Abstract
In this study, we show that NANOG, a master transcription factor, regulates S-phase entry in human embryonic stem cells (hESCs) via transcriptional regulation of cell cycle regulatory components. Chromatin immunoprecipitation combined with reporter-based transfection assays show that the C-terminal region of NANOG binds to the regulatory regions of CDK6 and CDC25A genes under normal physiological conditions. Decreased CDK6 and CDC25A expression in hESCs suggest that both CDK6 and CDC25A are involved in S-phase regulation. The effects of NANOG overexpression on S-phase regulation are mitigated by the down-regulation of CDK6 or CDC25A alone. Overexpression of CDK6 or CDC25A alone can rescue the impact of NANOG down-regulation on S-phase entry, suggesting that CDK6 and CDC25A are downstream cell cycle effectors of NANOG during the G1 to S transition.
Introduction
Nanog is a divergent homeodomain transcription factor that functions to maintain self-renewal of embryonic stem cells (ESCs; Chambers et al., 2003; Mitsui et al., 2003). Nanog expression is largely confined to the inner cell mass of human blastocysts (Hyslop et al., 2005) and is high in undifferentiated ESCs and embryonic carcinoma cells but down-regulated during ESC differentiation (Chambers et al., 2003; Armstrong et al., 2006). This has been attributed to the sequential methylation of CpG residues in the promoter region of NANOG (Deb-Rinker et al., 2005) as well as suppression by p53 and Tcf3, both of which were shown to bind to the Nanog promoter region (Lin et al., 2005; Pereira et al., 2006).
Overexpression of Nanog in mouse ESCs confers pluripotency independently of the leukemia inhibitory factor–STAT3 pathway (Chambers et al., 2003; Mitsui et al., 2003), whereas its down-regulation in mouse and human ESCs (hESCs) results in loss of pluripotency, reduction in cell growth, and differentiation toward extraembryonic lineages (Chambers et al., 2003; Mitsui et al., 2003; Hyslop et al., 2005; Zaehres et al., 2005). Elevated expression of Nanog has also been reported to result in clonal expansion of murine ESCs, the maintenance of Oct4 expression, and resistance to differentiation induced in monolayers (Chambers et al., 2003).
Large-scale studies have suggested that Nanog acts as a component of multiple protein complexes that are individually required for controlling the survival and differentiation of the inner cell mass in the embryo; some of the protein complexes are also putative Nanog and Oct4 targets as well as being their effectors in the pluripotency network (Boyer et al., 2005; Wang et al., 2006). Not surprisingly, transfection of OCT4, SOX2, NANOG, and LIN28 in human fibroblasts induces pluripotency, suggesting an important role for each of these factors in reprogramming the genome of somatic cells (Yu et al., 2007).
Despite these advances, very little is known about the role of NANOG in regulating ESC proliferation and survival. ESCs are characterized by rapid cell divisions, and their cell cycles have a rather large S phase and a truncated G1 phase (Burdon et al., 2002). They can proliferate without apparent limit and can be readily propagated, but very little is known about these unusual proliferative properties, their cell cycle structure, and how this affects the pluripotent phenotype. In this study, we sought to identify possible interactions between NANOG, one of the master pluripotency factors in ESCs, and cell cycle regulation.
To address how NANOG interacts with the cell cycle machinery, we constitutively overexpressed NANOG in hESCs. In this manuscript, we show that NANOG is able to enhance hESC proliferation while still maintaining the pluripotent phenotype. The results described in this work provide, for the first time, several lines of evidence that NANOG accelerates S-phase entry in hESCs by directly regulating at the transcriptional level two important cell cycle regulators: CDK6 and CDC25A.
Results
Generation and characterization of NANOG-overexpressing hESC clones
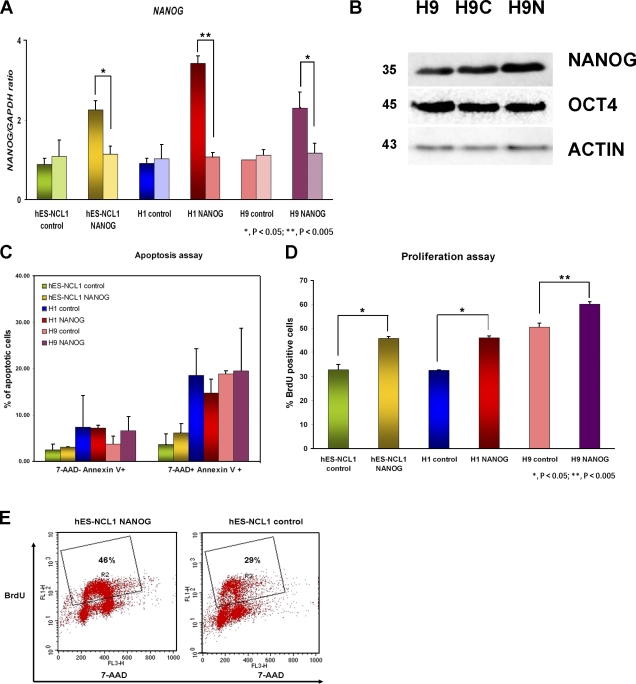
Three hESC lines, H1, hES-NCL1, and H9, were stably transfected with the empty pTP6 (Pratt et al., 2000) or pTP6-NANOG construct to generate three control sublines (H1 control, H9 control, and hES-NCL1 control) and three overexpressing sublines (H1 NANOG, H9 NANOG, and hES-NCL1 NANOG), respectively. Quantitative RT-PCR for the endogenous expression of NANOG showed no significant differences between clones generated from the same hESC line (Fig. 1 A). Total expression of NANOG analyzed by both quantitative RT-PCR (Fig. 1 A) and Western blotting (Fig. 1 B) revealed higher NANOG expression in NANOG-overexpressing sublines compared with the control, thus confirming gene overexpression. Each of the six sublines was subjected to karyotype analysis at 20 and 40 passages after transfection, and, in all cases, no chromosomal anomalies were detected (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1).
Figure 1.
Characterization of NANOG-overexpressing hESC sublines. (A) Quantitative RT-PCR for endogenous (right bars) and total (left bars) expression of NANOG in H1, H9, and hES-NCL1 sublines. The data represent the mean ± SEM (error bars) from four independent experiments. The value for the control clone from each hESC subline was set to 1, and all other values were calculated with respect to this. (B) Western blot showing NANOG and OCT4 expression in the H9 sublines. β-Actin was used as a loading control. H9, wild-type untransfected cells; H9C, H9 control subline; H9N, H9 NANOG-overexpressing subline. Molecular masses are indicated in kilodaltons. (C) Assessment of cell death in NANOG-overexpressing and control sublines by flow cytometry analysis. (D) Assessment of cell proliferation in NANOG-overexpressing and control sublines by flow cytometry after 45 min of BrdU incorporation. (C and D) The data represent the mean ± SEM from three independent experiments. (E) Flow cytometry images showing a higher percentage of proliferating cells (gate R2) in NANOG-overexpressing sublines compared with controls.
The NANOG-overexpressing sublines grew faster in culture and had to be subcultured more often than the respective controls. This could be caused by a decrease in apoptosis, an increase in hESC proliferation, or both. To distinguish these two possibilities, we performed annexin V staining to detect early (7-AAD− annexin V+) and late apoptotic (7-AAD+ annexin V+) cells (Fig. 1 C) and BrdU incorporation to detect DNA synthesizing/proliferating cells under the aforementioned culture conditions (Fig. 1, D and E). The results showed that overexpression of NANOG did not significantly affect the rate of apoptosis because percentages of apoptotic cells were similar to control sublines under normal culture conditions (Fig. 1 C). However, overexpression of NANOG increased the percentage of cells incorporating BrdU (Fig. 1, D and E) compared with the control sublines. It is of interest to note the variability between different hESC sublines with respect to the percentage of proliferating cells, with H9 being the most proliferative, which fits well with the shorter population doubling time compared with H1 and hES-NCL1 (unpublished data). Direct immunoblotting also indicated increased proliferating cell nuclear antigen expression, suggesting increased cell proliferation as a result of NANOG overexpression (unpublished data). Cell counting experiments over different periods of time showed a higher proliferation in the NANOG sublines compared with controls, which correlated to a shorter population doubling time observed across all of the NANOG sublines (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1).
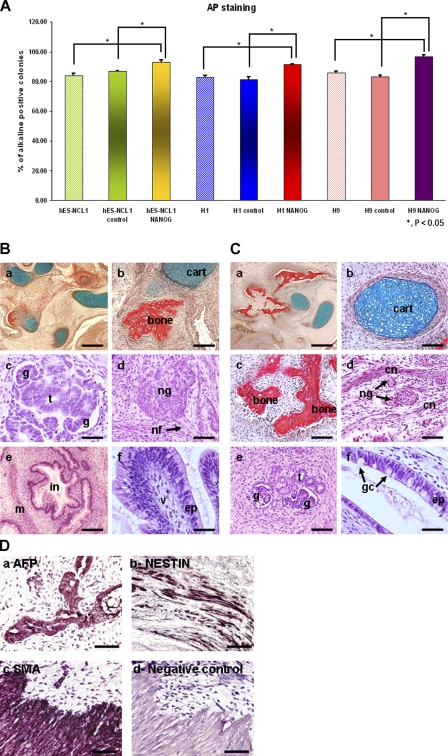
The NANOG-overexpressing sublines maintained typical hESC morphology in culture, whereas signs of spontaneous and typical differentiation were observed in the control sublines after 4–5 d in culture (unpublished data). AP staining analysis showed a small but significant increase in the number of AP-positive colonies in NANOG-overexpressing sublines compared with the controls (Fig. 2 A). This suggests that overexpression of NANOG results in the suppression of spontaneous differentiation that occurs during hESC culture. No changes in OCT4 expression were found by quantitative RT-PCR (not depicted) or direct immunoblotting (Fig. 1 B), corroborating data reported by Darr et al. (2006).
Figure 2.
Maintenance of pluripotency and differentiation capability of NANOG-overexpressing hESC sublines. (A) Assessment of pluripotency in NANOG-overexpressing and control sublines as well as wild-type cells by AP staining assay. The data represent the mean ± SEM (error bars) from three independent experiments. (B) Histological analysis of teratomas formed from grafted colonies of hESCs (hES-NCL1 NANOG) in SCID mice. (a) Low-power light micrograph showing heterogeneous structure within the body of the teratoma and the presence of a diverse range of different tissue types. (b) Cartilage (cart) and bone. (c) Kidney glomeruli (g) and associated tubules (t). (d) Large neural ganglion (ng) with connecting nerve fibers (nf) shown by a black arrow. (e) Transverse section through primitive intestine (in) with accompanying submucosal muscle layer (m). (f) Villous-type structure (v) lined with mucus-secreting cells resembling an intestinal epithelium (ep). Histological staining: Weigert's (a and b) and hematoxylin and eosin (c–f). Bars: (a) 400 μm; (b and e) 150 μm; (c, d, and f) 75 μm. (C) Histological analysis of teratomas formed from grafted colonies of hESCs (hES-NCL1 control) in SCID mice. (a) Low-power light micrograph showing heterogeneous structure within the body of the teratoma and the presence of a diverse range of different tissue types. (b) Cartilage. (c) Bone. (d) Small neural ganglia were clearly identifiable between layers of connective tissue (cn). (e) Kidney glomeruli and associated tubules. (f) Higher magnification image of a secretory cell resembling an intestinal epithelium producing mucus from goblet cells (gc) marked by two black arrows. Histological staining: Weigert's (a–c) and hematoxylin and eosin (d–f). Bars: (a) 400 μm; (b–e) 75 μm; (f) 25 μm. (D) Immunohistochemical analysis of teratomas formed from grafted colonies of hES–NCL1 NANOG in SCID mice. hES–NCL1 NANOG cells were found to be pluripotent, and the teratomas formed were composed of cell types from all three germ layers. (a) AFP (endoderm). (b) Nestin-positive cells (ectoderm). (c) SMA-positive cells (mesoderm). (d) Negative control. Bars, 150 μm.
We performed in vitro differentiation experiments using the embryoid body (EB) method to investigate whether NANOG overexpression interfered with the ability of hESCs to differentiate. The EBs obtained from NANOG sublines were indistinguishable from the controls at a morphological level (unpublished data). Recent studies in hESCs and primate ESCs have indicated that NANOG overexpression results in the enhancement of primitive ectoderm differentiation (Darr et al., 2006) and repression of primitive endoderm differentiation (Chen et al., 2006). The study performed in hESCs has used the H9 and H13 cell lines (Darr et al., 2006). To avoid duplicating findings, we focused our investigation on the impacts of NANOG overexpression on differentiation on the other two different cells lines, H1 and hES-NCL1. Our morphological observation and quantitative RT-PCR analysis showed no significant difference between each NANOG-overexpressing subline and controls in expression of the neuroepithelial marker (PAX6; Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1), mesodermal marker (BRACHYURY), endodermal marker (IHH), and trophoectoderm (CDX2) during the 21-d differentiation time course (not depicted). This notwithstanding, NANOG-overexpressing sublines showed an enhanced FGF5 peak expression (P < 0.05) compared with control sublines in which little or no expression of this gene was observed (Fig. S3 B), corroborating other published data reporting the enhancement of primitive ectoderm expression by NANOG (Darr et al., 2006). In addition, GATA4 expression was significantly repressed in NANOG sublines (P < 0.05) compared with the controls, suggesting that NANOG is likely to repress primitive endoderm differentiation (Fig. S3 C). The injection of NANOG and control hESCs into the testis of severe combined immunodeficient (SCID) mice resulted in the formation of teratomas (Fig. 2, B and C) comprised of cells from all three germ layers, indicating the pluripotent nature of the cells. The identity of endoderm, ectoderm, and mesoderm tissues within the tumors was confirmed by immunohistochemical staining with α-fetoprotein (AFP), NESTIN, and smooth muscle actin (SMA), respectively (Fig. 2 D).
NANOG accelerates S-phase entry
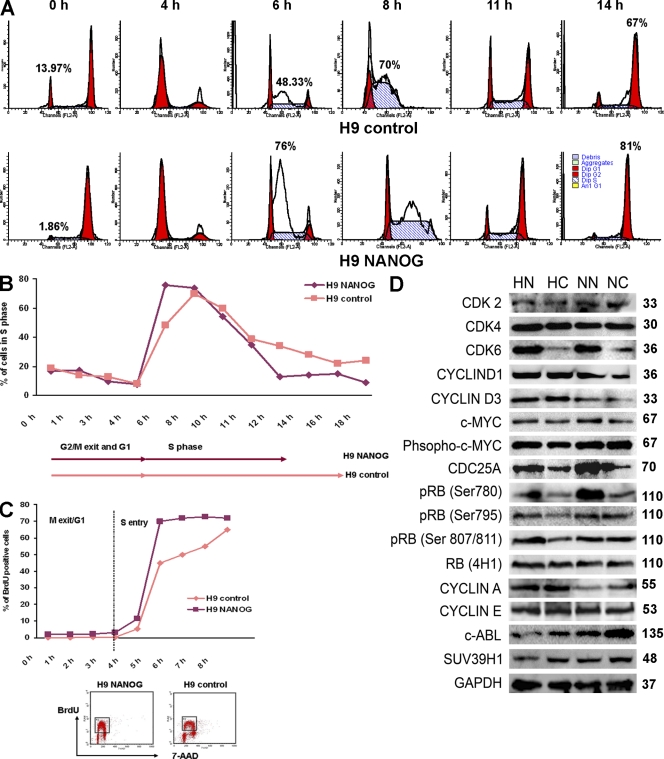
The increase in the number of cells incorporating BrdU in NANOG-overexpressing clones compared with controls is open to many explanations, and these include increased cell proliferation, slower progression through S phase, or transient stalling. To address some these issues, we synchronized human NANOG and control sublines by incubation with 200 ng/ml of the mitotic inhibitor nocodazole for 18 h. Flow cytometry analysis with cell surface markers associated with hESCs such as SSEA-4 and TRA-1-60 at 6 h upon release from nocodazole showed 10% and 8% more positive cells, respectively, in the NANOG sublines compared with controls, which fits well with the AP analysis shown in Fig. 2 A. In both cases, there were no significant changes in SSEA-4 or TRA-1-60 expression as a result of nocodazole treatment, thus indicating that nocodazole did not cause cell differentiation in either NANOG or control clones (unpublished data).
Upon release from inhibition, the hESC cell cycle distribution was investigated by flow cytometry every 1–2 h (Fig. 3, A and B). Analysis at the time of inhibitor release (0 h; Fig. 3 A) revealed that NANOG sublines showed a quicker cell cycle progression compared with controls because very few cells existed in G1 (1.86%) compared with control clones that showed 13.97% of cells still remaining in G1. Most importantly, 76% of the cells were in S phase within 6 h of release from the inhibitor in NANOG-overexpressing cells (Fig. 3 A). Control sublines showed a more gradual and slow entry into S phase, as only 48% and 70% were in S phase 6 h and 8 h after release from the inhibitor, respectively (Fig. 3, A and B). Although this analysis suggests that NANOG overexpression accelerates G1 to S transition, it is difficult to separate this event from S-phase progression. In view of this, we performed pulse-labeled BrdU experiments at different time points after nocodazole synchronization as described by Becker et al. (2006; Fig. 3 C). This analysis showed that in NANOG-overexpressing clones, most cells enter S phase 5 h after release from nocodazole treatment. In contrast, a more gradual and slower progression from G1 to S is observed in control clones (Fig. 3 C). Collectively, these data suggest that overexpression of NANOG shortens the time needed for S-phase entry.
Figure 3.
Changes in hESC cell cycle distribution as a result of NANOG overexpression. (A) Flow cytometry images showing movement of cells through the cell cycle after synchronization by nocodazole for 18 h assessed by propidium iodide staining. This figure represents an example of three independent experiments. (B) Chart representation of the fraction of cells in S phase assessed by propidium iodide staining over time after release from synchronization with nocodazole for 18 h. (C) Chart representation of the fraction of cells in S phase assessed by BrdU incorporation over time after release from synchronization with nocodazole for 18 h. The bottom panel shows flow cytometry images of BrdU-incorporating cells in NANOG and control H9 hESC sublines after 5 h of release from nocodazole. The dashed line represents the end of G1 and the start of S phase. (B and C) The figure represents an example of three independent experiments performed in H9 sublines. (D) Western blotting for expression of main components involved in G1 to S transition in NANOG-overexpressing and control clones. This summary is a representation of three independent experiments. GAPDH was used as a loading control. p16 and p15 were absent in all sublines, whereas p19 and p18 were expressed at very low but equal levels in NANOG-overexpressing and control clones (not depicted). Molecular masses are indicated in kilodaltons. HN, H1 NANOG; HC, H1 control; NN, hES-NCL1 NANOG; NC, hES-NCL1 control.
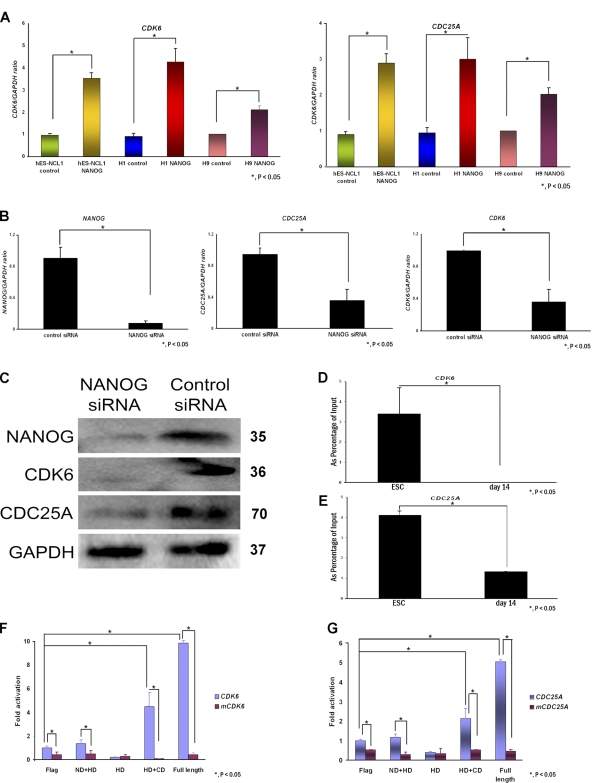
To investigate how NANOG regulates S-phase entry in hESCs, we first performed direct immunoblotting for the expression of key cell cycle components involved in the G1 to S transition in all NANOG-overexpressing and control sublines (Fig. 3 D and not depicted for H9 sublines). Most interestingly, we observed an increase in CDK6 and CDC25A in all NANOG-overexpressing sublines compared with controls (Fig. 3 D). It is important to point out that expression of other Cdks known to be important for the G1 to S progression such as CDK2, CDK4 (Fig. 3 D), and CDK1 (not depicted) were not changed as a result of NANOG overexpression. The aforementioned results were independently confirmed by hybridization of cell extracts from NANOG and control sublines to the Panorama antibody microarray that is designed to investigate several biological pathways, including cell cycle, signal transduction, apoptosis, cytoskeleton, etc. This analysis showed that CDK6 was the most up-regulated array target as a result of NANOG overexpression (3.43 ± 0.136 fold), whereas CDC25A occupied the 17th most up-regulated target (total of 42) showing, on average, a 2.3 ± 0.14-fold increase.
The C-terminal domain of NANOG is responsible for binding to CDK6 and CDC25A
To distinguish between transcriptional and posttranscriptional regulation, we performed quantitative RT-PCRs on components shown in Fig. 3 D and were able to confirm that CDK6 and CDC25A were transcriptionally activated in NANOG-overexpressing sublines compared with controls, suggesting that they might be direct transcriptional targets of NANOG (Fig. 4 A). We down-regulated NANOG expression in hESCs using RNA interference. More than 90% down-regulation of this gene was achieved 48 h after transfection (Fig. 4 B). Both CDK6 and CDC25A were down-regulated at the transcriptional and protein level (Fig. 4, B and C).
Figure 4.
CDK6 and CDC25A are NANOG transcriptional targets. (A) Quantitative RT-PCR analysis for the expression of CDK6 and CDC25A in NANOG-overexpressing and control sublines. The value for the control clone from each cell line was set to 1, and all other values were calculated with respect to this. The data represent the mean ± SEM (error bars) from three independent experiments. (B) Quantitative RT-PCR analysis for the expression of NANOG, CDK6, and CDC25A in hESC 42 h after the transfection of NANOG siRNA. The data represent the mean ± SEM from three independent experiments (one in each cell line: H1, H9, and hES-NCL1). The value for the control siRNA samples was set to 1, and all other values were calculated with respect to this. (C) Western blotting for NANOG, CDK6, and CDC25A in hES-NCL1 transfected with NANOG or control siRNA 42 h after the transfection of NANOG siRNA. GAPDH is used as a loading control. Molecular masses are indicated in kilodaltons. (D and E) Bar chart showing enrichment of CDK6 intragenic DNA region (intron 1) and CDC25A promoter fragment after ChIP with NANOG antibody in hESCs and day-14 differentiated sample from EBs. The data represent the mean ± SEM (error bars) from two experiments performed in the H1 cell line. (F and G) Bar chart showing activation of CDK6- and CDC25A-luciferase constructs upon transfection of different domains of NANOG. For each reporter construct, luciferase activities relative to pGL4 promoterless controls were determined, and data are represented as the fold change caused by the indicated NANOG expression relative to the nonmutated reporter cotransfected with the empty (Flag) control. mCDK6 or mCDC25A indicates the CDK6- or CDC25A-luciferase constructs with a mutated NANOG-binding site. Flag, DNA construct without NANOG cDNA; ND + HD, DNA construct containing the homeodomain and the N-terminal region of NANOG; HD, DNA construct containing the homeodomain region of NANOG; HD + CD, DNA construct containing the homeodomain and C-terminal region of NANOG; Full length, full-length NANOG cDNA. In both panels, the luciferase activity for cells transfected with the Flag construct and CDK6-luciferase (F) or CDC25A-luciferase construct (G) was set to 1, and all other values, including the luciferase activity achieved with mutated CDK6 and CDC25A constructs, were calculated with respect to that. The data represent the mean ± SEM.
Chromatin immunoprecipitation (ChIP) assays combined with quantitative PCR assays using primers that flanked a 580-bp region in intron 1 of CDK6 (suggested by Boyer et al. [2005] to be the NANOG-binding region; Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1) confirmed that NANOG does indeed bind to CDK6 (Fig. 4 D). The same assays, but using primers that flank the first 1.0 kb upstream of the transcription start sites of CDC25A, c-MYC, CDK2, and CDK4, revealed that NANOG also binds to the promoter region of CDC25A (Fig. 4 E and Fig. S4 A) but not of the other three targets (not depicted). ChIP assays were performed under identical conditions with various controls, including no input antibody, no input DNA, and a day-14 differentiated sample from EBs in which the expression of NANOG is detectable but low, as shown by recent work performed in our group (Neganova et al., 2008). The results corroborate, in part, the study by Boyer et al. (2005), who reported that NANOG and SOX2 bind to CDK6 but not to CDC25A and identified CDC25A as a transcriptional target of E2F4 but not of OCT4, SOX2, or NANOG.
A previous study (Mitsui et al., 2003) identified 5′-CGGACGCGCATTANGC-3′ as a NANOG consensus DNA binding sequence with the highest conservation observed for the tetranucleotide ATTA, which is common for DNA recognition sequences of many homeobox transcription factors. This consensus sequence was compared with the regulatory regions of CDK6 and CDC25A identified by Boyer et al. (2005) to identify NANOG binding domains that are shown in Fig. S4 A. PCR primers that flanked the NANOG binding domains were designed (Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1), and the corresponding fragments were inserted into the pGL4 luciferase reporter constructs (Fig. S4 B). Site-directed mutagenesis was performed, and the ATTA sequence was changed to AGGA to confirm the specificity of binding.
The aforementioned constructs (Fig. S4 B) were transfected in combination with a series of human NANOG domains (Fig. S4 C) in hESCs (Fig. 4, F and G) to identify which part of NANOG protein was important for CDK6 and CDC25A transactivation. The expression of NANOG constructs was tested by Western blotting (Fig. S4 D). The specificity of these interactions was confirmed by using CDK6 and CDC25A reporter constructs bearing mutations in the NANOG consensus binding sequence. The data obtained in hESCs showed that in the absence of exogenous NANOG expression (Flag-transfected group), the activity of luciferase reporter constructs driven by both CDC25A and CDK6 regulatory regions is significantly higher than the activity of mutated constructs, which precludes binding of endogenous NANOG. In addition, the data indicated that the C-terminal domain has a critical role in the transcriptional activation of CDK6 and CDC25A (Fig. 4, F and G), thus corroborating the data obtained by others (Oh et al., 2005). The specificity of the interaction between the C-terminal domain and regulatory regions of these two genes was confirmed by using CDK6 and CDC25A reporter constructs bearing mutations in the NANOG consensus binding sequence that showed significantly reduced transcriptional activity when compared with the wild-type constructs (Fig. 4, F and G).
CDK6 and CDC25A regulate S phase in hESCs
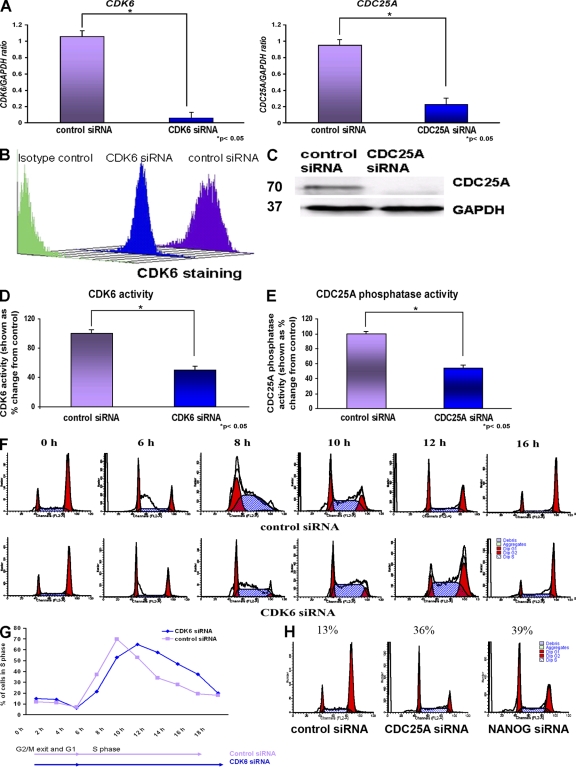
Our data have clearly shown that CDK6 and CDC25A are direct transcriptional targets of NANOG. However, the role of these two cell cycle regulators in hESCs has not been previously investigated. We set out to investigate this question using RNA interference. For each gene, two different mixed pools of siRNAs (targeted to different regions of each gene) were purchased from Invitrogen and Santa Cruz Biotechnology, Inc. (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1). For simplicity of presentation, the results from the application of mixed pools of three siRNAs obtained from Invitrogen are presented in this manuscript; however, similar results were obtained with the siRNAs obtained from Santa Cruz Biotechnology, Inc. (unpublished data). A 95 and 75% reduction was obtained at the transcriptional level for CDK6 and CDC25A, respectively, 42 h after transfection (Fig. 5 A). Both Western blotting and flow cytometry were used to confirm those findings at the protein level, and one example of each is presented in Fig. 5 (B and C). CDK6 kinase activity and CDC25A phosphatase activity were also reduced upon down-regulation of CDK6 and CDC25A, respectively (Fig. 5, D and E).
Figure 5.
CDK6 and CDC25A regulate S phase in hESCs. (A) Quantitative RT-PCR analysis for the expression of CDK6 and CDC25A in H1, H9, and hES-NCL1 cell lines 42 h after the transfection of CDK6 and CDC25A siRNA. The data represent the mean ± SEM (error bars) from three independent experiments (one in each cell line). The value for the control siRNA was set to 1, and all other values were calculated with respect to this. (B) Down-regulation of CDK6 by flow cytometry 42 h after the transfection of CDK6 siRNAs in hESCs (a representative example from the H9 line is shown). (C) Down-regulation of CDC25A by Western blotting 42 h after the transfection of CDC25A siRNAs (a representative example from the H9 line is shown). Molecular masses are indicated in kilodaltons. (D) Reduction in CDK6 kinase activity upon knockdown of CDK6. The value for the control siRNA was set to 100%, and all other values were calculated with respect to this. (E) Reduction in CDC25A phosphatase activity upon knockdown of CDC25A. The value for the control siRNA was set to 100%, and all other values were calculated with respect to this. (D and E) The data represent the mean ± SEM from three experiments performed in the H9 cell line. (F) Flow cytometry images showing movement of cells through the cell cycle after transfection of CDK6 siRNAs and synchronization by nocodazole for 18 h assessed by propidium iodide staining. (G) Chart representation of the fraction of cells in S phase over time after transfection of CDK6 siRNA and synchronization by nocodazole for 18 h assessed by propidium iodide staining. (H) Flow cytometry images showing retention of cells in G1 phase of the cell cycle after transfection of CDC25A siRNA and NANOG siRNA and synchronization by nocodazole for 18 h assessed by propidium iodide staining. (F–H) The figures represent an example of at least two independent experiments performed in the H9 subline.
To investigate changes in the cell cycle, the transfected hESCs were synchronized 24 h after transfection with nocodazole for an additional 18 h. Upon removal of the inhibitor, hESCs were analyzed by flow cytometry every 2 h. These experiments showed that down-regulation of CDK6 reduces the number of cells entering S phase and delays the S-phase completion by 2 h (Fig. 5, F and G). With this analysis, it is not possible to distinguish between S-phase entry and S-phase progression because any delays in S-phase entry would also delay S-phase progression. Notwithstanding this, we can conclude that, overall, CDK6 bears an impact on one or both of these events.
Flow cytometry analysis at removal of the inhibitor (0 h) showed that down-regulation of CDC25A causes a significant retention of cells in G1 compared with controls (Fig. 5 H), suggesting that CDC25A down-regulation prevents their S-phase entry. Further analysis at 2, 4, 6, and 8 h after release from the inhibitor showed similar results to those obtained at 0 h (unpublished data), suggesting that perhaps additional blocks at G2 to M and S phase were imposed upon CDC25A down-regulation, thus preventing further movement of cells into G2 to M and G1. Similar results at all time points examined (0, 2, 4, 6, and 8 h after release from the inhibitor) were obtained upon down-regulation of NANOG alone (Fig. 5 H).
NANOG regulates S-phase entry in hESCs via CDK6 and CDC25A
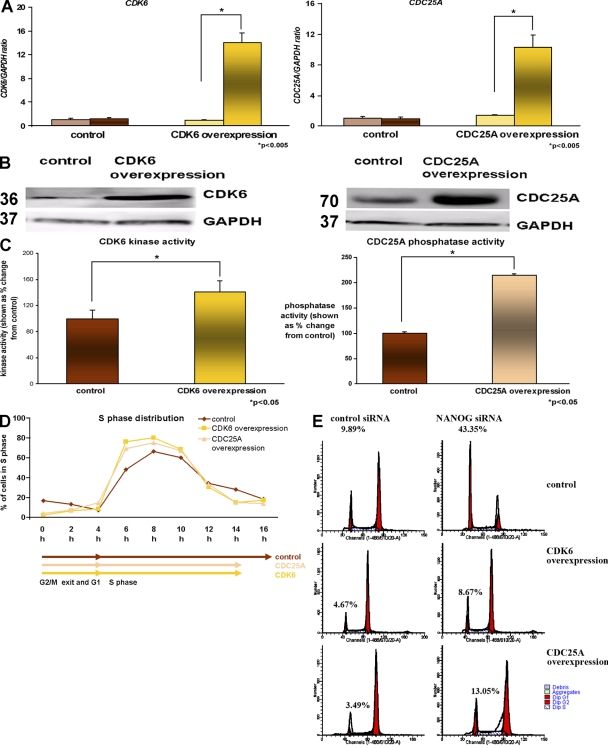
To investigate whether CDK6 and/or CDC25A overexpression alone was sufficient to substitute the role of NANOG in hESCs' S-phase entry, we created stable cell lines that overexpressed CDK6 and CDC25A. Quantitative RT-PCR (Fig. 6 A) and direct immunoblotting were used to confirm the overexpression of CDK6 and CDC25A (Fig. 6 B). A significant increase was observed in CDK6 kinase activity in the CDK6-overexpressing hESC sublines (Fig. 6 C). Similarly, a significant increase in phosphatase activity was observed in the CDC25A-overexpressing hESC sublines compared with controls (Fig. 6 C).
Figure 6.
Overexpression of CDK6 and CDC25A accelerates S-phase completion. (A) Quantitative RT-PCR for endogenous (left bars) and total (right bars) expression of CDK6 and CDC25A in the H9 hESC line. The data represent the mean ± SEM (error bars) from three independent experiments. The value for the control clone was set to 1, and all other values were calculated with respect to this. (B) Overexpression of CDC25A and CDC25A shown by Western blotting. GAPDH is used as a loading control (a representative example from the H9 line is shown). Molecular masses are indicated in kilodaltons. (C) Increase in CDK6 kinase activity and CDC25A phosphatase activity in CDK6- and CDC25A-overexpressing hESC sublines, respectively. The value for the control subline was set to 100%, and all other values were calculated with respect to this. The data represent the mean ± SEM from three experiments performed in the H9 cell line. (D) Chart representation of the fraction of cells in S phase over time in CDK6- and CDC25A-overexpressing hESC sublines assessed by propidium iodide staining (a representative example from the H9 subline is shown). (E) Flow cytometry images showing cell cycle distribution in H9 control as well as H9 CDK6 and H9 CDC25A after transfection of control siRNA and NANOG siRNA and synchronization by nocodazole for 18 h assessed by propidium iodide staining. This figure represents an example of at least two independent experiments performed in the H9 subline.
To investigate the effects of CDK6 and CDC25A overexpression, synchronization with nocodazole was performed for 18 h. We observed that a higher number of cells were found to enter S phase as a result of CDK6 and CDC25A overexpression (Fig. 6 D). In addition, the completion of S phase was shortened by 2 h in the CDK6- and CDC25A-overexpressing hESC sublines, thus indicating a role for both of these proteins in the regulation of S phase in hESCs.
To investigate whether CDK6 or CDC25A could substitute for NANOG in hESCs' S-phase entry, NANOG down-regulation was performed using RNA interference in CDK6- and CDC25A-overexpressing sublines (unpublished data). Quantitative RT-PCR analysis showed an 85–90% reduction in NANOG expression 42 h after siRNA transfection across the different hESC sublines (unpublished data). 24 h after siRNA transfection, the cells were synchronized with nocodazole for 18 h. Analysis of their cell cycle profile indicated a significant retention (43%) of hESCs in G1 as a result of NANOG knockdown, thus corroborating our previous results shown in Fig. 5 H. In contrast, a small number of cells, which was comparable with control transfections (8.67%), was present in G1 phase of the cell cycle in CDK6-overexpressing hESC sublines (Fig. 6 E), thus indicating that overexpression of CDK6 alone can rescue the effects of NANOG down-regulation in S-phase entry. The rescue effect was also observed in CDC25A-overexpressing hESC sublines, albeit at a lesser scale than CDK6-overexpressing sublines (Fig. 6 E). The difference in such results can be explained by changes in fold overexpression of the genes of interest between those sublines (14-fold CDK6 overexpression compared with 10-fold CDC25A overexpression). It is also likely that CDK6 enhances S-phase entry via an increase in CDK6 activity, whereas CDC25A could act by enhancing the activity of any G1-specific Cdks (CDK6, CDK4, or CDK2).
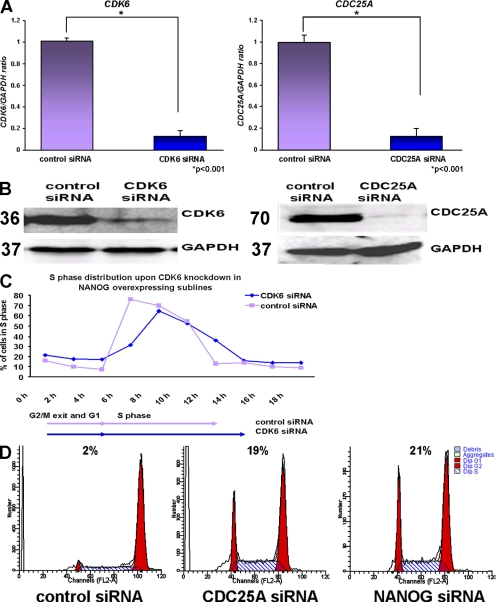
To investigate whether NANOG-mediated effects on S-phase regulation could be reversed by down-regulating CDK6 and CDC25A, we performed RNA interference experiments on NANOG-overexpressing sublines. An 86% and 88% reduction was obtained at the transcriptional level for CDK6 and CDC25A, respectively, 42 h after transfection (Fig. 7 A). Western blotting was used to confirm those findings at the protein level (Fig. 7 B).
Figure 7.
CDK6 and CDC25A are downstream effectors of NANOG. (A) Quantitative RT-PCR analysis showing down-regulation of CDK6 and CDC25A in H1 NANOG, H9 NANOG, and hES-NCL1 NANOG sublines 42 h after the transfection of CDK6 and CDC25A siRNA. The data represent the mean ± SEM (error bars) from three experiments (one in each subline). The value for the control-transfected sample (vector only) was set to 1, and all other values were calculated with respect to this. (B) Western blotting showing down-regulation of CDK6 and CDC25A in H1 NANOG sublines. Molecular masses are indicated in kilodaltons. (C) Chart representation of the fraction of cells in S phase over time after transfection of CDK6 siRNA and synchronization by nocodazole for 18 h in the H9 NANOG subline assessed by propidium iodide staining. This figure represents an example of at least two independent experiments carried in the H9 subline. (D) Flow cytometry images showing retention of cells in G1 phase of the cell cycle after transfection of CDC25A and NANOG siRNAs and synchronization by nocodazole for 18 h in the H9 NANOG subline assessed by propidium iodide staining. This figure represents an example of at least two independent experiments performed in the H9 subline.
To investigate changes in cell cycle, the hESCs were synchronized 24 h after transfection with CDK6 and CDC25A siRNAs with nocodazole for an additional 18 h. Upon removal of the inhibitor, hESCs were analyzed by flow cytometry every 2 h. These experiments showed that down-regulation of CDK6 in NANOG-overexpressing sublines lengthen the time needed for S-phase completion by 2 h compared with the control siRNA–transfected sample as well as reducing the numbers of hESCs entering S phase. Collectively, these data suggest that NANOG-accelerated S-phase entry and progression is in part mediated by CDK6, for its down-regulation alone reverses this effect, making NANOG cells comparable with controls.
Flow cytometry analysis of NANOG-overexpressing sublines that were transfected with CDC25A siRNA at the time of release from the inhibitor (0 h) showed a significant retention of cells in G1 (19%) compared with control (2%; Fig. 7 D). Similar results were obtained upon transfection of NANOG siRNA (Fig. 7 D). Repeated analysis at 2, 4, 6, and 8 h after release from the inhibitor showed similar results to those shown in Fig. 7 D for both NANOG and CDC25A knockdown experiments, suggesting likely additional blocks at S and G2 to M. Notwithstanding this, down-regulation of CDC25A in NANOG sublines results in a significant retention of cells in G1, supporting our hypothesis that NANOG regulation of S-phase entry is mediated by CDC25A.
Discussion
Recent publications suggest that combined overexpression of OCT4, SOX2, NANOG, and LIN28 is able to reprogram somatic cells to a pluripotent phenotype (Yu et al., 2007). One of the key features that distinguish somatic cells from ESCs is the length of the G1 phase of the cell cycle, which is much shorter in the latter (Becker et al., 2006). This means that during the transcription factor–mediated reprogramming, changes in expression or posttranslational modifications of cell cycle regulatory genes have to occur.
In this study, we sought to unravel the impacts of NANOG expression on hESCs and identify target genes that mediate its effects. We show that overexpression of NANOG in hESCs causes a significant increase in ESC proliferation while enhancing their pluripotent phenotype. Most importantly, overexpression of NANOG causes an increase in the numbers of cells entering S phase and shortens the time needed for S-phase entry. A combination of molecular assays indicated that the C-terminal region of NANOG binds to the regulatory regions of CDK6 and CDC25A. Overexpression of NANOG in hESCs results in a significant increase in the expression of CDK6 and CDC25A. Because NANOG overexpression results in the increased proportion of more pluripotent hESCs as suggested by our AP staining, it could be envisaged that an increase in CDK6 and CDC25A is achieved indirectly via increased homogeneity of hESC cultures on the proviso that CDK6 and CDC25A expression would be high in hESCs and down-regulated upon differentiation. However, this is not the case because CDK6 expression increases during hESC differentiation, whereas CDC25A shows a slight decrease in the first 3 d of differentiation followed by an increase at day 5 (Neganova et al., 2008). Together, these data suggest that NANOG has direct transcriptional effects on CDK6 and CDC25A.
Cdc25A phosphatase has been shown to control entry into and progression through S phase by removing inhibitory phosphates from cyclin E– and cyclin A–bound Cdk2 complexes. Published data have highlighted an important role for CDC25A in both mitotic entry in HeLa cells (Mailand et al., 2002) and S-phase progression in cancer cells (Lehmann and McCabe, 2007; Yao et al., 2007); however, very little is known about CDC25A function in ESCs. Our own data in hESCs have shown that CDC25A is present in hESCs and its expression peaks at G1 (Neganova et al., 2008). In addition, immunoprecipitation assays have shown that CDC25A is associated with active complexes formed between three G1 Cdks (CDK4, CDK6, and CDK2) and the respective cyclins (Neganova et al., 2008). Work described in this manuscript shows that one of the impacts of CDC25A down-regulation is the retention of cells at the G1-phase cycle in both NANOG and control sublines, which is indicative of its involvement in S-phase entry. In addition, its overexpression enhances the numbers of cells entering S phase and shortens the time interval needed for S-phase completion. Our current work has also indicated that down-regulation of CDK2 causes cell stalling at G1 phase of the cell cycle (Neganova et al., 2008), whereas down-regulation of CDK6 delays S-phase entry. The similarity in the phenotype and the results of the immunoprecipitation assays are suggestive of CDC25A acting through one or more G1-specific Cdk–cyclin complexes during S-phase entry. It now remains to be investigated how CDC25A is able to coordinate different functions during cell cycle progression and identify the target Cdk–cyclin complexes that are being regulated by CDC25A at different phases of the cell cycle. In particular, we are keen to investigate further the role of CDC25A and NANOG in S and G2 to M progression because the stalling of hESCs at S and G2 to M was also observed upon down-regulation of these two components. It is likely that more than one Cdk–cyclin complex is involved in this process, and, currently, our experiments are focused on the role of CDC2–CDK1 in hESC cell cycle regulation and likely association with CDC25A. It is interesting to point out that CDC2–CDK1 has also been identified as a transcriptional target of NANOG (Boyer et al., 2005); however, we were unable to identify any significant differences in CDK1 expression between NANOG and control clones (unpublished data).
Having established a role for CDK6 and CDC25A in cell cycle progression in hESCs, we sought to determine whether they were the downstream effectors of NANOG-mediated S entry and S progression effects. We were able to address this question by knockdown experiments in NANOG-overexpressing sublines and overexpression experiments. Data generated by an RNA interference approach clearly indicated that both CDK6 and CDC25A are involved in S-phase completion, for down-regulation of each component alone caused the lengthening of time needed for S phase and/or reduction in the numbers of cells able to enter S phase, thus reversing the effects of NANOG on S-phase entry. Most importantly, overexpression of either of these two genes alone rescued cell retention in G1 caused by NANOG down-regulation, suggesting that NANOG's effects on the S-phase entry of hESCs are mediated by these two cell cycle regulatory components. In this manuscript, we elucidated that NANOG has an effect on S-phase and G2 to M progression because NANOG down-regulation alone causes cell stalling in S and G2 to M in addition to G1. Our work on CDC25A also indicated that CDC25A itself has a function in S progression and G2 to M transition in hESCs. It is likely that other cell cycle regulatory components in addition to CDC25A mediate NANOG's effects on S-phase progression and G2 to M transition in hESCs. Large-scale ChIP–ChIP experiments have suggested that OCT4, SOX2, and NANOG regulate the expression of CDC7, which has been shown to be important for S-phase progression (Boyer et al., 2005). Therefore, it is important that the role of CDC7 is investigated in connection with NANOG in the S-phase progression in hESCs.
It is interesting to note that forced expression of Nanog in 3T3 cells and in a subset of T cells causes enhancement in cell growth as well as resulting in a transformed phenotype (Zhang et al., 2005; Piestun et al., 2006; Tanaka et al., 2007). Murine ESCs with high Nanog expression also show an increased expression of genes involved in cell cycle regulation, whereas cells with low Nanog expression have increased expression of cell cycle inhibitory genes (Singh et al. 2007). Together, these data suggest that Nanog possesses an oncogenic potential that might be related to the role it plays in germ cell tumors and to its function in self-renewal of ESCs. It remains to be investigated whether these oncogenic functions are related to its role in cell cycle regulation and in particular to the direct transactivation of CDK6 and CDC25A genes. This, of course, should be extended to the other two coregulators, OCT4 and SOX2, that occupy a substantial portion of the Nanog target genes and have been described to be involved in oncogenic transformations (de Jong and Looijenga, 2006; Rodriguez-Pinilla et al., 2007). Large-scale ChIP–ChIP experiments have suggested that all three factors regulate the expression of CDC7 shown to be important for S phase, whereas SOX2 and NANOG regulate the expression of CDC2–CDK1, which is crucial for G1 to S and G2 to M progression (Boyer et al., 2005). In addition, CYCLIN D1 and CDK4 have been shown to be transcriptional targets of SOX2 and OCT4, respectively (Boyer et al., 2005; Greco et al., 2007). A schematic summary of the published literature and the data generated from this manuscript is presented in Fig. 8. Collectively, our findings and those of others suggest that the regulation of cell cycle components is likely to be regulated by the important transcriptional network that controls pluripotency and self-renewal, and studies focused on these specific interactions will help us to understand the unusual cell cycle regulation in ESCs.
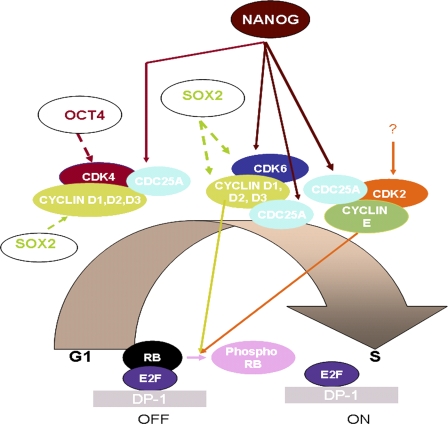
Figure 8.
Schematic presentation of G1 to S transition in hESCs showing the role of NANOG, CDK6, and CDC25A. A question mark is used to indicate lack of information of upstream regulators for CDK2. Data regarding the formation of active complexes between three G1-specific Cdks and CYCLIN D and E as well as CDC25A have been obtained from ongoing work in our group (Neganova et al., 2008). Continuous lines represent data obtained from this manuscript, whereas dashed lines indicate data obtained from literature (Boyer et al., 2005).
Materials and methods
Culture and differentiation of hESCs
hESCs were grown on mitotically inactivated mouse embryonic fibroblasts and passaged essentially as previously described (Stojkovic et al., 2004). EB differentiation was induced by harvesting hESCs with collagenase and culturing them in suspension in knockout DME (Invitrogen) containing 20% FCS (Hyclone), 1 mM l-glutamine (Invitrogen), 100 mM of nonessential amino acids (Invitrogen), 100 μM β-mercaptoethanol (Sigma-Aldrich), and 1% penicillin-streptomycin (Sigma-Aldrich). One to two passages before experiments, hESCs were transferred to Matrigel (BD)-coated plates with feeder-conditioned media as previously described (Stojkovic et al., 2004; Hyslop et al., 2005).
Karyotype analysis of hESCs
The karyotype of hESCs was determined by standard G-banding procedure.
Stable transfection of hESCs with the full-length cDNA of human NANOG, CDK6, and CDC25A
The full-length cDNA of human NANOG, CDK6, and CDC25A was isolated from cDNA of hESCs using the following oligonucleotides: NANOG forward, 5′-CATGAGTGTGGATCCAGCTTGT-3′; NANOG reverse, 5′-ATCTTCACACGTCTTCAGGTTG-3′; CDK6 forward, 5′-ACTGAATTCACCATGGAGAAGGACGGCCTGTG-3′; CDK6 reverse, 5′-ACTGAATTCTCAGGCTGTATTCAGCTCCGAG-3′; CDC25A forward, 5′-ACTGAATTCACCATGGAACTGGGCCCGGAG-3′; and CDC25A reverse, 5′-ACTGAATTCTCAGAGCTTCTTCAGACGACTG-3′. The cDNAs were cloned into the pTP6 vector. hESCs (H1, H9, and hES-NCL1) were plated on Matrigel-coated plates and cultured in the presence of feeder-conditioned media 4 d before transfection. The transfection of DNA was achieved using the Cell Line Nucleofector kit L (Amaxa) according to the manufacturer's instructions (program A-023). 2 d after the transfection, stable clones were selected using puromycin selection (0.8–1.2 μg/ml) for 10 d. Between 15 and 20 surviving colonies were pooled in each case, and the resulting subline from each cell line was expanded and named, for example, H1 NANOG, H9 NANOG, and hES-NCL1 NANOG. A similar procedure was performed after transfection of the empty vector. Each of the control sublines was named H1 control, H9 control, and hES-NCL1 control. All sublines were maintained with 0.6 μg/ml puromycin to ensure the maintenance of the transgene. Every 8–10 passages, quantitative RT-PCR and Western blot analysis were performed to confirm gene overexpression over time. For simplicity, in most figures, data from one or two overexpressing sublines or controls are shown.
Transient transfection of hESCs with CDK6 and CDC25A luciferase reporter constructs
hESCs were cultured under feeder-free conditions with feeder-conditioned media free of antibiotics for at least 4 d before transfections. hESCs were nucleofected simultaneously with firefly luciferase reporter constructs (6 μg in the case of CDK6 and CDC25A), a transfection control (0.6 μg in the case of the construct containing Renilla luciferase gene driven by the herpes simplex virus thymidine kinase promoter), and 6 μg NANOG cDNA (gift from J.-H. Kim, ChaBiotech Co. Ltd., Seoul, Republic of Korea) using the Cell Line Nucleofector kit L according to the manufacturer's instructions (program A-023). Site-directed mutagenesis for CDK6 and CDC25A luciferase constructs was performed using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. A similar procedure was followed for the transfection of site-directed mutagenesis constructs. After 24 h, cells were lysed using the lysis buffer provided in the Dual Luciferase Detection kit (Promega) according to the manufacturer's instructions. The firefly and Renilla luciferase activities were measured in turn using the LARII and Stop Glow solutions (Promega), and the ratio between the two was calculated.
siRNAs and transfection
siRNAs were obtained from Santa Cruz Biotechnology, Inc. and Invitrogen. The siRNA sequences are shown in Table S1. Transfection with scrambled control siRNAs with similar guanine-cytosine content to gene-specific siRNA sequences provided by the same company were used as a negative control. The transfection of siRNA into hESCs was performed using the high efficiency Cell Line Nucleofector kit L and 80 pmol siRNA (in 2 ml of medium) as outlined in the manufacturer's instructions (program A-023). 24 h after transfection, hESCs were synchronized in G2 to M by incubation in 200 ng/ml of a nocodazole-containing medium for 18 h. The cells were washed three times with normal medium and collected by Accutase (Millipore) treatment at various time points as indicated in the results section.
Flow cytometry analysis of hESCs
For the flow cytometry analysis, the hESCs were collected, processed, and analyzed as previously described (Armstrong et al., 2006).
Western blotting
Lysates were electrophoresed on a 10% SDS-PAGE gel and electrophoretically transferred to a polyvinylidene difluoride membrane (Hybond-P; GE Healthcare). Membranes were blocked in Tris-buffered saline with 5% milk and 0.1% Tween. The blots were probed with NANOG (1:1,000; R&D Systems), CDK4 (1:100; Santa Cruz Biotechnology, Inc.), CDK6 (1:100; Santa Cruz Biotechnology, Inc.), cyclin D1, D2, and D3 (1:100; all from Santa Cruz Biotechnology, Inc.), cyclin E, CDK2, CDC25A, c-ABL, retinoblastoma (phosphorylated or not), and c-Myc (1:100; all from Cell Signaling Technology), p15, p16, p18, p19, and Suv39H1 (1:100; all from Cell Signaling Technology), or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (1:2,000; Abcam) overnight and revealed with horseradish peroxidase–conjugated secondary antibodies, anti–goat (1:2,000; Dako), or anti–rabbit (1:20,000; GE Healthcare). Antibody–antigen complexes were detected using ECL reagent (GE Healthcare). Western blot images were acquired using a luminescent image analyzer (Fujifilm) and LAS-3000 software (Fujifilm). Protein molecular weights are indicated next to the image shown from the Western blotting.
Cell signaling assays
Panorama antibody microarray for cell signaling containing 224 different antibodies spotted in duplicate on nitrocellulose-coated glass was purchased from Sigma-Aldrich. 1 mg of NANOG-overexpressing or control subline cell extracts was collected, labeled with Cy3 and Cy5, respectively, and hybridized to the slides according to the manufacturer's instructions. Cy3 and Cy5 signals were read on the Gene Pix Pro 4.0 (MDS Analytical Technologies). The results from the NANOG sublines were analyzed together using the GeneSpring software (Agilent Technologies). Sample processing was performed using three normalization steps, which involved dye swap where necessary, the division of each spot by the control channel, and the normalization of each spot to the 50th percentile of the entire chip. Filter-on-confidence criteria was used to select the most significantly changed candidates (P < 0.05). A ratio of >1.0 indicates higher expression in both NANOG sublines compared with both respective control hESC sublines, and a ratio <1.0 indicates higher expression in control hESC control sublines compared with NANOG sublines.
LightCycler real-time PCR analysis
Quantitative RT-PCR analysis was performed using QuantiTect SYBR Green PCR Master Mix (QIAGEN) essentially as previously described (Boyer et al., 2005; Becker et al., 2006). The LightCycler experimental run protocol used was: PCR activation step (95°C for 15 min), amplification with data acquisition repeated 50 times (94°C for 15 s, annealing temperature for primers for 30 s, and 72°C for 20 s with a single fluorescence data collection), melting curve (60–95°C with a temperature transition rate of 0.1°C/s and continuous fluorescence data collection), and finally cooling to 40°C. The crossing point for each transcript was determined using the second derivative maximum method in the LightCycler software version 3.5.3 (Roche). The GAPDH crossing point for each sample was used as the internal control of these real-time analyses. The data were analyzed using the comparative threshold cycle method as described in the user bulletin for the LightCycler relative quantification software version 1.01 (Roche). For each gene, the control was set to one, and all other values were calculated with respect to this. PCR reactions were performed using the primers (final concentration of 0.5 μM) described in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1).
Apoptosis assay
Cells undergoing apoptosis can be enumerated using the annexin V–FITC apoptosis detection kit (BD). The protocol was performed in accordance with the manufacturer's instructions and, in brief, comprises the following steps. Cells were harvested using Accutase, washed twice with ice-cold phosphate-buffered saline, and counted. 105 cells were suspended in 100 μl of 1× binding buffer (supplied), and 5 μl annexin V–FITC and 5 μl propidium iodide solution were added. The mixture was vortexed gently and incubated for 15 min at room temperature in the dark. 400 μl of 1× binding buffer was added, and the cells were analyzed by flow cytometry (FACSCalibur; BD).
Measurement of cell proliferation using BrdU incorporation method
hESC proliferation was measured by incorporation of BrdU (Roche) into the genomic DNA during the S phase (DNA replication) of the cell cycle. hESCs were grown in a 4-well plate to day 2 and incubated in medium containing BrdU for 45 min at 37°C in a humidified atmosphere (5% CO2). Cells were fixed with ethanol and 50 mM glycine, pH 2.0, for 45 min at room temperature and denaturated in 4 M HCl for 10–20 min. Subsequent detection of BrdU was accomplished with antibodies for BrdU (1:5) according to the manufacturer's instructions and visualization at 488 nm using immunofluorescence microscopy. For flow cytometry assay, hESCs were incubated and processed with a BrdU Flow kit (BD) according to the manufacturer's protocol. Cells were stained with FITC or allophycocyanin anti-BrdU and 7-amino-actinomycin. Cells from the same population that were not BrdU labeled were used as a negative control. Flow cytometry analysis was performed using a FACSCalibur and CellQuest software (BD).
Cell cycle analysis
Cell cycle analysis was performed using the CycleTest Plus DNA reagent kit (BD). hESCs were harvested by Accutase treatment and counted with a hemocytometer. 500,000 cells were fixed, permeabilized, and stained in accordance with the manufacturers' instructions, and the sample was analyzed by flow cytometry using a FACSCalibur measuring FL2 area versus total counts. The data were analyzed using ModFit (Tree Star, Inc.) and FlowJo (Tree Star, Inc.) softwares to generate the percentages of cells in G1, S, and G2 to M phases of the cell cycle.
AP staining
The AP staining was performed using the Alkaline Phosphatase Detection kit (Millipore) according to the manufacturer's instructions. Cells were fixed in 90% methanol and 10% formamide for 2 min and washed with rinse buffer (20 mM Tris-HCl, pH 7.4, and 0.05% Tween 20) once. Staining solution (Naphthol/Fast Red Violet) was added to the wells, and plates were incubated in the dark for 15 min. The bright field images were obtained using a microscope (Axiovert; Carl Zeiss, Inc.) and AxioVision software (Carl Zeiss, Inc.).
ChIP experiments
ChIP assays were performed mainly as previously described (Atkinson et al., 2005). In brief, cells were harvested at 70–80% confluence, and ChIP was performed according to the manufacturer's instructions (Millipore). Sonication was optimized to produce chromatin fragments of 500–1,000 bp in length, and DNA from each immunoprecipitation was purified using the Qiaquick DNA Purification kit (QIAGEN). Also included in the experiment was a no antibody control immunoprecipitate to detect any background, and, if it was present, it was subtracted from each immunoprecipitate within that experiment. Pilot experiments performed with no antibody controls and irrelevant antibodies such as IgG revealed no significant differences; thus, no antibody controls were used in all ChIP experiments. Standard errors were generated for quantitative PCR reactions by reading each sample in triplicate. The sequences of the primers used for this purpose are given in Table S3.
Immunoprecipitation experiments
hESCs were washed with ice-cold PBS and lysed on ice for 30 min in radio immunoprecipitation assay buffer. Lysates were centrifuged at 14,000 g for 5 min. The supernatant from cell lysates was collected, and the protein concentration was measured using Bradford Reagent (Bio-Rad Laboratories). Protein G–agarose (PGA) beads were washed three times with PBS and incubated for 2 h in a rotor at 4°C in radio immunoprecipitation assay buffer with PMSF and protease inhibitor cocktail (Roche). 400 μg of protein recovered from cell supernatants was precleared with 20 μl PGA slurry for at least 2 h on a rotor at 4°C. PGA beads were removed by centrifugation at 14,000 g for 5 min at 4°C. Immunoprecipitation was performed by overnight incubation/rotation with 2 μg of mouse monoclonal anti-CDK2 antibody (D-12; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-CDK4 (C-22; Santa Cruz Biotechnology, Inc.), or rabbit polyclonal anti-CDK6 antibody (C-21; Santa Cruz Biotechnology, Inc.). A no antibody control was also included for each sample. After incubation, 20 μl of PGA beads was added to immunoprecipitated samples and returned to 4°C for 2 h with rotation. PGA beads with bound protein complexes were recovered by centrifugation at 14,000 g for 5 min, and beads were washed once with PBS and 0.2% Triton X-100 and twice with PBS. The sample was divided into two aliquots: one to be used for kinase assays and the second one for Western blotting. For the latter procedure, 40–60 μl of SDS sample buffer was added to the sample before boiling for 5 min. The samples were separated using denaturing acrylamide gels, and Western blotting was performed as indicated above.
Kinase activity assays
Kinase activity assays were performed using the PKLight Assay kit (LT07-500; Cambrex Bio Science Rockland, Inc.) according to the manufacturer's instructions. The PKLight Assay exploits the kinases' intrinsic ATPase activity, resulting in the cleavage of the γ-phosphate moiety of ATP and its subsequent insertion into the target substrate. This results in the phosphorylation of the substrate and the conversion of ATP to ADP. The PKLight Assay measures the consumption of ATP and is based on the bioluminescent measurement of the remaining ATP present in the wells after the kinase reaction. The bioluminescent signal of PKLight Assay is inversely proportional to kinase activity. Phosphorylation of Retinoblastoma or H1 was measured by incubating for 10 min at room temperature 20 μl of immunoprecipitation product for the kinase of interest (see previous section) with 1 mM ATP, kinase buffer (50 mM Tris, pH 7.5, and 5 mM MgCl2), and 5 mg/ml Retinoblastoma or H1 as substrate. 10 μl of kinase stop solution was added to each sample at room temperature for 10 min. Finally, 20 μl of ATP detection reagent was added to each sample at room temperature for 10 min, and the readings were taken using a luminometer. The difference in luminometer reading between the no antibody control and immunoprecipitation product containing the antibody was calculated. This figure, which is indicative of remaining ATP in the solution, was inversely correlated to the kinase activity. The kinase activity for the control sublines was set at 100%, and the respective values for the NANOG sublines were calculated with respect to that.
Phosphatase assays
These were performed using the SensoLyte fluorescein diphosphate (FDP) protein phosphatase assay kit (AnaSpec) according to the manufacturer's instructions. This kit provides a fluorogenic assay for measuring the activity of protein phosphatases such as tyrosine phosphatases and serine/threonine phosphatases that convert the FDP into fluorescein, which has a high extinction coefficient and emission quantum yield, therefore providing high assay sensitivity. Immunoprecipitations were performed using CDC25A antibody (F6; Santa Cruz Biotechnology, Inc.). The immunoprecipitation product was resuspended in 50 μl of phosphatase buffer (20 mM Tris-HCL, pH 8.3, 150 mM NaCL, 2 mM EDTA, 0.01% Triton X-100, 5 mM DTT, and 1 mg/ml BSA). 50 μl of a protein phosphatase–containing sample was mixed with 50 μl of FDP reaction solution. The reaction was incubated at 30°C for 30 min, and 50 μl of stop solution was added to stop the reaction. Fluorescence signal was measured using excitation/emission = 485 nm/538 nm. As a negative control, samples without phosphatase activity (distilled water) were used. The difference in fluorescence readings between the immunoprecipitation product and no antibody control was calculated to deduct background phosphatase activity. The phosphatase activity for the control sublines was set at 100%, and the respective values for the experiment sublines were calculated with respect to that.
Statistical analysis
Two-tailed pairwise Student's t test was used to analyze results obtained from two samples with one time point. Analysis of variance (single factor or two factors with replication) was used to compare multiple samples (at one time or several time points). The results were considered significant if P < 0.05.
Tumor formation in SCID mice
All procedures involving mice were performed in accordance with institutional guidelines and permission. Approximately 106 hESCs were injected into the testis of adult male SCID mice. After 70–90 d, mice were killed, and tissues were dissected, fixed in Bouins overnight, processed, and sectioned according to standard procedures and stained with either hematoxylin and eosin or Weiger's stain. Material for immunohistochemical analysis was fixed in 4% PFA (Sigma-Aldrich) in PBS (Cambrex Bio Science Rockland, Inc.) overnight, processed, and sectioned to 6 μm according to standard procedures. Sections were cleared using Histoclear (RA Lamb) and rehydrated, and endogenous hydrogen peroxide activity was blocked. Antigen retrieval was performed by microwaving (800 W) tissues in 10 mM of citrate buffer, pH 6 (citric acid [Sigma-Aldrich] and distilled H2O). Endogenous avidin/biotin activity was blocked using a blocking kit (Vector Laboratories). Sections were permeabilized (1% Triton X-100 [Thermo Fisher Scientific] and PBS solution) and blocked (5% normal goat serum [Invitrogen], 0.1% Triton X-100, and PBS), and sections were incubated with the following primary antibodies: AFP (1:100; Sigma-Aldrich), nestin (1:200; Millipore), and SMA (1:200; Sigma-Aldrich). Negative controls were performed with the omission of the primary antibody. A universal ABC detection kit (Vector Laboratories) with a purple-colored Vector VIP substrate (Vector Laboratories) was used to detect the primary antibodies. Sections were briefly counterstained using Mayer's hemalum and briefly blued using 4% alkaline alcohol (4% ammonia [Thermo Fisher Scientific] in 70% alcohol). Sections were dehydrated through a series of alcohols, cleared using Histoclear, and mounted using distyrene/plasticizer/xylene (RA Lamb).
Microscopy
Teratoma sections were visualized using a microscope (Diaphot 300; Nikon) with the following objectives: 4× NA 0.13, 10× NA 0.25, 20× NA 0.40, and 40× NA 1.3. Digital images were recorded using a digital camera (DXM1200; Nikon).
Online supplemental material
Fig. S1 shows karyotype analysis of H1 NANOG and hES-NCL1 NANOG clones after 20 passages in culture. Fig. S2 shows cell proliferation assessed by cell counting over three time points. Fig. S3 shows the maintenance of pluripotency and differentiation capability of NANOG-overexpressing hESC clones. Fig. S4 shows that the C-terminal domain of NANOG is responsible for transactivation of CDK6 and CDC25A. Table S1 shows the sequences of siRNAs used for the down-regulation of CDK6, CDC25A, and NANOG. Table S2 shows the sequences of oligonucleotides used for the quantitative RT-PCR analysis. Table S3 shows the sequences of oligonucleotides used for the quantitative PCR after ChIP experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801009/DC1.
Supplementary Material
Acknowledgments
We are grateful to Dr. Louise Hyslop, Dr. Rebecca Stewart, and Theresia Walter for the isolation of full-length NANOG cDNA and carrying out some of the transfections of full-length NANOG cDNA and control plasmid in hESCs. We also thank Ian Dimmick for help with setting up the BrdU flow cytometry analysis, Iliana Paraskevopoulou for carrying out the CDK1 expression analysis, Dr. Nick Allenby for help with the reading of antibody arrays, Jerome Evans for performing the karyotypic analysis, and Petra Stojkovic, Sun Yung, and Dennis Kirk for technical assistance.
This study was supported by Medical Research Council grant G0301182, Biotechnology and Biological Sciences Research Council grant BBS/B/14779, One North East Regional Development Agency, Sir James Knott Trust, and Life Knowledge Park. X. Zhang and I. Neganova performed most of the laboratory work, data collection, and interpretation; S. Przyborski, C. Yang, M. Cooke, S. Fenyk, S.P. Atkinson, G. Anyfantis, W.N. Keith, S.F. Hoare, and O. Hughes set up and performed some of the specialized laboratory techniques; T. Strachan and P.W. Hinds helped with manuscript writing and reagents; M. Stojkovic and P.W. Hinds provided vital reagents needed for this work; and L. Armstrong and M. Lako performed some of the laboratory work, designed the study, and wrote the manuscript.
M. Stojkovic's present address is Centro de Investigación Príncipe Felipe, 46012 Valencia, Spain.
Abbreviations used in this paper: AFP, α-fetoprotein; ChIP, chromatin immunoprecipitation; EB, embryoid body; ESC, embryonic stem cell; FDP, fluorescein diphosphate; GAPDH; glyceraldehyde 3-phosphate dehydrogenase; hESC, human ESC; PGA, protein G–agarose; SCID, severe combined immunodeficient; SMA, smooth muscle actin.
References
- Armstrong, L., O. Hughes, S. Yung, L. Hyslop, R. Stewart, I. Wappler, H. Peters, T. Walter, P. Stojkovic, J. Evans, et al. 2006. The role of PI3K/AKT, MAPK/ERK and NFkappabeta signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum. Mol. Genet. 15:1894–1913. [DOI] [PubMed] [Google Scholar]
- Atkinson, S.P., S.F. Hoare, R.M. Glasspool, and W.N. Keith. 2005. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 65:7585–7590. [DOI] [PubMed] [Google Scholar]
- Becker, K.A., P.N. Ghule, J.A. Therrien, J.B. Lian, J.L. Stein, A.J. van Wijnen, and G.S. Stein. 2006. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 209:883–893. [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., T.I. Lee, M.F. Cole, S.E. Johnstone, S.S. Levine, J.P. Zucker, M.G. Guenther, R.M. Kumar, H.L. Murray, R.G. Jenner, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon, T., A. Smith, and P. Savatier. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12:432–438. [DOI] [PubMed] [Google Scholar]
- Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 113:643–655. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Z. Du, and Z. Yao. 2006. Roles of the Nanog protein in murine F9 embryonal carcinoma cells and their endoderm-differentiated counterparts. Cell Res. 16:641–650. [DOI] [PubMed] [Google Scholar]
- Darr, H., Y. Mayshar, and N. Benvenisty. 2006. Over-expression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 133:1193–1201. [DOI] [PubMed] [Google Scholar]
- de Jong, J., and L.H. Looijenga. 2006. Stem cell marker OCT3/4 in tumor biology and germ cell tumor diagnostics: history and future. Crit. Rev. Oncog. 12:171–203. [DOI] [PubMed] [Google Scholar]
- Deb-Rinker, P., D. Ly, A. Jezierski, M. Sikorska, and P.R. Walker. 2005. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J. Biol. Chem. 280:6257–6260. [DOI] [PubMed] [Google Scholar]
- Greco, S.J., K. Liu, and P. Rameshwar. 2007. Functional similarities among genes regulated by OCT4 in human mesenchymal and embryonic stem cells. Stem Cells. 25:3143–3154. [DOI] [PubMed] [Google Scholar]
- Hyslop, L., M. Stojkovic, L. Armstrong, T. Walter, P. Stojkovic, S. Przyborski, M. Herbert, A. Murdoch, T. Strachan, and M. Lako. 2005. Down-regulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells. 23:1035–1043. [DOI] [PubMed] [Google Scholar]
- Lehmann, G.M., and M.J. McCabe. 2007. Arsenite slows S phase progression via inhibition of cdc25a dual specificity phosphatase gene transcription. Toxicol. Sci. 99:70–78. [DOI] [PubMed] [Google Scholar]
- Lin, T., C. Chao, S. Saito, S.J. Mazur, M.E. Murphy, E. Appella, and Y. Xu. 2005. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7:165–171. [DOI] [PubMed] [Google Scholar]
- Mailand, N., A.V. Podtelejnikov, A. Groth, M. Mann, J. Bartek, and J. Lukas. 2002. Regulation of G2/M events by CDC25A through phosphorylation-dependent modulation of its stability. EMBO J. 21:5911–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 113:631–642. [DOI] [PubMed] [Google Scholar]
- Neganova, I., X. Zhang, S.P. Atkinson, and M. Lako. 2008. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 10.1038/onc.2008.358. [DOI] [PubMed]
- Oh, J.H., H.J. Do, H.M. Yang, S.Y. Moon, K.Y. Cha, H.M. Chung, and J.H. Kim. 2005. Identification of a putative transactivation domain in human Nanog. Exp. Mol. Med. 37:250–254. [DOI] [PubMed] [Google Scholar]
- Pereira, L., F. Yi, and B.J. Merrill. 2006. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 26:7479–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piestun, D., B.S. Kochupurakkal, J. Jacob-Hirsch, S. Zeligson, M. Koudritsky, E. Domany, N. Amariglio, G. Rechavi, and D. Givol. 2006. Nanog transforms NIH3T3 cells and targets cell-type restricted genes. Biochem. Biophys. Res. Commun. 343:279–285. [DOI] [PubMed] [Google Scholar]
- Pratt, T., L. Sharp, J. Nichols, D.J. Price, and J.O. Mason. 2000. Embryonic stem cells and transgenic mice ubiquitously expressing a tau-tagged green fluorescent protein. Dev. Biol. 228:19–28. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pinilla, S.M., D. Sarrio, G. Moreno-Bueno, Y. Rodriguez-Gil, M.A. Martinez, L. Hernandez, D. Hardisson, J.S. Reis-Filho, and J. Palacios. 2007. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod. Pathol. 20:474–481. [DOI] [PubMed] [Google Scholar]
- Singh, A.M., T. Hamazaki, H.E. Hankowski, and N. Terada. 2007. A heterogenous expression pattern for Nanog in embryonic stem cells. Stem Cells. 25:2534–2542. [DOI] [PubMed] [Google Scholar]
- Stojkovic, M., M. Lako, P. Stojkovic, R. Stewart, S. Przyborski, L. Armstrong, J. Evans, M. Herbert, L. Hyslop, S. Ahmad, A. Murdoch, and T. Strachan. 2004. Derivation of human embryonic stem cells from day-8 blastocysts recovered after three-step in vitro culture. Stem Cells. 22:790–797. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., T. Era, S.I. Nishikawa, and S. Kawamata. 2007. Forced expression of Nanog in hematopoietic stem cells results in a {gamma}{delta}T cell disorder. Blood. 110:107–115. [DOI] [PubMed] [Google Scholar]
- Wang, J., S. Rao, J. Chu, X. Shen, D.N. Levasseur, T.W. Theunissen, and S.H. Orkin. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature. 444:364–368. [DOI] [PubMed] [Google Scholar]
- Yao, B., J. Fu, E. Hu, Y. Qi, and Z. Zhou. 2007. The Cdc25A is involved in S-phase checkpoint induced by benzo(a)pyrene. Toxicology. 237:210–217. [DOI] [PubMed] [Google Scholar]
- Yu, J., M.A. Vodyanik, K. Smuga-Otto, J. Antosiewicz-Bourget, J.L. Frane, S. Tian, J. Nie, G.A. Jonsdottir, V. Ruotti, R. Stewart, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318:1917–1920. [DOI] [PubMed] [Google Scholar]
- Zaehres, H., M.W. Lensch, L. Daheron, S.A. Stewart, J. Itskovitz-Eldor, and G.Q. Daley. 2005. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 23:299–305. [DOI] [PubMed] [Google Scholar]
- Zhang, J., X. Wang, B. Chen, G. Suo, Y. Zhao, Z. Duan, and J. Dai. 2005. Expression of Nanog gene promotes NIH3T3 cell proliferation. Biochem. Biophys. Res. Commun. 338:1098–1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.