Abstract
Sonic hedgehog (Shh) is an indispensable, extrinsic cue that regulates progenitor and stem cell behavior in the developing and adult mammalian central nervous system. Here, we investigate the link between the Shh signaling pathway and Hes1, a classical Notch target. We show that Shh-driven stabilization of Hes1 is independent of Notch signaling and requires the Shh effector Gli2. We identify Gli2 as a primary mediator of this response by showing that Gli2 is required for Hh (Hedgehog)-dependent up-regulation of Hes1. We also show using chromatin immunoprecipitation that Gli2 binds to the Hes1 promoter, which suggests that Hes1 is a Hh-dependent direct target of Gli2 signaling. Finally, we show that Shh stimulation of progenitor proliferation and cell diversification requires Gli2 and Hes1 activity. This paper is the first demonstration of the mechanistic and functional link between Shh, Gli, and Hes1 in the regulation of progenitor cell behavior.
Introduction
The neural retina represents an attractive model for investigating the molecular basis of progenitor cell proliferation and cell fate diversification in the central nervous system (CNS). The advantages of the retinal model system arise from the capacity for in vivo and in vitro analyses, the limited number of neurons and glial cell types comprising the mature retina, and the high degree of conservation of many developmental signaling mechanisms (Donovan and Dyer, 2005). The retinal cell types are derived in a temporal sequence from a common pool of multipotent progenitor cells (Young, 1985; Cepko et al., 1996). This conserved birth order is dependent on both intrinsic changes in competence of progenitor cells as well as cellular responses to environmental cues (Waid and McLoon, 1998; Belliveau et al., 2000; Zhang and Yang, 2001; Cayouette et al., 2003; Kim et al., 2005).
The Sonic hedgehog (Shh) pathway is a highly conserved cell extrinsic regulator of progenitor cell proliferation and diversification in many tissues, including the developing CNS and neural retina (Marti and Bovolenta, 2002; Dakubo and Wallace, 2004). Patched (Ptch) is the transmembrane receptor for Shh and normally antagonizes the activity of the transmembrane protein Smoothened (Smo), which is required for the activation of the Gli zinc finger transcription factors. Shh binding to Ptch alleviates the Ptch-mediated repression of Smo, allowing activation of Gli transcription factors and expression of target genes (Villavicencio et al., 2000).
In the mouse retina, Shh is secreted from postmitotic retinal ganglion cells (RGCs) and targets retinal progenitor cells (RPCs; Wang et al., 2005). Activation of the Shh pathway increases the proliferation of RPCs (Jensen and Wallace, 1997; Levine et al., 1997; Black et al., 2003; Moshiri and Reh, 2004; Moshiri et al., 2005; Wang et al., 2005), whereas conditional inactivation of Shh results in decreased numbers of progenitor cells, confirming a role for Shh in RPC proliferation (Wang et al., 2005). Genetic ablation of Shh in the embryonic mouse retina also results in increased RGC production, revealing a role for Shh signaling in cell fate regulation (Wang et al., 2005). During later stages of retinal development, loss of Shh signaling results in a reduction of Müller glial cells and bipolar neurons, which is only restored with Shh pathway activation, indicating a potential instructive role for Shh in specifying cell fate (Wang et al., 2002). Few Gli target genes important for these Shh-induced cellular responses have been identified. Cyclin D1, the major D-type cyclin expressed in the retina, is a reported target of Shh signaling during retinal development (Wang et al., 2005; Locker et al., 2006). However, loss of Cyclin D1 in the retina does not recapitulate the cell fate changes observed with loss of Shh signaling (Ma et al., 1998), which indicates that other unidentified targets of Shh/Gli signaling are necessary for establishing Shh-dependent effects.
Hes1 is a basic helix-loop-helix (bHLH) repressor and functions as a target of Notch signaling, a pathway that plays a key role in maintaining neural progenitor identity (Kageyama et al., 2005). There are several redundant functions of Shh and Hes1 during CNS and retinal development that suggest convergence of these pathways. For example, Hes1 has been implicated in regulating cell cycling in the chick retina and Müller cell development in the mouse retina (Furukawa et al., 2000; Takatsuka et al., 2004; Hashimoto et al., 2006). Also, the retinas of Hes1 and Shh mutants are phenotypically similar, as both are characterized by an increased production of RGCs, precocious cell cycle exit, and depletion of RPCs (Takatsuka et al., 2004; Wang et al., 2005). In contrast, conditional Notch1 mouse mutants are characterized by a propensity to develop cone photoreceptors without an increase in RGC development (Jadhav et al., 2006; Yaron et al., 2006). The differing phenotypes resulting from loss of Notch and Hes1 in the retina suggest that Hes1 may have Notch-independent roles in retinal development. Furthermore, preliminary observations indicate that Shh signaling may influence the maintenance of Hes1 expression in the retina (Wang et al., 2005).
Here, we establish a novel, Shh-dependent regulatory mechanism for controlling neural progenitor cell behavior. Inhibition of Hes1 activity results in a decrease in RPC proliferation as well as a disruption of neuronal cell development in response to Shh pathway activation. Furthermore, the increased proliferation characterizing PtchlacZ+/− retinas is rescued in compound PtchlacZ+/−Hes1+/− heterozygous mice, which suggests that Hes1 is epistatic to Ptch and is required to potentiate the proliferative response induced by the Shh pathway in vivo. We show that modulation of Hes1 by Shh requires signaling through the activator Gli2 and is independent of the Notch pathway. Finally, chromatin immunoprecipitation (ChIP) analysis suggests that Hes1 is a Shh-dependent, direct transcriptional target of Gli2. Thus, we have identified a novel mechanism linking Shh, Gli2, and Hes1 that is important for controlling neural progenitor cell proliferation.
Results
Hes1 is activated by Shh signaling in a temporally regulated manner
To investigate the Shh dependence of Hes gene expression in RPCs, we used retina organ cultures (explants) derived from postnatal mice (Fig. 1 a). Key features of normal retinal development, including critical cell fate decisions, are recapitulated in retinal cell explant cultures (Zhang et al., 2002). We have shown previously that Shh induction of target genes is abolished in postnatal mouse retinal explants due to the death of Shh-secreting RGCs because of a lack of trophic support from target tissues in the CNS (Wang et al., 2002). We treated retinal explants with a Smo agonist (Frank-Kamenetsky et al., 2002), which restores Hedgehog (Hh) target gene expression (Wang et al., 2005), and analyzed Hes induction by quantitative RT-PCR (RT-qPCR). Activation of the Shh pathway in retinal explants derived from postnatal mice results in an ∼20-fold induction of Hes1 mRNA (Fig. 1 b). Interestingly, Shh signaling in explants cultured from embryonic day 14 (E14) retinas resulted in only a twofold induction of Hes1 mRNA, which indicates temporal regulation of the magnitude of Hes1 expression by the Shh pathway. The modest induction of Hes1 at E14 is not caused by a lack of progenitor cell competence to respond to Shh because the Hh target gene, Gli1, is potently activated by Shh in both E14 and postnatal retinal explants (Fig. 1 b). These data indicate that Hes1 is inducible by Shh signaling at developmental stages when Shh is regulating both RGC development and RPC proliferation. A similar temporal regulatory pattern was observed for Hes5 in response to Shh signaling. Shh activation resulted in a fivefold induction of Hes5 mRNA in postnatal retinal explants, whereas no significant induction was observed in E14 explants (Fig. 1 b). Shh pathway activation also results in stabilization of Hes1 protein (Fig. 1 c), whereas Hes1 protein was undetectable in untreated explants devoid of Shh ligand, which indicates that an active Hh pathway is necessary for the maintenance of Hes1 in RPCs.
Figure 1.
Shh is required to maintain Hes1 protein and mRNA in postnatal retinal explants. (a) Diagram of the retinal explant culture method. Once the retina is surgically detached from the lens and surrounding ocular tissues, it is flattened by making four incisions and cultured on a membrane in the presence of a Smo agonist to activate the Hh signaling pathway. A cross section of a postnatal retinal explant is shown demonstrating Gli1 transcript expression in the Hh-responsive progenitor cells of the neuroblast region. The RGC layer is comprised of a population of postmitotic neurons that are not responsive to Hh signaling. Bar, 100 μm. (b) Retinal explants were treated with and without a Smo agonist at E14 (n = 3) and P0 (n = 3) for 3 d in culture and analyzed for Hes1, Hes5, and Gli1 mRNA by RT-qPCR. Values represent fold mRNA induction in Smo agonist–treated explants relative to untreated explants. Error bars represent SEM. *, P < 0.05. (c) Western blot for Hes1 from P0 retinal explants cultured for 3 d from untreated and Smo agonist–treated explants; β-tubulin protein level was used as a loading control.
Shh activation of Hes1 and Hes5 does not require the active NICD
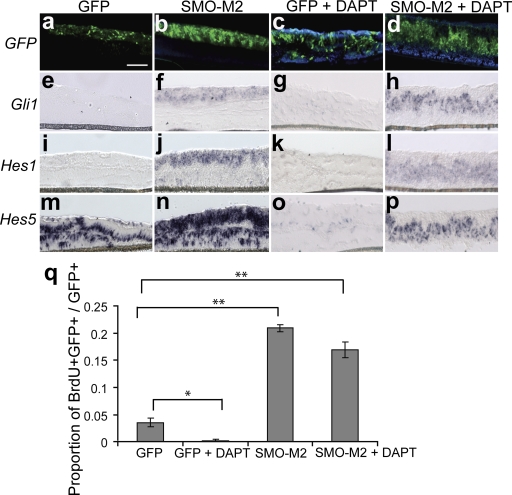
Notch is a transmembrane protein and requires cleavage by a γ-secretase complex to free its active intracellular domain (NICD) and induce target genes including Hes1 and Hes5 (Kageyama and Ohtsuka, 1999). To address whether Shh induction of Hes1 and Hes5 requires Notch signaling, we used a widely used chemical inhibitor of the γ-secretase complex, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT), to inhibit propagation of the Notch pathway (Geling et al., 2002). Hes5, which is robustly expressed in untreated retinal explants, was used as a read-out for Notch signaling. Hes5 was down-regulated in DAPT-treated retinal explants, which verifies Notch inactivation with DAPT treatment (Fig. 2, m and o) and is consistent with previous reports showing the Notch dependence of Hes5 expression in RPCs (Nelson et al., 2007). Activation of the Hh pathway was accomplished using a constitutively active allele of Smo (Smo-M2; Xie et al., 1998). Transfection of RPCs was accomplished using electroporation of transgenes in retinal explants. It has been previously found that transfection efficiency of postmitotic cells in electroporated retinal explants is very low, which indicates that dividing cells are the primary targets for electroporation (Matsuda and Cepko, 2004). Retinal explants were coelectroporated with Smo-M2 and pUb-GFP to localize the transfected cells, and simultaneously treated with DAPT for Notch pathway inactivation. In situ hybridization (ISH) for Gli1 confirmed Shh pathway activation in response to Smo-M2 (Fig. 2, f and h). Induction of Shh signaling with SMO-M2 resulted in a cell-autonomous increase in Hes1 and Hes5 mRNA in the presence of DAPT, which indicates that Shh activation of these genes is independent of the NICD (Fig. 2, l and p). It is also noteworthy that control retinal explants electroporated with pUb-GFP, which do not exhibit Shh signaling, do not have detectable Hes1 expression (Fig. 2 i). Because endogenous Hes5 expression is lost with DAPT treatment, this suggests that Notch signaling is active in control retinal explants yet insufficient to maintain Hes1 expression.
Figure 2.
Shh activation of Hes1 and Hes5 is independent of Notch signaling. Retinas at P0 were electroporated with SMO-M2 cotransfected with pUB-GFP or pUB-GFP alone and cultured for 3 d with DAPT or DMSO control. (a–d) GFP fluorescence localizes the transfected cells. ISH was performed for Gli1 (e–h), Hes1 (i–l), and Hes5 (m–p). Differences in the localization of transfected cells within the explants are caused by folding and twisting during tissue processing. Bars, 100 μm. (q) Retinal explants (P0 + 3 days in vitro [DIV]) were electroporated with Smo-M2/pUb-GFP, treated with DAPT, dissociated, and scored for the proportion of BrdU+GFP+/GFP+ cells. The magnitude of Smo-M2–induced proliferation is not changed with DAPT treatment. Error bars represent SEM. *, P < 0.05; **, P < 0.005.
We also tested whether Shh activation can induce cellular proliferation in RPCs independently of Notch signaling. Retinal explants were electroporated with SMO-M2, treated with DAPT, dissociated, and scored for BrdU incorporation. Antagonizing Notch activity did not significantly affect the proliferative effect of Shh signaling, which indicates that Shh can regulate progenitor cell behavior in a Notch-independent manner (Fig. 2 q).
Differential mechanism for the induction of Hes1 and Hes5 by Shh signaling
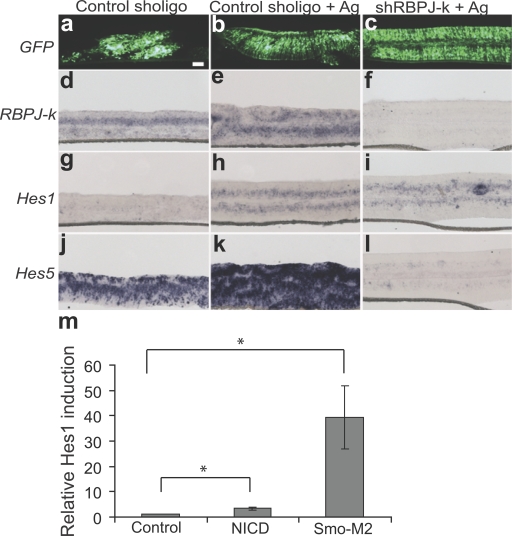
Although the previous experiment (Fig. 2) demonstrates that Shh can regulate Hes1 independently of NICD activity, it does not address whether other downstream effectors of the Notch pathway are responsible for mediating Shh-dependent Hes1 expression. We next asked whether the induction of Hes1 by the Shh pathway requires the activity of RBPJ-κ by investigating Hes1 induction in the context of RBPJ-κ knockdown. To control for the specificity of the shRBPJ-κ, we show that Notch (NICD; Nofziger et al., 1999)-mediated activation of a Notch reporter in retinal explants is abrogated by coexpression of shRBPJ-κ (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200805155/DC1). Retinal explants from postnatal day 0 (P0) mice were electroporated with an shRBPJ-κ plasmid and cotransfected with pUb-GFP to localize the transfected cohort of cells. Gli1 is induced in the presence of the control and shRBPJ-κ vectors, which indicates activation of the Shh pathway (unpublished data). ISH analysis revealed a marked reduction in RBPJ-κ mRNA in transfected cells, which was also associated with a cell-autonomous reduction of Hes5 mRNA, which provides additional confirmation of the specificity of the knockdown (Fig. 3, f and l). ISH for Hes1 demonstrates that Hes1 is induced in RPCs by the Smo agonist despite RBPJ-κ knockdown (Fig. 3 i). However, the Shh pathway is unable to induce Hes5 in cells expressing the shRBPJ-κ construct (Fig. 3 l). This result infers a differential mechanism for the modulation of Hes1 and Hes5 by Shh signaling. The induction of Hes1 via Shh is independent of the Notch signaling pathway, whereas induction of Hes5 requires functional RBPJ-κ signaling.
Figure 3.
RBPJ-κ signaling is not required for Shh induction of Hes1 but is necessary for Shh induction of Hes5. Retinal explants were electroporated with shRBPJ-κ or a control short hairpin plasmid at P0 and cultured for 4 d with or without a Smo agonist. (a–c) GFP fluorescence localizes the transfected cells. ISH was performed for RBPJ-κ (d–f), Hes1 (g–i), and Hes5 (j–l). Bar, 100 μm. (m) Retinal explants were coelectroporated with an empty vector control, NICD, or Smo-M2 and pUB-GFP, and cultured for 3 d, then Hes1 expression was analyzed by RT-qPCR. Values represent the relative induction of Hes1 expression normalized to GFP. Error bars represent SEM. *, P < 0.005.
Activated Notch signaling is a weak regulator of Hes1 in RPCs
Because Shh-mediated induction of Hes1 is independent of Notch signaling, we wanted to compare the efficiency of the Shh and Notch pathways in activating Hes1 in RPCs. The active NICD, Smo-M2, or an empty-vector control were coelectroporated with pUB-GFP in retinal explant cultures. Hes1 expression was analyzed using RT-qPCR and normalized to GFP to account for variations in transfection efficiency. Activation of the Hh pathway resulted in a 10-fold greater Hes1 induction compared with Notch pathway activation (Fig. 3 m), which indicates that Hes1 expression in RPCs is primarily responsive to the Shh pathway when compared with Notch signaling.
Shh induction of Hes1 requires signaling through activator Gli2
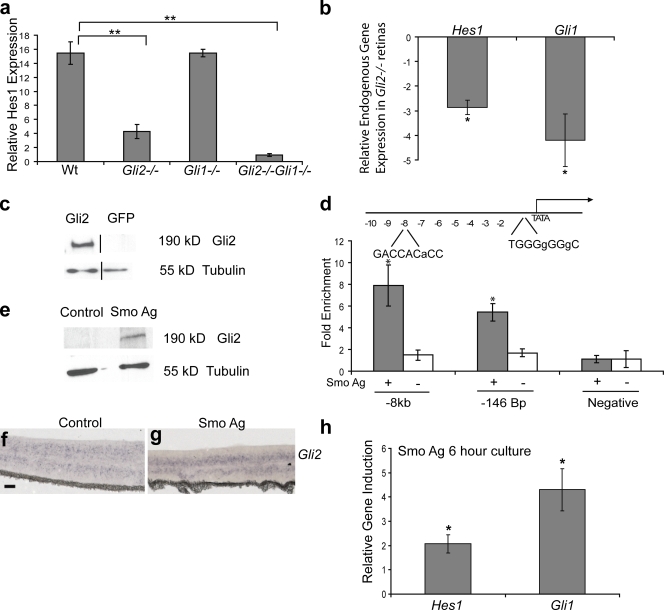
The major downstream activators of the Shh pathway are the Gli1 and Gli2 transcription factors, which activate target gene expression in response to Smo signaling (Park et al., 2000; Bai et al., 2002). Gli1 and Gli2 are also expressed in the Hh-responsive neuroblast region of the developing retina. To evaluate the role of activator Gli function in the regulation of Hes1, we generated Gli1−/−, Gli2−/−, and Gli1−/−Gli2−/− mutant mice. Retinal explants derived from these mice were cultured in the presence of Smo agonist and analyzed by RT-qPCR for induction of Hes1 mRNA. The Hh-mediated induction of Hes1 was reduced fourfold in Gli2−/− explants compared with wild-type explants, whereas loss of Gli1 signaling did not effect Hes1 induction (Fig. 4 a). These data indicate that Gli2 is the predominate factor responsible for Hes1 regulation downstream of Shh activation. However, Hes1 induction was completely attenuated in retinal explants cultured from Gli1−/−Gli2−/− compound mutant mice in response to Hh pathway activation, which indicates that Gli1 can minimally compensate for Hes1 activation in the absence of Gli2 signaling (Fig. 4 a). To establish a link between Gli2 signaling and Hes1 expression in vivo, we analyzed endogenous levels of Hes1 in acutely dissected Gli2−/− and wild-type E18 retinas (Fig. 4 b). Hes1 expression is down-regulated nearly threefold in Gli2−/− retinas compared with wild-type retinas, which establishes Gli2 as an important regulator of Hes1 expression in vivo.
Figure 4.
Shh induction of Hes1 requires Gli2. (a) Retinal explants were cultured from wild-type (Wt; n = 5), Gli1−/− (n = 3), Gli2−/− (n = 6), and Gli2−/−Gli1−/− (n = 3) mice with or without a Smo agonist at E18 for 3 d, then analyzed for Hes1 expression by RT-qPCR. Values represent fold mRNA induction in Smo agonist–treated explants relative to untreated explants. (b) RT-qPCR on acutely dissected retinas from E18 wild-type (n = 5) and Gli2−/− (n = 5) animals. Values represent fold mRNA induction in Gli2−/− retinas compared with the wild type. The black lines in the Western blot indicate that intervening lanes have been spliced out. (c) Western blot analysis of protein lysates from Myc-Gli2–transfected or control COS cells blotted with an anti-Gli2 antibody. The β-tubulin protein level was used as a loading control. (d) Schematic of the 10-kb region of the Hes1 promoter. The Gli2-binding sites are indicated with the mismatched nucleotides relative to the ideal Gli consensus sequence in small letters. ChIP reveals enrichment of Gli2 at sites −7,808 bp and −146 bp upstream of the transcription start site in the Hes1 promoter in retinal explants treated with a Smo agonist. No enrichment of Gli2 was detected at these sites in untreated retinal explants. Association of Gli2 at a region of the Hes1 promoter that does not contain a Gli consensus sequence was used as a negative control. (e) Western blot analysis for Gli2 on retinal explants treated with or without a Smo agonist (P0 + 3 DIV). The β-tubulin protein level was used as a loading control. (f and g) Retinal explants cultured with or without a Smo agonist for 3 DIV and subjected to ISH for Gli2. Bars, 100 μm. (h) Retinal explants (P0) were cultured with (n = 5) or without a Smo agonist (n = 4) for 6 h and analyzed for Hes1 and Gli1 expression by RT-qPCR. Values represent fold mRNA induction in Smo agonist–treated explants relative to untreated explants. Error bars represent SEM. *, P < 0.05; **, P < 0.001.
Because the induction of Hes1 by Shh does not require signaling through Notch but does require Gli2, we addressed whether Hes1 is a direct transcriptional target of Gli2. To study Hes1 regulation in vitro, we generated a luciferase reporter containing the Hes1 promoter and 10 kb of additional upstream sequence. The Hes1 reporter is not sufficient to mimic endogenous Hes1 activity by Hh signaling, which could indicate that we have not identified all of the relevant regulatory sequences or that the function of Gli2 in this context will not be revealed in simple reporter assays with nonchromatinized substrates (Kleinjan and van Heyningen, 2005; Ni et al., 2008). We therefore examined the possibility of Shh-mediated regulation of Hes1 in the context of native chromatin by performing ChIP. These experiments allowed us to determine whether Gli2 binds the Hes1 promoter in a physiologically relevant context. Candidate Gli consensus sequences were identified within a 10-kb region of the Hes1 promoter and analyzed for association with Gli2 in control and Smo agonist–treated retinal explants. Validation of the specificity of the Gli2 antibody used for ChIP analysis was tested by Western blotting using COS cells transfected with a full-length Gli2 expression plasmid or a GFP control plasmid (Fig. 4 c). Physical association of Gli2 at the Hes1 promoter was detected at two putative Gli consensus sequences located at −146 bp and −7,808 bp upstream of the transcription start site (Fig. 4 d). Enrichment for Gli2 at the Hes1 promoter was only detected in Smo agonist–treated explants, with no enrichment of Gli2 detected in untreated explants, which suggested that Hes1 is a Shh-dependent direct transcriptional target of Gli2 (Fig. 4 d).
The observation that Gli2 binds the Hes1 promoter only in the presence of Shh signaling may be attributed to the stability of Gli protein (Huntzicker et al., 2006; Pan et al., 2006). To test this hypothesis, we investigated whether we can detect endogenous Gli2 protein in RPCs in the absence of Hh signaling. Western blot analysis revealed that Gli2 protein is only detected in cultures with an active Hh pathway despite the presence of Gli2 transcript in untreated retinal explants (Fig. 4, e–g). This data indicates that Hh signaling is necessary for the stability of Gli2 protein in RPCs and accounts for the Hh-dependent binding of Gli2 to the Hes1 promoter.
To further explore the relationship between Shh signaling and Hes1 regulation, we examined the kinetics of Hes1 induction in retinal explants. Retinal explants treated with a Smo agonist for 6 h exhibited a significant induction of Hes1 compared with untreated explants, strengthening the evidence of a direct relationship between Hes1 and Hh signaling (Fig. 4 h). This induction of Hes1 is not caused by decay of Hh signaling in untreated explants because retinal explants cultured for 6 h do not exhibit a significant decrease in levels of Gli1 or Hes1 when compared with acutely dissected retinas (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200805155/DC1).
Hes1 is necessary for Shh-mediated proliferation and cell fate specification in RPCs
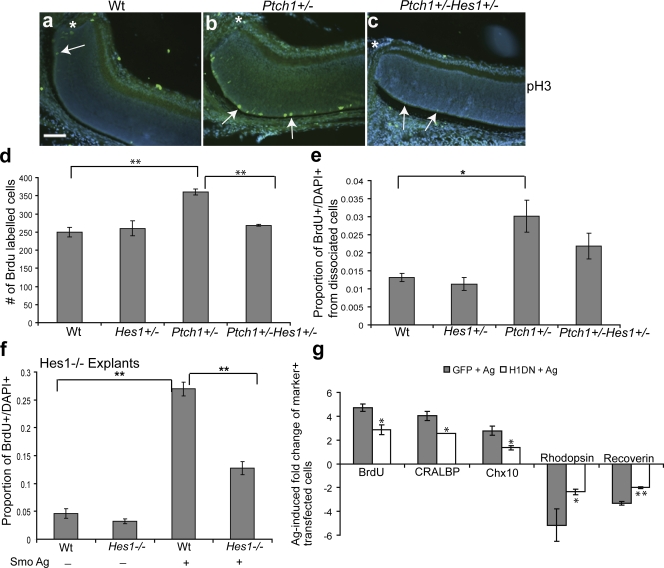
Because we have demonstrated a novel mechanism for Hes1 regulation by Shh-Gli2 signaling, we wanted to examine the physiological significance of Hes1 as a Shh target gene. To investigate this, we used PtchlacZ+/− mice, which exhibit constitutive Hh pathway activation that results in delayed cell cycle exit in the central retina of postnatal mice (Black et al., 2003; Moshiri and Reh, 2004). To directly evaluate whether Hes1 is necessary for the Hh-mediated proliferation of progenitor cells in vivo, we generated PtchlacZ+/−Hes1+/− compound heterozygous mice. We chose to work with mice heterozygous for Ptch because Ptch−/− mutants exhibit early embryonic lethality before retinal development (Goodrich et al., 1997). Immunohistochemistry (IHC) for the mitotic marker pH3 revealed that mitotic cells were reduced in the compound heterozygous retinas (PtchlacZ+/−Hes1+/−) compared with retinas from PtchlacZ+/− mice (Fig. 5, a–c). Furthermore, quantification of BrdU incorporation from retinal sections of PtchlacZ+/−Hes1+/− double heterozygous animals revealed a significant reduction in the total number of cells in S phase compared with PtchlacZ+/− retinas, which demonstrates that Hes1 is a mediator of Shh-dependent proliferation in the retina in vivo (Fig. 5 d). Also, quantification of the proportion of BrdU-labeled cells from dissociated retinas revealed a significant increase in proliferation in PtchlacZ+/− retinas compared with the wild type; however, there was no significant increase in proliferation in PtchlacZ+/−Hes1+/− retinas (Fig. 5 e). This data provides novel evidence for an in vivo genetic interaction between Hh signaling and Hes1 in the regulation of progenitor cell proliferation.
Figure 5.
Shh-mediated RPC proliferation and cell fate specification requires Hes1. (a–c) In vivo anti-pH3 staining of the central retina adjacent to the optic nerve (asterisks) in P5 wild-type (Wt), PtchlacZ+/−, and PtchlacZ+/−Hes1+/− retinas. Arrows indicate pH3-positive cells. Note that pH3+ cells in the vicinity of the optic nerve are rare in Wt and compound heterozygous mice. Bar, 100 μm. (d) Quantitative analysis of BrdU incorporation in vivo from P5 Wt (n = 3), Hes1+/− (n = 3), PtchlacZ+/− (n = 3), and PtchlacZ+/−Hes1+/− (n = 6) retinas. Values represent the mean number of BrdU-positive cells counted from three sections per animal. (e) Quantification of the proportion of BrdU+ cells in single-cell dissociates from the retinas of Wt (n = 5), Hes1+/− (n = 3), PtchlacZ+/− (n = 8), and PtchlacZ+/−Hes1+/− (n = 7) retinas at P5. (f) Retinal explants from Hes1−/− (n = 3) or Wt (n = 3) animals were treated with a Smo agonist for 3 d, dissociated, and scored for the proportion of BrdU+DAPI+ cells. (g) Quantitative analysis for BrdU, CRALBP, Chx10, rhodopsin, and recoverin-positive cells in Smo agonist–treated P0 retinal explants electroporated with GFP and Hes1DN. Values are based on scoring marker+ cells among the transfected cohort in dissociates from retinal explants and represent the fold induction of double-positive (marker+GFP+) cells in GFP + Ag and Hes1DN + Ag cultures compared with double-positive cells in GFP-transfected untreated explants. There is no difference in proliferation or cell type composition in GFP and Hes1DN-transfected cells in untreated explants. Error bars represent SEM. *, P < 0.05; **, P < 0.01.
We also investigated the proliferative response induced by Shh in Hes1−/− mutant retinal explants. Loss of Hes1 resulted in a significant decrease in Shh-mediated BrdU incorporation compared with wild-type explants (Fig. 5 f). To study whether acute inhibition of Hes1 activity antagonizes progenitor cell proliferation in response to Shh, we antagonized Hes1 activity using a previously characterized Hes1DN construct that carries three point mutations in the basic DNA-binding domain that interfere with its DNA-binding activity (Strom et al., 1997). The Hes1DN protein will dimerize with wild-type Hes1 and HesR proteins to form transcriptionally inactive complexes. Retinal explants were electroporated with the Hes1DN construct and cultured in the presence of a Smo agonist for 3 d in vitro. The explants were dissociated by enzymatic digestion followed by IHC for cell type–specific markers and scored for marker+ cells among the transfected cohort of cells, which was identified by GFP fluorescence. Normally, Smo agonist treatment of control electroporated RPCs results in an increased proportion of dividing progenitors compared with control explants (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200805155/DC1). BrdU incorporation in response to the agonist was reduced by 50% in the Hes1DN-expressing cohort compared with the GFP-expressing cells (Fig. 5 g), which indicates that Hes1 is required for Shh-mediated proliferation. This data also reinforces the hypothesis that Hes1 is a Notch-independent target of Shh, as inhibition of Notch signaling did not compromise Shh-induced proliferation (Fig. 2 q).
The Shh pathway also promotes the development of Müller glia and bipolar cells at the expense of rod photoreceptors (Fig. S3; Wang et al., 2005; Yu et al., 2006). As Hes1 has been implicated as a regulatory factor in promoting bipolar and Müller cell specification (Tomita et al., 1996; Furukawa et al., 2000; Takatsuka et al., 2004), we investigated whether Hes1 was required for the acquisition of specific cell fates downstream of Shh signaling. Retinal explants were electroporated with the Hes1DN construct or GFP, cultured with the Smo agonist for 7 d in vitro, and dissociated and scored using IHC for specific cell type markers. Significantly, there was a 50% reduction in the proportion of Müller (anti-cellular retinaldehyde-binding protein [CRALBP]) and bipolar cells (anti–Chx10) in agonist-treated Hes1DN-expressing cells compared with agonist-treated GFP-expressing cells (Fig. 5 g). The reduction in cells with a bipolar and Müller cell identity in the Hes1DN cohort was associated with an increase in the proportion of cells positive for rod photoreceptor markers rhodopsin and recoverin. Therefore, the Müller- and bipolar-promoting effects of Shh require Hes1.
Loss of Gli2 represses proliferation and cell fate specification in response to Shh signaling, similar to inhibition of Hes1
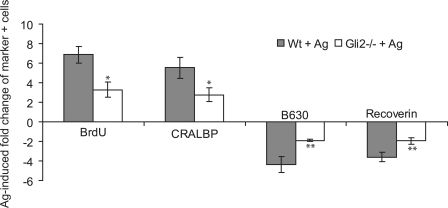
Because Gli2 is necessary for the induction of Hes1 by Shh signaling, we asked whether loss of Gli2 phenocopies the effect of Hes1 inhibition in the context of an activated Shh pathway. Gli2−/− mutant explants at E18 were cultured with the Smo agonist and analyzed for proliferation and the development of specific cell types. Proliferation was attenuated in Gli2−/−-treated explants relative to wild type–treated explants after 3 d (Fig. 6). Müller and bipolar cell development was also reduced in Gli2−/−-treated explants cultured for 7 d compared with control explants (Fig. 6), which demonstrates that Gli2 is required for Shh effects on cell type development. In contrast, Hh-mediated proliferation was normal in the absence of Gli1 correlating with normal Hes1 induction (unpublished data).
Figure 6.
Gli2 is required for the Shh effects on proliferation and cell fate. Retinal explants were cultured from wild-type (Wt; n = 3) and Gli2−/− (n = 3) mice at E18 for 3 d in culture with or without a Smo agonist. IHC was performed on dissociated cells using anti-BrdU, anti-CRALBP, anti-rhodopsin, and anti-recoverin antibodies. Values represent the fold induction of positive cells in Wt + Ag or Gli2−/− + Ag cultures compared with nontreated explants. Error bars represent SEM. *, P < 0.05; **, P < 0.01.
Discussion
In this study, we sought to identify the molecular mechanisms that Shh utilizes to regulate CNS progenitor cell behavior. We have used the neural retina as a model for CNS development to evaluate the mechanism and function of Hes1 as a putative Shh target gene. Hes1 is a key target of the Notch pathway, and its role during development is normally associated with activated Notch signaling. However, Hes1 mutant retinas do not mimic Notch1 mutants, which suggests that Hes1 may have Notch-independent roles in retinal development (Takatsuka et al., 2004; Jadhav et al., 2006; Yaron et al., 2006). This idea is corroborated by the persistent expression of Hes1 in Notch1 and RBPJ-κ mutant embryos and the identification of other factors capable of activating Hes1 (de la Pompa et al., 1997; Furukawa et al., 2000; Stockhausen et al., 2005; Nguyen et al., 2006; Ingram et al., 2008; Nakazaki et al., 2008). Also, inhibition of the Notch pathway in chick retinal explants results in a weak reduction of Hes1 expression and a much more potent reduction in Hes5 levels, which is further evidence for Notch-independent Hes1 regulation (Nelson et al., 2007). Shh treatment of cerebellar granule precursors results in induction of Hes1, which suggests a more general role for Shh in the regulation of Hes1 in CNS development (Solecki et al., 2001). Here, we show that Gli2 is a novel regulator of Hes1 expression and that Hes1 is a novel mediator of Shh-mediated proliferation in neural progenitors. Surprisingly, we have observed a differential mechanism for Shh-mediated regulation of Hes1 and Hes5. Induction of Hes5 is independent of Notch; however, it requires signaling by RBPJ-κ, suggesting that RBPJ-κ can function independently of Notch downstream of Shh activation. To date, Notch-independent RBPJ-κ activity has only been reported in Drosophila melanogaster mechanoreceptor physiology, mouse pancreas development, and the specification of GABAergic neurons (Barolo et al., 2000; Beres et al., 2006; Hori et al., 2008).
The requirement for both Shh- and Notch-driven regulation of Hes1 in progenitor cells may be explained by several hypotheses. First, Shh and Notch could be targeting different progenitor populations. A recent study has shown that the activated NICD is heterogeneously expressed in subsets of progenitors in the mouse retina (Nelson et al., 2007), which implies that not all progenitors are responsive to activated Notch signaling. Second, progenitor cells are sensitive to Hes1 dosage; therefore, Notch and Shh signaling may be required to achieve the spectrum of Hes1 levels needed for cell fate specification and proliferation. For example, Hes1+/− retinas exhibit accelerated rod photoreceptor differentiation without the proliferative or RGC phenotype characterizing Hes1−/− retinas (Takatsuka et al., 2004). Also, the decision to adopt an RGC fate is dependent on levels of proneuronal bHLHs NGN2 and Math5 as well as Hes1, with high levels of Hes1 antagonizing Math5 expression and function and thereby inhibiting the RGC fate (Matter-Sadzinski et al., 2005). Mechanistically, oscillation in Hes1 expression (Hirata et al., 2002) could be one way to achieve functionally relevant modulations in Hes1 levels in progenitor cells. Notch-induced Hes1 oscillation is required for the maintenance of neural progenitors (Shimojo et al., 2008), and it is conceivable that the effects of Hh signaling on Hes1 expression could be mediated by a similar mechanism. Further analysis is necessary to determine the mode of Hes1 expression in the context of Hh pathway activation.
We have identified a direct interaction of Gli2 and Hes1 at two Gli consensus sites in the Hes1 promoter. In these studies, we obtained strong evidence for a direct effect because ChIP analysis using primary RPCs revealed Shh-dependent recruitment of Gli2 to the Hes1 promoter in vivo. These findings, coupled with our strong genetic evidence linking Shh to Hes1, provide the first example of an interaction between Gli2 signaling and Hes1 expression, and the first example of the importance of this mechanism in the regulation of progenitor cell proliferation. Our findings also raise the possibility that Gli2-dependent regulation of proliferation in other tissues could function with a similar mechanism (Matise et al., 1998; Corrales et al., 2004; Palma and Ruiz i Altaba, 2004; Hutchin et al., 2005; Hu et al., 2006; Zhang et al., 2008). In addition to activator Gli1 and Gli2 function, the Gli3 transcription factor is also a mediator of the Shh pathway, and it functions to repress target genes in the absence of Shh signaling. Because loss of function of both Gli1 and Gli2 completely attenuates Shh-mediated induction of Hes1, it appears that derepression of Gli3 is not a primary regulator in this context.
Hh control of Hes1 appears to be an evolutionary conserved signaling mechanism involved in widespread tissue patterning. Hh drives expression of the D. melanogaster homologue of Hes1, hairy, along the dorsal/ventral axis of the leg imaginal disc to negatively regulate the development of sensory cell fates (Hays et al., 1999). The induction of hairy in response to Hh requires Cubitus interruptus (Ci), the D. melanogaster homologue of Gli, and dorsal/ventral expression of hairy is lost in Smo-deficient clones (Hays et al., 1999). Patterning of the retinal field during development of the D. melanogaster compound eye also reveals Notch-independent regulation of hairy by Hh (Fu and Baker, 2003). In this system, both Hh and Notch negatively regulate hairy expression to promote a wave of photoreceptor differentiation.
In this study, we have elucidated a novel mechanism for Shh-controlled progenitor cell behavior. Our observation that Gli2 occupies the Hes1 promoter is one of the first identified direct relationships between Gli2 and a target gene in neural progenitor cells. This study provides a mechanistic link between Shh-Gli2 signaling and Hes1 in regulating the proliferation of RPCs, thereby shedding light on a new means of manipulating Shh-induced cellular responses.
Materials and methods
Transgenic mice
Several transgenic mouse lines were used in this study. PtchlacZ+/− mice (Goodrich et al., 1997) were obtained from the Jackson Laboratory and maintained on a C57Bl6 background; Gli1+/− and Gli2+/− mice (obtained from A. Joyner, Sloan-Kettering Institute, New York, NY; Mo et al., 1997; Park et al., 2000), and Hes1+/− (Ishibashi et al., 1995) were maintained on a CD1 background. PtchlacZ+/−were crossed with Hes1+/− mice to generate double heterozygous mice (PtchlacZ+/−Hes1+/−). Gli1+/− and Gli2+/− strains were mated to generate double heterozygous animals, Gli1+/−Gli2+/−, which were subsequently crossed to give double homozygous null mice Gli1−/−Gli2−/−. Unless otherwise stated, retinal explants were derived from CD1 (the Jackson Laboratory) wild-type mice.
Cell culture, retinal explants, and BrdU labeling
Mouse strains were continuously mated or time-mated to generate specimens of the appropriate age, with the day of the vaginal plug designated day 0 of gestation. Retinal explants were prepared as described previously (Wang et al., 2005). Retinal explant medium was supplemented with 10 nM of Smo agonist (Ag1.10; a kind gift from Curis, Inc.; Frank-Kamenetsky et al., 2002) or 10 μM DAPT (Millipore), then cultured at 8% CO2 and 37°C. Selected explants were labeled with 10 μM BrdU for the last 6 h of culture to identify cells in S phase. Explants were either fixed in a 4% paraformaldehyde phosphate buffer for 1 h, transferred to a 30% sucrose/PBS solution overnight, and embedded in 1:1 optimal cutting temperature/30% sucrose/PBS mixture or dissociated into single cells with trypsin (Sigma-Aldrich) and plated onto Superfrost slides (Sigma-Aldrich) for quantitative analysis as described previously (Wang et al., 2005). COS cells were cultured in 10% FBS DME and transfected with full-length Myc-tagged Gli2 (a gift from H. Sasaki, RIKEN Center for Developmental Biology, Chuo-ku, Kobe, Japan; Sasaki et al., 1999) or pUb-GFP (a gift from T. Matsuda, Harvard Medical School, Boston, MA) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Histology, IHC, and ISH
IHC or ISH was performed as described previously (Jensen and Wallace, 1997; Wallace, 1999; Dakubo et al., 2003). Antibodies used in this study include rabbit polyclonal anti-CRALBP (a kind gift from J. Saari, University of Washington, Seattle, WA), mouse monoclonal anti-BrdU (BD), mouse monoclonal anti-rhodopsin (Röhlich et al., 1989), rabbit polyclonal anti-recoverin (Millipore), sheep polyclonal anti-Chx10 (a gift from R. Bremner, Toronto Western Research Institute, Toronto, Ontario, Canada), rabbit polyclonal phosphohistone H3 (Millipore), and rabbit polyclonal anti-GFP (Invitrogen). Secondary antibodies include donkey anti–goat IgG Cy3 (Invitrogen), goat anti–rabbit IgG FITC (Invitrogen), goat anti–mouse IgG Cy3 (Jackson ImmunoResearch Laboratories), and goat anti–rabbit IgG Cy3 (Jackson ImmunoResearch Laboratories). The antisense riboprobes used for ISH include Gli1 (a gift from A. Joyner), RBPJ-κ (a gift from T. Hongo, Kyoto University, Kyoto, Japan), Gli2 (a gift from H. Sasaki), Hes1, and Hes5. Bright field images were analyzed using an Axioplan microscope and captured with an Axiovision camera (2.05; both from Carl Zeiss, Inc.). Pictures were taken at magnifications of 10× (NA 0.30) and 20× (NA 0.05; both from Carl Zeiss, Inc.). Fluorescent images were analyzed using an Axiocam microscope (HRm) and captured with an Axioimager camera (M1; both from Carl Zeiss, Inc.). Florescent images were taken at 20× (0.8 NA). All images were processed using Photoshop CS2 (Adobe).
In vitro electroporation
Electroporation was performed on retinal explants based on the protocol from Matsuda and Cepko (2004). Explanted retinas were electroporated (ECM 830; BTX Harvard Apparatus) in a 2-mm gap cuvette (VWR) with 0.5–1.5 μg/μl of plasmid DNA in endotoxin-free TE buffer with a 10:1 ratio of plasmid DNA/pUb-GFP or pUb-GFP alone. The DNA plasmids used in this study include: SMO-M2 (a gift from G. Fishell, New York University Langone Medical Center, New York, NY), an activated NICD (a gift from G. Weinmaster, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA), LacZ 2.1 double-stranded shcontrol (Invitrogen), shRBPJ-k (Invitrogen), and Hes1DN.
Western blotting
Protein was extracted from Smo agonist–treated and nontreated retinal explant cultures using RIPA buffer (125 mM Tris-HCL, 2% SDS + protease inhibitor cocktail), and Western blotting performed as described previously (Dakubo et al., 2008). Protein samples were probed with 1:1,500 rabbit polyclonal anti-Hes1 (a gift from N. Brown, University of Cincinnati College of Medicine, Cincinnati, OH), 1:200 dilution of goat polyclonal anti-Gli2 (sc-20291; Santa Cruz Biotechnology, Inc.), 1:500 dilution of rabbit polyclonal Gli2 (Abcam), or 1:50 dilution of mouse monoclonal antibody E7 ascites (Developmental Studies Hybridoma Bank). Secondary antibodies used include goat anti–rabbit IgG HRP (1:5,000; Sigma-Aldrich), donkey anti–goat HRP (1:3,000; Santa Cruz Biotechnology, Inc.), and sheep anti–mouse IgG HRP (1:3,000; Sigma-Aldrich).
RT-qPCR
RNA was harvested from Smo agonist–treated and nontreated retinal explants using Trizol (1 explant per 1 ml of Trizol). cDNA was synthesized using 2 μg of total RNA with the Invitrogen kit according to the manufacturer's instructions. qPCR was performed using 1 μl of cDNA with Brillant SyBr Green mastermix (Agilent Technologies) according to the manufacturer's instructions, with the exception that the reaction was scaled down to a total volume of 25 μl. Primers (200 nM) used include Hes1 (forward, 5′-AAAGACGGCCTCTGAGCACA-3′; reverse, 5′-TCATGGCGTTGATCTGGGTCA-3′), Hes5 (forward, 5′-AAGAGCCTGCACCAGGACTA-3′; reverse, 5′-CGCTGGAAGTGGTAAAGCA-3′), and 18S (forward, 5′-CGGCTACCACATCCAAGG-3′; reverse, 5′-CTGGAATTACCGCGGCT-3′), and GFP (forward, 5′-CGTCGCCGTCCAGCTCGACCAG-3′; reverse, 5′-CATGGTCCTGCTGGAGTTCGTG-3′), Gli1 (forward, 5′-CACTACCTGGCCTCACACCT-3′; reverse, 5′-GTACTCGGTTCGGCTTCTCC-3′). The PCR reaction was performed on a MX3000P (Stratagene) with 40 amplification cycles. Changes in gene expression were quantified based on the 2ΔCt value normalized to 18S. Normalization to GFP was used to standardize for transfection efficiency in electroporated retinal explants. Statistical significance was determined using a two-tailed student's t test.
ChIP
Candidate Gli consensus sequences were identified as GACCACCCA or TGGGTGGTC (Lai et al., 2004), and primers were designed to amplify regions of genomic DNA that contain at least a seven-base match within a 10-kb region of the Hes1 promoter. CD1 retinal explants treated with or without a Smo agonist for 3 d were fixed in cold 4% paraformaldehyde PBS solution (two explants per condition) for 30 min. The DNA was sheared to less than 1 kb by sonication. ChIP was performed using the EZ ChIP kit (Millipore) according to the manufacturer's instructions. Immunoprecipitations were performed using 10 μl of a goat anti-Gli2 polyclonal antibody (Santa Cruz Biotechnology, Inc.) or an irrelevant antibody of the same species (goat anti-Brn3b). DNA was analyzed using qPCR with 2 μl of DNA with Brilliant SyBr green mastermix (Stratagene) and 200 nM of Hes1 primers (−7,808 bp site: forward, 5′-CAGTGCTACAGACCACACAGG-3′; and reverse, 5′-AGAACGTGACATCGGCTTTC-3′; −146 bp site: forward, 5′-TCCTTTTGATTGACGTTGTAGC-3′; and reverse, 5′-CCCAAACTTTCTTTCCCACA-3′), with an annealing temperature of 60°C on a MX3000P for 40 cycles. A primer set (forward, 5′-TTGAGGGTTTTTGTTTTGTTTTG-3′; reverse, 5′-CGGTTGCTTTTTAAACAGTGG-3′) spanning a region of the Hes1 promoter without a Gli consensus sequence was used as a negative control. The Ct values were expressed relative to unprecipitated input chromatin and fold enrichment were calculated by 2ΔCt, where ΔCt = Ct (anti-Gli2 – anti-control Brn3b). Standard deviation was calculated based on three independent experiments, and significance was calculated using a two-tailed student's t test.
Online supplemental material
Fig. S1 shows knockdown of Hes1 reporter activity in response to shRBPJ-κ. Fig. S2 confirms that Hes1 and Gli1 levels are maintained in retinal explant cultures after a culture period of 6 h. Fig. S3 shows the proportions of retinal cell types in response to Smo Ag treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200805155/DC1.
Supplementary Material
Acknowledgments
We are grateful to Izhar Livnebar for technical advice on ChIP; and R. Bremner, Ruth Slack, Lynn Megeney, and Ilona Skerjanc for critical reading of the manuscript.
Dana Wall is a recipient of a Canadian Institutes of Health Research/CNIB studentship award. This work was supported by an operating grant to V. Wallace from the Canadian Cancer Society (National Cancer Institute of Canada grant 016435).
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; CNS, central nervous system; CRALBP, cellular retinaldehyde-binding protein; DAPT, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; DIV, days in vitro; E, embryonic day; Hh, Hedgehog; IHC, immunohistochemistry; ISH, in situ hybridization; P, postnatal day; Ptch, Patched; RGC, retinal ganglion cell; RPC, retinal progenitor cell; RT-qPCR, quantitative RT-PCR; Shh, Sonic hedgehog; Smo, Smoothened.
References
- Bai, C.B., W. Auerbach, J.S. Lee, D. Stephen, and A.L. Joyner. 2002. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 129:4753–4761. [DOI] [PubMed] [Google Scholar]
- Barolo, S., R.G. Walker, A.D. Polyanovsky, G. Freschi, T. Keil, and J.W. Posakony. 2000. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 103:957–969. [DOI] [PubMed] [Google Scholar]
- Belliveau, M.J., T.L. Young, and C.L. Cepko. 2000. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. J. Neurosci. 20:2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres, T.M., T. Masui, G.H. Swift, L. Shi, R.M. Henke, and R.J. MacDonald. 2006. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 26:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, G.C., C.J. Mazerolle, Y. Wang, K.D. Campsall, D. Petrin, B.C. Leonard, K.F. Damji, D.G. Evans, D. McLeod, and V.A. Wallace. 2003. Abnormalities of the vitreoretinal interface caused by dysregulated Hedgehog signaling during retinal development. Hum. Mol. Genet. 12:3269–3276. [DOI] [PubMed] [Google Scholar]
- Cayouette, M., B.A. Barres, and M. Raff. 2003. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron. 40:897–904. [DOI] [PubMed] [Google Scholar]
- Cepko, C.L., C.P. Austin, X. Yang, M. Alexiades, and D. Ezzeddine. 1996. Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. USA. 93:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales, J.D., G.L. Rocco, S. Blaess, Q. Guo, and A.L. Joyner. 2004. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 131:5581–5590. [DOI] [PubMed] [Google Scholar]
- Dakubo, G.D., and V.A. Wallace. 2004. Hedgehogs and retinal ganglion cells: organizers of the mammalian retina. Neuroreport. 15:479–482. [DOI] [PubMed] [Google Scholar]
- Dakubo, G.D., Y.P. Wang, C. Mazerolle, K. Campsall, A.P. McMahon, and V.A. Wallace. 2003. Retinal ganglion cell-derived sonic hedgehog signaling is required for optic disc and stalk neuroepithelial cell development. Development. 130:2967–2980. [DOI] [PubMed] [Google Scholar]
- Dakubo, G.D., S.T. Beug, C.J. Mazerolle, S. Thurig, Y. Wang, and V.A. Wallace. 2008. Control of glial precursor cell development in the mouse optic nerve by sonic hedgehog from retinal ganglion cells. Brain Res. 1228:27–42. [DOI] [PubMed] [Google Scholar]
- de la Pompa, J.L., A. Wakeham, K.M. Correia, E. Samper, S. Brown, R.J. Aguilera, T. Nakano, T. Honjo, T.W. Mak, J. Rossant, and R.A. Conlon. 1997. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 124:1139–1148. [DOI] [PubMed] [Google Scholar]
- Donovan, S.L., and M.A. Dyer. 2005. Regulation of proliferation during central nervous system development. Semin. Cell Dev. Biol. 16:407–421. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky, M., X.M. Zhang, S. Bottega, O. Guicherit, H. Wichterle, H. Dudek, D. Bumcrot, F.Y. Wang, S. Jones, J. Shulok, et al. 2002. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., and N.E. Baker. 2003. Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development. 130:5229–5239. [DOI] [PubMed] [Google Scholar]
- Furukawa, T., S. Mukherjee, Z.Z. Bao, E.M. Morrow, and C.L. Cepko. 2000. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 26:383–394. [DOI] [PubMed] [Google Scholar]
- Geling, A., H. Steiner, M. Willem, L. Bally-Cuif, and C. Haass. 2002. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3:688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, L.V., L. Milenkovic, K.M. Higgins, and M.P. Scott. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 277:1109–1113. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T., X.M. Zhang, B.Y. Chen, and X.J. Yang. 2006. VEGF activates divergent intracellular signaling components to regulate retinal progenitor cell proliferation and neuronal differentiation. Development. 133:2201–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, R., K.T. Buchanan, C. Neff, and T.V. Orenic. 1999. Patterning of Drosophila leg sensory organs through combinatorial signaling by hedgehog, decapentaplegic and wingless. Development. 126:2891–2899. [DOI] [PubMed] [Google Scholar]
- Hirata, H., S. Yoshiura, T. Ohtsuka, Y. Bessho, T. Harada, K. Yoshikawa, and R. Kageyama. 2002. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 298:840–843. [DOI] [PubMed] [Google Scholar]
- Hori, K., J. Cholewa-Waclaw, Y. Nakada, S.M. Glasgow, T. Masui, R.M. Henke, H. Wildner, B. Martarelli, T.M. Beres, J.A. Epstein, et al. 2008. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 22:166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M.C., R. Mo, S. Bhella, C.W. Wilson, P.T. Chuang, C.C. Hui, and N.D. Rosenblum. 2006. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development. 133:569–578. [DOI] [PubMed] [Google Scholar]
- Huntzicker, E.G., I.S. Estay, H. Zhen, L.A. Lokteva, P.K. Jackson, and A.E. Oro. 2006. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 20:276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchin, M.E., M.S. Kariapper, M. Grachtchouk, A. Wang, L. Wei, D. Cummings, J. Liu, L.E. Michael, A. Glick, and A.A. Dlugosz. 2005. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 19:214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, W.J., K.I. McCue, T.H. Tran, A.R. Hallahan, and B.J. Wainwright. 2008. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 27:1489–1500. [DOI] [PubMed] [Google Scholar]
- Ishibashi, M., S.L. Ang, K. Shiota, S. Nakanishi, R. Kageyama, and F. Guillemot. 1995. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9:3136–3148. [DOI] [PubMed] [Google Scholar]
- Jadhav, A.P., H.A. Mason, and C.L. Cepko. 2006. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 133:913–923. [DOI] [PubMed] [Google Scholar]
- Jensen, A.M., and V.A. Wallace. 1997. Expression of Sonic hedgehog and its putative role as a precursor cell mitogen in the developing mouse retina. Development. 124:363–371. [DOI] [PubMed] [Google Scholar]
- Kageyama, R., and T. Ohtsuka. 1999. The Notch-Hes pathway in mammalian neural development. Cell Res. 9:179–188. [DOI] [PubMed] [Google Scholar]
- Kageyama, R., T. Ohtsuka, J. Hatakeyama, and R. Ohsawa. 2005. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 306:343–348. [DOI] [PubMed] [Google Scholar]
- Kim, J., H.H. Wu, A.D. Lander, K.M. Lyons, M.M. Matzuk, and A.L. Calof. 2005. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 308:1927–1930. [DOI] [PubMed] [Google Scholar]
- Kleinjan, D.A., and V. van Heyningen. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76:8–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, K., M.J. Robertson, and D.V. Schaffer. 2004. The sonic hedgehog signaling system as a bistable genetic switch. Biophys. J. 86:2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, E.M., H. Roelink, J. Turner, and T.A. Reh. 1997. Sonic hedgehog promotes rod photoreceptor differentiation in mammalian retinal cells in vitro. J. Neurosci. 17:6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker, M., M. Agathocleous, M.A. Amato, K. Parain, W.A. Harris, and M. Perron. 2006. Hedgehog signaling and the retina: insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 20:3036–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., D. Papermaster, and C.L. Cepko. 1998. A unique pattern of photoreceptor degeneration in cyclin D1 mutant mice. Proc. Natl. Acad. Sci. USA. 95:9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, E., and P. Bovolenta. 2002. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends Neurosci. 25:89–96. [DOI] [PubMed] [Google Scholar]
- Matise, M.P., D.J. Epstein, H.L. Park, K.A. Platt, and A.L. Joyner. 1998. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 125:2759–2770. [DOI] [PubMed] [Google Scholar]
- Matsuda, T., and C.L. Cepko. 2004. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 101:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter-Sadzinski, L., M. Puzianowska-Kuznicka, J. Hernandez, M. Ballivet, and J.M. Matter. 2005. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 132:3907–3921. [DOI] [PubMed] [Google Scholar]
- Mo, R., A.M. Freer, D.L. Zinyk, M.A. Crackower, J. Michaud, H.H. Heng, K.W. Chik, X.M. Shi, L.C. Tsui, S.H. Cheng, et al. 1997. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 124:113–123. [DOI] [PubMed] [Google Scholar]
- Moshiri, A., and T.A. Reh. 2004. Persistent progenitors at the retinal margin of ptc+/− mice. J. Neurosci. 24:229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri, A., C.R. McGuire, and T.A. Reh. 2005. Sonic hedgehog regulates proliferation of the retinal ciliary marginal zone in posthatch chicks. Dev. Dyn. 233:66–75. [DOI] [PubMed] [Google Scholar]
- Nakazaki, H., A.C. Reddy, B.L. Mania-Farnell, Y.W. Shen, S. Ichi, C. McCabe, D. George, D.G. McLone, T. Tomita, and C.S. Mayanil. 2008. Key basic helix-loop-helix transcription factor genes Hes1 and Ngn2 are regulated by Pax3 during mouse embryonic development. Dev. Biol. 316:510–523. [DOI] [PubMed] [Google Scholar]
- Nelson, B.R., B.H. Hartman, S.A. Georgi, M.S. Lan, and T.A. Reh. 2007. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol. 304:479–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, B.C., K. Lefort, A. Mandinova, D. Antonini, V. Devgan, G. Della Gatta, M.I. Koster, Z. Zhang, J. Wang, A. Tommasi di Vignano, et al. 2006. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 20:1028–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z., M. Abou El Hassan, Z. Xu, T. Yu, and R. Bremner. 2008. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 9:785–793. [DOI] [PubMed] [Google Scholar]
- Nofziger, D., A. Miyamoto, K.M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 126:1689–1702. [DOI] [PubMed] [Google Scholar]
- Palma, V., and A. Ruiz i Altaba. 2004. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 131:337–345. [DOI] [PubMed] [Google Scholar]
- Pan, Y., C.B. Bai, A.L. Joyner, and B. Wang. 2006. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 26:3365–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.L., C. Bai, K.A. Platt, M.P. Matise, A. Beeghly, C.C. Hui, M. Nakashima, and A.L. Joyner. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 127:1593–1605. [DOI] [PubMed] [Google Scholar]
- Röhlich, P., G. Adamus, J.H. McDowell, and P.A. Hargrove. 1989. Binding pattern of anti-rhodopsin monoclonal antibodies to photoreceptor cells: an immunocytochemical study. Exp. Eye Res. 49:999–1013. [DOI] [PubMed] [Google Scholar]
- Sasaki, H., Y. Nishizaki, C. Hui, M. Nakafuku, and H. Kondoh. 1999. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 126:3915–3924. [DOI] [PubMed] [Google Scholar]
- Shimojo, H., T. Ohtsuka, and R. Kageyama. 2008. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 58:52–64. [DOI] [PubMed] [Google Scholar]
- Solecki, D.J., X.L. Liu, T. Tomoda, Y. Fang, and M.E. Hatten. 2001. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 31:557–568. [DOI] [PubMed] [Google Scholar]
- Stockhausen, M.T., J. Sjolund, and H. Axelson. 2005. Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp. Cell Res. 310:218–228. [DOI] [PubMed] [Google Scholar]
- Strom, A., P. Castella, J. Rockwood, J. Wagner, and M. Caudy. 1997. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 11:3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka, K., J. Hatakeyama, Y. Bessho, and R. Kageyama. 2004. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 1004:148–155. [DOI] [PubMed] [Google Scholar]
- Tomita, K., M. Ishibashi, K. Nakahara, S.L. Ang, S. Nakanishi, F. Guillemot, and R. Kageyama. 1996. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 16:723–734. [DOI] [PubMed] [Google Scholar]
- Villavicencio, E.H., D.O. Walterhouse, and P.M. Iannaccone. 2000. The sonic hedgehog-patched-gli pathway in human development and disease. Am. J. Hum. Genet. 67:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waid, D.K., and S.C. McLoon. 1998. Ganglion cells influence the fate of dividing retinal cells in culture. Development. 125:1059–1066. [DOI] [PubMed] [Google Scholar]
- Wallace, V.A. 1999. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 9:445–448. [DOI] [PubMed] [Google Scholar]
- Wang, Y.P., G. Dakubo, P. Howley, K.D. Campsall, C.J. Mazarolle, S.A. Shiga, P.M. Lewis, A.P. McMahon, and V.A. Wallace. 2002. Development of normal retinal organization depends on Sonic hedgehog signaling from ganglion cells. Nat. Neurosci. 5:831–832. [DOI] [PubMed] [Google Scholar]
- Wang, Y., G.D. Dakubo, S. Thurig, C.J. Mazerolle, and V.A. Wallace. 2005. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 132:5103–5113. [DOI] [PubMed] [Google Scholar]
- Xie, J., M. Murone, S.M. Luoh, A. Ryan, Q. Gu, C. Zhang, J.M. Bonifas, C.W. Lam, M. Hynes, A. Goddard, et al. 1998. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 391:90–92. [DOI] [PubMed] [Google Scholar]
- Yaron, O., C. Farhy, T. Marquardt, M. Applebury, and R. Ashery-Padan. 2006. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 133:1367–1378. [DOI] [PubMed] [Google Scholar]
- Young, R.W. 1985. Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 353:229–239. [DOI] [PubMed] [Google Scholar]
- Yu, C., C.J. Mazerolle, S. Thurig, Y. Wang, M. Pacal, R. Bremner, and V.A. Wallace. 2006. Direct and indirect effects of hedgehog pathway activation in the mammalian retina. Mol. Cell. Neurosci. 32:274–282. [DOI] [PubMed] [Google Scholar]
- Zhang, J., B. Wang, Z. Xiao, Y. Zhao, B. Chen, J. Han, Y. Gao, W. Ding, H. Zhang, and J. Dai. 2008. Olfactory ensheathing cells promote proliferation and inhibit neuronal differentiation of neural progenitor cells through activation of Notch signaling. Neuroscience. 153:406–413. [DOI] [PubMed] [Google Scholar]
- Zhang, S.S., X.Y. Fu, and C.J. Barnstable. 2002. Tissue culture studies of retinal development. Methods. 28:439–447. [DOI] [PubMed] [Google Scholar]
- Zhang, X.M., and X.J. Yang. 2001. Regulation of retinal ganglion cell production by Sonic hedgehog. Development. 128:943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.