Abstract
Schizosaccharomyces pombe cdc15 homology (PCH) family members participate in many cellular processes by bridging the plasma membrane and cytoskeleton. Their F-BAR domains bind and curve membranes, whereas other domains, typically SH3 domains, are expected to provide cytoskeletal links. We tested this prevailing model of functional division in the founding member of the family, Cdc15, which is essential for cytokinesis in S. pombe, and in the related PCH protein, Imp2. We find that the distinct functions of Imp2 and Cdc15 are SH3 domain independent. However, the Cdc15 and Imp2 SH3 domains share an essential role in recruiting proteins to the contractile ring, including Pxl1 and Fic1. Together, Pxl1 and Fic1, a previously uncharacterized C2 domain protein, add structural integrity to the contractile ring and prevent it from fragmenting during division. Our data indicate that the F-BAR proteins Cdc15 and Imp2 contribute to a single biological process with both distinct and overlapping functions.
Introduction
For cytokinesis to occur, an actomyosin ring composed of a wide variety of conserved proteins must form and associate with the plasma membrane as an anchored yet dynamic lattice (for reviews see Glotzer, 2005; Wolfe and Gould, 2005; Barr and Gruneberg, 2007). Not only must the cytokinetic ring be associated firmly with the membrane upon ring formation, but rearrangements in both protein composition and membrane curvature drive ring constriction and ultimately abscission. Although great progress has been made in identifying the protein composition of the contractile ring, the interactions between proteins and between specific proteins and the plasma membrane are still being elucidated.
Schizosaccharomyces pombe is an ideal model system in which to study the assembly and dynamics of the cytokinetic ring. Indeed, genetic screens revealed key players, including Mid1 (Chang et al., 1996; Sohrmann et al., 1996), which binds to the plasma membrane and directs medial placement of the ring (Celton-Morizur et al., 2004). Other conserved proteins also participate in the first steps of ring assembly, including the heavy and light chains of nonmuscle myosin II, actin, an IQGAP (Rng2), tropomyosin, profilin, a formin, and two proteins with a membrane-binding F-BAR domain, Cdc15 and Imp2 (for review see Wolfe and Gould, 2005). Several of these components organize into cortical nodes directed by Mid1 before contractile ring formation (Wu et al., 2006), and models for the first steps of ring assembly have been formulated focusing on the coalesce of the nodes (Vavylonis et al., 2008) or, alternatively, proceeding through a leading cable of filamentous actin emanating from a spotlike concentration of formin (Chang, 1999; Arai and Mabuchi, 2002; Kamasaki et al., 2007). However, many questions remain regarding how these first components organize the nascent ring, anchor the ring to the membrane, and recruit dozens of additional proteins as mitosis proceeds. Furthermore, the reorganization inherent in contractile ring constriction and abscission has not been determined either in terms of protein–protein or protein–membrane interactions. Because a global proteome localization study indicated that >200 proteins localize to the site of division at some stage during cytokinesis (Matsuyama et al., 2006), the complexity of the process has only begun to be unraveled.
Cdc15 is one of the first proteins observed at the incipient ring (Wu et al., 2003). It is the founding member of the S. pombe cdc15 homology (PCH) family of proteins, which are typified by the conserved domain architecture of an N-terminal F-BAR domain and usually a C-terminal SH3 domain (for reviews see Lippincott and Li, 2000; Chitu and Stanley, 2007). Although Cdc15 plays an essential role in cytokinesis (Fankhauser et al., 1995), other PCH family members play diverse roles not only in cytokinesis but also in endocytosis, motility, and neural morphogenesis. Because most described PCH proteins bind and curve the cell membrane through their F-BAR domains (Itoh et al., 2005; Tsujita et al., 2006) and recruit protein-binding partners through SH3 domains (see table in Chitu and Stanley, 2007), it is reasonable to predict that PCH proteins achieve scaffolding functions through a division of labor between the two domains. However, we previously found that Cdc15 interacts with two key elements regulating the medial filamentous actin cytoskeleton, formin Cdc12 and type I myosin Myo1, through its F-BAR domain (Carnahan and Gould, 2003), leaving the role of its SH3 domain unclear. Another PCH family member in S. pombe, Imp2, also localizes to the contractile ring and functions in cytokinesis (Demeter and Sazer, 1998). With no binding partners identified for Imp2, it is unclear whether the roles played by Cdc15 and Imp2 overlap in any way.
In this study, we examine how the SH3 domains of Cdc15 and Imp2 contribute to contractile ring assembly. Surprisingly, the endogenous Cdc15ΔSH3 truncation fulfills the essential role of Cdc15, and imp2ΔSH3 causes no detectable phenotype on its own. Still, truncation of the Cdc15 SH3 domain results in significant cytokinetic defects. We identified two binding partners for the Cdc15 SH3 domain, Pxl1 and a novel C2 domain protein, Fic1. We also exposed a functional redundancy between the SH3 domains of Cdc15 and the PCH family member Imp2 in recruitment of contractile ring components and an additional cooperation between proteins such as Fic1 and Pxl1 recruited by these SH3 domains. The shared function of the Cdc15 and Imp2 SH3 domains is essential for cytokinesis. Consistent with this functional redundancy, replacement of the Cdc15 SH3 domain with that of Imp2 rescues most of the cytokinetic defects of cdc15ΔSH3. Our data indicate that the overlapping function accomplished by the SH3 domains of Imp2 and Cdc15 in contractile ring stabilization is separate from their distinct SH3-independent functions.
Results
The SH3 domain of Cdc15 plays a nonessential role in cytokinesis
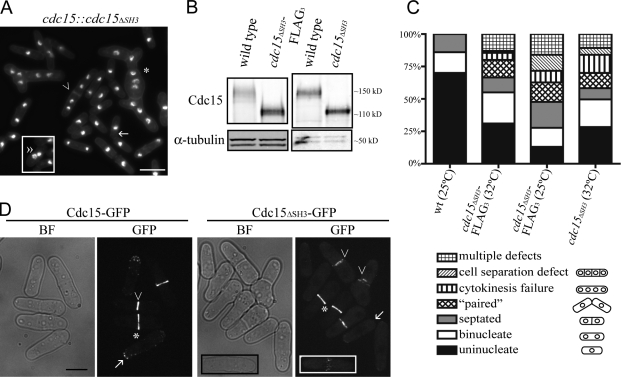
Preliminary data suggested that the Cdc15 SH3 domain was not essential for Cdc15 function (Carnahan and Gould, 2003), but to confirm this observation, we constructed a truncation of cdc15+ at its endogenous locus. Cdc15ΔSH3, representing amino acids 1–752, performs the essential function of Cdc15 (Fig. 1 A and not depicted), and the levels of truncated Cdc15 were found to be near those of wild type (Fig. 1 B and Fig. S1 D, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). However, although viable, 20–35% of cdc15ΔSH3 cells exhibited significant cytokinesis defects (Fig. 1, A and C). Additionally, >10% of cells displayed multiple defects. These defects were similarly severe when the truncated protein was tagged with a variety of epitopes and in cells expressing a more precise truncation of the SH3 domain at amino acid 869 (cdc15ΔSH3869; Fig. S1, A and B). Thus, the different cdc15ΔSH3 alleles were used interchangeably in further experiments.
Figure 1.
cdc15ΔSH3 cells are viable but exhibit cytokinesis defects. (A) cdc15ΔSH3 cells were grown to midlog phase, fixed, and stained with DAPI and methyl blue to visualize DNA and cell walls, respectively. The asterisk indicates a cell with both cell separation and cytokinesis defects, the arrow marks a “paired” cell, the arrowhead denotes a cell with impaired cell separation, and the double arrowhead marks a cell with a “kissing nuclei” phenotype. The boxed area is from a separate micrograph but was included to demonstrate all of the phenotypes scored. (B) Denatured lysates were prepared from wild-type, cdc15ΔSH3, and cdc15ΔSH3-FLAG3 cells grown at 32°C and immunoblotted with anti-Cdc15 serum or anti–α-tubulin for loading control. (C) Wild-type, cdc15ΔSH3-FLAG3, and cdc15ΔSH3 cells were grown at the temperatures indicated, fixed, and imaged as in A (n = 200). (D) cdc15-GFP and cdc15ΔSH3-GFP cells were grown to midlog phase at 25°C and imaged. Arrows indicate interphase cells, asterisks indicate contractile rings that have not begun constriction, and arrowheads mark constricting rings. The insets show a cell in which nodes of Cdc15ΔSH3 are present. BF, bright field. Bars: (A) 10 μm; (D) 5 μm.
cdc15ΔSH3 cells exhibit contractile ring defects
Because cdc15ΔSH3 cells exhibited cytokinetic defects, we reasoned that the contractile ring was defective in these cells and examined whether Cdc15 localization was affected. Cdc15-GFP is detected at cell tips during interphase and at the contractile ring during mitosis first in nodes and cables, which later coalesce (Carnahan and Gould, 2003; Wu et al., 2006). Cdc15ΔSH3-GFP and Cdc15ΔSH3869-GFP localized to nodes and rings (Fig. 1 D and Fig. S1 C) but localized to fewer and fainter spots at cell tips. Known Cdc15 binding partners Myo1 and Cdc12 (Carnahan and Gould, 2003) also localized to contractile rings in cdc15ΔSH3 cells (Fig. S1, F and G), affirming that Cdc15ΔSH3 localization to the ring and recruitment of its known binding partners remained superficially unaffected. However, Cdc15ΔSH3-GFP and Cdc15ΔSH3869-GFP rings appeared less well organized than full-length Cdc15-GFP during constriction (Fig. 1 D and Fig. S1 C).
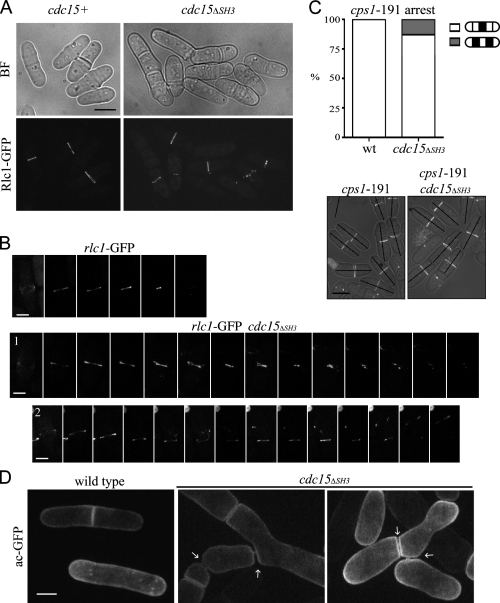
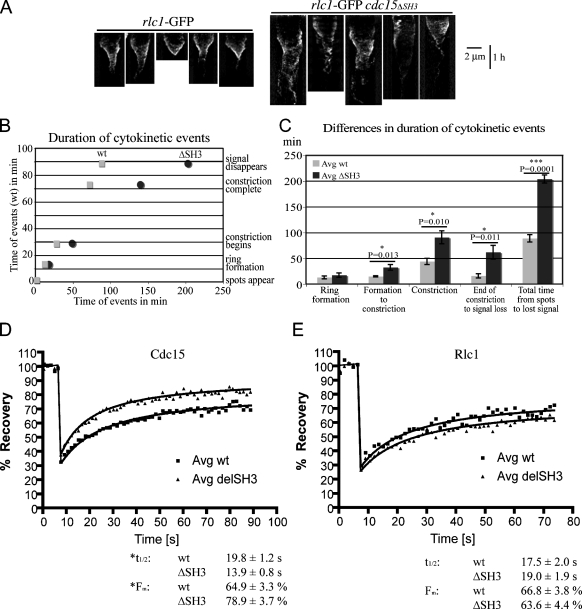
To examine contractile rings in cdc15ΔSH3 cells directly, we imaged the regulatory light chain for type II myosin, Rlc1-GFP. Many Rlc1-GFP rings appeared fragmented or frayed in cdc15ΔSH3 cells, especially once constriction had begun, which is similar to Cdc15ΔSH3-GFP rings (Fig. 2 A). To define the contractile ring defects in these cells, we imaged rlc1-GFP and cdc15ΔSH3-FLAG3 rlc1-GFP cells by time-lapse microscopy (Fig. 2 B and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). Kymographs from these videos revealed that rings constricted unsteadily in the mutant background relative to wild-type cells (Fig. 3 A). To quantify the defects, the timing of cytokinetic events was measured in five cells per genotype and averaged (Fig. S1 H). Although ring formation proceeded normally, cdc15ΔSH3 cells took significantly longer to complete cytokinesis, specifically taking longer to initiate constriction, complete constriction, and clear the Rlc1-GFP signal from the newly formed septum (Fig. 3, B and C). Because the cells chosen for analysis were those that successfully completed cytokinesis and because cells with rings that moved along the longitudinal axis were excluded (Fig. 2 B, cell 2), the overall defect in the cdc15ΔSH3 population was even more severe.
Figure 2.
cdc15ΔSH3 cells show defects in contractile ring dynamics. (A) rlc1-GFP and rlc1-GFP cdc15ΔSH3-FLAG3 cells were grown and imaged as in Fig. 1 D. BF, bright field. (B) rlc1-GFP and cdc15ΔSH3-FLAG3 rlc1-GFP cells were imaged live at 25°C every 2.5 min (Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). Montages of cells completing cytokinesis are shown. Each image is 12.5 min apart. (C) cps1-191 rlc1-GFP and cps1-191 cdc15ΔSH3-FLAG3 rlc1-GFP cells were arrested and imaged. In the images, cells were divided into fifths, and the location of the contractile ring was quantified (n = 100). wt, wild type. (D) acyl-GFP was expressed in wild-type or cdc15ΔSH3-FLAG3 cells and imaged live. Membrane bridges remaining after cytokinesis are marked with arrows. Bars: (A and C) 5 μm; (B) 2 μm; (D) 3 μm.
Figure 3.
Dynamics of rings and ring components in cdc15ΔSH3. (A) Kymographs were created from lines drawn across the division site in five videos for each rlc1-GFP and cdc15ΔSH3-FLAG3 rlc1-GFP. Time is depicted on the vertical scale bar. (B) Contractile ring event timing for each cell in A was determined and averaged (n = 5 each; Fig. S1 H, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). Timing of cytokinesis in wild-type (wt) cells was plotted on the y axis, and the time taken for corresponding events in wild-type and cdc15ΔSH3-FLAG3 cells was plotted on the x axis. (C) The time taken for wild-type and cdc15ΔSH3-FLAG3 cells to complete stages of cytokinesis was determined for each genotype, and differences were determined by Student's t tests. P-values and SEM (error bars) are included. (D) FRAP measurements of Cdc15-GFP or Cdc15ΔSH3-GFP signals are shown (n = 24 each). t1/2 and mobile fraction values were calculated from best-fit curves and are shown below. The difference in t1/2 is significant (P = 0.046), as is the difference in Fm (P = 0.0009). R2 values for wild-type and cdc15ΔSH3 curves are 0.9753 and 0.9752, respectively. (E) FRAP measurements of Rlc1-GFP in cdc15+ or cdc15ΔSH3-FLAG3 backgrounds (n = 28 each). t1/2 and mobile fraction values were calculated from best-fit curves. The differences in t1/2 and Fm are not significant (P = 0.651 and P = 0.589, respectively). R2 values for wild-type and cdc15ΔSH3 curves are 0.9365 and 0.9518, respectively.
The contractile ring appeared to be displaced from the cell middle in some cdc15ΔSH3 cells. To evaluate this phenotype more closely, we examined the placement of Rlc1-GFP rings in a cps1-191 mutant that arrests septation and blocks cells with medially placed rings (Le Goff et al., 1999; Liu et al., 1999). In cdc15+, all rings in the cps1-191 arrest localized medially, but in the cdc15ΔSH3 background, rings in 13% of cells localized outside the middle fifth of the cell (Fig. 2 C), indicating a defect in ring anchoring.
We used a membrane marker, acyl-GFP, to determine whether abscission was slow. Membrane bridges were never visible in wild-type cells but were visible in many cdc15ΔSH3 cells (Fig. 2 D). Live cell imaging demonstrated that these bridges persisted for hours (unpublished data), suggesting that cdc15ΔSH3 cells are defective not only in the timing of contractile ring events but also in the abscission of the membrane connection between mother and daughter cells. The thick septum visible around each bridge is consistent with perturbation of the actin cytoskeleton (Ishijima et al., 1999).
Finally, we investigated the dynamics of Cdc15-GFP and Cdc15ΔSH3-GFP using FRAP. Cdc15ΔSH3-GFP recovered with a significantly shorter t1/2 than Cdc15-GFP and with a significantly higher mobile fraction, together indicating that Cdc15ΔSH3-GFP is more mobile at the contractile ring (Fig. 3 D and Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). FRAP of Rlc1-GFP in cdc15+ and cdc15ΔSH3 cells did not reveal significantly different dynamics (Fig. 3 E and Fig. S2 B), indicating that although the cdc15ΔSH3 mutation affects contractile ring organization, it does not globally alter ring component mobility. Consistent with these analyses, cdc15ΔSH3 synthetically interacts with a wide variety of cell cycle and contractile ring mutants (Fig. S1 I). Collectively, these results indicate that the SH3 domain of Cdc15 is involved in contractile ring anchoring, stability, and constriction.
Cdc15 SH3 domain binds to C2 domain protein Fic1
We next sought to identify binding partners for the Cdc15 SH3 domain. SH3 domains typically interact with proline-rich regions of target proteins, especially the motif PXXP (for review see Mayer, 2001). We could not demonstrate Cdc15 interaction with Vrp1 or Wsp1 (unpublished data), which were likely candidates based on previously investigated PCH family members (Wu et al., 1998; Qualmann and Kelly, 2000; Naqvi et al., 2001; Ho et al., 2004). Some SH3 domains that contain a phenylalanine residue at position 75 bind ubiquitin or ubiquitinated proteins (Stamenova et al., 2007). Although the SH3 domain of Cdc15 has the corresponding residue, we did not detect binding between this domain and ubiquitin (Fig. S2 D).
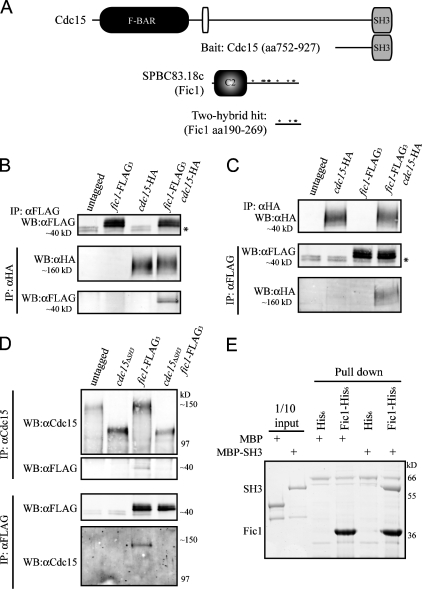
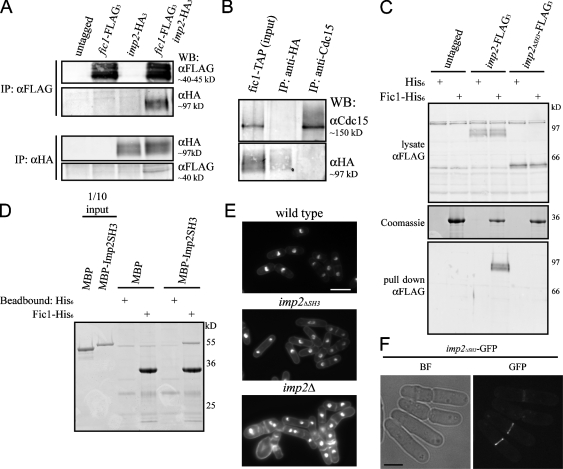
We then conducted a yeast two-hybrid screen using amino acids 752–927 of Cdc15 as bait and an S. pombe cDNA library as prey (Fig. 4 A). One positive clone was isolated from the screen, and a BLAST (basic local alignment search tool) search using the recovered sequence revealed that the plasmid contained base pairs 568–807 of SPBC83.18c cDNA. This fragment encodes amino acids 190–269 of the protein, which includes four PXXP motifs (Fig. 4 A, asterisks). The isolated plasmid showed a positive interaction when directly tested with the bait plasmid and also a shorter portion of Cdc15, containing only amino acids 843–927 (Fig. S2, E, F, and H).
Figure 4.
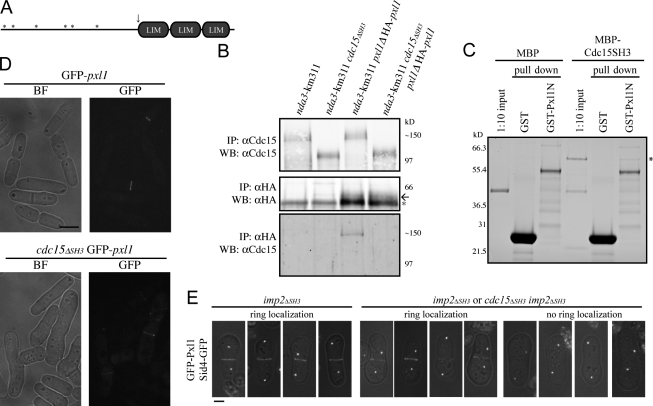
Cdc15 SH3 domain is necessary and sufficient for interaction with C2 domain protein Fic1. (A) Schematic representations of the domains of Cdc15 (top) and Fic1 (bottom). Domains are as indicated, except the PEST domain of Cdc15 is the white oval, and PXXP motifs in Fic1 are indicated with asterisks. Fragments of cdc15 and fic1 from the two-hybrid screen are depicted. (B and C) Coimmunoprecipitation of endogenously tagged Fic1-FLAG3 and Cdc15-HA3 from asynchronous cell lysate. Asterisks indicate a background band present in FLAG immunoblots. (D) Coimmunoprecipitations were performed in which Cdc15 or Cdc15ΔSH3 was isolated using anti-Cdc15 serum, or Fic1-FLAG3 was isolated with the anti-FLAG antibody, and bound proteins were identified by immunoblotting with either the anti-FLAG antibody or anti-Cdc15 serum. (E) Recombinant MBP or MBP-Cdc15SH3 was purified and incubated with recombinant bead-bound His6 or Fic1-His6. Beads were washed, and bound proteins were run on SDS-PAGE and detected by Coomassie staining. One tenth of each MBP protein input is shown as a control. IP, immunoprecipitation; WB, Western blot.
The SPBC83.18c gene encodes a C2 domain–containing protein and exhibits homology to the essential Saccharomyces cerevisiae gene INN1 and Cryptococcus neoformans cts1, which have established roles in cytokinesis (Fig. S2 G; Fox et al., 2003; Sanchez-Diaz et al., 2008). Because the S. pombe protein binds Cdc15, we named it “15-interacting C2 domain” protein, Fic1.
To verify that Fic1 indeed interacts with Cdc15 in vivo, reciprocal coimmunoprecipitation experiments were performed in which either endogenously tagged Cdc15-HA3 or Fic1-FLAG3 was immunoprecipitated and the other protein was detected by immunoblotting (Fig. 4, B and C). Fic1-FLAG3 did not coimmunoprecipitate Cdc15ΔSH3 (Fig. 4 D), indicating that the SH3 domain of Cdc15 is necessary for its interaction with Fic1. Additionally, an in vitro binding assay showed that Fic1-His6 and maltose-binding protein (MBP)–Cdc15SH3 interact directly (Fig. 4 E), demonstrating that the SH3 domain of Cdc15 is sufficient for the interaction. Quantitative in vitro binding revealed that Fic1 and the SH3 domain of Cdc15 bind with a Kd of ∼3 μM (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1), which falls within the range (0.27 to >20 μM) of other SH3 domain Kd measurements (Landgraf et al., 2004; Haynes et al., 2007).
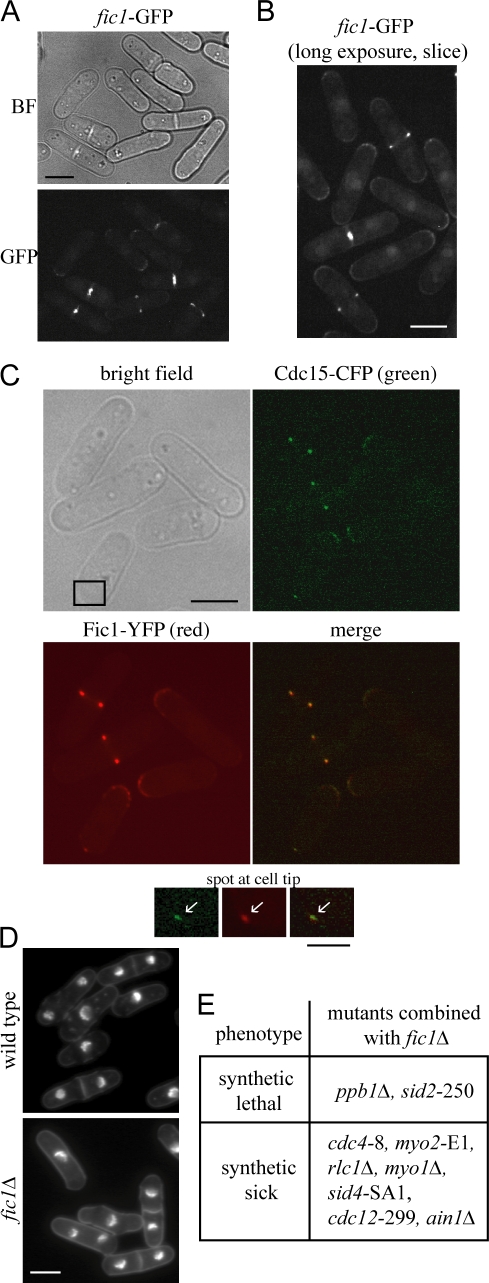
Fic1 is a contractile ring protein with a minor role in cytokinesis
In live cell microscopy, endogenously tagged Fic1-GFP localizes to cell tips and nuclei in interphase cells and in mitotic cells concentrates at the contractile ring, where it remains during ring constriction (Fig. 5, A and B). Localization of Fic1-GFP in cells also producing Sid4-GFP, a marker of the spindle pole body (Chang and Gould, 2000), revealed that Fic1-GFP arrives at the cytokinetic ring before anaphase and is not seen at nodes (Fig. S3 B). Imaging of a fic1-YFP cdc15-CFP strain revealed that Cdc15 and Fic1 colocalize in both contractile rings and in spots at cell tips (Fig. 5 C). Fic1 localized to the ring and cell tips independently of the SH3 domain of Cdc15 (Fig. S3 C), suggesting that in these cells, Fic1 is able to localize properly through direct membrane binding or interaction with another protein.
Figure 5.
Fic1 localizes to the contractile ring and plays a nonessential role in cytokinesis. (A and B) fic1-GFP cells were imaged live. In B, one 0.5-μm slice was taken through cell middles to allow imaging of fainter structures. BF, bright field. (C) cdc15-CFP fic1-YFP cells were imaged through one 0.5-μm plane. Bright field, CFP (green), YFP (red), and merged images are shown. Insets (boxed area) show colocalization of Cdc15 and Fic1 at cell tips (arrows). (D) Wild-type and fic1Δ cells were stained as in Fig. 1 A. Quantitation of cell phenotypes is shown in Fig. S3 E (available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). (E) fic1Δ was crossed with several mutants or deletions in cytokinesis genes to determine synthetic interactions. Bars: (A–D) 5 μm; (C, inset) 2 μm.
fic1Δ cells are viable and behave as wild type, as indicated by overall morphology (Fig. 5 D and Fig. S3 E), doubling time, and Cdc15-GFP localization (Fig. S3 D). Time-lapse imaging of Rlc1-GFP in fic1Δ cells also demonstrated that cytokinesis in these cells is essentially unaltered (unpublished data). Visualization of the acyl-GFP marker in fic1Δ cells did not reveal a defect in abscission (Fig. S3 F). However, fic1Δ interacted negatively with several cytokinesis mutants and was synthetically lethal with mutants of two cytokinesis regulators, calcineurin homologue ppb1 and septation signaling kinase sid2 (Fig. 5 E). Our genetic analyses indicate that although Fic1 is not essential, it does play a role in cytokinesis.
Fic1 interacts with PCH family member Imp2
Because Fic1 localized correctly in the absence of the Cdc15 SH3 domain, we sought to identify additional Fic1 binding partners that could recruit it to the ring. We isolated endogenously expressed Fic1–tandem affinity purification (TAP) on IgG-coated Dynabeads from lysate derived from cells arrested in mitosis because of a mutation in β-tubulin, nda3-km311 (Hiraoka et al., 1984). Copurified proteins were identified by liquid chromatography tandem mass spectrometry (MS; Fig. S3 G). Cdc15 was identified with the highest sequence coverage (75.8%), whereas the only other ring component in the top 25 hits was PCH family member Imp2 (61.5%). Coimmunoprecipitation of Imp2-HA3 and Fic1-FLAG3 verified their in vivo interaction (Fig. 6 A). Fic1-FLAG3 did not coimmunoprecipitate the PCH family member Bzz1-HA3 (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1), indicating that Fic1 does not bind SH3 domains or PCH family members nonspecifically.
Figure 6.
Fic1 binds the SH3 domain of PCH family member Imp2. (A) Coimmunoprecipitations of Imp2-HA3 and Fic1-FLAG3 were performed as in Fig. 4 (B and C). IP, immunoprecipitation. (B) Fic1-TAP–containing complexes were purified from nda3-km311 imp2-HA3 cells using IgG beads followed by TEV cleavage. These complexes were immunoprecipitated using either an anti-HA antibody or anti-Cdc15 serum, and bound proteins were detected by immunoblotting. WB, Western blot. (C) Lysates from wild-type, imp2-FLAG3, or imp2ΔSH3-FLAG3 cells were incubated with recombinant bead-bound His6 or Fic1-His6. Bound proteins were detected by immunoblotting. (D) An in vitro binding experiment was performed with MBP-Imp2SH3 and Fic1-His6 as in Fig. 4 E. (E) Wild-type, imp2ΔSH3-HA3, and imp2Δ cells were fixed and imaged as in Fig. 1 A (quantitation in Fig. S4 D, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1). (F) Imp2ΔSH3-GFP was visualized in live cells. BF, bright field. Bars: (E) 10 μm; (F) 5 μm.
Because C2 domains sometimes dimerize in crystal structures (Sutton and Sprang, 1998; Liu et al., 2006), and because it is unknown whether PCH family members can form heterodimers, we examined whether Fic1, Cdc15, and Imp2 were present in one complex. We purified Fic1-TAP from mitotic lysate containing Imp2-HA3 and cleaved Fic1 complexes from beads using tobacco etch virus (TEV) protease. Two fractions were then immunoprecipitated with anti-HA antibody or anti-Cdc15 serum, and each immunoprecipitation was immunoblotted to test the presence or absence of Cdc15 and Imp2-HA3 (Fig. 6 B). Imp2-HA3 complexes did not coimmunoprecipitate Cdc15 or vice versa, indicating that Fic1 binds the two PCH family members in separate complexes.
To determine whether the SH3 domain of Imp2 mediates its binding to Fic1, we created an imp2ΔSH3 strain (Fig. S4 B), and either full-length Imp2 or Imp2ΔSH3 was pulled from the lysate using recombinant bead-bound Fic1-His6 (Fig. 6 C). Fic1-His6 pulled full-length Imp2 but not Imp2ΔSH3 from cell lysate, indicating that the SH3 domain of Imp2 is essential for its interaction with Fic1. In an in vitro binding assay, the SH3 domain of Imp2 was also sufficient to bind recombinantly produced Fic1 directly (Fig. 6 D) and bound Fic1 with a Kd of ∼0.9 μM (Fig. S4 C).
Next, we examined the imp2ΔSH3 strain to determine whether it exhibited any growth defects. Although imp2Δ cells display severe cytokinesis defects that become lethal at higher temperatures (Demeter and Sazer, 1998), imp2ΔSH3-HA3 cell morphology was indistinguishable from wild type at all tested temperatures (Fig. 6 E and Fig. S4 D). Imp2ΔSH3-GFP localized to the contractile ring (Fig. 6 F), and Rlc1-GFP localization in the imp2ΔSH3 background was normal (Fig. S4 E). No membrane bridges were visible in imp2ΔSH3 cells, indicating no abscission defect (Fig. S4 F). However, the imp2ΔSH3 mutation showed negative genetic interactions with several mutations or deletions in other known contractile ring components or regulators (Fig. S4 H). Collectively, these results indicate that although the SH3 domain of Imp2 plays a minor role during cytokinesis, Imp2ΔSH3 fulfills the major function of Imp2.
Cdc15 and Imp2 SH3 domains play a cooperative, essential role in cytokinesis
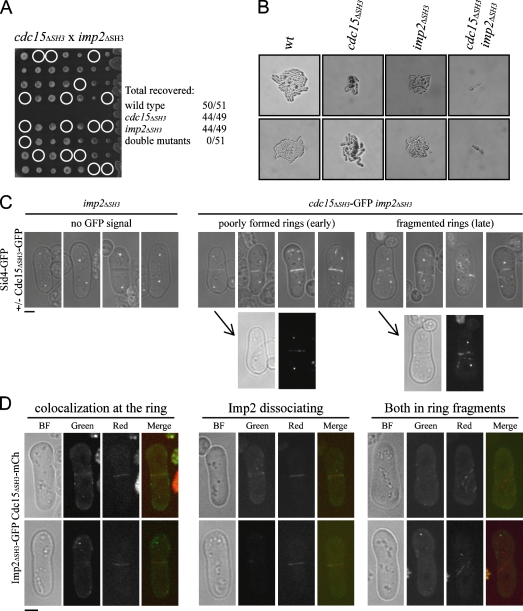
We speculated that the SH3 domains of Cdc15 and Imp2 might be acting cooperatively to recruit binding partners like Fic1 to the contractile ring. Based on this hypothesis, we tested whether the cdc15ΔSH3 and imp2ΔSH3 truncations were synthetically lethal and did not recover any double mutants at any tested temperature (Fig. 7 A and Fig. S1 E), and double mutants died before the first cell division (Fig. 7 B). To examine the defect of the double mutation, we crossed single mutants carrying fluorescent labels and germinated the resultant spores in the presence of hygromycin to select for the imp2ΔSH3 allele. Cdc15ΔSH3-GFP localizes transiently to the contractile ring in the double mutant, but later, especially in cells with the faint beginning of a septum, these rings fragment (Fig. 7 C). Imp2ΔSH3-GFP colocalizes with Cdc15ΔSH3-mCherry in some cells, but this colocalization decreases as first Imp2ΔSH3 and then Cdc15ΔSH3 leave the medial region of the cell in spots and ring fragments (Fig. 7 D).
Figure 7.
SH3 domains of Cdc15 and Imp2 function cooperatively in cytokinesis. (A) cdc15ΔSH3-FLAG–KanR and imp2ΔSH3-FLAG–HygR cells were crossed, and tetrads were pulled. Circles indicate missing double mutants, and the recovery rates are listed. (B) Images were taken for several resulting tetrads, and single colonies of each genotype are shown. wt, wild type. (C) Spores of the indicated genotypes were germinated overnight and imaged in the first cell division. Bright field images are overlaid onto GFP images. Below, bright field (BF) and GFP images of two germinating spores before (left) and after (right) initiation of septation are shown separately. (D) Spores were germinated as in C and imaged for both Imp2ΔSH3-GFP and Cdc15ΔSH3-mCherry localization. Bars, 2 μm.
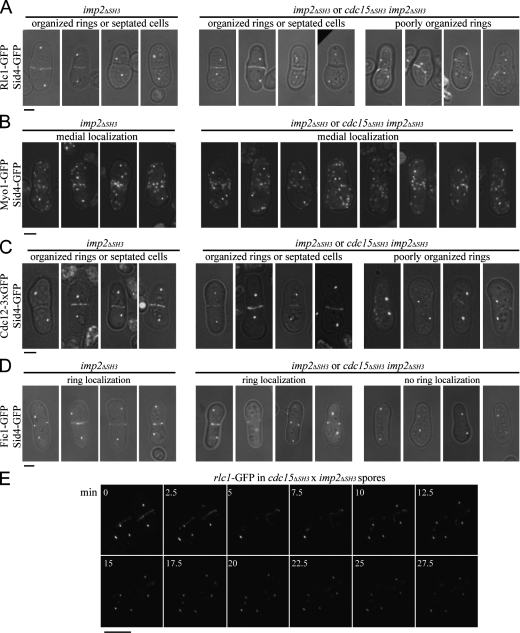
To examine more closely the stage of cytokinetic failure in these double mutants, we crossed cdc15ΔSH3 and imp2ΔSH3 strains carrying sid4-GFP and rlc1-GFP and grew the resultant spores in the presence of hygromycin. In parallel, we germinated imp2ΔSH3 spores as a control for the single mutant phenotype. In the mixture of imp2ΔSH3 and imp2ΔSH3 cdc15ΔSH3 germinated spores, approximately half of the cells with two spindle pole bodies showed normal contractile ring organization or complete septa, as seen in the imp2ΔSH3 single mutant (Fig. 8 A). The other half had very poorly organized rings, including disconnected wisps of ring material in the majority of cells and fragmented or maloriented rings in the remainder (Fig. 8 A). To make certain that there was not a defect in the localization of known binding partners Myo1 and Cdc12, these proteins were also visualized in the double mutant. Myo1-GFP localized medially in single and double mutants (Fig. 8 B). Cdc12-GFP3 localized to contractile rings in single mutants and appeared at ring fragments in the double mutant background, suggesting that its localization to the ring parallels that of Cdc15 and Rlc1 (Fig. 8 C).
Figure 8.
Contractile ring defects of cdc15ΔSH3 imp2ΔSH3 spores. (A–D) Spores of the indicated genotypes were germinated in the presence of hygromycin overnight and imaged as in Fig. 7 C. (E) Images were taken at 150-s intervals from a video of Rlc1-GFP and Sid4-GFP in the cdc15ΔSH3 imp2ΔSH3 background. Time on the montage is indicated in minutes. Bars: (A–D), 2 μm; (E) 5 μm.
To determine whether SH3 domain binding factors fail to localize to the medial region of the cell in the imp2ΔSH3 cdc15ΔSH3 mutants, we performed a similar spore germination experiment with fic1-GFP sid4-GFP cells. Approximately half of the germinating spores with two spindle pole bodies showed Fic1-GFP at well-formed contractile rings, as seen in the single imp2ΔSH3 mutant (Fig. 8 D). The other half of germinating spores with two spindle pole bodies showed no Fic1-GFP in the medial region of the cell and often showed cortical localization of Fic1-GFP at one tip (Fig. 8 D), demonstrating that the SH3 domains of Imp2 and Cdc15 are essential for recruiting binding partners, including but not limited to Fic1, to the cytokinetic ring.
To observe the dynamics of contractile ring failure in the double SH3 mutant, we imaged Rlc1-GFP and Sid4-GFP spores live through time-lapse microscopy (Fig. 8 E and Video 3). In the two cells depicted, contractile rings form but fray and disassemble, suggesting that although incipient rings form, they are inherently unstable.
Pxl1 is recruited to the medial ring in an SH3-dependent manner
Because the phenotypes observed for cdc15ΔSH3 and the double SH3 deletions were much more severe than fic1Δ, we sought other binding partners for the Cdc15 SH3 domain. Paxillin-related protein Pxl1 localizes to the contractile ring, coimmunoprecipitates with Cdc15, and contains six PXXP motifs in its N-terminal half, whereas pxl1Δ cells show contractile ring fraying reminiscent of the cdc15ΔSH3 phenotype (Fig. 9 A; Ge and Balasubramanian, 2008; Pinar et al., 2008). We found that Cdc15 coimmunoprecipitates HA-Pxl1 in an SH3-dependent manner from mitotically arrested cells (Fig. 9 B). MBP-Cdc15SH3 also bound to GST-Pxl1 (N terminus) in vitro (Fig. 9 C), indicating that the SH3 domain of Cdc15 is sufficient to mediate its direct interaction with Pxl1.
Figure 9.
Pxl1 binds the SH3 domain of Cdc15 and functions in parallel with Fic1. (A) Diagram of Pxl1 to scale. LIM domains are shown as gray ovals, and PXXP motifs are depicted as asterisks. The site of the truncation used in the in vitro binding assay is indicated with an arrow. (B) Coimmunoprecipitation of Cdc15 with HA-Pxl1 was performed as in Fig. 4 D. The asterisk indicates a background band, and the arrow marks the HA-Pxl1 bands. IP, immunoprecipitation; WB, Western blot. (C) in vitro binding assay was performed as in Fig. 4 E. Binding is indicated with an asterisk. (D) GFP-pxl1 and cdc15ΔSH3 GFP-pxl1 strains were grown at 25°C and imaged live. (E) Spores were germinated and imaged as in Fig. 8 (A–D). BF, bright field. Bars, 5 μm.
GFP-Pxl1 localized to the medial ring normally in the cdc15ΔSH3 mutant (Fig. 9 D) but failed to localize in the cdc15ΔSH3 imp2ΔSH3 spores (Fig. 9 E), suggesting that like Fic1, Pxl1 is recruited to the medial ring by the two SH3 domains. This result suggests that the unraveling of the contractile ring in cdc15ΔSH3 imp2ΔSH3 cells might be brought about by the loss of at least two stabilizing protein–protein and possibly protein–membrane interactions. Consistent with this model, pxl1Δ and fic1Δ are synthetically lethal when combined (Fig. S5, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1).
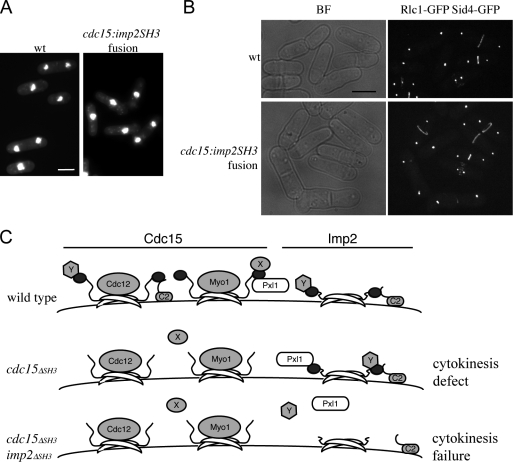
Finally, a fusion was generated in which the SH3 domain of Cdc15 was seamlessly replaced with that of Imp2 (cdc15-imp2SH3) at the cdc15 genomic locus. Remarkably, the cdc15-imp2SH3 phenotype was nearly the same as wild type (Fig. 10 A and Fig. S5 C), and contractile rings in cdc15-imp2SH3 cells did not show significant defects (Fig. 10 B). Together, these data indicate that (a) the Imp2 SH3 domain can fulfill most of the roles of the Cdc15 SH3 domain, and (b) the cdc15ΔSH3 phenotype might largely be the result of a stoichiometric reduction in available SH3-binding spots at the ring rather than the failure of a specific component to be recruited.
Figure 10.
The Imp2 SH3 domain can fulfill most functions of the Cdc15 SH3 domain. (A) The SH3 domain of Cdc15 was replaced with that of Imp2 at the endogenous cdc15 locus. Cells were grown and stained as in Fig. 1 A. (B) Rlc1-GFP and Sid4-GFP were imaged live in wild-type (wt) and cdc15-imp2SH3 backgrounds. BF, bright field. (C) Cdc15 and Imp2 are depicted as dimers at the membrane; F-BAR domains are shown as crescents, and SH3 domains are shown as dark ovals. In wild-type cells, both Cdc15 and Imp2 bind Fic1 (the C2 domain–containing protein) and possibly other proteins (Y). Cdc15 binds to Pxl1 as well. In the absence of the SH3 domain of Cdc15, some proteins (Fic1, Pxl1, and Y) continue to localize to the contractile ring through Imp2, whereas other Cdc15-specific proteins (X) fail to localize. In the absence of the SH3 domains of either Imp2 or Cdc15, none of these proteins localize properly, resulting in cytokinetic failure. Bars, 5 μm.
Discussion
The majority of proteins known to associate with PCH family members do so through SH3 domain interactions (see table in Chitu and Stanley, 2007). By examining the role of the SH3 domain in the founding member of this family, S. pombe Cdc15, we determined that this domain plays a supportive but nonessential role in cytokinesis. Similarly, the SH3 domain of Imp2 is clearly not the most important domain for Imp2 function. Unexpectedly, however, we discovered that the SH3 domains of Cdc15 and Imp2 cooperate in recruiting contractile ring components that are, in total, required for ring anchoring, stability, and constriction. The partial redundancy in function revealed in this study might be an explanation for why siRNA screens in human and Drosophila melanogaster cells have yet to identify F-BAR proteins as important players in the cell division process in those organisms (Echard et al., 2004; Eggert et al., 2004; Kittler et al., 2007). Functional redundancy in the cell division process might be expected, as a robustly constructed and well-tethered contractile ring would decrease the likelihood of errors during cytokinesis.
What proteins are recruited through the Cdc15 and Imp2 SH3 domains? We have identified two here, Pxl1 and Fic1, but there are most likely others. Biochemical purification of Cdc15 SH3 domain–interacting proteins identified a dozen candidates that contain multiple PXXP motifs and localize to the contractile ring and/or cell tips (unpublished data). Some nonoverlapping partners might also exist based on our findings that Imp2 (but not Cdc15) can be pulled from lysates by recombinant ubiquitin, presumably through its SH3 domain (Fig. S2 D), that a small percentage (2.3%) of cdc15-imp2SH3 fusion–expressing cells still show cytokinetic defects, and that Fic1 binds the two SH3 domains with somewhat different affinities. On this note, it will be informative to compare the overall binding specificities of these two SH3 domains even though the SH3 domain of Imp2 seems largely capable of replacing that of Cdc15. Clearly, the next step in expanding our understanding of how a mature contractile ring is constructed is to characterize other binding partners for the Cdc15 SH3 domain, to determine whether each of these proteins binds to Imp2, and also to investigate whether other domains within the SH3-interacting proteins provide additional protein-tethering and/or membrane-binding capabilities.
What roles do SH3-binding proteins play at the contractile ring? Consistent with previous work emphasizing a role for Cdc15 in anaphase (Wachtler et al., 2006), loss of just the Cdc15 SH3 domain results in cells taking longer to undergo constriction and abscission. This late role for the SH3 domain of Cdc15 suggests that some of its binding partners might play roles in ring remodeling or vesicle trafficking. Fic1 might contribute to contractile ring stability by reinforcing interaction between the ring and membrane through its C2 domain; this model is supported by evidence that the fic1 homologue in C. neoformans, cts1, binds phospholipids (Fox et al., 2003). If the Fic1 C2 domain binds membranes, this would mean that Cdc15 and Imp2 bind the plasma membrane directly through their N-terminal F-BAR domains and indirectly through their SH3 domains. We note that a significant fraction of Cdc15 is immobile in the ring as determined by FRAP analysis (Fig. 3 D; Clifford et al., 2008) and that this property of Cdc15 is compromised with the loss of the SH3 domain. Higher order redundancy of membrane tethering at the contractile ring might secure the ring, especially when forces increase during constriction. S. cerevisiae Inn1 tethers the contractile ring to the plasma membrane (Sanchez-Diaz et al., 2008), and this tethering may be accomplished through the F-BAR protein Hof1 as a result of their interaction in yeast two-hybrid assays (Ito et al., 2001; Tong et al., 2002). Unlike Fic1, Inn1 plays an essential role in S. cerevisiae cytokinesis. One possibility for differences in the relative requirements for fic1 and INN1 might be that the actomyosin ring is connected to the cell cortex by many more proteins in S. pombe than in S. cerevisiae. Fic1 shares some similarity to human MCTP1/2 (26% and 27%, respectively) and TOLLIP1; although these proteins have no described role in cytokinesis, it is intriguing to think that they (and/or another of the 343 C2 domain–containing human proteins) might be acting orthologously to Fic1, Inn1, and Cts1. Also, although no clear paralogue of Fic1 exists in S. pombe, 5 of its 15 C2 domain–containing proteins localize to the contractile ring or septum (Matsuyama et al., 2006), raising the possibility that they might act in concert with Fic1 during S. pombe cytokinesis.
The N-terminus of Pxl1 has been shown previously to target it to the contractile ring (Pinar et al., 2008). This is consistent with our findings that the N terminus of Pxl1 binds the SH3 domain of Cdc15 and that Pxl1 requires SH3 domains to localize. Paxillins are large scaffolding proteins important for recruiting other adaptors and enzymes to focal adhesions (for review see Schaller, 2001), and, therefore, it is likely that once Pxl1 has been recruited through SH3 domain interactions, it complexes with yet other proteins to anchor and extend the contractile ring. Interestingly, paxillin was recently found to be essential for mammalian cytokinesis (Shafikhani et al., 2008).
When are SH3 domain–interacting proteins recruited by Cdc15 and Imp2? Imp2 localizes to the contractile ring before anaphase onset but does not localize to nodes (Demeter and Sazer, 1998; unpublished data), suggesting that its recruitment to the contractile ring occurs after that of Cdc15. Likewise, neither Pxl1 nor Fic1 localizes to nodes (Fig. 5, A and B; Ge and Balasubramanian, 2008), indicating that SH3 domain binding partners are not recruited concomitantly with Cdc15. We note that although cdc15ΔSH3 imp2ΔSH3 cells form nascent contractile rings, rings were not observed in any cdc15ΔSH3 imp2ΔSH3 cells beginning to septate. We speculate that when forces on the ring increase at the onset of constriction and when scaffolding protein Mid1 leaves the contractile ring (Sohrmann et al., 1996), the integration of additional proteins might be especially important to prevent ring collapse. What regulates the timing of recruitment of these proteins? Given that Cdc15, Imp2, Fic1, and Pxl1 are all phosphoproteins (Fankhauser et al., 1995; unpublished data), we suspect that phosphoregulation is involved. Also, among the many proteins recruited to the site of cell division are other modifiers of protein function, including ubiquitinating and deubiquitinating enzymes (Hermand et al., 2003; Matsuyama et al., 2006).
Importantly, in addition to exposing the shared role of the SH3 domains of Cdc15 and Imp2, we have shown that Cdc15ΔSH3 can perform the essential function of Cdc15; our previous observation that Cdc12 and Myo1 are recruited by the F-BAR domain of Cdc15 (Carnahan and Gould, 2003) might be key to its essential role in contractile ring assembly. It is noteworthy that Imp2 cannot compensate for this essential role, even when overproduced (Demeter and Sazer, 1998). Imp2ΔSH3 also fulfilled the major cytokinetic role of Imp2, suggesting that its key function in cytokinesis (Demeter and Sazer, 1998) is also performed by another domain. Based on the presence of F-BAR domains, one common function of the Cdc15 and Imp2 N termini is likely to be inducing or maintaining membrane curvature. That two PCH family members contribute to cytokinesis with a common SH3 domain function raises the question of whether their F-BAR domains cause similar or distinct membrane curvatures. If the curvatures generated by the two F-BAR domains differ, perhaps it is important to balance their presence temporally to achieve membrane remodeling events key to successful cytokinesis.
In light of the unexpected functional overlap between the SH3 domains of Cdc15 and Imp2 as well as the distinction in function between their F-BAR and SH3 domains, it will be interesting to consider for other PCH family proteins (a) to what extent SH3 domains are required for endogenous functions, (b) to what extent SH3 domains of proteins in this family act redundantly, and (c) whether F-BAR domains are involved in additional protein–protein interactions. Because some family members such as FCHO1/2 and PSTPIP2 and alternatively spliced versions of Cip4 lack SH3 domains entirely (Wu et al., 1998; Dombrosky-Ferlan et al., 2003; Katoh and Katoh, 2004), it would not be surprising to find that many PCH proteins engage in protein–protein interactions mediated by their F-BAR domains or else the gulf of protein sequence between F-BAR and SH3 domains, the function of which is undefined in any family member. The shared essential function of the SH3 domains of Cdc15 and Imp2 shown in this study, combined with the observation that many PCH family members in higher eukaryotes bind the same proteins through their SH3 domains (for review see Chitu and Stanley, 2007) suggest a degree of cooperation between PCH family members as a whole that might be necessary for the completion of a variety of biological processes that involve membrane curvature remodeling.
Materials and methods
Yeast strains, media, and genetic methods
Strains (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1) were grown in yeast extract (YE) media (Moreno et al., 1991) except for the large-scale purification, which was grown in 2 liters of 4× YE media. nda3-km311 cells were grown at 32°C and arrested at 17°C for 7 h. cdc15, fic1, imp2, and bzz1 were tagged endogenously at the 3′ termini with GFP, CFP, YFP, TAP, FLAG3, HA3, mCherry, or linker-GFP (Sandblad et al., 2006) as previously reported (Bahler et al., 1998). A lithium acetate method was used for all yeast cell transformations (Keeney and Boeke, 1994). Strain construction and tetrad analysis were accomplished through standard methods. Strains were also supplied by F. Chang (Columbia Medical Center, New York, NY), M. Balasubramanian (Temasek Life Sciences Laboratory, Singapore), David Kovar (University of Chicago, Chicago, IL), and Pilar Pérez (Universidad de Salamanca, Salamanca, Spain).
Truncations of cdc15 and imp2 were generated by PCR-mediated insertions of stop codons or epitope tags after amino acids 752 and 869 of Cdc15 and 499 of Imp2 (Bahler et al., 1998). For integration at the endogenous locus, pIRT2 cdc15ΔSH3 or cdc15-imp2SH3 with 5′ and 3′ noncoding regions was used to rescue cdc15Δ. Integrants were recovered based on resistance to 5-fluorourotic acid, and correct integration was verified by PCR. fic1+ was deleted by homologous recombination as previously described (Bahler et al., 1998).
For spore germination experiments, mating colonies were digested with glusulase in water. Spores were allowed to recover in YE at 32°C before overnight incubation at 25°C in the presence or absence of 50–100 μg/ml hygromycin.
Molecular biology methods
To integrate cdc15ΔSH3, the 5′ noncoding region of cdc15 was amplified by PCR from genomic DNA using oligonucleotides containing 5′ Sph1 and 3′ Pst1 sites and cloned into pIRT2 (Hindley et al., 1987), and an Nde1 site was introduced at the end of the flank immediately before the Pst1 site by site-directed mutagenesis. All oligonucleotides were purchased from Integrated DNA Technologies. The 3′ noncoding region was amplified by PCR from genomic DNA with oligonucleotides containing 5′ BamHI and 3′ Sma1 sites. Finally, a cdc15 cDNA (encoding amino acids 1–752 plus a stop codon) with 5′ Nde1 and 3′ BamH1 site was cloned into the new vector using these sites. For integration of cdc15-imp2SH3, DNA encoding amino acids 610–670 of Imp2 was cloned behind the codon for amino acid 869 of Cdc15, and the restriction site was removed by site-directed mutagenesis. The aforementioned pIRT2/flanks construct was used as a backbone.
cdc15 (amino acids 752–927) was amplified by PCR from a cDNA-containing plasmid and cloned into pGBT and pMAL2c vectors using EcoRI and BamHI sites. imp2 (amino acids 584–670) was amplified by PCR from a cDNA library and subcloned into pMAL2c using BamHI and Not1 sites. fic1+ was amplified from genomic DNA and cloned into pSK, and its intron was removed using a QuikChange Site-Directed Mutagenesis kit (Agilent Technologies). fic1+ cDNA was then subcloned using Ase1 sites at both ends into a bacterial expression vector that produced a C-terminal His6-tagged protein. DNA encoding amino acids 1–257 of Pxl1 was amplified from genomic DNA, and introns were removed by site-directed mutagenesis. The resulting fragment was cloned into pGEX-2T using BamHI restriction sites. All clones were sequenced in their entirety to ensure that mutations had not been introduced. Vectors to express ubiquitin tagged with GST at both N (GST-ubiquitin) and C (ubiquitin-GST) termini were a gift from L. Hicke (Northwestern University, Evanston, IL; Stamenova et al., 2007).
Yeast two-hybrid screen
For construction of the S. pombe cDNA library, mRNA was purified from wild-type cells, and cDNA was synthesized with the ZAP-cDNA kit (Agilent Technologies) using random primers. The resulting cDNA was ligated with EcoRI adaptors, and only 0.5–1.5-kb-long cDNAs were ligated into pGAD424 at its EcoRI site. The resulting plasmids were transformed and pooled, producing an amplified library of 108 cfu/ml.
Yeast two-hybrid assays were performed as described previously (James et al., 1996). S. cerevisiae strain PJ69-4A containing the pGBT-cdc15SH3 plasmid was transformed with the pGAD cDNA library and interacting pairs selected on media lacking histidine; plasmids were isolated as described previously (Robzyk and Kassir, 1992).
Protein methods
Cell pellets were frozen in a dry ice/ethanol bath and lysed by bead disruption in NP-40 lysis buffer in either native or denaturing conditions as previously described (Gould et al., 1991), except with the addition of 0.1–0.5 mM diisopropyl fluorophosphate (Sigma-Aldrich). Proteins were immunoprecipitated by anti-HA (12CA5) or anti-FLAG (M2; Sigma-Aldrich) antibodies or a serum raised against GST-Cdc15 (amino acids 1–405; VU326; Cocalico Biologicals). Western blot analysis was performed as previously described (Wolfe et al., 2006) except that secondary antibodies were conjugated to Alexa Fluor 680 (Invitrogen) or IRDye800 (LI-COR Biosciences) and visualized using an Odyssey machine (LI-COR Biosciences).
For purification of Fic1-TAP, cells were lysed as described previously (Tasto et al., 2001), and protein was purified on tosylactivated Dynabeads (Invitrogen) coated with rabbit IgG (MP Biomedicals). Purified complexes were washed thoroughly and eluted with 0.5 M NH4OH and 0.5 mM EDTA or by TEV protease (Invitrogen) cleavage at 18°C for 1 h for use in immunoprecipitation experiments. For MS analysis, proteins were resuspended in 100 mM NH4HCO3 and 8 M urea, reduced and alkylated with Tris (2-carboxyethyl) phosphine and iodoacetamide, and digested with sequencing-grade trypsin (Promega) after decreasing to 2 M urea. MS was performed as previously described (McDonald et al., 2002) with the following modifications. Peptides were loaded onto columns with a pressure cell and were separated and analyzed by three-phase multidimensional protein identification technology on a linear trap quadrupole instrument (Thermo Electron). An autosampler (FAMOS) was used for 12 salt elution steps, each with 2 μl ammonium acetate. Each injection was followed by elution of peptides with a 0–40% acetonitrile gradient except the first and last injections, in which a 0–90% acetonitrile gradient was used. Eluted ions were analyzed by one full precursor MS scan (400–2,000 mass-to-charge ratio) and four tandem MS scans of the most abundant ions detected in the precursor MS scan under dynamic exclusion. The S. pombe database was searched using the SEQUEST algorithm, and results were processed using the CHIPS program (jointly developed by the Vanderbilt University Mass Spectrometry Research Center and University of Arizona). Filter settings for peptides were Xcorr ≥ 1.8 for singly charged, Xcorr ≥ 2.5 for doubly charged, and Xcorr ≥ 3.3 for triply charged.
Recombinant proteins were produced in chemically competent BL21 cells and purified on GST-Bind Resin (EMD), amylose beads (New England Biolabs, Inc.), or His-Bind resin (EMD) according to the manufacturers' protocols. For in vitro binding assays, recombinant proteins were incubated together for 1 h at 4°C. For lysate binding assays, cell lysates were incubated with bead-bound protein for 1 h at 4°C. In both cases, beads were washed thoroughly, and proteins were resolved by SDS-PAGE for Coomassie staining or Western blot analysis. Binding affinity experiments were performed as described previously (Disanza et al., 2004).
Microscopy methods
Cells in midlog growth were imaged live at 25°C using a spinning disk confocal microscope (Ultraview LCI; PerkinElmer) with a 100× NA 1.40 Plan-Apochromat oil immersion objective and a 488-nm argon ion laser (GFP and YFP), a 442-nm helium cadmium laser (CFP), or a 594-nm helium neon laser (mCherry). Images were captured on a charge-coupled device camera (Orca-ER; Hamamatsu Photonics) and processed using Metamorph 7.1 software (MDS Analytical Technologies). Z-section slices were 0.5 μm. For time-lapse imaging, cells on YE agar pads were sealed using Valap (a Vaseline, lanolin, and paraffin mixture). Images were captured at 150-s intervals, and kymographs were created with Metamorph 7.1 software. Differences were determined by Student's t tests. For induction of acyl-GFP, cells were grown without thiamine for 20 h before imaging. For cps1-191 arrest, cells were incubated at 36°C for 4 h before imaging.
Alternatively, cells were fixed in 70% ethanol and stained with DAPI (Chang et al., 2001) and 1 mg/ml methyl blue. Cells were imaged using a microscope (Axioskop II; Carl Zeiss, Inc.) with a 100× NA 1.40 Plan-Apochromat oil immersion objective and a halogen lamp. Images were processed using OpenLab 4.0.3 software (PerkinElmer). Images of colonies were captured with a camera (PowerShot SD750; Canon) through a microscope (Universal; Carl Zeiss, Inc.) with a 20× NA 0.32 objective.
FRAP experiments were performed as described previously (Clifford et al., 2008) except that a square (11 × 11 U) region was bleached. Data were analyzed using laser-scanning microscopy software (Carl Zeiss, Inc.) and averaged for plotting on Prism 4.0c software (GraphPad Software, Inc.). Best-fit curves were generated and used to obtain measurements of mobile fraction (Fm) and t1/2. Fm was calculated as the amount recovered to Bmax as a percentage of the bleached signal. A Student's t test was used to evaluate whether results were significantly different for Fm and t1/2.
Other methods
Sequence alignments were formulated at http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html (Corpet, 1988).
Online supplemental material
Fig. S1 shows that the Cdc15 SH3 domain plays a role in cytokinesis. Fig. S2 shows the SH3 domain binding partner that was identified. Fig. S3 shows a characterization of Fic1. Fig. S4 shows a characterization of the interaction between Fic1 and Imp2. Fig. S5 shows the genetic interaction between pxl1Δ and fic1Δ and characterization of cdc15-imp2SH3 fusion. Video 1 shows Rlc1-GFP live cell imaging. Videos 2 and 3 show Rlc1-GFP live cell imaging in the cdc15ΔSH3 mutant and cdc15ΔSH3 imp2ΔSH3 double mutant backgrounds, respectively. Table S1 shows the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806044/DC1.
Acknowledgments
We would like to thank Liping Ren for her technical assistance, Drs. Fred Chang and Linda Hicke for plasmids used in this study, and Drs. Mohan Balasubramanian, David Kovar, and Pilar Pérez for sharing strains. We would also like to thank Dr. Dannel McCollum, Dr. Erfei Bi, and members of the Gould laboratory for critically reading this manuscript and for many helpful discussions. We are also grateful to the Vanderbilt University Medical Center Cell Imaging Shared Resource Core for training.
R.H. Roberts-Galbraith has been supported by National Science Foundation fellowship DGE-02387141. This work was supported by the Howard Hughes Medical Institute, of which K.L. Gould is an investigator.
Abbreviations used in this paper: MBP, maltose-binding protein; MS, mass spectrometry; PCH, S. pombe cdc15 homology; TAP, tandem affinity purification; TEV, tobacco etch virus; YE, yeast extract.
References
- Arai, R., and I. Mabuchi. 2002. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 115:887–898. [DOI] [PubMed] [Google Scholar]
- Bahler, J., J.Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie III, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., and U. Gruneberg. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860. [DOI] [PubMed] [Google Scholar]
- Carnahan, R.H., and K.L. Gould. 2003. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 162:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur, S., N. Bordes, V. Fraisier, P.T. Tran, and A. Paoletti. 2004. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol. Cell. Biol. 24:10621–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F. 1999. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr. Biol. 9:849–852. [DOI] [PubMed] [Google Scholar]
- Chang, F., A. Woollard, and P. Nurse. 1996. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109:131–142. [DOI] [PubMed] [Google Scholar]
- Chang, L., and K.L. Gould. 2000. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA. 97:5249–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, L., J.L. Morrell, A. Feoktistova, and K.L. Gould. 2001. Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol. Cell. Biol. 21:6681–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu, V., and E.R. Stanley. 2007. Pombe Cdc15 homology (PCH) proteins: coordinators of membrane-cytoskeletal interactions. Trends Cell Biol. 17:145–156. [DOI] [PubMed] [Google Scholar]
- Clifford, D.M., B.A. Wolfe, R.H. Roberts-Galbraith, W.H. McDonald, J.R. Yates III, and K.L. Gould. 2008. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J. Cell Biol. 181:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter, J., and S. Sazer. 1998. imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J. Cell Biol. 143:415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza, A., M.F. Carlier, T.E. Stradal, D. Didry, E. Frittoli, S. Confalonieri, A. Croce, J. Wehland, P.P. Di Fiore, and G. Scita. 2004. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat. Cell Biol. 6:1180–1188. [DOI] [PubMed] [Google Scholar]
- Dombrosky-Ferlan, P., A. Grishin, R.J. Botelho, M. Sampson, L. Wang, W.A. Rudert, S. Grinstein, and S.J. Corey. 2003. Felic (CIP4b), a novel binding partner with the Src kinase Lyn and Cdc42, localizes to the phagocytic cup. Blood. 101:2804–2809. [DOI] [PubMed] [Google Scholar]
- Echard, A., G.R. Hickson, E. Foley, and P.H. O'Farrell. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14:1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert, U.S., A.A. Kiger, C. Richter, Z.E. Perlman, N. Perrimon, T.J. Mitchison, and C.M. Field. 2004. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., A. Reymond, L. Cerutti, S. Utzig, K. Hofmann, and V. Simanis. 1995. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 82:435–444. [DOI] [PubMed] [Google Scholar]
- Fox, D.S., G.M. Cox, and J. Heitman. 2003. Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryot. Cell. 2:1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, W., and M.K. Balasubramanian. 2008. Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Mol. Biol. Cell. 19:1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M. 2005. The molecular requirements for cytokinesis. Science. 307:1735–1739. [DOI] [PubMed] [Google Scholar]
- Gould, K.L., S. Moreno, D.J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10:3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, J., B. Garcia, E.J. Stollar, A. Rath, B.J. Andrews, and A.R. Davidson. 2007. The biologically relevant targets and binding affinity requirements for the function of the yeast actin-binding protein 1 Src-homology 3 domain vary with genetic context. Genetics. 176:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermand, D., S. Bamps, L. Tafforeau, J. Vandenhaute, and T.P. Makela. 2003. Skp1 and the F-box protein Pof6 are essential for cell separation in fission yeast. J. Biol. Chem. 278:9671–9677. [DOI] [PubMed] [Google Scholar]
- Hindley, J., G. Phear, M. Stein, and D. Beach. 1987. Sucl+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol. Cell. Biol. 7:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka, Y., T. Toda, and M. Yanagida. 1984. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 39:349–358. [DOI] [PubMed] [Google Scholar]
- Ho, H.Y., R. Rohatgi, A.M. Lebensohn, M. Le, J. Li, S.P. Gygi, and M.W. Kirschner. 2004. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 118:203–216. [DOI] [PubMed] [Google Scholar]
- Ishijima, S.A., M. Konomi, T. Takagi, M. Sato, J. Ishiguro, and M. Osumi. 1999. Ultrastructure of cell wall of the cps8 actin mutant cell in Schizosaccharomyces pombe. FEMS Microbiol. Lett. 180:31–37. [DOI] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., K.S. Erdmann, A. Roux, B. Habermann, H. Werner, and P. De Camilli. 2005. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 9:791–804. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki, T., M. Osumi, and I. Mabuchi. 2007. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 178:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, M., and M. Katoh. 2004. Identification and characterization of human FCHO2 and mouse Fcho2 genes in silico. Int. J. Mol. Med. 14:327–331. [PubMed] [Google Scholar]
- Keeney, J.B., and J.D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 136:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler, R., L. Pelletier, A.K. Heninger, M. Slabicki, M. Theis, L. Miroslaw, I. Poser, S. Lawo, H. Grabner, K. Kozak, et al. 2007. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9:1401–1412. [DOI] [PubMed] [Google Scholar]
- Landgraf, C., S. Panni, L. Montecchi-Palazzi, L. Castagnoli, J. Schneider-Mergener, R. Volkmer-Engert, and G. Cesareni. 2004. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff, X., A. Woollard, and V. Simanis. 1999. Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet. 262:163–172. [DOI] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li. 2000. Involvement of PCH family proteins in cytokinesis and actin distribution. Microsc. Res. Tech. 49:168–172. [DOI] [PubMed] [Google Scholar]
- Liu, J., H. Wang, D. McCollum, and M.K. Balasubramanian. 1999. Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 153:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., X. Song, D. He, C. Komma, A. Kita, J.V. Virbasius, G. Huang, H.D. Bellamy, K. Miki, M.P. Czech, and G.W. Zhou. 2006. Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2alpha. J. Biol. Chem. 281:4254–4260. [DOI] [PubMed] [Google Scholar]
- Matsuyama, A., R. Arai, Y. Yashiroda, A. Shirai, A. Kamata, S. Sekido, Y. Kobayashi, A. Hashimoto, M. Hamamoto, Y. Hiraoka, et al. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24:841–847. [DOI] [PubMed] [Google Scholar]
- Mayer, B.J. 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114:1253–1263. [DOI] [PubMed] [Google Scholar]
- McDonald, W.H., R. Ohi, D.T. Miyamoto, T.J. Mitchison, and J.R. Yates III. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219:245–251. [Google Scholar]
- Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823. [DOI] [PubMed] [Google Scholar]
- Naqvi, S.N., Q. Feng, V.J. Boulton, R. Zahn, and A.L. Munn. 2001. Vrp1p functions in both actomyosin ring-dependent and Hof1p-dependent pathways of cytokinesis. Traffic. 2:189–201. [DOI] [PubMed] [Google Scholar]
- Pinar, M., P.M. Coll, S.A. Rincón, and P. Pérez. 2008. Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol. Biol. Cell. 19:1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann, B., and R.B. Kelly. 2000. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J. Cell Biol. 148:1047–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk, K., and Y. Kassir. 1992. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20:3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz, A., V. Marchesi, S. Murray, R. Jones, G. Pereira, R. Edmondson, T. Allen, and K. Labib. 2008. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat. Cell Biol. 10:395–406. [DOI] [PubMed] [Google Scholar]
- Sandblad, L., K.E. Busch, P. Tittmann, H. Gross, D. Brunner, and A. Hoenger. 2006. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 127:1415–1424. [DOI] [PubMed] [Google Scholar]
- Schaller, M.D. 2001. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 20:6459–6472. [DOI] [PubMed] [Google Scholar]
- Shafikhani, S.H., K. Mostov, and J. Engel. 2008. Focal adhesion components are essential for mammalian cell cytokinesis. Cell Cycle. 7:2868–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann, M., C. Fankhauser, C. Brodbeck, and V. Simanis. 1996. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 10:2707–2719. [DOI] [PubMed] [Google Scholar]
- Stamenova, S.D., M.E. French, Y. He, S.A. Francis, Z.B. Kramer, and L. Hicke. 2007. Ubiquitin binds to and regulates a subset of SH3 domains. Mol. Cell. 25:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R.B., and S.R. Sprang. 1998. Structure of the protein kinase Cbeta phospholipid-binding C2 domain complexed with Ca2+. Structure. 6:1395–1405. [DOI] [PubMed] [Google Scholar]
- Tasto, J.J., R.H. Carnahan, W.H. McDonald, and K.L. Gould. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 18:657–662. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., B. Drees, G. Nardelli, G.D. Bader, B. Brannetti, L. Castagnoli, M. Evangelista, S. Ferracuti, B. Nelson, S. Paoluzi, et al. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 295:321–324. [DOI] [PubMed] [Google Scholar]
- Tsujita, K., S. Suetsugu, N. Sasaki, M. Furutani, T. Oikawa, and T. Takenawa. 2006. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 172:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis, D., J.Q. Wu, S. Hao, B. O'Shaughnessy, and T.D. Pollard. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100. [DOI] [PubMed] [Google Scholar]
- Wachtler, V., Y. Huang, J. Karagiannis, and M.K. Balasubramanian. 2006. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol. Biol. Cell. 17:3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B.A., and K.L. Gould. 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15:10–18. [DOI] [PubMed] [Google Scholar]
- Wolfe, B.A., W.H. McDonald, J.R. Yates III, and K.L. Gould. 2006. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell. 11:423–430. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q., J.R. Kuhn, D.R. Kovar, and T.D. Pollard. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q., V. Sirotkin, D.R. Kovar, M. Lord, C.C. Beltzner, J.R. Kuhn, and T.D. Pollard. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., D. Dowbenko, and L.A. Lasky. 1998. PSTPIP 2, a second tyrosine phosphorylated, cytoskeletal-associated protein that binds a PEST-type protein-tyrosine phosphatase. J. Biol. Chem. 273:30487–30496. [DOI] [PubMed] [Google Scholar]