Abstract
Drosophila melanogaster β4GalNAcTB mutant flies revealed that this particular N-acetylgalactosaminyltransferase is predominant in the formation of lacdiNAc (GalNAcβ1,4GlcNAc)-modified glycolipids, but enzymatic activity could not be confirmed for the cloned enzyme. Using a heterologous expression cloning approach, we isolated β4GalNAcTB together with β4GalNAcTB pilot (GABPI), a multimembrane-spanning protein related to Asp-His-His-Cys (DHHC) proteins but lacking the DHHC consensus sequence. In the absence of GABPI, inactive β4GalNAcTB is trapped in the endoplasmic reticulum (ER). Coexpression of β4GalNAcTB and GABPI generates the active enzyme that is localized together with GABPI in the Golgi. GABPI associates with β4GalNAcTB and, when expressed with an ER retention signal, holds active β4GalNAcTB in the ER. Importantly, treatment of isolated membrane vesicles with Triton X-100 disturbs β4GalNAcTB activity. This phenomenon occurs with multimembrane-spanning glycosyltransferases but is normally not a property of glycosyltransferases with one membrane anchor. In summary, our data provide evidence that GABPI is required for ER export and activity of β4GalNAcTB.
Introduction
Glycosylation in the secretory pathway is a complex process in which hundreds of glycosyltransferases are involved (Taniguchi et al., 2002). Many glycosyltransferases appear in gene families specified mainly by the nature of the nucleotide sugar donor (Coutinho et al., 2003). Within a given family, individual glycosyltransferases differ regarding the recognized acceptor structures. To understand and modulate cellular glycosylation pathways, it is important to know how this substrate specificity is generated (de Graffenried and Bertozzi, 2004).
Important in this respect is the observation that acceptor specificity in many glycosyltransferases is not restricted to recognition of one or a few specifically linked monosaccharides. Some protein-specific glycosyltransferases obtain additional selectivity by recognizing specific peptide motifs in the acceptor. A classic example is the N-acetylgalactosaminyltransferase (GalNAcT), which modifies glycoprotein hormones with high selectivity (Smith and Baenziger, 1988).
Some glycosyltransferases require other proteins that are not part of the acceptor structure for their specific activity. β1,4-galactosyltransferase (β4GalT) acts on terminally positioned N-acetylglucosamine (GlcNAc) residues conjugated to proteins or lipids. Its specificity changes if it builds a complex with α-lactalbumin. In the complex, free glucose is used as an acceptor, and lactose is formed (Brew et al., 1968). In the case of core 1 β3-galactosyltransferase (C1β3GalT), a molecular chaperone called Cosmc, with specificity for this single client, is required for folding and transportation to the Golgi (Ju and Cummings, 2002, 2005; Ju et al., 2008). Also, for O-mannosylation, two proteins, POMT1 and POMT2, are required (Manya et al., 2004). However, in this case, a two-protein enzymatic complex is proposed. The same is true in heparin sulfate biosynthesis in which two different exostosins are required for efficient biosynthesis (McCormick et al., 2000).
For several glycosyltransferases involved in glycolipid biosynthesis, data indicate that factors other than the enzyme and the acceptor substrate play a role. This is the case for β4GalT-V and -VI, which are homologues of the β4GalT mentioned in the previous paragraph and of Drosophila melanogaster β4GalNAcTB (the subject of this study). Under in vitro conditions, β4GalT-V and -VI transfer galactose (Gal) into β1-4 linkage to terminally expressed GlcNAc residues on glycoproteins (van Die et al., 1999; Guo et al., 2001). However, their involvement in the biosynthesis of lactosyl ceramide (Cer) by Gal transfer onto glucosyl Cer has been demonstrated as well (Nomura et al., 1998; Sato et al., 2000; Kolmakova and Chatterjee, 2005). At least in the case of galactosyltransferase V, this latter activity depends on the enzyme's anchorage in the membrane (van Die et al., 1999; Sato et al., 2000). Other enzymes involved in glycolipid biosynthesis have been shown to exhibit very low (de Vries et al., 1995; Zhu et al., 1998; Togayachi et al., 2001) or no (Steffensen et al., 2000; Schwientek et al., 2002) activity if expressed as soluble proteins. In general, very little is known about how lipid acceptors are recognized by glycosyltransferases. However, it has been suggested that a membrane-bound activator protein is required to present glycolipid acceptors to the modifying glycosyltransferases (Ramakrishnan et al., 2002). This hypothesis is substantiated by analogy to the lysosomal sphingolipid degradation machinery in which the sphingolipid activator protein presents the glycolipid substrates to glycosidases (Kolter and Sandhoff, 2005).
In this study, we describe a novel mechanism of glycosyltransferase maturation and functionalization for the glycolipid-specific β4GalNAcTB from Drosophila. This enzyme, which has been described as an inactive homologue of β4GalNAcTA in a previous study (Haines and Irvine, 2005), is a member of the invertebrate branch of the β4GalT family involved in the biosynthesis of the lacdiNAc (GalNAcβ1,4GlcNAc) epitope (Kawar et al., 2002; Vadaie et al., 2002; Haines and Irvine, 2005; Stolz et al., 2008). Because β4GalNAcT had not been cloned when this study was started, we searched for the corresponding activity using expression cloning (Bakker et al., 1997, 2005; Münster et al., 1998). In a heterologous approach, a cDNA library from Drosophila was used for expression in CHO cells, whereas formation of the lacdiNAc epitope was monitored with a specific monoclonal antibody (van Remoortere et al., 2000). As will be demonstrated in this study, the expression of two cDNA clones was required to install the functionally active enzyme.
Results
Expression cloning of a Drosophila β4GalNAcT
Other than in many invertebrates, the lacdiNAc element has been identified on only a few glycoconjugates in mammals (Sato et al., 2003). We ascertained that CHO cells are negative for lacdiNAc. Considering that in mammalian cells terminal GlcNAc residues are recognized by several galactosyltransferases, we additionally hypothesized that signals in the complementation cloning approach could be improved by the use of CHO Lec8 cells. They lack the Golgi UDP-Gal transporter and, consequently, show drastically reduced incorporation of Gal in glycans (Deutscher and Hirschberg, 1986). A cDNA library was constructed from Drosophila, subdivided into pools, and, in an established sibling selection procedure (Bakker et al., 1997), used to search for clones that rendered cells positive for lacdiNAc. Cell surface lacdiNAc expression was monitored with antibody 259-2A1, which was originally raised from Schistosoma mansoni–infected mice (van Remoortere et al., 2000). In this procedure, it became obvious that two cDNA clones were required for the expression of the lacdiNAc epitope. Although clone one was a member of the β1,4GalT family (van Die et al., 1997), the second clone (flybase CG17257) encoded a type III membrane protein that was related to a gene family referred to as Asp-His-His-Cys (DHHC) proteins (Mitchell et al., 2006). This protein was termed β4GalNAcTB pilot (GABPI) to describe its crucial role in generating a functionally active β4GalNAcTB as shown in the following experiments. The identified β4GalNAcT was identical to the inactive β4GalNAcTB recently cloned by Haines and Irvine (2005) in a homology-based approach. In agreement with this study, we found no in vivo activity for full-length β4GalNAcTB expressed in mammalian cells. It is remarkable that expression cloning identified β4GalNAcTB and not the homologous β4GalNAcTA, which was shown in an earlier study (Haines and Irvine, 2005) to be an active enzyme. To resolve the controversial finding, β4GalNAcTA was cloned by PCR and expressed in comparison with β4GalNAcTB–GABPI in CHO and HEK293 cells. These experiments demonstrated that the cell surface lacdiNAc expression detected with antibody 259-2A1 as a result of β4GalNAcTA was much lower than the lacdiNAc formation after the combined expression of β4GalNAcTB and GABPI. Thus, the data demonstrate the existence of two functionally active β4GalNAcTs (β4GalNAcTA and β4GalNAcTB) in Drosophila of which β4GalNAcTB needs the cooperation of GABPI. Because CHO cells demonstrated a low tolerance to expression of the Drosophila N-acetylgalactosamine (GalNAc) transferases and HEK293 cells turned out to be a more suitable expression system, subsequent experiments were performed exclusively in HEK293 cells.
β4GalNAcTB specifically modifies glycolipids
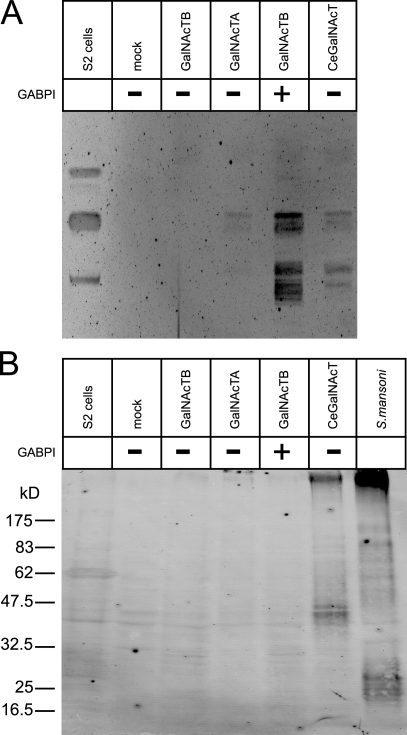
Despite elaborated analyses of Drosophila glycoproteins (North et al., 2006), the lacdiNAc structure has so far only been found as a modification of glycolipids (Seppo et al., 2000). With both cloned enzymes at hand, we evaluated the question of acceptor specificity. HEK293 cells were transfected with β4GalNAcTA, β4GalNAcTB, or the combination β4GalNAcTB–GABPI and analyzed for the presence of lipid- and protein-bound lacdiNAc using TLC followed by immunooverlay (Fig. 1 A) and Western blotting (Fig. 1 B), respectively. In both systems, Drosophila S2 cells, which are naturally positive for the antibody epitope, and HEK293 cells transfected with Caenorhabditis elegans β4GalNAcT (Kawar et al., 2002) were used as controls. Although expression of the C. elegans enzyme confirmed the availability of β4GalNAcT acceptors on proteins, the absence of specific signals in both HEK293 cells transfected with the β4GalNAcTs and in S2 cells confirmed the earlier observations in flies. In contrast, immunostaining of the lipid extracts resulted in positive signals for S2 cells as well as for HEK293 cells transfected with the β4GalNAcTB–GABPI pair. Expression of β4GalNAcTB alone was not sufficient to produce a signal, whereas faint signals were reliably obtained with β4GalNAcTA. It is important to mention that lipid specificity is preserved, although the glycolipid acceptor structures are different in Drosophila and HEK293 cells.
Figure 1.
Drosophila-derived β4GalNAcTs are specific for glycolipids. (A) Glycolipid extracts from S2 cells and HEK293 cells after transfection with empty vector (mock) and GalNAcTs were separated on TLC and immunooverlayed with antibody 259-2A1. (B) Protein extracts from cells as described in A were analyzed by Western blotting with 259-2A1. A protein extract from S. mansoni eggs was loaded as a control.
In vitro activity of β4GalNAcTB–GABPI is detergent sensitive
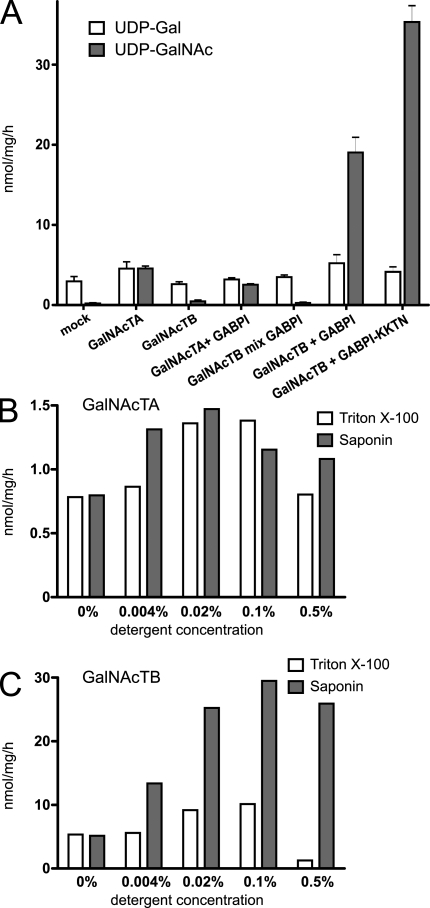
The observations that β4GalNAcTB is inactive if it is separately expressed in a heterologous cell system or if it is tested as a recombinant soluble protein (Haines and Irvine, 2005) encouraged further analyses to examine at which step β4GalNAcTB and GABPI interact with each other in the biosynthesis. In the first experiment, it was established that a soluble secreted construct of β4GalNAcTB was still inactive when coexpressed with GABPI. Subsequently, we wondered whether the two proteins expressed in separate cells have the capacity to form an active enzyme. Microsomal fractions of HEK293 cells transfected with either β4GalNAcTB or GABPI were isolated, mildly treated with detergent (saponin 0.01%), and functionally tested in mixtures. The assay system used to follow β4GalNAcT activity was adapted from an established assay (Palcic et al., 1988). In this assay system, [3H]UDP-GalNAc is the donor, and GlcNAc–p-nitrophenyl (pNP) is the acceptor substrate. No GalNAc transfer was measured in mixed vesicles (Fig. 2 A), whereas controls with microsomes of β4GalNAcTA– or β4GalNAcTB–GABPI-transfected HEK293 cells were active after identical treatment with detergent. In accordance with lacdiNAc formation in intact cells, β4GalNAcTB was not active when expressed alone, but it showed higher activity than β4GalNAcTA when expressed in combination with GABPI.
Figure 2.
Activity of β4GalNAcTB is disrupted by Triton X-100. (A) Microsomal preparations of HEK293 transfected with β4GalNAcTs and GABPI as indicated were assayed for activity with GlcNAc-pNP as an acceptor and [3H]UDP-GalNAc or [3H]UDP-Gal (endogenous activity as an internal control) as donor substrates. Each value represents the mean of three independent vesicle preparations with standard deviation. GalNAcTB mix GABPI indicates that the proteins were expressed separately but mixed afterward for assays. (B and C) Microsomal fractions of HEK293 cells transfected with β4GalNAcTA (B) or the combination β4GalNAcTB–GABPI (C) were treated with Triton X-100 and saponin in various concentrations and assayed for GalNAcT activity as in A.
In contrast to β4GalNAcTA, the β4GalNAcTB activity strongly depended on the detergent used. Only background levels were measured if membranes were treated with Triton X-100 (Fig. 2 B) or NP-40 (not depicted) at 0.5%, which is routinely used in glycosyltransferase assays. The milder detergent saponin increased activity over a wide concentration range (Fig. 2 B). The rather low activity measured in the absence of detergent was probably a result of limited transport of the substrates over the vesicle membranes. As saponin is known to perforate and not disrupt membranes (Schulz, 1990), these data suggest that the maintenance of protein complexes in intact membrane patches is required for β4GalNAcTB activity.
ER export of β4GalNAcTB requires GABPI
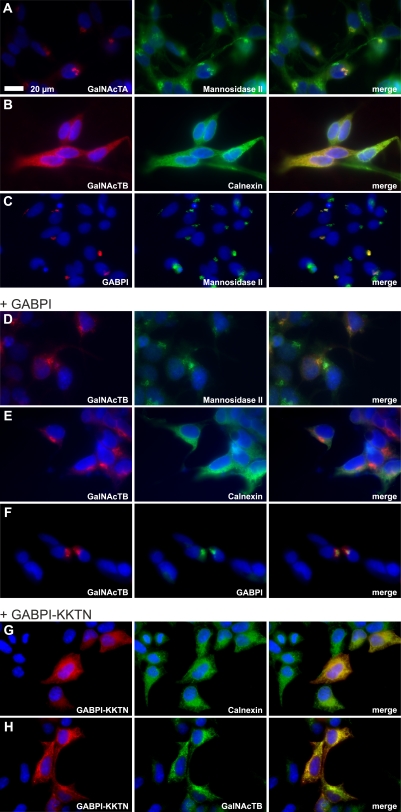
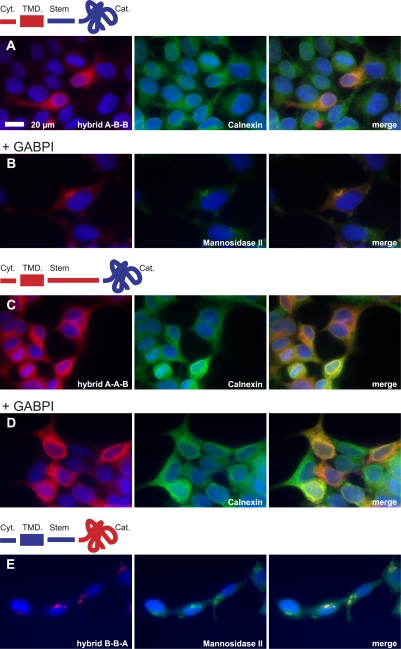
The data presented so far for the interaction between β4GalNAcTB and GABPI are highly reminiscent of the interactions between the human C1β3GalT generating the T antigen (core 1 O-glycan Galβ1-3GalNAcα1-Ser/Thr) and its client-specific molecular chaperone, Cosmc (Ju and Cummings, 2002). Cosmc supports functional folding of C1β3GalT in the ER but then dissociates and releases C1β3GalT (Ju et al., 2002b; Ju et al., 2008). Therefore, the following experiments addressed the subcellular localization of GABPI and β4GalNAcTs. Flag-β4GalNAcTs and Myc-GABPI were separately expressed in HEK293 cells and, after selection of stable clones, were detected by indirect immunofluorescence. Flag-β4GalNAcTA and Myc-GABPI colocalized with the Golgi marker α-mannosidase II (Fig. 3, A and C). Only the signal generated by Flag-β4GalNAcTB overlapped with the ER marker calnexin (Fig. 3 B). However, when GABPI was cotransfected (Fig. 3, D–F), the immunofluorescence images showed a clear shift of β4GalNAcTB to the Golgi. Moreover, as shown in Fig. 3 F, GABPI and β4GalNAcTB colocalized in this compartment. This experiment demonstrated that ER export of β4GalNAcTB needs piloting by GABPI, which by itself is an autonomous protein fully equipped with the information required for folding and transport to the Golgi.
Figure 3.
The Golgi resident protein GABPI pilots β4GalNAcTB to the Golgi apparatus. The subcellular localization of β4GalNAcTs and GABPI was analyzed with N-terminally tagged proteins expressed in HEK293 cells. (A–C) Flag-β4GalNAcTA, -B, and Myc-GABPI were visualized by indirect immunofluorescence (red). Subcellular compartments were labeled with anti–α-mannosidase II (Golgi) or calnexin (ER; green). (D–F) Flag-β4GalNAcTB (red) coexpressed with Myc-GABPI (F, green). (G) Subcellular localization of Myc-GABPI containing an N-terminal KKTN sequence. (H) Flag-β4GalNAcTB (green) coexpressed with Myc-GABPI–KKTN (red). Nuclei were stained with Hoechst 33258 (blue).
As GABPI moves with β4GalNAcTB, the question was raised whether both proteins remain associated in the Golgi. To answer this question, GABPI was tagged with a C-terminal KKTN dilysine signal (Zerangue et al., 2001), which retains proteins in the ER. GABPI was indeed successfully localized in the ER using this approach (Fig. 3 G). More importantly, β4GalNAcTB was also retained in the ER in cells expressing KKTN-tagged GABPI (Fig. 3 H). In vitro enzymatic activity of β4GalNAcTB was about two times as high as the nonretained construct (Fig. 2 A, right bars), and cell surface lacdiNAc was also detectable in these cells. Although the latter probably required cycling to the Golgi of at least part of the enzyme, ER retention might have allowed a higher protein expression level that was enzymatically active in vitro. Together, these data demonstrated that the DHHC family–related protein has, in contrast to Cosmc, functions that go beyond those of a client-specific chaperone.
Depletion of GABPI in Drosophila S2 cells delocalizes GalNAcTB and reduces lacdiNAc-containing glycolipid formation
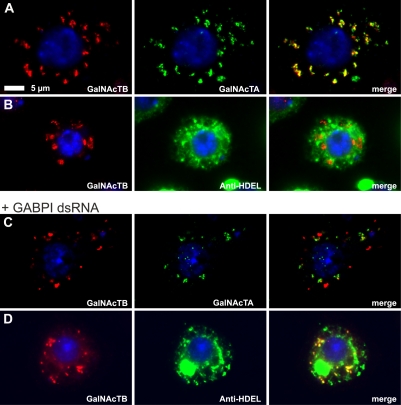
To additionally evaluate the influence of GABPI on β4GalNAcTB localization in the natural environment, RNAi experiments were performed. S2 cells were transiently transfected with N-terminally tagged β4GalNAcTs, and localization of the enzymes was monitored. As shown in Fig. 4 A, both β4GalNAcTs were colocalized in vesicular structures presumed to be the Golgi. β4GalNAcTB did not show any overlap with the ER-specific antibody anti-HDEL (Fig. 4 B). Incubation of cells with double-stranded RNA (dsRNA; Clemens et al., 2000) corresponding to a central coding region of GABPI dissected the HA-β4GalNAcTB signal from Flag-β4GalNAcTA (Fig. 4 C) and shifted the signal to structures that are part of the ER (Fig. 4 D).
Figure 4.
Knockdown of GABPI in S2 cells interferes with Golgi localization of β4GalNAcTB. (A–D) S2 cells after 3 d of culture in the absence (A and B) or presence (C and D) of dsRNA directed against GABPI were transiently transfected with Flag-β4GalNAcTA and HA-β4GalNAcTB. The Flag and HA epitopes were detected 2 d after transfection using respective antibodies, and the ER was marked with anti-HDEL. Nuclei were stained with Hoechst 33258.
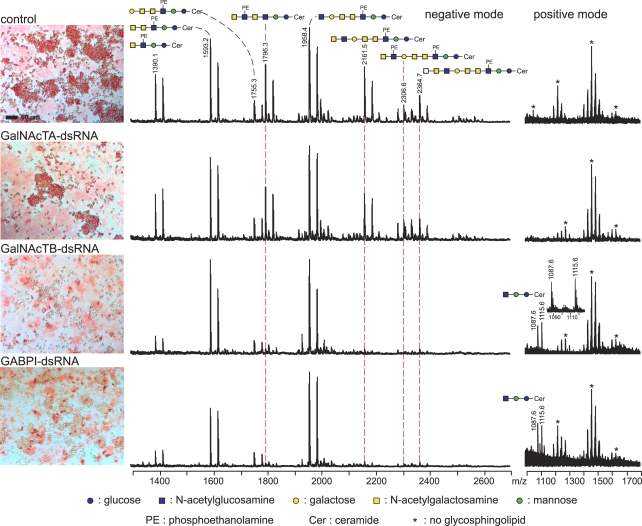
To answer whether the RNAi-induced redistribution of the enzyme is also followed by a change in activity, a second knockdown experiment was performed in which dsRNAs were designed to down-regulate β4GalNAcTA, β4GalNAcTB, or GABPI. S2 cells were cultured for 3 d in the absence or presence of dsRNA, after which the expression of lacdiNAc was displayed by immunocytochemistry and matrix-assisted laser desorption/ionization (MALDI) time of flight (TOF) mass spectrometry (MS), as illustrated in Fig. 5. The intense staining of control cells with antibody 259-2A1 was in accordance with the detection of lacdiNAc-containing glycolipid structures by negative-ion mode MALDI-TOF-MS. β4GalNAcTA knockdown did not change the signal pattern in comparison with control cells, whereas depletion of either β4GalNAcTB or GABPI had comparably strong effects on lacdiNAc expression. In the negative-ion mode, reduction of glycolipids carrying lacdiNAc repeats was accompanied by an enrichment of GlcNAcβ,3Galβ,3GalNAcα,4GalNAcβ,4(PE-6)GlcNAcβ,3Manβ,4GlcβCer species with a molecular mass of 1,958.4 D, an acceptor structure for β4GalNAcT. The positive-ion mode analyses clearly demonstrated the accumulation of a second β4GalNAcT acceptor structure, GlcNAcβ,3Manβ,4GlcβCer, having a molecular mass of 1,087.6 D. Changes in the glycolipid structures are very similar to changes observed in β4GalNAcTB knockout flies (Stolz et al., 2008) and thus are not further addressed in this paper. In addition, a new glycolipid species carrying lacdiNAc repeats with a molecular mass of 1,796.3 D has been identified and characterized by MALDI-TOF/TOF-MS (Table S1 and Figs. S1 and S2, available at http://www.jcb.org/cgi/content/full/jcb.200801071/DC1) as well as two extended species (Table S1 and Fig. S3). In summary, the results presented in Figs. 4 and 5 allow the conclusions that (a) β4GalNAcTB is the major lacdiNAc-synthesizing enzyme in S2 cells as it is in the fly, (b) GABPI enables Golgi targeting of β4GalNAcTB, and (c) β4GalNAcTB is essentially required to convey functionality.
Figure 5.
Knockdown of GABPI in S2 cells abrogates β4GalNAcTB activity. S2 cells before and after dsRNA down-regulation of β4GalNAcTs and GABPI as indicated were stained with antibody 259-2A1 to display lacdiNAc structures on the surface. Extracted glycolipid fractions were analyzed by MALDI-TOF-MS in negative- and positive-ion mode. Glycolipid species carrying multiple lacdiNAc structures that show differential expression are connected by dashed red lines. The positive-mode spectra show accumulation of the β4GalNAcT trisaccharide precursor.
Pull down of the complex formed between β4GalNAcTB and GABPI
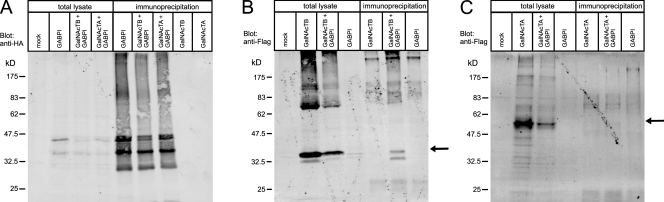
Because the data shown so far consistently argue for the existence of β4GalNAcTB and GABPI as a complex in the Golgi, we examined this contact in pull-down experiments. HEK293 cells transiently transfected with cDNA constructs encoding Myc-GABPI–HA and Flag-β4GalNAcTA and -B were lysed with buffer containing 1% NP-40. The anti-HA antibody 12CA5 coupled to Sepharose beads was used to precipitate Myc-GABPI–HA. To control the expression of recombinant proteins, total cell lysates were analyzed by Western blotting in parallel to precipitated proteins (Fig. 6). Both proteins were well expressed, as shown in the total cell lysates, whereas only Flag-β4GalNAcTB and not Flag-β4GalNAcTA was precipitated via Myc-GABPI–HA. This provided additional evidence for a tight interaction between β4GalNAcTB and GABPI.
Figure 6.
Myc-GABPI–HA coprecipitates Flag-β4GalNAcTB. HEK293 cells transiently transfected with Myc-GABPI–HA and Flag-β4GalNAcTA or Flag-β4GalNAcTB as indicated were lysed and precipitated with mouse monoclonal anti-HA (12CA5) coupled to Sepharose A beads. (A–C) Precipitated proteins as well as total cell lysates were displayed on Western blots using rabbit anti-HA (A) or mouse anti-Flag antibody (B and C). In spite of similar expression levels seen in total cell lysates for the Flag-tagged β4GalNAcTs, immunoprecipitation of Myc-GABPI–HA pulled down only Flag-β4GalNAcTB. Arrows indicate the running position of Flag-β4GalNAcTB and Flag-β4GalNAcTA in B and C, respectively.
The DHHC family–related protein GABPI is not an acyltransferase
Characterized DHHC protein family members are palmitoyltransferases in which the cysteine residue in the conserved DHHC motif is essential for activity (Lobo et al., 2002; Roth et al., 2002; Valdez-Taubas and Pelham, 2005). GABPI, in contrast to all mammalian and the other Drosophila members in the family, has exchanged this motif from DHHC to DHHS. This was already an argument against its function as acyltransferase. To validate this assumption, a series of mutants was constructed with which a potential involvement of the DHHS motif in GABPI functions could be tested. The ability of GABPI to install a functional β4GalNAcTB was not abolished by reconstruction of the DHHC motif, by replacement of the serine by alanine, or by successive replacement to AAAA (unpublished data). In addition, the critical cysteine residue (C29) that may serve as acyl residue acceptor in β4GalNAcTB was mutated. Again, no effect on GABPI–β4GalNAcTB Golgi localization and activity was found (unpublished data).
The stem region of β4GalNAcTB is needed for activation by GABPI
Because the β4GalNAcTs isolated from Drosophila are highly homologous proteins, it was of relevance to identify primary sequence elements responsible for the strict GABPI dependency of β4GalNAcTB. The aligned primary sequences indicated the stem region to be the domain of highest variability. Consequently, hybrids were made by domain swapping, as shown in Fig. 7. The chimera in which cytoplasmic and transmembrane domains of β4GalNAcTA were added to stem and catalytic regions of β4GalNAcTB (hybrid A-B-B) remained GABPI dependent for Golgi localization (Fig. 7, A and B) and activity (not depicted). However, additional replacement of the stem region destroyed activation by GABPI. The resulting protein was inactive and retained in the ER (Fig. 7, C and D). Because the stem region in β4GalNAcTA is considerably longer than in β4GalNAcTB, additional constructs were prepared in which the size was trimmed from the N and C termini to the exact length of the β4GalNAcTB stem region. All constructs remained inactive (unpublished data), allowing the conclusion that information contained in the stem region of β4GalNAcTB is essential for its function. In contrast, β4GalNAcTA remained Golgi localized and active independently of GABPI when fused to the cytoplasmic and stem region of β4GalNAcTB (construct B-B-A; Fig. 7 E). This is in agreement with the fact that the catalytic domain of β4GalNAcTA can be produced as soluble enzyme and, therefore, is an independent active entity.
Figure 7.
Protein domains of β4GalNAcTB involved in the interaction with GABPI. Hybrids of Flag-β4GalNAcTA (red) and -B (blue) were cloned by domain swapping of the cytoplasmic (Cyt) and transmembrane domain (TMD), stem region, and catalytic domain (Cat) as illustrated and expressed in HEK293 with and without GABPI. The intracellular localization was analyzed by indirect immunofluorescence using α-mannosidase II and calnexin as markers for Golgi and ER, respectively.
Discussion
Using a heterologous expression cloning approach, we isolated β4GalNAcTB as the major enzyme responsible for the biosynthesis of lacdiNAc structures in Drosophila. In this study, this enzyme was demonstrated to depend on the cooperation of a multimembrane-spanning protein related to the DHHC protein family. To point out the complexity of its involvement in forming a functionally active β4GalNAcTB, it was called GABPI. GABPI was cloned simultaneously to β4GalNAcTB in a classical expression cloning approach. This demonstrates the power of this technique, which exclusively screens for activity. The fact that the second β4GalNAc transferase in Drosophila, β4GalNAcTA (Haines and Irvine, 2005), was not detected in the expression cloning approach is a result of the much lower activity of this enzyme, which we confirmed in in vitro and in cellular test systems as well as on the systemic level (Stolz et al., 2008). Although β4GalNAcTA has been shown to act on protein acceptors in vitro (Sasaki et al., 2007), the low activity of β4GalNAcTA in HEK293 cells observed in this study does not allow a conclusion on the nature of the acceptor. Obviously, β4GalNAcTB is strictly lipid specific. This specificity is remarkable because glycan structures added to lipid anchors are different between mammals and flies. This suggests that selectivity is at least partly established through the lipid anchors. GABPI might be involved in the lipid specificity, but β4GalNAcTB also requires GABPI for in vitro activity with the synthetic acceptor substrate GlcNAc-pNP. In particular, these types of small hydrophobic aglycon-linked monosaccharide acceptors usually overcome the restricted specificity of glycosyltransferases; even the glycoprotein hormone-specific β4GalNAcT is reactive with such acceptors (Smith and Baenziger, 1988).
Trials to assemble an active enzyme by combining vesicle preparations containing β4GalNAcTB and GABPI separately failed. This was also the case for the O-mannosyltransferases (Manya et al., 2004). This shows that GABPI and β4GalNAcTB do not act in a sequential reaction mechanism. Combined with the experiments in which it was shown that β4GalNAcTB remains in the ER in an inactive state if expressed alone and can only reach the Golgi in the presence of GABPI, it can be concluded that interaction between β4GalNAcTB and GABPI most likely starts in the ER and requires coexpression of the two proteins. Most importantly, GABPI is an autonomous protein equipped with all of the information needed for Golgi destination. In this respect, GABPI seems to be different from Cosmc, the client-specific molecular chaperone required to activate C1β3GalT. A soluble, active form of recombinant C1β3GalT can be produced (Ju et al., 2002a), although Cosmc is not associated with this enzyme (Ju and Cummings, 2002). Purified rat liver C1β3GalT was also devoid of Cosmc (Ju et al., 2002b). According to the classical definition of a chaperone, Cosmc releases an active C1β3GalT (Ju et al., 2002b, 2008). In contrast, GABPI moves with β4GalNAcTB to the Golgi and retains β4GalNAcTB in the ER if it is retained itself. This and the fact that the proteins can be coimmunoprecipitated argue for a stable complex of both.
An insertion of the complex in an intact membrane patch is indispensable for functionality. Proof of this is provided by the fact that β4GalNAcTB activity tested with GlcNAc-pNP was almost completely abolished after addition of Triton X-100 or NP-40. Although these are rather mild detergents that normally do not dissociate protein complexes, their presence interferes with membrane integrity. This in turn may cause deformation of associated complexes. In contrast, saponin, which only perforates membranes, most likely increased activity by allowing the substrates to enter the vesicles without disturbing the proper embedding of the enzyme in the membrane. Detergent sensitivity is a property of many mannosyltransferases in the ER (Schutzbach, 1997), including the protein O-mannosyltransferase complex (POMT1 and POMT2), which is inactivated by Triton X-100 (Manya et al., 2004), and egghead, the mannosyltransferase acting two steps upstream of β4GalNAcTB in Drosophila glycolipid biosynthesis (Wandall et al., 2003). These enzymes are multitransmembrane-spanning proteins. Glycosyltransferases of the Golgi containing one transmembrane domain are usually not sensitive to detergents. As β4GalNAcTB is a typical member of the Golgi type II transmembrane glycosyltransferases, the observed detergent sensitivity is expected to be conveyed by disturbance of GABPI or the GABPI–β4GalNAcTB complex.
In line with the experiments in HEK293 cells, dsRNA-induced knockdown of GABPI in Drosophila S2 cells separated β4GalNAcTB from β4GalNAcTA, depleted cell surface expression of the lacdiNAc epitope, and provoked an accumulation of the β4GalNAcT glycolipid acceptor structures. These effects observed at the cellular level were exactly phenocopied in a Drosophila mutant with an inactivated β4GalNAcTB gene (Stolz et al., 2008).
The knowledge that all functionally characterized DHHC family proteins are palmitoyltransferases prompted experiments designed to determine whether GABPI could function as an acyltransferase. All residues critical for a potential acyltransferase activity in GABPI (Mitchell et al., 2006) as well as the only cysteine residue that may serve as acyl acceptors in β4GalNAcTB were point mutated. None influenced the functionality of GABPI or activity of β4GalNAcTB. The functionally crucial cysteine in the name-giving DHHC motif is exchanged by serine in GABPI, which argues against its function as a palmitoyltransferase.
In experiments aimed at understanding how β4GalNAcTB and GABPI interact, we demonstrated that the selectivity with which GABPI activates β4GalNAcTB and not the highly homologous β4GalNAcTA is attributed to a structural element in the stem region. However, this area cannot be the solely responsible element. Additional sequences in the catalytic domain must be involved in determining GABPI dependency.
The exact function of GABPI in priming activity of β4GalNAcTB in the Golgi is difficult to address. However, several glycosyltransferases acting exclusively in the glycolipid biosynthetic pathways need membrane anchorage and cannot be expressed as soluble recombinant proteins (Amado et al., 1998; Steffensen et al., 2000; Schwientek et al., 2002). One of these enzymes is brainiac (Schwientek et al., 2002), a β3GlcNAc transferase acting right upstream of β4GalNAcTB in glycolipid biosynthesis of Drosophila. The factors determining membrane dependency of brainiac are not yet identified. Because we found the product of brainiac accumulated in S2 cells treated with RNAi against GABPI (Fig. 5), an involvement of GABPI for brainiac function in vivo is not likely. However, because mammalian lipid-modifying enzymes have been suggested to form multienzyme complexes (Giraudo and Maccioni, 2003), GABPI, being an essential part of β4GalNAcTB, might be an anchor position in the pathway without being essential for the activity of all enzymes. A striking parallel exists between β4GalNAcTB and β4GalT-V and -VI described in the Introduction. These mammalian galactosyltransferases are members of the same gene family and are essentially dependent on membrane contact for transfer of Gal onto glucosyl Cer (van Die et al., 1999; Sato et al., 2000). As soluble enzymes, β4GalT-V and -VI recognize terminal GlcNAc residues instead of glucose. Therefore, it can be speculated that these enzymes require a cofactor similar to GABPI, which mediates glycolipid acceptor recognition. Orthologues of GABPI are found in arthropod and vertebrate species but not in nematodes, indicating that GABPI homologues might play a role in higher eukaryotes as well. In summary, it can be concluded that the identification of GABPI reveals a novel mechanism to generate specificity in the complex glycosylation pathway.
Materials and methods
Expression cloning
A cDNA library from Drosophila larval poly(A)+ RNA (Clontech Laboratories, Inc.) was constructed in pCMV-Script using the pCMV-ScriptXR cDNA library construction kit (Agilent Technologies). The library was divided into pools of 10,000 independent clones and used for expression cloning after the sibling selection strategy described previously (Bakker et al., 1997, 2005). The CHO cell line Lec8 (Deutscher and Hirschberg, 1986) grown in α-MEM supplemented with 10% FCS (both obtained from Biochrom AG) was used as the host. Pools or clones of the cDNA library were transfected into Lec8 cells using Metafectane (Biontex). After 2 d, cells grown in 6-well plates were fixed with 1.5% glutaraldehyde, incubated with the antilacdiNAc monoclonal antibody 259-2A1 (van Remoortere et al., 2000) followed by HRP-conjugated goat anti–mouse antibody (Jackson ImmunoResearch Laboratories), and detected by tyramide signal amplification using biotin-tyramide (Speel et al., 2006), streptavidin-AP (Invitrogen), and Fast-Red (Sigma-Aldrich) as chromogenic substrate.
Plasmid constructs
All tagged mammalian expression constructs were made in pcDNA3 (Invitrogen). Myc-GABPI (flybase gene number CG17257) contains an N-terminal Myc tag (MAQKLISEEDLNLRPLE [antibody-bound sequence underlined]) and Myc-GABPI–HA, an additional C-terminal HA tag (SRYPYDVPDYASL). Flag-β4GalNAcTB (CG14517), Flag-β4GalNAcTA (CG8536), and C. elegans GalNAcT (Kawar et al., 2002) contain N-terminal Flag tags (MDYKDDDDKGS). The Myc-GABPI–KKTN construct was cloned by PCR using Myc-GABPI as a template. For expression in Drosophila S2 cells, Flag-β4GalNAcTA and HA-β4GalNAcTB (N-terminal HA tag; MYPYDVPDYAGS) were cloned in pIB/V5-His (Invitrogen). Hybrids of β4GalNAcTA and β4GalNAcTB are identified by a three-letter code, whereby the first letter indicates the cytoplasmic plus transmembrane region, the second letter indicates the stem region, and the third letter indicates the catalytic domain (e.g., A-B-B). Borders between the three regions are after amino acids 29 and 135 in β4GalNAcTA and after 33 and 65 in β4GalNAcTB. Flag- or Myc-tagged constructs were used for all experiments unless indicated.
Preparation of ER and Golgi fractions from transfected HEK293 cells
HEK293 cells were grown in DME/HAM's F-12 supplemented with 10% FCS (both obtained from Biochrom AG). Cells transiently transfected as described in the Expression cloning section for CHO cells were washed with PBS and collected by centrifugation (5 min at 1,500 g). The cell pellets from three 175-cm2 plates (9 × 107 cells) were resuspended in 7 ml of lysis buffer (10 mM Hepes-Tris, pH 7.4, 0.8 M sorbitol, and 1 mM EDTA) containing an EDTA-free protease inhibitor mixture (Roche). After 10 strokes in a Dounce homogenizer, the lysate was centrifuged (10 min at 1,500 g). The supernatant was collected, and the pellet was subjected to a second homogenization/centrifugation round. The ER/Golgi-rich fraction was obtained by centrifugation of the combined supernatants at 100,000 g for 1 h. Pelleted vesicles were resuspended in 500 μl of assay buffer (0.1 M MOPS, pH 7.5) and 20-μl aliquots kept at −80°C. Protein concentrations were determined using a bicinchoninic acid kit (Thermo Fisher Scientific).
In vitro β4GalNAcT assays
Standard enzyme assays were performed with 20 μl of the ER/Golgi preparations in 50 μl of assay buffer (0.1 MOPS, pH 7.5, 20 mM MnCl2, 10 mM ATP, 100 mM GalNAc, 0.1% BSA, and 0.01% saponin). Therefore, 20-μl aliquots of the ER/Golgi vesicle preparation were supplemented to obtain the appropriate buffer composition and 0.5 mM of the radio-labeled nucleotide sugars UDP-6[3H]Gal (specific activity of 32 Bq/nmol; GE Healthcare) or UDP-1[3H]GalNAc (specific activity of 36 Bq/nmol [PerkinElmer]; diluted with cold nucleotide sugars [Sigma-Aldrich]). Reactions were started by adding the acceptor substrate GlcNAc-pNP (Sigma-Aldrich) at 1 mM and were incubated for 2 h at 28°C. Control samples were incubated in the absence of GlcNAc-O-pNP and subtracted from measured values. Reactions were stopped by addition of 1 ml of ice-cold water, and products were isolated on columns (Sep Pak Plus C18; Waters Corporation) as described previously (Palcic et al., 1988). The elutes were dried and counted in 2 ml of scintillation cocktail (Luma Safe Plus; Lumac LSC). Incorporated radioactivity was measured in a counter (LS 6500; Beckman Coulter).
Analyses of glycosphingolipids and proteins from transfected HEK293 cells
Transiently transfected HEK293 cells were washed with PBS, scraped off the plates, and collected by centrifugation (10 min at 1,500 g). Drosophila S2 cells were harvested by centrifugation and extracted in the same way. The cell pellets (107 cells) were resuspended in 300 μl of water and sonicated for 5 min in a bath sonicator. 2-propanol and hexane were added to obtain a solvent ratio of 55:25:20 (2-propanol/hexane/water), and the mixtures were sonicated again for 5 min. Samples were centrifuged for 10 min at 1,500 g, and supernatants were dried under nitrogen. The extracts were resuspended in chloroform/methanol/water (3:47:48) and desalted by reverse-phase chromatography (Sep Pak Plus C18 columns; Williams and McCluer, 1980). The eluted glycosphingolipids were dried under nitrogen, and one fourth of each sample was spotted onto a TLC plate (Nano-Durasil-20; Macherey-Nagel) and developed in running solvent composed of chloroform/methanol/0.25% aqueous KCl (5:4:1). For immunostaining, the silica plate was fixed in 0.1% polyisobutylmethylacrylate (Sigma-Aldrich) in aceton. The plate was blocked overnight with 1% BSA in TBS at 4°C followed by incubation with primary antibody (mouse antilacdiNAc 259-2A1) for 2 h at room temperature and with secondary antibody goat anti–mouse IRDye 800 (LI-COR Biosciences) for 30 min. After washing, the plate was analyzed on an infrared imaging system (Odyssey; LI-COR Biosciences).
Protein samples for Western blotting were isolated from the same cells by dissolving 107 cells in 750 μl of lysis buffer (2 mM EDTA, 50 mM Tris-HCl, pH 8.0, 1 mM MgCl2, and 1% NP-40 supplemented with a protease inhibitor mixture [Roche]) and analyzing 20 μl of these samples by standard Western blotting techniques. The blot was incubated with the same antibodies as the TLC plate and analyzed in the same way.
Subcellular localization studies by immunofluorescence
Subcellular localizations of recombinant Flag-β4GalNAcTA, Flag-β4GalNAcTB, and Myc-GABPI were performed with stably transfected HEK293 cells (without selecting clones). Therefore, transfected cells were cultured for 3 wk in the presence of G-418 (EMD). For staining, cells were seeded onto glass coverslips, fixed in 4% PFA, and permeabilized for 30 min with 0.1% saponin in PBS containing 0.1% BSA. Samples were incubated with the respective primary antibodies (anti-Flag tag M5, anti-Flag tag F7425, anti-HA tag 12CA5 or anti-Myc tag 9E10, and rabbit anti–α-mannosidase II or -calnexin as a Golgi or ER marker) for 1.5 h at room temperature. After three washings (in PBS, 0.1% BSA, and 0.1% Tween 20), cells were incubated with anti–mouse IgCy3 and anti–rabbit IgG Alexa Fluor 488 for 1 h at room temperature. After staining with the nuclear dye (Hoechst 33258; Hoechst Pharmaceuticals), the slides were washed with water, mounted (Dako), and analyzed under a microscope (Axiovert 200M; Carl Zeiss, Inc.) using a Plan Apochromat 63×/1.40 oil differential interference contrast objective (M27; Carl Zeiss, Inc.) at room temperature. Images (1,388 × 1,040 pixels) were taken using a camera (AxioCam MRm; Carl Zeiss, Inc.) and Axiovision 4.4 software (Carl Zeiss, Inc.). Images taken in an automatic exposure setting with filter sets for Hoechst 33258, Alexa Fluor 488, and Cy3 were converted in blue, green, and red, respectively; intensities were adapted to be equal for the three colors, and images were reduced to 600 dots per inch for display in Figs. 3 and 7.
Knockdown experiments in Drosophila S2 cells
dsRNA was made using the MEGAscript T7 transcription kit (Applied Biosystems). Each primer used in the PCR contained a 5′ T7 RNA polymerase–binding site followed by sequences specific for the target genes: GABPI (5′-CCGGCACCTCCAATTTTCTTTC-3′ and 5′-GTCCATATCCCCCACCTCGTCA-3′), β4GalNAcTA (5′-ATGTACCTCTTCACCAAGGCGA-3′ and 5′-ATAACCAATGTTCATCATGGCA-3′), and β4GalNAcTB (5′-TCAACTTTTCCTGCCAACAATG-3′ and 5′-ACCACGCCGCCGAAAAGACC-3′).
Drosophila Schneider (S2) cells were grown in Schneider's Drosophila medium (Invitrogen) supplemented with 10% FCS and 4 mM l-glutamine (Biochrom AG). For RNAi knockdown experiments (Clemens et al., 2000), 106 cells were plated per 6 wells in serum-free medium, and dsRNA of β4GalNAcTA, βGalNAcTB, and/or GABPI was added directly to the media in a final concentration of 37 nM (15 μg). After 30 min at room temperature, 2 ml of Schneider's medium containing FCS was added, and incubation was continued for 3 d at 27°C. For the immunocytochemical analysis of surface-expressed lacdiNAc structures, the protocol described in the Expression cloning section for CHO cells was used. Light microscopic images of Fig. 5 were taken using the aforementioned microscope and software using a camera (AxioCam MRc; Carl Zeiss, Inc.) and a Plan Apochromat 10×/0.45 objective (Ph1M27; Carl Zeiss, Inc.). To determine the subcellular localization of β4GalNAcTA and β4GalNAcTB in GABPI dsRNA–treated cells, the N-terminally Flag- and HA-tagged enzymes were transiently transfected with Fugene (Roche) into cultures that had been treated for 3 d with dsRNA. The day after transfection, cells were washed with serum-free medium, and RNAi treatment was repeated with a concentration of 18.5 nM dsRNA. 2 d after transfection, cells were transferred to concanavalin A–coated coverslips for 1 h, fixed in 4% PFA, and further processed as described in the previous section for HEK293 cells except that 0.1% saponin was kept in all incubation and washing solutions. Mouse anti-Flag M5 (Sigma-Aldrich) in combination with rabbit anti-HA (Sigma-Aldrich) was used to visualize the tagged β4GalNAc transferases, whereas the ER was stained with mouse anti-HDEL (Santa Cruz Biotechnology, Inc.). Secondary antibodies used were goat anti–mouse, Alexa Fluor 488, and goat anti–rabbit Cy3. Fluorescent images were made using a microscope (Axiovert 200M) as for the aforementioned HEK293 cells except that the ApoTome mode was used and five images were averaged. Fig. 4 shows 330 × 236-pixel sections of the original images. In addition, glycosphingolipid extracts (prepared as described in Analyses of glycosphingolipids and proteins…) from S2 cells before and after dsRNA treatment were analyzed by MALDI-TOF-MS in a TOF/TOF mass spectrometer (Ultraflex II; Bruker Daltonics) as described previously (Wuhrer and Deelder, 2005; Stolz et al., 2008).
Immunoprecipitation
Transiently transfected HEK293 cells were lysed for 30 min at 4°C using 750 μl of lysis buffer (2 mM EDTA, 50 mM Tris-HCl, pH 8.0, 1 mM MgCl2, and 1% NP-40 supplemented with protease inhibitor mixture). After centrifugation for 30 min at 12,000 g, anti-HA antibody 12CA5 coupled to Sepharose A beads was added to supernatants and incubated for 3 h at 4°C on a rotating wheel. Immunocomplexes were pelleted by centrifugation (300 g for 5 min) and washed twice with 50 mM Tris-HCl, pH 8.0, and 1% NP-40, twice with 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 1% NP-40, and once with the first washing buffer. Immunoprecipitated proteins were separated in SDS-PAGE, blotted onto polyvinylidene difluoride membranes (Waters Corporation), and stained with mouse anti-Flag M5 or rat anti-HA antibody.
Online supplemental material
Table S1 shows newly registered zwitterionic glycosphingolipid species. Fig. S1 shows negative-mode MALDI-TOF-MS of S2 cell glycosphingolipids. Fig. S2 shows MALDI-TOF/TOF-MS fragmentation analysis of two zwitterionic glycolipid species containing lacdiNAc tandem repeats. Fig. S3 shows MALDI-TOF/TOF-MS analysis of two zwitterionic glycolipid species. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801071/DC1.
Supplementary Material
Acknowledgments
We would like to thank Drs. Françoise Routier and Birgit Lüttig for critical reading of the manuscript.
Supporting financial resources for this study were obtained from the Hannover Medical School bonus system Leistungsorientierte Mittel and Regenerative Biology to Reconstructive Therapy, a Cluster of Excellence financed by the Deutsche Forschungsgemeinschaft.
Abbreviations used in this paper: Cer, ceramide; dsRNA, double-stranded RNA; GABPI, β4GalNAcTB pilot; Gal, galactose; GalNAc, N-acetylgalactosamine; GalNAcT, N-acetylgalactosaminyltransferase; GlcNAc, N-acetylglucosamine; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; pNP, p-nitrophenyl; TOF, time of flight.
References
- Amado, M., R. Almeida, F. Carneiro, S.B. Levery, E.H. Holmes, M. Nomoto, M.A. Hollingsworth, H. Hassan, T. Schwientek, P.A. Nielsen, et al. 1998. A family of human β3-galactosyltransferases. Characterization of four members of a UDP-galactose:β-N-acetyl-glucosamine/β-N-acetyl-galactosamine β-1,3-galactosyltransferase family. J. Biol. Chem. 273:12770–12778. [DOI] [PubMed] [Google Scholar]
- Bakker, H., I. Friedmann, S. Oka, T. Kawasaki, N. Nifant'ev, M. Schachner, and N. Mantei. 1997. Expression cloning of a cDNA encoding a sulfotransferase involved in the biosynthesis of the HNK-1 carbohydrate epitope. J. Biol. Chem. 272:29942–29946. [DOI] [PubMed] [Google Scholar]
- Bakker, H., F. Routier, S. Oelmann, W. Jordi, A. Lommen, R. Gerardy-Schahn, and D. Bosch. 2005. Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology. 15:193–201. [DOI] [PubMed] [Google Scholar]
- Brew, K., T.C. Vanaman, and R.L. Hill. 1968. The role of α-lactalbumin and the A protein in lactose synthetase: a unique mechanism for the control of a biological reaction. Proc. Natl. Acad. Sci. USA. 59:491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, J.C., C.A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B.A. Hemmings, and J.E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho, P.M., E. Deleury, G.J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307–317. [DOI] [PubMed] [Google Scholar]
- de Graffenried, C.L., and C.R. Bertozzi. 2004. The roles of enzyme localisation and complex formation in glycan assembly within the Golgi apparatus. Curr. Opin. Cell Biol. 16:356–363. [DOI] [PubMed] [Google Scholar]
- de Vries, T., C.A. Srnka, M.M. Palcic, S.J. Swiedler, D.H. van den Eijnden, and B.A. Macher. 1995. Acceptor specificity of different length constructs of human recombinant α1,3/4-fucosyltransferases. Replacement of the stem region and the transmembrane domain of fucosyltransferase V by protein A results in an enzyme with GDP-fucose hydrolyzing activity. J. Biol. Chem. 270:8712–8722. [DOI] [PubMed] [Google Scholar]
- Deutscher, S.L., and C.B. Hirschberg. 1986. Mechanism of galactosylation in the Golgi apparatus. A Chinese hamster ovary cell mutant deficient in translocation of UDP-galactose across Golgi vesicle membranes. J. Biol. Chem. 261:96–100. [PubMed] [Google Scholar]
- Giraudo, C.G., and H.J. Maccioni. 2003. Ganglioside glycosyltransferases organize in distinct multienzyme complexes in CHO-K1 cells. J. Biol. Chem. 278:40262–40271. [DOI] [PubMed] [Google Scholar]
- Guo, S., T. Sato, K. Shirane, and K. Furukawa. 2001. Galactosylation of N-linked oligosaccharides by human β-1,4-galactosyltransferases I, II, III, IV, V, and VI expressed in Sf-9 cells. Glycobiology. 11:813–820. [DOI] [PubMed] [Google Scholar]
- Haines, N., and K.D. Irvine. 2005. Functional analysis of Drosophila β1,4-N-acetlygalactosaminyltransferases. Glycobiology. 15:335–346. [DOI] [PubMed] [Google Scholar]
- Ju, T., and R.D. Cummings. 2002. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. USA. 99:16613–16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, T., and R.D. Cummings. 2005. Protein glycosylation: chaperone mutation in Tn syndrome. Nature. 437:1252. [DOI] [PubMed] [Google Scholar]
- Ju, T., K. Brewer, A. D'Souza, R.D. Cummings, and W.M. Canfield. 2002. a. Cloning and expression of human core 1 β1,3-galactosyltransferase. J. Biol. Chem. 277:178–186. [DOI] [PubMed] [Google Scholar]
- Ju, T., R.D. Cummings, and W.M. Canfield. 2002. b. Purification, characterization, and subunit structure of rat core 1 β1,3-galactosyltransferase. J. Biol. Chem. 277:169–177. [DOI] [PubMed] [Google Scholar]
- Ju, T., R.P. Aryal, C.J. Stowell, and R.D. Cummings. 2008. Regulation of protein O-glycosylation by the endoplasmic reticulum–localized molecular chaperone Cosmc. J. Cell Biol. 182:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawar, Z.S., I. van Die, and R.D. Cummings. 2002. Molecular cloning and enzymatic characterization of a UDP-GalNAc:GlcNAc(β)-R β1,4-N-acetylgalactosaminyltransferase from Caenorhabditis elegans. J. Biol. Chem. 277:34924–34932. [DOI] [PubMed] [Google Scholar]
- Kolmakova, A., and S. Chatterjee. 2005. Platelet derived growth factor recruits lactosylceramide to induce cell proliferation in UDP Gal:GlcCer: β1→4Galactosyltransferase (GalT-V) mutant Chinese hamster ovary cells. Glycoconj. J. 22:401–407. [DOI] [PubMed] [Google Scholar]
- Kolter, T., and K. Sandhoff. 2005. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21:81–103. [DOI] [PubMed] [Google Scholar]
- Lobo, S., W.K. Greentree, M.E. Linder, and R.J. Deschenes. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268–41273. [DOI] [PubMed] [Google Scholar]
- Manya, H., A. Chiba, A. Yoshida, X. Wang, Y. Chiba, Y. Jigami, R.U. Margolis, and T. Endo. 2004. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. USA. 101:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, C., G. Duncan, K.T. Goutsos, and F. Tufaro. 2000. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl. Acad. Sci. USA. 97:668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.A., A. Vasudevan, M.E. Linder, and R.J. Deschenes. 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47:1118–1127. [DOI] [PubMed] [Google Scholar]
- Münster, A.K., M. Eckhardt, B. Potvin, M. Mühlenhoff, P. Stanley, and R. Gerardy-Schahn. 1998. Mammalian cytidine 5′-monophosphate N-acetylneuraminic acid synthetase: a nuclear protein with evolutionarily conserved structural motifs. Proc. Natl. Acad. Sci. USA. 95:9140–9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., M. Takizawa, J. Aoki, H. Arai, K. Inoue, E. Wakisaka, N. Yoshizuka, G. Imokawa, N. Dohmae, K. Takio, et al. 1998. Purification, cDNA cloning, and expression of UDP-Gal: glucosylceramide β-1,4-galactosyltransferase from rat brain. J. Biol. Chem. 273:13570–13577. [DOI] [PubMed] [Google Scholar]
- North, S.J., K. Koles, C. Hembd, H.R. Morris, A. Dell, V.M. Panin, and S.M. Haslam. 2006. Glycomic studies of Drosophila melanogaster embryos. Glycoconj. J. 23:345–354. [DOI] [PubMed] [Google Scholar]
- Palcic, M.M., L.D. Heerze, M. Pierce, and O. Hindsgaul. 1988. The use of hydrophobic synthetic glycosides as acceptors in glycosyltransferase assays. Glycoconj. J. 5:49–63. [Google Scholar]
- Ramakrishnan, B., E. Boeggeman, and P.K. Qasba. 2002. β-1,4-galactosyltransferase and lactose synthase: molecular mechanical devices. Biochem. Biophys. Res. Commun. 291:1113–1118. [DOI] [PubMed] [Google Scholar]
- Roth, A.F., Y. Feng, L. Chen, and N.G. Davis. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, N., H. Yoshida, T.J. Fuwa, A. Kinoshita-Toyoda, H. Toyoda, Y. Hirabayashi, H. Ishida, R. Ueda, and S. Nishihara. 2007. Drosophila β1,4-N-acetylgalactosaminyltransferase-A synthesizes the LacdiNAc structures on several glycoproteins and glycosphingolipids. Biochem. Biophys. Res. Commun. 354:522–527. [DOI] [PubMed] [Google Scholar]
- Sato, T., S. Guo, and K. Furukawa. 2000. Involvement of recombinant human β-1, 4-galactosyltransferase V in lactosylceramide biosynthesis. Res. Commun. Biochem. Cell Mol. Biol. 4:3–10. [Google Scholar]
- Sato, T., M. Gotoh, K. Kiyohara, A. Kameyama, T. Kubota, N. Kikuchi, Y. Ishizuka, H. Iwasaki, A. Togayachi, T. Kudo, et al. 2003. Molecular cloning and characterization of a novel human β1,4-N-acetylgalactosaminyltransferase, β4GalNAc-T3, responsible for the synthesis of N,N′-diacetyllactosediamine, GalNAc β1-4GlcNAc. J. Biol. Chem. 278:47534–47544. [DOI] [PubMed] [Google Scholar]
- Schulz, I. 1990. Permeabilizing cells: some methods and applications for the study of intracellular processes. Methods Enzymol. 192:280–300. [DOI] [PubMed] [Google Scholar]
- Schutzbach, J.S. 1997. The role of the lipid matrix in the biosynthesis of dolichyl-linked oligosaccharides. Glycoconj. J. 14:175–182. [DOI] [PubMed] [Google Scholar]
- Schwientek, T., B. Keck, S.B. Levery, M.A. Jensen, J.W. Pedersen, H.H. Wandall, M. Stroud, S.M. Cohen, M. Amado, and H. Clausen. 2002. The Drosophila gene brainiac encodes a glycosyltransferase putatively involved in glycosphingolipid synthesis. J. Biol. Chem. 277:32421–32429. [DOI] [PubMed] [Google Scholar]
- Seppo, A., M. Moreland, H. Schweingruber, and M. Tiemeyer. 2000. Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. Eur. J. Biochem. 267:3549–3558. [DOI] [PubMed] [Google Scholar]
- Smith, P.L., and J.U. Baenziger. 1988. A pituitary N-acetylgalactosamine transferase that specifically recognizes glycoprotein hormones. Science. 242:930–933. [DOI] [PubMed] [Google Scholar]
- Speel, E.J., A.H. Hopman, and P. Komminoth. 2006. Tyramide signal amplification for DNA and mRNA in situ hybridization. Methods Mol. Biol. 326:33–60. [DOI] [PubMed] [Google Scholar]
- Steffensen, R., K. Carlier, J. Wiels, S.B. Levery, M. Stroud, B. Cedergren, S.B. Nilsson, E.P. Bennett, C. Jersild, and H. Clausen. 2000. Cloning and expression of the histo-blood group Pk UDP-galactose: Ga1β-4G1cβ1-cer α1, 4-galactosyltransferase. Molecular genetic basis of the p phenotype. J. Biol. Chem. 275:16723–16729. [DOI] [PubMed] [Google Scholar]
- Stolz, A., N. Haines, A. Pich, K.D. Irvine, C.H. Hokke, A.M. Deelder, R. Gerardy-Schahn, M. Wuhrer, and H. Bakker. 2008. Distinct contributions of β4GalNAcTA and β4GalNAcTB to Drosophila glycosphingolipid biosynthesis. Glycoconj. J. 25:167–175. [DOI] [PubMed] [Google Scholar]
- Taniguchi, N., K. Honke, and M. Fukuda. 2002. Handbook of Glycosyltransferases and Related Genes. Springer-Verlag New York Inc., New York. 670 pp.
- Togayachi, A., T. Akashima, R. Ookubo, T. Kudo, S. Nishihara, H. Iwasaki, A. Natsume, H. Mio, J. Inokuchi, T. Irimura, et al. 2001. Molecular cloning and characterization of UDP-GlcNAc:lactosylceramide β1,3-N-acetylglucosaminyltransferase (β3Gn-T5), an essential enzyme for the expression of HNK-1 and Lewis X epitopes on glycolipids. J. Biol. Chem. 276:22032–22040. [DOI] [PubMed] [Google Scholar]
- Vadaie, N., R.S. Hulinsky, and D.L. Jarvis. 2002. Identification and characterization of a Drosophila melanogaster ortholog of human β1,4-galactosyltransferase VII. Glycobiology. 12:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas, J., and H. Pelham. 2005. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 24:2524–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Die, I., H. Bakker, and D.H. van den Eijnden. 1997. Identification of conserved amino acid motifs in members of the β1→4-galactosyltransferase gene family. Glycobiology. 7:v–viii. [DOI] [PubMed] [Google Scholar]
- van Die, I., A. van Tetering, W.E. Schiphorst, T. Sato, K. Furukawa, and D.H. van den Eijnden. 1999. The acceptor substrate specificity of human β4-galactosyltransferase V indicates its potential function in O-glycosylation. FEBS Lett. 450:52–56. [DOI] [PubMed] [Google Scholar]
- van Remoortere, A., C.H. Hokke, G.J. van Dam, I. van Die, A.M. Deelder, and D.H. van den Eijnden. 2000. Various stages of schistosoma express Lewis(x), LacdiNAc, GalNAcß1-4 (Fucα1-3)GlcNAc and GalNAcß1-4(Fucα1-2Fucα1-3)GlcNAc carbohydrate epitopes: detection with monoclonal antibodies that are characterized by enzymatically synthesized neoglycoproteins. Glycobiology. 10:601–609. [DOI] [PubMed] [Google Scholar]
- Wandall, H.H., J.W. Pedersen, C. Park, S.B. Levery, S. Pizette, S.M. Cohen, T. Schwientek, and H. Clausen. 2003. Drosophila egghead encodes a β1,4-mannosyltransferase predicted to form the immediate precursor glycosphingolipid substrate for brainiac. J. Biol. Chem. 278:1411–1414. [DOI] [PubMed] [Google Scholar]
- Williams, M.A., and R.H. McCluer. 1980. The use of Sep-Pak C18 cartridges during the isolation of gangliosides. J. Neurochem. 35:266–269. [DOI] [PubMed] [Google Scholar]
- Wuhrer, M., and A.M. Deelder. 2005. Negative-mode MALDI-TOF/TOF-MS of oligosaccharides labeled with 2-aminobenzamide. Anal. Chem. 77:6954–6959. [DOI] [PubMed] [Google Scholar]
- Zerangue, N., M.J. Malan, S.R. Fried, P.F. Dazin, Y.N. Jan, L.Y. Jan, and B. Schwappach. 2001. Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc. Natl. Acad. Sci. USA. 98:2431–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G., M.L. Allende, E. Jaskiewicz, R. Qian, D.S. Darling, C.A. Worth, K.J. Colley, and W.W. Young Jr. 1998. Two soluble glycosyltransferases glycosylate less efficiently in vivo than their membrane bound counterparts. Glycobiology. 8:831–840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.