSD) of 0.35Å, which suggests that this region remains static during channel gating. Major structural changes appear to occur in the pore lining inner helices just below the selectivity filter. The inner helices in the NaK structure appear to be nearly straight, reminiscent of the KcsA structure, whereas those in NaKNΔ19 appear to develop a bend (about 34°, Fig. 1d) that splays the intracellular gate wide open as seen for the MthK open pore structure[4]. This inner helix bending occurs at residue Gly87 in NaK, shown by sequence alignment (Supplementary Fig. 1) to be the conserved gating hinge glycine seen in MthK and other selective and non-selective cation channels. In addition to a simple bending motion, the inner helices also undergo a 45° clockwise twist around their helical axis as viewed from intracellular side (Fig. 1d). Along with inner helix bending, the outer helices tilt tangentially in the same direction by about 11° without any twisting motion (Fig. 1c).
Figure 1.
Structure of NaKNΔ19 in an open conformation. (a) Overall structure of NaKNΔ19 with the front subunit removed for clarity. Selectivity filter residues, Gly87 and Phe92 are colored yellow, red and green, respectively. M1=outer helix, M2=inner helix, and P=pore helix b) Superimposition of the structures of NaKNΔ19 (blue) and NaK (red, PDB code 2AHY) with its M0 helix removed, viewed from the plane of the membrane with proximal and distal subunits removed and c) from the intracellular side. d) Conformational change of NaK inner helices from the closed (red cylinder) to open state (blue cylinder) involves a 34° bending and a 45° twisting around the helical axis. e) Comparison of NaK with KcsA (left) and MthK (right). Superimposition of the closed NaK structure (red, PDB code 2AHY) with KcsA (yellow, PDB code 1K4C) viewed from the intracellular side along 4-fold axis and the equivalent superimposition of the open NaKNΔ19 structure (blue) with the MthK pore (orange, PDB code1LNQ) are shown.
Structural conservation in the open and closed NaK channel pores
Despite the structural difference between the selectivity filters of NaK and K+ channels, their overall tertiary structures, comprising the inner and outer helices along with the pore helix, are virtually identical. The availability of both closed and open conformation structures of the NaK channel at high resolution allows us to compare and analyze the global arrangement of these three structural elements in NaK with those in K+ channels structures of equivalent pore conformations. Figure 1e shows such a comparison of the closed and open structures of NaK with KcsA and MthK, respectively. In the closed conformation, the inner and pore helices of NaK superimpose very well with those of KcsA with a main chain RMSD of 0.73 Å, whereas the respective outer helices are slightly shifted translationally (Fig.1e, left). In the open conformation, all three components of NaKNΔ19 and MthK (outer, inner, and pore helices) superimpose very well with a main chain RMSD of 0.74 Å (Fig. 1e, right). Considering the fact that these four structures are from three different channels crystallized in different conditions, the observed structural and positional conservation of the pore-lining inner helices is a clear indication of conserved gating mechanics in tetrameric cation channels. Since the major conformational changes upon channel gating occur within the inner helices in both NaK and K+ channels, the position of outer helices does not seem to require the same level of conservation as their pore lining counterparts. This was also seen in a structural comparison of the KcsA and Kir channels where their respective outer helices did not superimpose well compared to the inner helices [9].
Inter/intra subunit interactions in the closed and open NaK channel
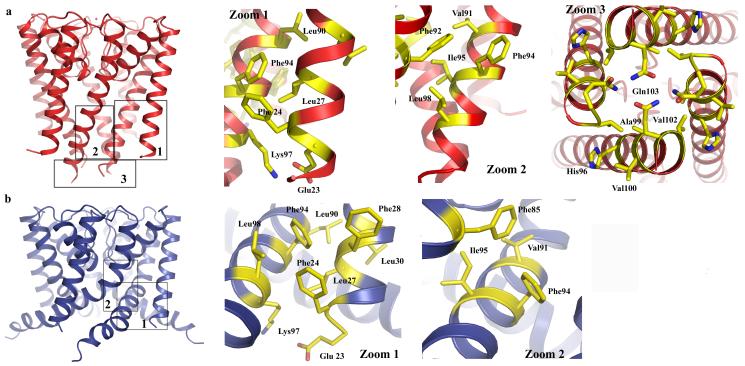
The determination of high resolution structures for both the open and closed conformation of the NaK channel allows us, for the first time, to visualize the rearrangement of intra and inter-subunit protein contacts within the same channel pore upon gating. The intra-subunit interaction between inner and outer helices remains similar between closed (Fig. 2a, zoom 1) and open (Fig. 2b, zoom 1) conformations as a consequence of the concurrent movement of both helices and inner helix twisting. However, the inter-subunit interactions between neighboring inner helices at the bundle crossing seen in the closed NaK structure (Fig. 2a, zoom 3) are disrupted upon pore opening. It is also interesting to note that a major change in inter-subunit interaction occurs at the region just above the bundle crossing. In the closed conformation, Phe92 from each inner helix forms contacts with a hydrophobic patch on the opposite face of Phe92 from the neighboring inner helix formed by Val91, Phe94, Ile95 and Leu98 (Fig. 2a zoom 2). Inner helix bending in the open conformation causes Phe92 to swing away and point its side chain towards the central ion conduction pathway. However, the hydrophobic patch also slides along the neighboring inner helix by two helical turns and forms new van der Waals contacts with Phe85 (Fig. 2b, zoom 2).
Figure 2.
Analysis of inter and intra-subunit interactions in the closed (a) and open (b) structures of the NaK channel. The numbers in the boxed regions in both structures correspond to the zoom-in panels 1-3 for the closed structure and 1-2 for the open NaK structure. Zoom 1 in both cases highlights intra-subunit interactions between outer (M1) and inner (M2) helices. Zoom 2 highlights inter-subunit interactions between inner helices of two neighboring subunits. Zoom 3 shows an intracellular view of the interactions involved in the bundle crossing formation in the closed structure.
Effect of ion conduction pathway size on channel conduction
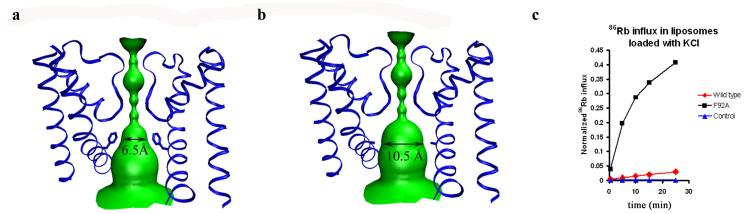
In the closed NaK channel, side chains from residues Ile95, Ala99 and Gln103 at or above the bundle crossing form several constriction points along the ion conduction pathway, with the intracellular gate almost completely sealed off by the side chain of Gln103 (Fig. 2a, zoom 3). The bending of inner helices upon channel opening disrupts this bundle crossing. However, a new constriction point is created by the side chain of residue Phe92 (Fig. 1a and Fig. 3a). It is interesting to note that residue positions equivalent to Phe92 in NaK are occupied by small side chain amino acids in most other cation channels, most notably alanine as in MthK where it forms the narrowest part of the open pore [4](Supplementary Fig. 1). By roughly orienting its aromatic ring towards the central ion conduction pathway, Phe92 forms a constriction point in the open NaK pore with an ion pathway diameter of about 6.5 Å (Fig. 3a), which is narrower than what is seen in the open MthK channel (∼ 9.5 Å) and is likely to lower ion conduction rates. Indeed, the structure of a Phe92Ala mutation shows an ion conduction pathway comparatively less hindered by steric barriers (Fig. 3b) with a diameter of 10.5 Å at the constriction point and expectedly leads to increased accumulation of Rb86 in liposomes (Fig. 3c). Although the effects of this mutation on increasing channel open probability in NaK cannot be discounted based on results from the 86Rb flux assay, our recent electrophysiological study on equivalent mutations in MthK shows an obvious change in ion conduction rates (Y. Li, S. Ye, and Y.J, unpublished data). This supports the suggestion that a small side chain residue at this position is important in allowing for a wider ion passageway and easier access to the selectivity filter in cation channels as previously proposed[4].
Figure 3.
Effect of the size of the ion conduction pathway size on ion conduction in NaK. a) Surface representation of the ion conduction pathway of the NaKNΔ19 open pore and b) its Phe92Ala mutant. c) 86Rb flux assay showing time dependent Rb+ influx in liposomes reconstituted with NaKNΔ19 (Red trace) and the Phe92Ala mutant (black trace). Both proteins were reconstituted in liposomes loaded with KCl at the same concentration. Liposomes with no reconstituted protein were used as a control (blue trace).
Discussion
In this study we present a high resolution structure of the NaKNΔ19 channel in an open conformation. A comparison of this open state structure with that of the previously determined closed state shows that the region surrounding the selectivity filter, the top 1/3 of the pore at towards the extracellular side, remains static during channel gating. In contrast, major conformational changes occur in the inner helices at residue Gly87 just below the filter. The structure similarity between NaKNΔ19 and MthK pore points to similar pore opening mechanics in these two channels, specifically, utilization of a conserved glycine residue as a gating hinge to allow for inner helix bending. This glycine hinge has also been functionally shown to be important for gating in other tetrameric cation channels, such as voltage-gated K+ and Na+ channels [12-15], although these channel pores may not open as wide as what is observed in MthK or NaK.
Even though the intra- and inter-subunit interactions in the open state of NaK appear to be less extensive than those in the closed state, mainly due to the disruption of the helix bundle crossing, a number of factors could contribute to the stabilization of the open state captured in the crystal. First, the M0 helices are absent in NaKNΔ19. In the closed NaK structure, four M0 helices form a cuff surrounding the bundle crossing and could prevent the inner helices from splaying open, a spatial hindrance absent in the truncated construct. Indeed, NaKNΔ19 does functionally display substantially higher ion conduction rates compared to the full length channel in 86Rb flux assays[11], which most likely stems from a higher channel open probability. Second, the use of detergents instead of lipids in the purification and crystallization procedures, in other words the absence of a lipid environment in the crystal, could allow for more conformational freedom for the channel. Finally, protein packing in the crystal could be a main factor contributing to the open state. Two kinds of inter-tetramer packing interactions were observed in the NaKNΔ19 crystal (Supplementary Fig. 2). One is the anti-parallel hydrophobic packing between outer helices from two neighboring channel tetramers (Supplementary Fig. 2, squared region). The other is hydrophilic contacts between the C-terminal end of the inner helix and the extracellular surface of a neighboring tetramer (Supplementary Fig 2, circled regions). These hydrophilic contacts are predominantly hydrogen bonding in nature and some are mediated by water molecules. Since both kinds of inter-tetramer packing interactions are quite extensive, we believe this is the dominant force stabilizing NaKNΔ19 in the open conformation. Despite the possibility of structural distortion due to crystal packing, the fact that NaKNΔ19 and MthK, two different crystals of different proteins, share virtually the same structure is a clear indication that NaKNΔ19 does represent an open pore structure.
KcsA has been one of the most well studied K+ channels in regards to its gating properties, specifically the physical movement of its intracellular gate [16-24]. The global conformational changes observed between the open and closed conformation structures of NaK match quite well with those observed in KcsA. In a recent study using single KcsA tetramers labeled with gold nano crystals, a global twisting motion of +/- 40° was observed for inner helices in channels undergoing gating transitions[24]. Such a movement can be easily mapped on to the NaK structures. By using the Cα of Ala99 as an example, the movement of residues at the C-terminal part of inner helices between closed and open state results in a rotation of about 42° relative to the central axis (Supplementary Fig. 3a), equivalent to the global twisting motion reported for the KcsA pore.
It is also important to note that some of the cross subunit (diagonal) distances calculated between residues in the open and closed conformation of NaK are quite different from those reported in a gating study on KcsA using Electron Paramagnetic Resonance (EPR) spectroscopic methods [17, 18]. For example, the distance between two spin-labeled Ala109s in KcsA shows a decrease of about 9 Å from closed to open states in the EPR measurement, whereas the diagonal distance between the Cβs of the equivalent residues (Lys97) in NaK show an increase of 5.5 Å from closed to open state based on our crystal structures. We believe this discrepancy arises mainly from contributions of the spin label arm length, which can extend up to 9 Å from the Cβ of the labeled residue and the paramagnetic center in the nitroxide moiety [25], rather than from differences in gating mechanics. As shown in Supplementary Fig 3b, in the closed state of NaK, Lys97 is positioned at the distal face of the inner helix relative to the central axis of the pore. A spin label at this position likely has the nitroxide moiety extended away from the central axis leading to an overestimation of the diagonal distance. On the other hand, Lys97 in the open state would be oriented more towards the ion conduction pathway as a consequence of inner helix bending and twisting and the spin label at this position will have the nitroxide moiety extending closer to the central axis leading to an underestimation of the diagonal distance. If we measure the diagonal distance between the amine atoms of Lys97s instead of Cßs as a simple approximation of the distance between the paramagnetic centers, a decrease of 7 Å is observed (from 29 Å in the closed state to 22 Å in the open state), which matches well with that observed in the EPR measurement of KcsA labeled at the equivalent residue (Ala109). Here we only consider one residue as an example instead of mapping and comparing every relevant NaK residue with corresponding KcsA residues in the EPR studies. The important conclusion that arises, however, is that the structure of KcsA in its open state would likely be similar to that of NaKNΔ19 or MthK and the structures of NaK and NaKNΔ19 represent the general structures of the tetrameric cation channel pore in closed and open states, respectively.
Methods
Protein expression and purification
We cloned the gene encoding the NaKNΔ19 channel, a truncated form of the NaK channel lacking the N-terminal 19 residues, into the PQE60 expression vector between NcoI and BamHI restriction endonuclease sites with a thrombin cleavage site between a C-terminal hexahistidine sequence and the channel. The Phe92 to Ala mutant was generated using the Strategene Quick Change II site directed mutagenesis kit. The channels were expressed in E. coli XL1 Blue or SG13009 cells by induction (at A600 ∼ 0.8) with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 25°C for 20 hours. Cells were harvested and lysed in a solution of 50 mM Tris·HCl, pH 8.0, and 100 mM NaCl. Expressed protein was then extracted from the cell lysate for 3 hours at room temperature in the above solution by adding 40 mM n-decyl-β-D-maltoside (DM) and purified on a Talon Co2+ affinity column (Clontech). The column was first washed with 20 mM Tris·HCl, pH 8.0, 100 mM NaCl and 5 mM DM, then with the above buffer containing 15 mM imidazole to remove non-specifically bound proteins. Finally, the NaKNΔ19 channel was eluted with 300 mM imidazole. After elution, the protein was incubated overnight at room temperature (∼ 20 °C) in the presence of 1 unit of thrombin per 2 mg of protein to remove the hexahistidine tag, and then further purified on a Superdex-200 (10/30) column in a solution of 20 mM Tris·HCl, pH 8.0, 100 mM NaCl and 5 mM DM.
Crystallization and structure determination
Purified proteins were concentrated to 12-20 mg ml-1 using an Amicon Ultra centrifugal filtration device (50kD MW cutoff) and crystallized using the sitting drop vapor diffusion method by mixing equal volumes of concentrated protein and well solution. The highest resolution (1.6Å) structure used in this discussion was determined from crystal grown over a well solution containing 55-70% (v/v) (±)-2-Methyl-2,4-pentanediol (MPD), 1 mM CaCl2 and 100 mM Glycine pH 9.5. All crystals were frozen in liquid propane with the crystallization solution serving as the cryo-protectant. Crystals were of space group I4 with unit cell dimensions around a=b=68 Å, c = 89 Å, α=β=γ=90° and contained 2 molecules per asymmetric unit. The four-fold axis of the channel tetramer coincided with the crystallographic tetrad.
X-ray data were collected at the Advanced Photon Source (APS) Beamlines 19-ID/BM and 23-ID. Data processing and scaling was performed using the HKL2000 software [26]. The structures were determined by molecular replacement using Molrep in the CCP4 suite [27] using the region surrounding the filter of the full length NaK structure (residues 30 to 90, PDB code 2AHY) as an initial search model followed by repeated cycles of model building in O [28] and refinement in CNS [29]. 95.1 % of the residues were in most favored regions and 4.9 % in additionally allowed regions on a Ramachandran plot. Some discontinuous electron density can be attributed to bound lipid, detergent, or other solvent molecules and was assigned as water molecules for simplicity. Detailed data collection and refinement statistics are listed in Table I.
Table 1.
Data collection and refinement statistics (molecular replacement)
| NaKNΔ19 100mM Na+/1mM Ca2+ | |
|---|---|
| Data collection | |
| Space group | |
| Cell dimensions | |
| a, b, c (Å) | 68.030, 68.030, 89.264 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50-1.6 |
| Rsym or Rmerge | 4.2 (48.4) |
| I / σI | 48 (1.5) |
| Completeness (%) | 94.9 (62.4) |
| Redundancy | 6.7(2.3) |
| Refinement | |
| Resolution (Å) | 50-1.6 |
| No. reflections | 25448 |
| Rwork / Rfree | |
| No. atoms | |
| Protein | 1462 |
| Ligand/ion | 23 |
| Water | 152 |
| B-factors | |
| Protein | 35.629 |
| Ligand/ion | 42.35 |
| Water | 55.228 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0090 |
| Bond angles (°) | 1.299 |
All structure figures were generated in Pymol (www.pymol.org) with the exception of the surface renderings of the ion conduction pathways of NaKNΔ19 and its F92A mutant, which were calculated in the program HOLE [30] and rendered in VMD [31].
86Rb flux assay
All channel proteins used in the flux assay were purified in the detergent DM and reconstituted into lipid vesicles composed of 1-palmitoyl-2-oleoyl-phosphatidylethanolamine (POPE, 7.5 mg ml-1) and 1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG, 2.5 mg ml-1) at a protein/lipid ratio of 5-10 μg mg-1 using the same method as described[32, 33] with the following modifications: 10 mM DM was used to solubilize the lipid and dialysis (against a reconstitution buffer of 10 mM Hepes, pH 7.4, 400 mM NaCl or KCl, and 4 mM NMG) was used to slowly remove the detergent from the detergent/lipid/protein mixture. The reconstituted liposome samples were kept at -80°C in 100 ul aliquots.
The 86Rb flux assay was performed following virtually the same procedures as described[33]. Liposomes were thawed and sonicated in a bath sonicator for 30 seconds before the assay. To remove extra-liposomal salt (NaCl or KCl), 95 ul samples were passed through a pre-spun Sephadex G-50 fine gel filtration column (1.5 ml bed volume in a 5 ml disposable spin column) swollen in 400 mM Sorbitol, 10 mM Hepes, pH 7.4 and 4 mM NMG. 160 ul of liposome samples collected after this buffer exchange step were added to 320 ul 86Rb flux buffer (400 mM Sorbitol, 10 mM Hepes, pH 7.4, 4 mM NMG, 50 uM NaCl or KCl, and 5 uM 86RbCl). 75ul of this reaction mixture were passed through another pre-spun gel filtration column as described above to eliminate extraliposomal 86Rb at the desired time points. For normalization, 1ug ml-1 (final) valinomycin was added to the mixture for an additional 2 minutes before taking the final reading after the last gel filtration step. The final eluate was mixed with 10 ml scintillation cocktail and its radioactivity measured in a scintillation counter
Supplementary Material
Acknowledgments
Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US Department of Energy, Office of Energy Research. We thank the beamline staff for assistance in data collection. This work was supported by Howard Hughes Medical Institute and by grants from the NIH/NIGMS (RO1 GM079179), David and Lucile Packard Foundation and McKnight Endowment for Neuroscience. A.A. was supported by National Institutes of Health Training Grant T32 GM008297.
Footnotes
Competing interests statement: The authors declared no competing interests.
Coordinates. Protein Data Bank: The atomic coordinates and structure factors of NaKNΔ19 have been deposited (accession number 3E86).
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd edition ed. Sinauer Associates, Inc.; Sunderland, MA: 2001. [Google Scholar]
- 2.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417(6888):515–22. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, et al. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414(6859):43–8. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417(6888):523–6. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 5.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 6.Long SB, et al. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168):376–82. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 7.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 8.Kuo A, et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300(5627):1922–6. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 9.Nishida M, et al. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. Embo J. 2007;26(17):4005–15. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam A, Shi N, Jiang Y. Structural insight into Ca2+ specificity in tetrameric cation channels. Proc Natl Acad Sci U S A. 2007;104(39):15334–9. doi: 10.1073/pnas.0707324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi N, et al. Atomic structure of a Na+- and K+-conducting channel. Nature. 2006;440(7083):570–4. doi: 10.1038/nature04508. [DOI] [PubMed] [Google Scholar]
- 12.Jin T, et al. The (beta)gamma subunits of G proteins gate a K(+) channel by pivoted bending of a transmembrane segment. Mol Cell. 2002;10(3):469–81. doi: 10.1016/s1097-2765(02)00659-7. [DOI] [PubMed] [Google Scholar]
- 13.Magidovich E, Yifrach O. Conserved gating hinge in ligand- and voltage-dependent K+ channels. Biochemistry. 2004;43(42):13242–7. doi: 10.1021/bi048377v. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron. 2004;41(6):859–65. doi: 10.1016/s0896-6273(04)00116-3. [DOI] [PubMed] [Google Scholar]
- 15.Ding S, et al. Investigating the putative glycine hinge in Shaker potassium channel. J Gen Physiol. 2005;126(3):213–26. doi: 10.1085/jgp.200509287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5(6):459–69. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 17.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285(5424):73–8. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 18.Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001;8(10):883–7. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 19.Kelly BL, Gross A. Potassium channel gating observed with site-directed mass tagging. Nat Struct Biol. 2003;10(4):280–4. doi: 10.1038/nsb908. [DOI] [PubMed] [Google Scholar]
- 20.Zimmer J, Doyle DA, Grossmann JG. Structural characterization and pH-induced conformational transition of full-length KcsA. Biophys J. 2006;90(5):1752–66. doi: 10.1529/biophysj.105.071175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwamoto M, et al. Surface structure and its dynamic rearrangements of the KcsA potassium channel upon gating and tetrabutylammonium blocking. J Biol Chem. 2006;281(38):28379–86. doi: 10.1074/jbc.M602018200. [DOI] [PubMed] [Google Scholar]
- 22.Baker KA, et al. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14(11):1089–95. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi K, et al. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282(20):15179–86. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu H, et al. Global twisting motion of single molecular KcsA potassium channel upon gating. Cell. 2008;132(1):67–78. doi: 10.1016/j.cell.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 25.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci U S A. 1995;92(18):8239–43. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, et al. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65(6):2455–60. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 32.Heginbotham L, et al. Single streptomyces lividans K(+) channels: functional asymmetries and sidedness of proton activation. J Gen Physiol. 1999;114(4):551–60. doi: 10.1085/jgp.114.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. J Gen Physiol. 1998;111(6):741–9. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.