Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis in some but not all breast cancer cell lines. Breast cancers can be divided into those which express the estrogen (ER) and progesterone (PR) receptors, those with HER-2 amplification, and those without expression of ER, PR, or HER-2 amplification (referred to as basal or triple-negative breast cancer). We tested a panel of 20 breast cancer cell lines representing the different types of breast cancer to evaluate if the molecular phenotype of the breast cancer cells determined their response to TRAIL. The most striking finding was that eight of eleven triple-negative cell lines are sensitive to TRAIL-mediated apoptosis. The eight TRAIL-sensitive triple-negative cell lines have a mesenchymal phenotype while the three TRAIL-resistant triple-negative cell lines have an epithelial phenotype. Two of five cell lines with HER-2 amplification were sensitive to TRAIL and none of the five ER positive cell lines were sensitive. RNAi-mediated knockdown of TRAIL receptor expression demonstrated that TRAIL Receptor 2 (TRAIL-R2) mediates the effects of TRAIL, even when both TRAIL-R1 and TRAIL-R2 are expressed. Finally, inhibition of EGFR, expressed in both TRAIL-sensitive and TRAIL-resistant triple-negative breast cancer cell lines, using a small molecule tyrosine kinase inhibitor (AG1478) enhanced TRAIL-induced apoptosis in TRAIL-sensitive cell lines but did not convert resistant cells into TRAIL-sensitive cells. Together, these findings suggest that a subset of triple-negative breast cancer, those with mesenchymal features, may be the most likely to benefit from TRAIL targeted therapy. These findings could form the basis to select breast cancer patients for clinical trials of TRAIL-R2 ligands.
Keywords: TRAIL, apoptosis, basal breast cancer, triple-negative breast cancer
Introduction
Activation of the death receptors by the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) has been shown to induce apoptosis selectively in a variety of cancer cell lines including breast cancer cell lines, but not in normal cells [1–3]. Also, animal studies have shown that TRAIL can induce regression of cancer xenografts without toxicity to normal tissues [3]. The selective induction of apoptosis in cancer cells, but not in normal cells, has prompted clinical studies of TRAIL and agonistic antibodies as cancer therapy. However, we have shown previously that the majority of breast cancer cell lines are resistant to TRAIL-mediated apoptosis [1]. In fact, of the breast cancer cell lines we tested for TRAIL sensitivity, only the MDA-MB-231 breast cancer cell line is sensitive to TRAIL-induced apoptosis [1]. Studies in breast cancer cell lines have failed to identify clearly the major determinants of TRAIL sensitivity.
Breast cancer is a heterogeneous group of diseases. Approximately 60–70% of breast cancers express estrogen receptors (ER) and/or progesterone receptors (PR) and approximately 20–30% of breast cancers have amplified HER-2 and express high levels of the HER-2 protein [4]. Molecularly targeted therapies that inhibit the estrogen pathway or that target amplified HER-2 are effective in the treatment of breast cancer in patients whose tumors express these targets [4]. In approximately 15–20% of patients with breast cancer, the tumors do not express ER or PR and do not have amplification of HER-2 [4]. These tumors are called triple-negative breast cancer and patients with these tumors have a poor prognosis [4]. There is no clinically validated, molecularly targeted therapy for these patients and they can be treated only with chemotherapy [4]. Thus, identification of novel, molecularly targeted therapies for triple-negative breast cancer would be of great benefit.
In the work presented here, we investigated whether the molecular phenotype of breast cancer determined the response to TRAIL-induced apoptosis. We found that triple-negative breast cancer cells, and particularly those with mesenchymal features, are preferentially killed by TRAIL. Further, we found that TRAIL-induced apoptosis is mediated predominantly by TRAIL Receptor 2 (TRAIL-R2).
Materials and methods
Cell lines
The AU565, BT474, BT549, HCC38, HCC1500, HCC1954, MB231, MB436, MB453, MB468, MCF7, SKBR3, T47D, and ZR75-1 cell lines were obtained from ATCC (Manassas, VA, USA). BT20, HCC1937, HS578T, and MB157 were obtained from Reinhard Ebner (Avalon Pharmaceuticals; Germantown, MD, USA). SUM149 and SUM159 were obtained from Steve Ethier (Karmanos Cancer Institute, Detroit, MI, USA). All cell lines were grown in RPMI 1640 supplemented with 10% fetal calf serum, 100 units/ml of penicillin, and 100 units/ml of streptomycin.
Cell culture and MTS assays
The GST-TRAIL construct and the isolation of recombinant GST-TRAIL fusion protein have been previously described [1]. The GST-TRAIL protein (at 0.5mg/mL in culture media) was stored at −70°C in aliquots until used. TRAIL-mediated cytotoxcity was assessed by the MTS assay using Cell Titer 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA ) as previously described [5]. All MTS measurements were done in replicates of six wells and each experiment was carried out at least three times. Results are given as the mean +/− the standard error of the mean (SE) for at least three independent experiments.
The tetra-peptide pan-caspase inhibitor Z-VAD-fmk (Biomol International, Plymouth Meeting, PA) was resuspended in DMSO at a concentration of 10 mM. Z-VAD-fmk was added to cells at a final concentration of 30 μM 30 min prior to any treatments. Control cells were incubated with DMSO at the same concentration as Z-VAD-fmk-treated cells. Cell viability was analyzed by the MTS assay after 16 h incubation with TRAIL.
The EGFR tyrosine kinase inhibitor AG1478 (EMD Biosciences, San Diego, CA) was reconstituted in DMSO (Sigma-Aldrich Co., St. Louis, MO) at 10 mM. Cells were incubated with the inhibitor or an equivalent amount of DMSO at a final concentration of 30 μM for 4 h prior to addition of TRAIL.
Apoptosis and caspase assays
Apoptosis was assessed in TRAIL-treated cells using the Cell Death Detection ELISAplus kit (Roche Diagnostic Corp, Indianapolis, IN). Briefly, 105 cells were plated in 6 well plates and allowed to adhere overnight. TRAIL was added at 1 μg/ml for 4 h after which media was removed and cells were lysed with 200 μl of lysis buffer following the instructions supplied with the kit. Mono- and oligonucleosomes in the cytoplasmic fraction of cell lysates, indicative of fragmented DNA, were measured using the protocol supplied with the kit.
Caspase activation by TRAIL was assessed using the Caspase-Glo® 3/7 Assay Kit (Promega, Madison, WI, USA). 1×104 cells were plated in 96 well microtiter plates, allowed to adhere overnight, and then treated with TRAIL at the indicated concentrations for 4 h. Cells were lysed and caspase activity was measured in the cell lysates using the protocol supplied with Thermo kit. Luminescence was measured on a LabSystems Fluoroscan Ascent Fluorometer (Thermo LabSystems, Milford, MA, USA).
Isolation and analysis of protein lysates
Cells (8–10 ×105) were plated onto 20 cm diameter tissue culture dishes and grown to 60–80% confluence. Cell lysates were prepared and immunoblots performed as previously described [1]. The following rabbit polyclonal antibodies were used for immunoblotting: anti-caspase-3 (#9662; Cell Signaling, Danvers, MA, USA); anti-EGFR (#2232; Cell Signaling, Danvers, MA, USA); anti-ErbB-2 (#RB-103-P; Lab Vision Corp. Neomarkers, Freemont, CA, USA); anti-TRAIL-R1 (#556554; BD Pharmingen, San Diego, CA, USA); and anti-TRAIL-R2 (#2019; ProSci, Poway, CA, USA). The following mouse monoclonal antibodies were used for immunoblotting: anti-caspase-8 (#05-477; Upstate Biotechnology, Lake Placid, NY, USA); anti-E-cadherin (#610181; BD Transduction Laboratories, San Jose, CA, USA); anti-estrogen receptor (#MS-1071; Lab Vision Corp. Neomarkers, Freemont, CA, USA); anti-tubulin (#T9026; Sigma, St. Louis, MO, USA); anti-TRAIL-R3 (#SC-65309; Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-TRAIL-R4 (#SC-65310; Santa Cruz Biotechnology, Santa Cruz, CA,USA); anti-vimentin (#550513; BD Pharmingen, San Diego, CA, USA). Rabbit polyclonal anti-moeisin antibody was graciously provided by Dr. Steven Shaw. Mouse monoclonal anti-EGFR (#GR01; EMD Biosciences, San Diego, CA, USA) was used to immunoprecipitate the EGFR. HRP-conjugated mouse monoclonal anti-phosphotyrosine (#16-184; Upstate Biotechnology, Lake Placid, NY) was used to evaluate EGFR tyrosine phosphorylation in EGFR precipitates.
RNA interference (RNAi)
siRNAs were purchased from Dharmacon (Lafayette, CO, USA) for TRAIL-R1 (ON-TARGETplus SMARTpool #L-008090-00), TRAIL-R2 (ON-TARGETplus SMARTpool #L-004448-00), vimentin (ON-TARGETplus SMARTpool #L-003551-00), moesin (ON-TARGETplus SMARTpool #L-011732-00), E-cadherin ON-TARGETplus SMARTpool #L-003877-00), and a non-targeting siRNA control pool(ON-TARGETplus SMARTpool control #D-001810-10). Cells were transfected in solution with siRNA at a final concentration of 50 nM. The stock siRNA at 20 μM was diluted into 3.5 ml RPMI media and incubated at room temperature for 10 min in 50 ml tubes. 49 μl of Oligofectamine (Invitrogen Corp, Carlsbad, CA, USA) was added to the siRNA and incubated at room temperature for 15 min. 3.5 ml of cells at 2×105/ml in RPMI supplemented with 10% FCS (without antibiotics) were added to the siRNA/Oligofectamine solution, mixed, and then 100 μl of the solution was plated in each of 60 wells in a 96 well tissue culture plate. The remaining transfection mixture was plated in 6 well plates for protein analysis. TRAIL was added to the 96 well plates at 48 h post-transfection and MTS reagent was added at 72 h post-transfection for viability measurements. Cells plated in the 6 well plates were harvested for protein analysis at 72 h post-transfection as described above.
Microarray analysis
Published microarray data representing 51 breast cancer cell lines [6] were downloaded from the caArray website (http://caArray.nci.nih.gov). All analyses were performed using software from the Bioconductor project [7]. Raw CEL files were normalized using RMA from the affy software package (http://www.bioconductor.org). Genes significantly correlated with TRAIL sensitivity (measured at 1 μg/ml) were chosen by linear regression using the Limma Bioconductor package [8]. For plotting, TRAIL-sensitive cell lines were defined as having an IC50 ≤ 1 μg/ml. P-values were corrected for multiple comparisons [9].
Statistics
Statistical comparison of mean values was performed using the paired and unpaired Student’s t-tests. Associations between TRAIL sensitivity and phenotype, molecular classification based on array, or protein expression data were analysed by Pearson’s Chi-squared test with Yates’ continuity correction using the R statistics (http://www.R-project.org). All p-values are two tailed.
Results
TRAIL preferentially kills triple-negative breast cancer cells with mesenchymal features
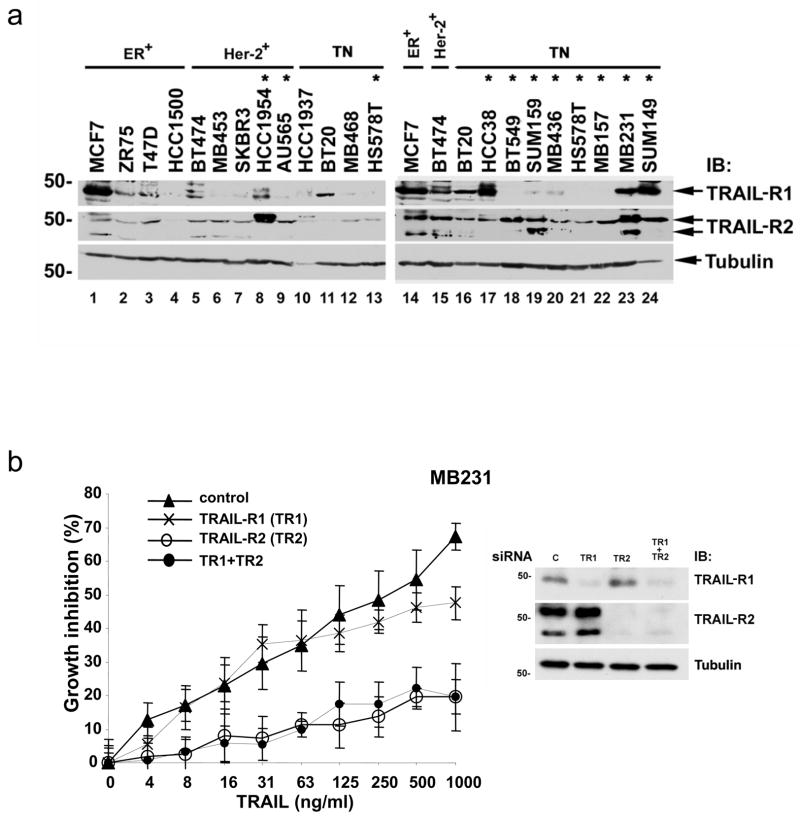
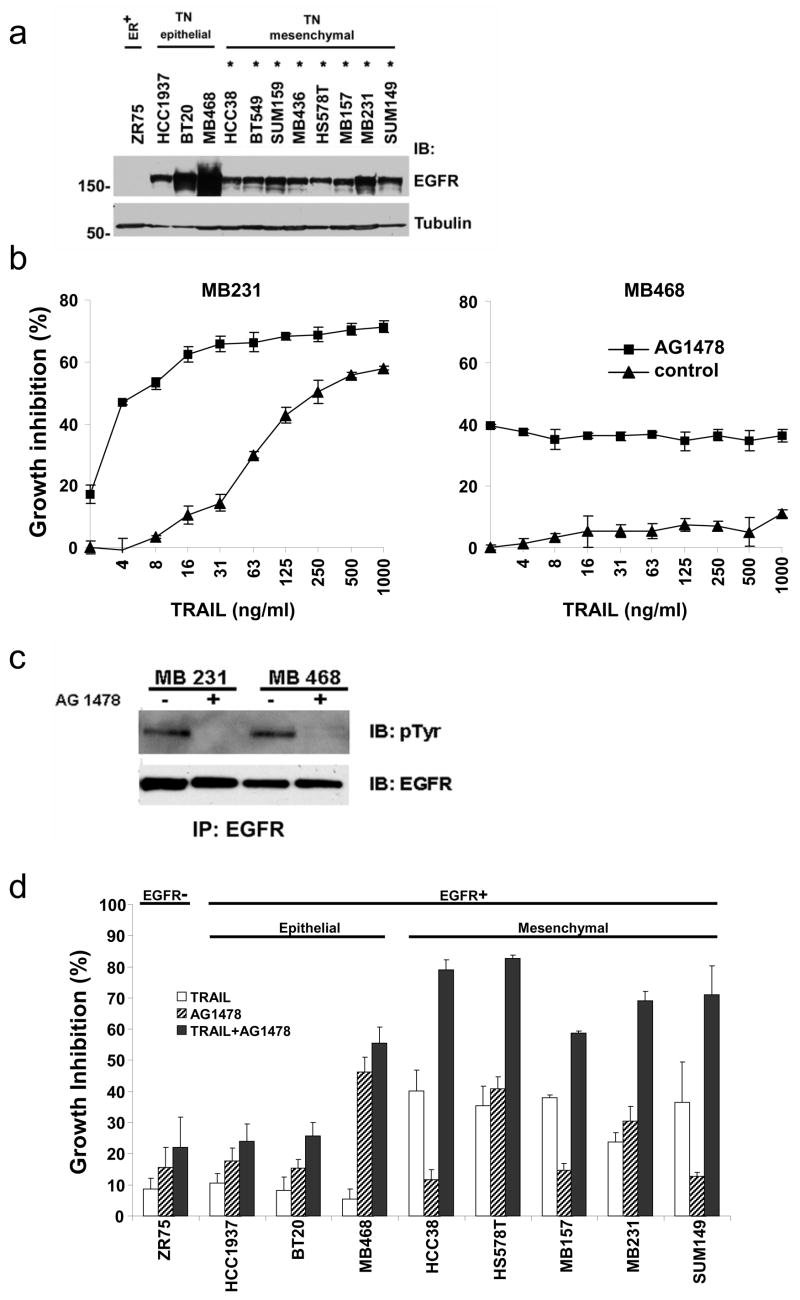
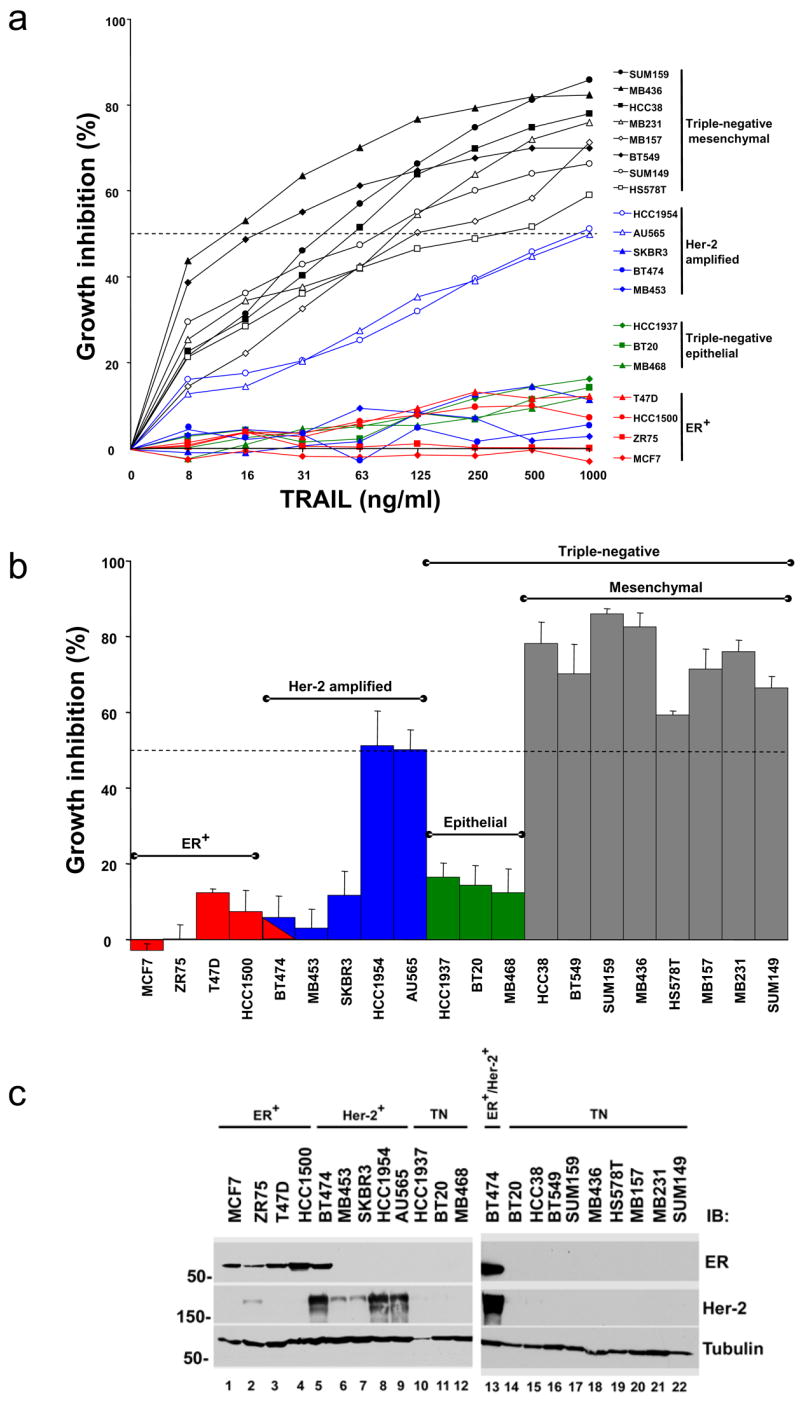
Previous work has found that most breast cancer cell lines are resistant to TRAIL-mediated toxicity [1]. However, only a limited number of breast cancer cell lines have been tested systematically for TRAIL sensitivity. To correlate the phenotype of the cancer cells with TRAIL sensitivity, we treated a panel of 20 breast cancer cell lines with TRAIL (Fig. 1a, b). These cell lines represented the three major classifications of breast cancer: ER positive, HER-2 amplified, and triple-negative cancers (Fig. 1c). One cell line, BT454, expresses both ER and amplified HER-2 (Fig. 1c, lanes 5 and 13). Expression of ER and HER-2 for all of these cell lines is consistent with the phenotype of the original tumors and with previously published data (http://www.atcc.org and [10, 11]). Published data have demonstrated that PR was expressed in several of the ER positive cell lines but was not expressed in any of the ER negative cell lines (http://www.atcc.org and [6]). Ten of the cell lines showed significant toxicity over the range of TRAIL concentrations tested (Fig. 1a, b). The most striking result is that eight of eleven triple-negative breast cancer cell lines were sensitive to TRAIL-induced toxicity (Fig. 1a, 1b). The IC50 for the sensitive triple-negative cell lines ranged from 16–250 ng/ml. Also, TRAIL induced significant toxicity in two of the five HER-2 amplified cell lines tested (HCC1954 and AU565). The IC50 for these cell lines (~1000 ng/ml) was higher than the IC50 in the triple-negative breast cancer cell lines. None of the five ER expressing cell lines were inhibited by TRAIL by more than 10–15% (this includes the BT474 cell line which is both ER positive and HER-2 amplified).
Fig. 1.
TRAIL-induced toxicity in breast cancer cell lines. (a) Cell lines were incubated with TRAIL at the indicated concentrations for 16 h. Cell viability was measured by the MTS assay. The data represent mean growth inhibition as a percentage of untreated cells for three experiments with each cell line. Percentage growth inhibition was calculated as [1-(MTS reading in treated cells/MTS reading in control cells)] X 100. Black lines represent mesenchymal triple-negative cell lines, green lines represent epithelial triple-negative cell lines; blue lines represent HER-2 amplified cell lines, and red lines represent ER positive cell lines (see text for discussion). The horizontal dashed line indicates 50% growth inhibition. (b) The data from the dose curve of TRAIL toxicity with each cell line at the highest TRAIL concentration (1 μg/ml) is replotted with the cells grouped by phenotype as indicated along the top of the graph and color coded as above. The data represent mean growth inhibition +/− SE as a percentage of untreated cells for three experiments with each cell line calculated as above. The dashed line indicates 50% growth inhibition. (c) Lysates prepared from the cell lines shown above were immunoblotted for expression of ER and HER-2 as indicated to the right of the panels. ER expressing (ER+), HER-2 amplified (Her-2+), and triple-negative cells (TN) are indicated along the top of the blot. The lysates were run on two separate gels. BT474 was run on the second gel as an internal positive control for ER expression and HER-2 amplification. Tubulin is included as a loading control. MW is indicated in kDa to the left of the panels.
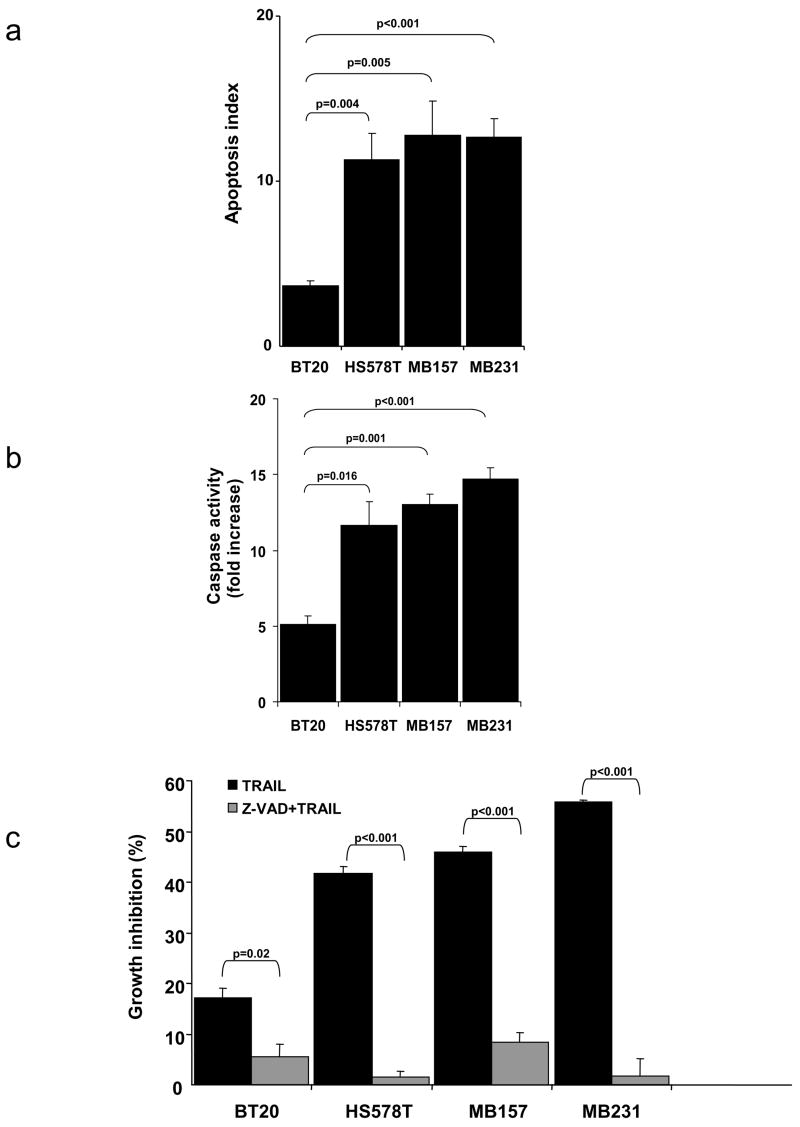
To assess if the growth inhibition measured in the MTS assays described in Fig. 1 was due to apoptosis, an assay to measure DNA fragmentation was performed (Fig. 2a). The TRAIL-sensitive triple-negative cell lines (HS578T, MB157, and MB231) showed >10 fold increase in the amount of fragmented DNA upon treatment with TRAIL (Fig. 2a). In contrast, the TRAIL-resistant triple-negative cell line, BT20, showed only an ~3 fold increase in fragmented DNA (consistent with the modest growth inhibition shown in Fig. 1). The increased DNA fragmentation seen in the sensitive cell lines was statistically greater than that seen in the BT20 cell line. TRAIL induces apoptosis by a caspase-dependent mechanism [12]. Treatment of TRAIL-sensitive cells (HS578T, MB157, and MB231) results in approximately a 12–14 fold increase in caspase activity (Fig. 2b). In contrast, the relatively TRAIL-insensitive BT20 cell line showed significantly less activation of caspases upon TRAIL treatment (Fig. 2b). Inhibition of caspase activation using the pan-caspase inhibitor, Z-VAD-fmk, markedly reduced the toxicity of TRAIL in both the TRAIL-sensitive and relatively TRAIL-insensitive cell lines (Fig. 2c). Together, the results demonstrate that the growth inhibition of the cells by TRAIL shown in the MTS assay is due to caspase-mediated apoptosis.
Fig. 2.
TRAIL induces caspase dependent apoptosis in breast cancer cells. (a) Apoptosis was measured in cells treated with or without 1 μg/ml of TRAIL for 4 h using the Cell Death Detection ELISAplus kit as described in the methods. The apoptosis index was calculated as [ELISA reading in the presence of TRAIL/ELISA reading in untreated cells] for each cell line. Data represent the mean apoptosis index +/− SE for four experiments with each cell line. The p values for the apoptosis index of the TRAIL-sensitive cells compared by two tailed t-tests to the apoptosis index of the BT-20 cell line are shown above the bars. (b) Caspase activation was measured in cells treated with or without TRAIL (1 μg/ml) for 4 h using the Caspase-Glo® 3/7 Assay Kit. Data represent the mean fold increase in caspase activity +/− SE for three experiments compared to untreated cells in the indicated cell lines. The p values for the increase in caspase activation in the TRAIL-sensitive cells compared by two tailed t-tests to the increase in caspase activation in the BT-20 cell line are shown above the bars. (c) Cells were preincubated with the caspase inhibitor Z-VAD-fmk (Z-VAD) and then treated with or without TRAIL. Cell viability was measured by an MTS assay. The data represent mean growth inhibition as a percentage of untreated cells of three experiments +/− SE in each cell line. Percentage growth inhibition was calculated as above. The p values comparing the MTS assay results in the presence and absence of Z-VAD-fmk by two tailed t-test are shown above the bars.
TRAIL-induced apoptosis is mediated by TRAIL-R2
To determine if expression of TRAIL receptors accounts for the differences in sensitivity to TRAIL-induced apoptosis, we analyzed protein expression of the agonist TRAIL receptors. There was no correlation between TRAIL sensitivity and expression of the death-inducing TRAIL receptors (TRAIL-R1 and TRAIL-R2) (Fig. 3a). Since the expression of TRAIL-R1 and TRAIL-R2 did not clearly correlate with TRAIL sensitivity, selective knockdown of each receptor using siRNA was performed to see which receptor(s) mediates TRAIL-induced apoptosis. We first tested MB231 as this cell line expresses high levels of TRAIL-R1 and TRAIL-R2 (Fig. 3a). The loss of TRAIL-R2 inhibited the toxic effects of TRAIL over a wide range of TRAIL concentrations while loss of TRAIL-R1 had little or no effect (Fig. 3b). Loss of both TRAIL-R1 and TRAIL-R2 was similar to loss of TRAIL-R2 alone (Fig. 3b). Similar results were obtained in two additional triple-negative cell lines (MB157 and HCC38) and in a TRAIL sensitive HER-2 amplified cell line (HCC1954) (Fig. 3c). Thus TRAIL-R2 is the predominant mediator of the effects of TRAIL in breast cancer cells.
Fig. 3.
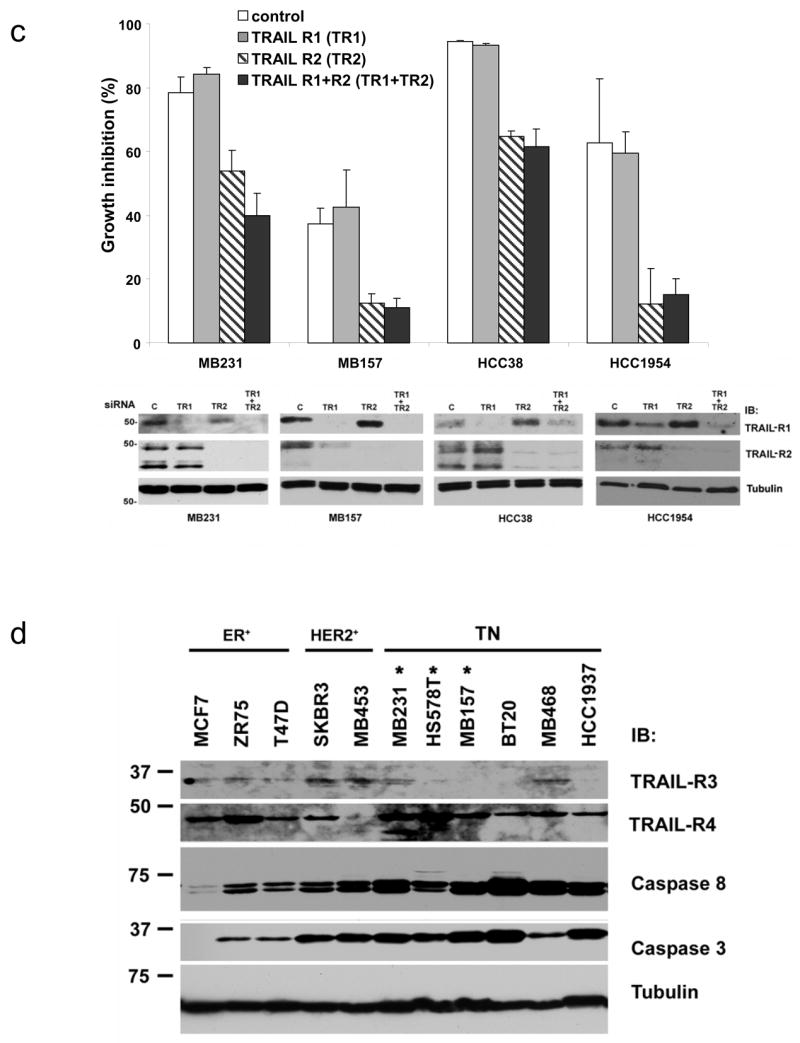
Expression and functional analysis of TRAIL pathway components in cell lines. Lysates prepared from the cell lines tested for TRAIL sensitivity in Fig. 1 were immunoblotted for expression of the proteins as indicated to the right of the panels. (a) Expression of TRAIL-R1 and TRAIL-R2 in the cell lines. ER expressing (ER+), HER-2 amplified (Her-2+), and triple-negative cells (TN) are indicated along the top of the blot. Cell lines marked with an asterisk were sensitive to TRAIL-induced cell death. The lysates were run on two gels. As internal controls on the immunoblots, at least one representative cell line from each phenotype was run on each gel (HS578T on the left panels and MCF7, BT474, and BT20 on the right panel). (b) MB231 was transfected with siRNA targeting TRAIL-R1 (TR1), TRAIL-R2 (TR2), both (TR1+TR2) or a nontargeting siRNA control. Transfected cells were assessed for TRAIL sensitivity over a wide range of TRAIL doses using an MTS assay as in Fig. 1. Data are the mean +/− SD for a representative experiment. Degree of protein knock down is shown in the immunoblot to the right. (c) MB231, MB157, HCC38 and HCC1954 were transfected with siRNA as above and assessed for TRAIL-sensitivity by MTS assay. Data are the mean +/− SD for a representative experiment in each cell line showing only the highest dose of TRAIL (1 μg/ml). Degree of protein knock down is shown in the immunoblot below the MTS data for each cell line. (d) Expression of TRAIL-R3, TRAIL-R4, caspase 8, and caspase 3 for representative TRAIL-resistant and TRAIL-sensitive cell lines. ER expressing (ER+), HER-2 amplified (Her-2+), and triple-negative cells (TN) are indicated along the top of the blot. Cell lines marked with an asterisk are sensitive to TRAIL-induced cell death. Tubulin is included as a loading control and MW is indicated in kDa to the left of the panels.
No correlation was found between TRAIL sensitivity and expression of the TRAIL decoy receptors (TRAIL-R3 and TRAIL-R4), c-FLIP, caspase 8, or caspase 3 (Fig. 3d). Similarly, cDNA expression profiling revealed no correlation between TRAIL sensitivity and expression of any genes implicated as regulators of the TRAIL pathway including TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4, cFLIP, caspases, Bcl2 family members, IAPs, and survivin (data not shown).
Mesenchymal markers predict TRAIL sensitivity in triple-negative cell lines
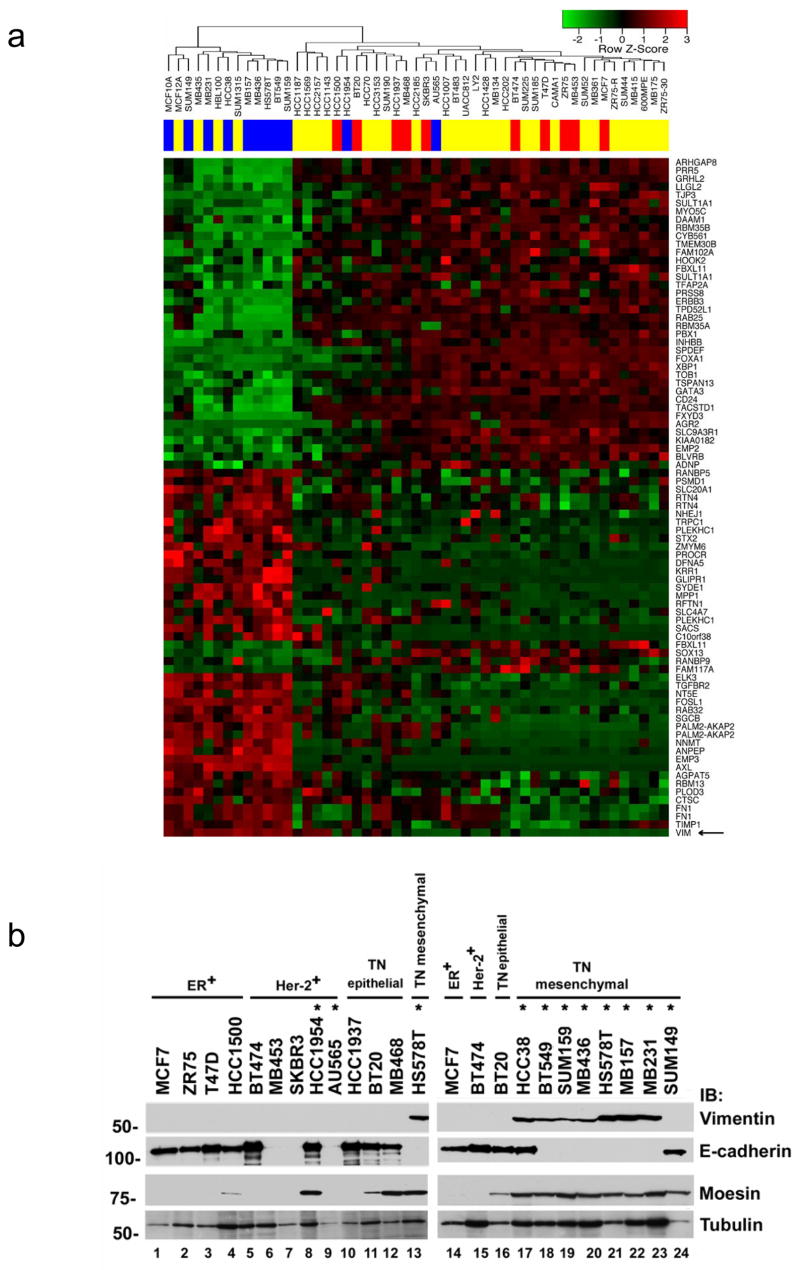
Recent work has classified 51 breast cancer cell lines based on transcriptional profiling [6]. In that work, breast cancer cell lines could be classified into two main groups, luminal and basal. The basal group was further subdivided into basal “A” (BaA) and basal “B” (BaB) groups. The BaB cell lines were distinguished based on expression of mesenchymal markers such as the cytoskeletal protein vimentin. In the published paper, the hormone receptor positive cell lines all clustered with the luminal cells. The HER-2 amplified cell lines were divided between the luminal and BaA groups. The triple-negative cell lines were divided between the BaA and BaB groups, with the majority of triple-negative cell lines classified as mesenchymal or BaB. Based on this work, we found that all of the TRAIL-sensitive triple-negative cell lines we tested are classified as BaB or mesenchymal cells (shown in black in Fig. 1a and gray in Fig. 1b). In contrast, the three TRAIL-resistant triple-negative cell lines are classified as epithelial or BaA (shown in green in Fig. 1a, b).
To identify determinants of TRAIL-sensitivity, we sought to define the genes most highly correlated with TRAIL sensitivity using the published cDNA microarray data (Fig. 4a, Supplemental Table 1, and [6]). Using these genes to visualize the gene expression pattern, this analysis clustered the eight TRAIL-sensitive triple-negative cell lines together in one arm of the dendogram (Fig. 4a, shown in blue on the upper left side of the dendogram). Strikingly, the eight TRAIL-sensitive triple-negative breast cancer cell lines clustered together with other triple-negative cell lines that have a mesenchymal (or BaB) gene expression profile (Fig. 4a and [6]). Interestingly, the immortalized, but non-transformed MCF10A cell line clusters with the mesenchymal (or BaB) cell lines by this analysis and is TRAIL-sensitive ([1] and data not shown). The TRAIL-resistant cell lines clustered together with the BaA and luminal cell lines from the published analysis (Fig. 4a, right side of dendogram). This group included all of the ER positive cell lines and the TRAIL-resistant HER-2 amplified cell lines. The two TRAIL-sensitive HER-2 amplified cell lines (HCC1937 and AU565) are clustered with the TRAIL-resistant cells by this analysis (Fig. 4a, right side of dendogram). Importantly, the TRAIL-resistant triple-negative breast cancer cell lines were identified as BaA cell lines in the previous work and cluster with the resistant cells in our analysis (Fig. 4a, shown in red on the upper right side of the dendogram). A number of cell lines that we did not test that cluster with the resistant cells have been reported as TRAIL-resistant [2]. This includes the ER positive cell line SUM52 and the HER-2 amplified cell lines SUM185, SUM190, and SUM225 [2].
Fig. 4.
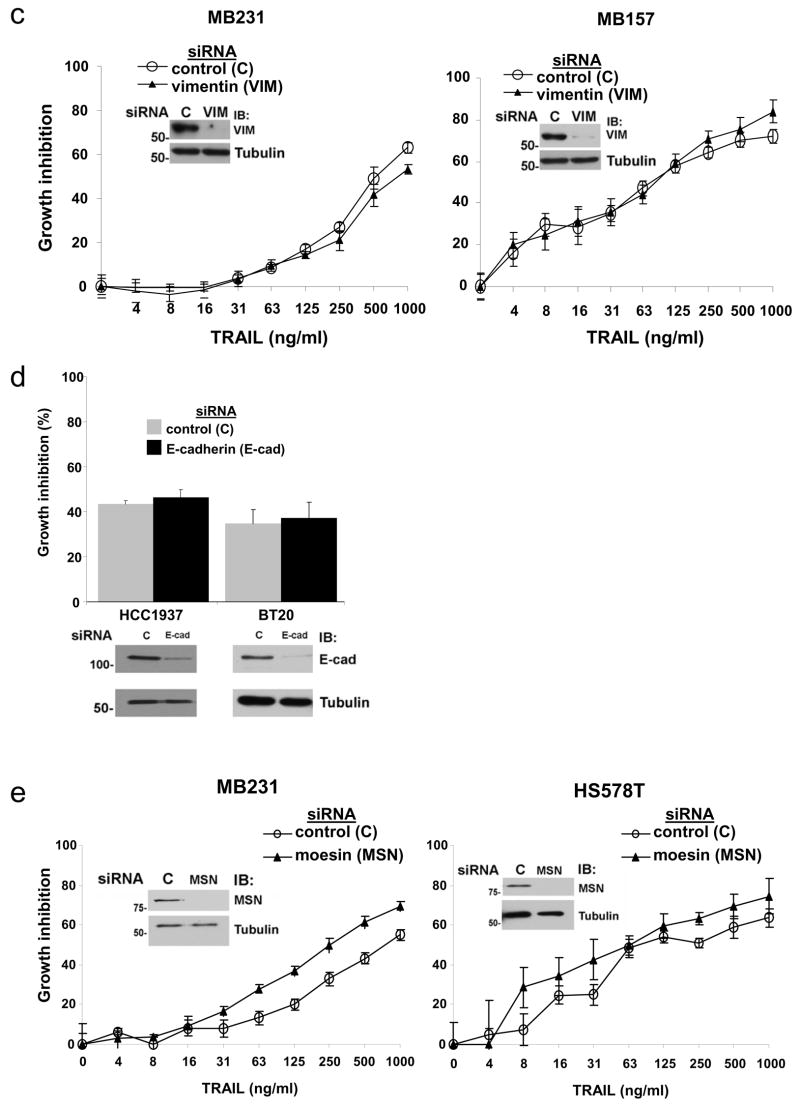
Triple-negative breast cancer cells with a mesenchymal phenotype are sensitive to TRAIL. (a) Supervised clustering analysis, performed with the most significant genes after regression analysis, of the microarray gene expression data are shown for the cell lines tested for TRAIL sensitivity along with additional cell lines. In this analysis we included the 20 breast cancer cell lines we tested for TRAIL sensitivity as well as the MCF10A cell line and found a set of 77 unique genes (83 probesets) that had a false discovery rate less than 2%. These genes were used to cluster the data for all of the cell lines analyzed in the previous study. Cell lines are shown along the top of the figure. The color bar indicates TRAIL-sensitive cells in blue, TRAIL-resistant cells in red, and untested cells in yellow. Genes are listed to the right of the figure. Red color indicates expression greater than the average expression for the entire sample set and green color indicates expression lower than the average for the entire sample set. The color key is shown in the upper right. (b) Protein expression in cell lines. Lysates prepared from the cell lines tested for TRAIL sensitivity in Fig. 1 were immunoblotted for expression of the proteins as indicated to the right of the panels. ER expressing (ER+), HER-2 amplified (Her-2+), and triple-negative cells (TN) are indicated along the top of the blot. Mesenchymal triple-negative and epithelial triple-negative phenotypes are indicated along the top of the blots. Cell lines marked with an asterisk are sensitive to TRAIL-induced cell death. The cell lysates were run on two separate gels. As internal controls on the immunoblots, at least one representative cell line from each phenotype was run on each gel (HS578T on the left panel and MCF7, BT474, and BT20 on the right panel). Tubulin is included as a loading control. MW is indicated in kDa to the left of the panels. (c) The TRAIL-sensitive triple-negative cell lines MB231 and MB157 were transfected with siRNA targeting vimentin or a nontargeting siRNA control. Transfected cells were assessed for TRAIL sensitivity over a wide range of TRAIL doses using an MTS assay as in Fig. 1. Data are the mean +/− SD for a representative experiment. Degree of protein knock down is shown in the immunoblots within each panel. (d) TRAIL-resistant triple-negative cell lines, HCC1937 and BT20, were transfected with siRNA targeting E-cadherin or a nontargeting siRNA control. Transfected cells were assessed for TRAIL sensitivity over a wide range of TRAIL doses using an MTS assay as in Fig. 1. Data show only the highest TRAIL concentration (1 μg/ml) and are the mean +/− SD for a representative experiment. Degree of protein knock down is shown in the immunoblots below the graph. (e) TRAIL-sensitive triple-negative cell lines, MB231 and HS578T, were transfected with siRNA targeting moesin or a nontargeting siRNA control. Transfected cells were assessed for TRAIL sensitivity over a wide range of TRAIL doses using an MTS assay as in Fig. 1. Data are the mean +/− SD for a representative experiment. Degree of protein knock down is shown in the immunoblots within each panel.
Based on the mRNA expression analysis, vimentin was expressed in nine of the 10 TRAIL-sensitive cell lines (Fig. 4a, arrow). These included all of the triple-negative cell lines that cluster with the mesenchymal group and one of the HER-2 amplified cell lines. Vimentin expression is high in mesenchymal cells and low in epithelial cells [13]. In contrast, E-cadherin is expressed at high levels in epithelial cells and at low levels in mesenchymal cells [13]. To confirm the cDNA microarray data, we performed immunoblots to examine protein expression of vimentin and E-cadherin in the 20 cell lines. Six of eight of the TRAIL-sensitive triple-negative cell lines express high levels of vimentin protein and low levels of E-cadherin protein - consistent with a mesenchymal phenotype (Fig. 4b, lanes 18–23). One of the TRAIL-sensitive triple-negative cell lines (HCC38) expresses high levels of both vimentin and E-cadherin proteins (Fig. 4b, lane 17). Only one TRAIL-sensitive triple-negative cell line (SUM149) does not express vimentin protein and has high levels of E-cadherin (Fig. 4b, lane 24). In contrast, all of the TRAIL-resistant triple-negative cell lines express high levels of E-cadherin and no vimentin, consistent with an epithelial phenotype (Fig. 4b, lanes 10–12, 16). Within the triple-negative breast cancer cell lines, seven of seven that express vimentin protein are TRAIL-sensitive and three of four cell lines that do not express vimentin protein are TRAIL-resistant. Thus, within the triple-negative breast cancer cell lines, mesenchymal features (in particular vimentin expression) are strong predictors of TRAIL sensitivity. All of the TRAIL-resistant ER positive cell lines express high levels of E-cadherin and no vimentin protein (Fig. 4b, lanes 1–5, 14–15) The HER-2 amplified cell lines do not express vimentin protein, including the two TRAIL-sensitive HER-2 amplified cell lines (Fig. 4b, lanes 5–9, 15). The expression of E-cadherin is more variable in these HER-2 amplified cell lines but does not correlate with TRAIL sensitivity (Fig. 4b, lanes 5–9, 15).
To test if vimentin or E-cadherin proteins are mechanistic determinants of sensitivity or resistance to TRAIL, respectively, we used specific siRNAs to knock down the expression of each of these molecules and tested the effect on TRAIL-induced toxicity (Fig. 4c, d). Loss of vimentin did not decrease the effect of TRAIL in the sensitive MB231 cells nor in MB157 cells (Fig. 4c). Loss of E-cadherin did not increase the sensitivity to TRAIL in the resistant HCC1937 cells and BT20 cells (Fig. 4d). Also, reintroduction of E-cadherin into the TRAIL-sensitive MB231 cell line did not alter the sensitivity to TRAIL (data not shown).
Recently moesin protein expression has been identified as a marker of triple-negative breast cancers by immunohistochemical analysis of primary tumors and cell lines [14, 15]. We therefore measured moesin expression by immunoblot in the cell lines. Expression of moesin was seen in all of the TRAIL-sensitive triple-negative breast cancer cell lines and in one of the two TRAIL-sensitive HER-2 amplified cell lines (Fig. 4b, lanes 8, 13, 17–24). Thus, nine of ten TRAIL-sensitive cell lines express moesin protein. Only three of the ten TRAIL–resistant cell lines (HCC1500, BT20, MB468) express moesin (Fig 4b, lanes 4, 11, 12 and 16). Thus, moesin expression is also correlated with TRAIL sensitivity. However, siRNA knockdown of moesin expression in TRAIL-sensitive MB231 and HS578T cells did not decrease the effects of TRAIL in these cells (Fig. 4e).
Together, these data suggest that vimentin expression, E-cadherin expression, and moesin expression may be markers of TRAIL sensitivity but do not regulate TRAIL sensitivity. Using Pearson’s Chi-squared test with Yates’ continuity correction TRAIL-sensitivity was strongly associated with mesenchymal classification on microarray (p-value = 0.0014), vimentin expression (p-value = 0.005), and moesin expression (p = 0.02). Triple-negative classification (based on the lack of hormone receptors and lack of amplification of HER2) showed a borderline significant association with TRAIL-sensitivity (p-value = 0.07). E-cadherin expression showed a borderline significant statistical association with TRAIL resistance (p-value = 0.07).
Inhibition of EGFR increases TRAIL-sensitivity in mesenchymal but not in epithelial triple-negative breast cancer cells
EGFR expression is another marker of triple-negative breast cancer cells [16, 17]. All of the triple-negative breast cancer cell lines we examined express high levels of EGFR (Fig. 5a). Previous reports have described EGFR activity promoting resistance to TRAIL and EGFR inhibition increasing sensitivity to TRAIL [18–20]. Thus, we investigated whether inhibition of the EGFR by a small molecule tyrosine kinase inhibitor (AG1478) enhances TRAIL-mediated apoptosis in triple-negative breast cancer cells. This inhibitor is a relatively selective inhibitor of EGFR and has an ~10,000 fold lower IC50 for EGFR compared to other related tyrosine kinases such as Her2 [21]. The MB231 cell line is TRAIL-sensitive, but the addition of AG1478 results in maximal toxicity at significantly lower doses of TRAIL (Fig. 5b, left panel). The TRAIL-resistant MB468 cell line expresses high levels of EGFR and growth of this cell line is significantly inhibited by AG1478 (Fig. 5b, right panel). However, the combination of AG1478 and TRAIL in the MB468 cell line did not result in significantly increased toxicity compared to AG1478 alone (Fig. 5b, right panel). These experiments were performed in the absence of exogenous EGF ligand. To test whether the EGFR is active in the absence of exogenous ligand, the EGFR was immunoprecipitated and immunoblotted with anti-phosphotyrosine antibody. In both MB231 and MB468, there was detectable EGFR activity in the absence of exogenous ligand that is inhibited by incubation with AG1478 (Fig. 5c). We then tested the ability of AG1478 to enhance TRAIL sensitivity in a panel of cells including the EGFR negative cell line (ZR75), the three TRAIL-resistant, EGFR-expressing epithelial triple-negative breast cancer cell lines (HCC1937, BT20, and MB468), and several of the TRAIL-sensitive, EGFR-expressing mesenchymal triple-negative breast cancer cell lines (HCC38, HS578T, MB157, MB231, and SUM149) (Fig. 5d). The combination of TRAIL and AG1478 was significantly more toxic to mesenchymal triple-negative breast cancer cells than either agent alone (compare black bars in Fig. 5d to white or striped bars). This was true over a wide range of TRAIL concentrations and was due to an increase in caspase-mediated apoptosis (data not shown). Importantly, the combination of TRAIL and AG1478 was not significantly more toxic to the epithelial triple-negative cell lines than AG1478 alone (Fig. 5d; HCC1937, BT20, MB468). This was observed over a wide range of doses of both TRAIL and AG1478 (Fig. 4b and data not shown). The addition of AG1478 to TRAIL did not induce significantly more toxicity in EGFR-negative cells such as ZR75 (Fig. 5d).
Fig. 5.
EGFR inhibition enhances TRAIL sensitivity in mesenchymal triple-negative cell lines. (a) Expression of EGFR was measured in one ER+ cell line (ZR75) and in the triple-negative cell lines by immunoblot. Tubulin is included as a loading control. MW is indicated in kDa to the left of the panels. (b) MB231 or MB468 cells were preincubated with AG1478 (30 μM) or vehicle for 4 h and then assessed for TRAIL sensitivity over a wide range of TRAIL doses using an MTS assay as in Fig. 1. Data are the mean +/− SD for a representative experiment. (c) The EGFR was immunoprecipitated from MB231 and MB468 cells that had been incubated with AG1478 (30 μM) or vehicle for 4 h and immunoblotted for phosphotyrosine (pTyr) or total EGFR as indicated. (d) Cell lines were preincubated with AG1478 (30 μM) for 4 h and then assessed for TRAIL sensitivity over a wide range of TRAIL doses using an MTS assay as in Fig. 1. Data are the mean +/− SE for at least three experiments with each cell line. Results are shown for TRAIL at 100 ng/ml.
Discussion
The results presented here using breast cancer cell lines identify differential sensitivity to TRAIL-mediated apoptosis based on the molecular phenotype of the breast cancer cells. The majority of triple-negative breast cancer cell lines, and particularly all of those with a mesenchymal phenotype, are TRAIL-sensitive (Fig. 1, 4). The three TRAIL-resistant triple-negative breast cancer cell lines do not have a mesenchymal phenotype, both based on our expression analysis of vimentin and E-cadherin and the published cDNA microarray expression data (Fig. 4b and [6]). Of note, two of the three TRAIL-resistant triple-negative breast cancer cell lines (MB468 and BT20) have amplification of the EGFR and thus may represent a unique subset of triple-negative breast cancers [22, 23].
The mesenchymal cytoskeletal protein, vimentin, was expressed predominantly in the majority of the TRAIL-sensitive triple-negative cell lines. The expression of vimentin protein is highly predictive of TRAIL-sensitivity among the triple-negative breast cancer cell lines (seven of seven vimentin protein positive cell lines were TRAIL-sensitive). In contrast, only one of the four triple-negative breast cancer cell lines that do not express vimentin protein is sensitive to TRAIL-induced apoptosis (i.e., SUM149). This cell line expresses vimentin mRNA and clusters with the mesenchymal cell lines by transcriptional profiling (Fig. 4a and [6]).
While previous transcriptional profiling data has identified triple-negative breast cancer cell lines with a mesenchymal phenotype, this classification has not been identified in transcriptional profiling of primary breast cancer samples [6, 24, 25]. The reason for this discrepancy is not clear. These mesenchymal cell lines may represent a transformation that occurs in culture due, for example, to the lack of interactions with the tumor stroma. Alternatively, these cells may be a minor population that is better able to grow under cell culture conditions. However, immunohistochemical analysis of primary tumors has identified tumors in which the cancer cells express vimentin [17, 26, 27]. In the largest study (>2000 samples), approximately 14% of breast tumors expressed vimentin in the tumor cells [27]. Also, in this study, 35% of the ER negative tumors were vimentin positive [27]. The enrichment of vimentin-positive tumors among the ER negative tumors is consistent with vimentin expression in triple-negative tumors. However, this work did not simultaneously characterize HER-2 amplification and so the tumors can not be accurately classified as triple-negative [27]. Two small studies demonstrated vimentin expression in 17 of 18 and 4 of 11 triple-negative breast cancer samples [17, 26]. In one of these studies, all of the samples had been classified as basal-type tumors by microarray analysis and the vimentin staining observed was strong and diffuse in the tumors [17]. Thus, vimentin-positive triple-negative tumors are a clinical entity and not a cell culture artifact.
Recently moesin has been identified as a marker of triple-negative breast cancer cells in primary breast cancer samples and breast cancer cell lines [14, 15]. This protein is a member of the ezrin, radixin, moesin family of proteins which play important roles in cytoskeletal function, cell adhesion, and motility [28]. In our work, moesin protein was expressed in all of the mesenchymal TRAIL-sensitive triple-negative cell lines and in one of the two TRAIL- sensitive HER-2 amplified cell lines. Thus 9 of 10 TRAIL-sensitive cell lines express moesin - making moesin a potential biomarker for TRAIL sensitivity.
Together, these data suggest that characterization of vimentin and moesin expression by immunohistochemistry may be useful biomarkers for TRAIL sensitivity. Measurements of vimentin and moesin mRNA and/or protein expression could be incorporated readily into future clinical trials of TRAIL agonists to investigate whether these proteins have predictive value for TRAIL response in patients with breast cancer.
The mechanism underlying the differential TRAIL sensitivity of the mesenchymal triple-negative breast cancer cell lines is unknown. siRNA knockdown experiments indicate that while vimentin, moesin, and E-cadherin protein expression are predictive of TRAIL sensitivity or resistance, they do not regulate TRAIL function (Fig. 4c, d, e). Recent analysis in pancreatic cancer, colorectal cancer, non-small-cell lung cancer, and melanoma cell lines has identified low expression of O-glycosylation genes as a potential mechanism of TRAIL-resistance [29]. However our gene expression analysis did not find a correlation between the genes described in that work for O-glycosylation and TRAIL sensitivity in the breast cancer cell lines (unpublished observation). Our analysis of gene expression identified 77 genes that classified the cell lines into two clusters based on TRAIL sensitivity. None of these genes have been implicated as regulators of TRAIL activity (Fig. 4a). Several of the genes that are expressed at a higher level in the TRAIL-sensitive triple-negative cell lines have been described previously in breast cancer. For example, the receptor tyrosine kinase Axl, also preferentially expressed in the mesenchymal breast cancer cells (Fig. 4a and [30]), has been implicated in angiogenesis and in breast cancer tumorigenesis [31]. The membrane protein EMP3 was found highly expressed in the mesenchymal cell lines but its function is unknown [32]. PROCR has been described as expressed in breast cancer cells with a “stem cell” phenotype [33]. Interestingly, the mesenchymal breast cancer cell lines have characteristics consistent with putative breast cancer “stem cells” (e.g., high CD44 and low CD24 expression) (Fig. 4a and [6]). A role for these proteins in TRAIL activity has not been studied.
We also investigated inhibition of the EGFR as a means to overcome TRAIL resistance in the epithelial triple-negative breast cancer cell lines. EGFR is frequently expressed in triple-negative breast cancer and we found that it was expressed in all of the triple-negative breast cancer cell lines tested whether they are TRAIL-sensitive or resistant ([16, 17] and Fig. 5a). While we found that EGFR inhibition can enhance TRAIL-mediated apoptosis in EGFR-expressing breast cancer cells that are already sensitive to TRAIL (i.e., the mesenchymal triple-negative breast cancer cell lines), EGFR inhibition does not overcome the resistance in the epithelial triple-negative breast cancer cell lines (Fig. 5b and d).
The patients with triple-negative breast cancer have no clinically validated molecularly targeted therapies and have a poor prognosis relative to those patients having other breast cancer subtypes [4]. The data here provide a strong rationale for testing TRAIL receptor agonists in this population - especially those with tumors that have mesenchymal features (i.e., vimentin expression). In addition, combination of EGFR inhibitors with TRAIL may be particularly effective for the treatment of triple-negative breast cancers with mesenchymal features.
HER-2 amplified tumors represent a second population that may be sensitive to TRAIL. However, only two of five cell lines tested here with HER-2 amplification are sensitive to TRAIL-induced apoptosis and three additional lines reported in the literature are resistant to TRAIL [2]. In addition, the IC50 in these cell lines is higher than the IC50 in the sensitive triple-negative cell lines (Fig. 1). These cell lines cluster with the resistant cell lines by transcriptional profiling and they do not express the vimentin protein (Fig. 4). Thus, they do not have mesenchymal markers. One of the TRAIL-resistant HER-2 amplified cell lines (BT474) also expresses ER, and all of the ER-positive cell lines tested are resistant to TRAIL-mediated apoptosis (Fig. 1). Thus, ER expression appears to identify TRAIL-resistant cell lines. It is not clear what distinguishes the other TRAIL-resistant HER-2 amplified cell lines from the two TRAIL-sensitive HER-2 amplified cell lines. Interestingly, our previous work suggests that TRAIL-induced apoptosis could be enhanced in the resistant cell lines with HER-2 amplification (e.g., SKBR3 and MB453) when TRAIL was combined with trastuzumab pre-treatment [5]. Thus, tumors with HER-2 amplification may benefit from combined molecularly targeted therapies of TRAIL and trastuzumab.
Expression levels of TRAIL receptors do not predict whether a cell will be sensitive or resistant to TRAIL (Fig. 3). TRAIL agonists which target both TRAIL receptors and those which selectively target either TRAIL-R1 or TRAIL-R2 are currently in phase I and phase II clinical trials. Previous work has demonstrated that TRAIL induces apoptosis predominantly via TRAIL-R1 in some tumors and via TRAIL-R2 in others [34–36]. Using RNAi to selectively reduce expression of either TRAIL-R1 or TRAIL-R2, our data indicate that TRAIL-R2 is the predominant death-inducing receptor in the sensitive breast cancer cell lines (in both the TRAIL-sensitive triple-negative and HER-2 amplified cells (Fig. 3b, c). Thus, agonists that activate both TRAIL-R1 and TRAIL-R2 or those that selectively activate TRAIL-R2 (but not those that selectively activate TRAIL-R1) should be tested in breast cancer patients.
Overall, our work suggests that TRAIL receptor agonists will have differential effects on breast cancer tumors with different molecular phenotypes. This work suggests that patients with triple-negative tumors with mesenchymal features, a group of patients with a poor prognosis, are most likely to benefit from TRAIL receptor agonists. Importantly, these data provide a basis for designing clinical studies using these agents in triple-negative breast cancer patients.
Supplementary Material
List of genes with a false discovery rate of 2% for the regression analysis of TRAIL sensitivity and gene expression ranked from most to least significant. Column headings are, from left to right, official gene symbol, gene description, Entrez Gene ID, Affymetrix probeset ID, t-statistic, and false discovery rate (adj.P.val). A positive t-statistic is associated with genes that are higher in TRAIL sensitive cell lines. The raw expression data described in Neve et al. [6] was downloaded from http://caArray.nci.nih.gov.
Acknowledgments
We thank Steven Shaw for helpful discussion about moesin and for providing the anti-moesin antibody. We thank Jeffrey Rubin for his careful review of this manuscript and thoughtful comments.
Funding: This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
S Davis and JG Pumphrey contributed equally to this work.
Contributor Information
Monzur Rahman, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892.
Sean R. Davis, Genetics Department, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892
Janet G. Pumphrey, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892
Jing Bao, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892.
Marion M. Nau, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892
Paul S. Meltzer, Genetics Department, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892
Stanley Lipkowitz, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892.
References
- 1.Keane MM, Ettenberg SA, Nau MM, Russell EK, Lipkowitz S. Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer Res. 1999;59(3):734–741. [PubMed] [Google Scholar]
- 2.Chinnaiyan A, Prasad U, Shankar S, Hamstra DA, Shanaiah M, Chenevert TL, Ross BD, Rehemtulla A. Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci U S A. 2000;97(4):1754–1759. doi: 10.1073/pnas.030545097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis- inducing ligand in vivo. Nat Med. 1999;5(2):157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 4.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23(29):7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 5.Cuello M, Ettenberg SA, Clark AS, Keane MM, Posner RH, Nau MM, Dennis PA, Lipkowitz S. Down-regulation of the erbB-2 receptor by trastuzumab (herceptin) enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB-2. Cancer Res. 2001;61(12):4892–4900. [PubMed] [Google Scholar]
- 6.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth GK. In: Limma: linear models for microarray data., in Bioinformatics and computational biology solutions using R and Bioconductor 2005. Gentleman RC, et al., editors. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a pratical and powerful approach to multiple testing. Journal Of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 10.Ethier SP. Human breast cancer cell lines as models of growth regulation and disease progression. J Mammary Gland Biol Neoplasia. 1996;1(1):111–21. doi: 10.1007/BF02096306. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan L, Van Weelden K, Ammerman C, Ethier SP, Welsh J. SUM-159PT cells: a novel estrogen independent human breast cancer model system. Breast Cancer Res Treat. 1999;58(3):193–204. doi: 10.1023/a:1006331716981. [DOI] [PubMed] [Google Scholar]
- 12.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2(6):420–30. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 13.Weinberg RA. The biology of cancer. Garland Science; New York: 2007. [Google Scholar]
- 14.Charafe-Jauffret E, Monville F, Bertucci F, et al. Moesin expression is a marker of basal breast carcinomas. Int J Cancer. 2007;121(8):1779–85. doi: 10.1002/ijc.22923. [DOI] [PubMed] [Google Scholar]
- 15.Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, Slamon DJ. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105(3):319–26. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 16.Korsching E, Packeisen J, Agelopoulos K, et al. Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest. 2002;82(11):1525–33. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- 17.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–71. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 18.Gibson EM, Henson ES, Haney N, Villanueva J, Gibson SB. Epidermal growth factor protects epithelial-derived cells from tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by inhibiting cytochrome c release. Cancer Res. 2002;62(2):488–496. [PubMed] [Google Scholar]
- 19.Gibson S, Tu S, Oyer R, Anderson SM, Johnson GL. Epidermal growth factor protects epithelial cells against Fas-induced apoptosis. Requirement for Akt activation. J Biol Chem. 1999;274(25):17612–17618. doi: 10.1074/jbc.274.25.17612. [DOI] [PubMed] [Google Scholar]
- 20.Park SY, Seol DW. Regulation of Akt by EGF-R inhibitors, a possible mechanism of EGF-R inhibitor-enhanced TRAIL-induced apoptosis. Biochem Biophys Res Commun. 2002;295(2):515–518. doi: 10.1016/s0006-291x(02)00719-2. [DOI] [PubMed] [Google Scholar]
- 21.Levitzki A, Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science. 1995;267(5205):1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 22.Filmus J, Pollak MN, Cailleau R, Buick RN. MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochem Biophys Res Commun. 1985;128(2):898–905. doi: 10.1016/0006-291x(85)90131-7. [DOI] [PubMed] [Google Scholar]
- 23.Lebeau J, Goubin G. Amplification of the epidermal growth factor receptor gene in the BT20 breast carcinoma cell line. Int J Cancer. 1987;40(2):189–91. doi: 10.1002/ijc.2910400210. [DOI] [PubMed] [Google Scholar]
- 24.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–84. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 25.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umemura S, Takekoshi S, Suzuki Y, Saitoh Y, Tokuda Y, Osamura RY. Estrogen receptor-negative and human epidermal growth factor receptor 2-negative breast cancer tissue have the highest Ki-67 labeling index and EGFR expression: gene amplification does not contribute to EGFR expression. Oncol Rep. 2005;14(2):337–43. [PubMed] [Google Scholar]
- 27.Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, Woelfle U, Rau T, Sauter G, Heukeshoven J, Pantel K. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res. 2005;11(22):8006–14. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 28.Hughes SC, Fehon RG. Understanding ERM proteins--the awesome power of genetics finally brought to bear. Curr Opin Cell Biol. 2007;19(1):51–6. doi: 10.1016/j.ceb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Wagner KW, Punnoose EA, Januario T, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13(9):1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 30.Meric F, Lee WP, Sahin A, Zhang H, Kung HJ, Hung MC. Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res. 2002;8(2):361–7. [PubMed] [Google Scholar]
- 31.Holland SJ, Powell MJ, Franci C, et al. Multiple roles for the receptor tyrosine kinase axl in tumor formation. Cancer Res. 2005;65(20):9294–303. doi: 10.1158/0008-5472.CAN-05-0993. [DOI] [PubMed] [Google Scholar]
- 32.Evtimova V, Zeillinger R, Weidle UH. Identification of genes associated with the invasive status of human mammary carcinoma cell lines by transcriptional profiling. Tumour Biol. 2003;24(4):189–98. doi: 10.1159/000074429. [DOI] [PubMed] [Google Scholar]
- 33.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Kelley RF, Totpal K, Lindstrom SH, Mathieu M, Billeci K, Deforge L, Pai R, Hymowitz SG, Ashkenazi A. Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J Biol Chem. 2005;280(3):2205–12. doi: 10.1074/jbc.M410660200. [DOI] [PubMed] [Google Scholar]
- 35.MacFarlane M, Kohlhaas SL, Sutcliffe MJ, Dyer MJ, Cohen GM. TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res. 2005;65(24):11265–70. doi: 10.1158/0008-5472.CAN-05-2801. [DOI] [PubMed] [Google Scholar]
- 36.van der Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A, Serrano L, Quax WJ. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci U S A. 2006;103(23):8634–9. doi: 10.1073/pnas.0510187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes with a false discovery rate of 2% for the regression analysis of TRAIL sensitivity and gene expression ranked from most to least significant. Column headings are, from left to right, official gene symbol, gene description, Entrez Gene ID, Affymetrix probeset ID, t-statistic, and false discovery rate (adj.P.val). A positive t-statistic is associated with genes that are higher in TRAIL sensitive cell lines. The raw expression data described in Neve et al. [6] was downloaded from http://caArray.nci.nih.gov.