Abstract
Nuclear pore complexes (NPCs) mediate all nucleocytoplasmic traffic and provide docking sites for the spindle assembly checkpoint (SAC) protein Mad1p. Upon SAC activation, Mad1p is recruited onto kinetochores and rapidly cycles between NPCs and kinetochores. We examined the mechanism of Mad1p movement onto kinetochores and show that it is controlled by two components of the nuclear transport machinery, the exportin Xpo1p and Ran–guanosine triphosphate (GTP). Mad1p contains a nuclear export signal (NES) that is recognized by Xpo1p. The NES, Xpo1p, and RanGTP are all required for Mad1p recruitment onto kinetochores in checkpoint-activated cells. Consistent with this function, Xpo1p also accumulates on kinetochores after SAC activation. We have also shown that Xpo1p and RanGTP are required for the dynamic cycling of Mad1p between NPCs and kinetochores in checkpoint-arrested cells. These results reveal an important function for Xpo1p in mediating intranuclear transport events and identify a signaling pathway between kinetochores and NPCs.
Introduction
Nuclear pore complexes (NPCs) control macromolecular movement across the nuclear envelope. The gateways formed by NPCs selectively allow soluble nuclear transport factors (many referred to as karyopherins [kaps] or importins and exportins) and their cargoes to partition across the nuclear envelope (for review see Hetzer et al., 2005). Importins bind to cargoes containing NLSs, whereas exportins bind to nuclear export signals (NESs). A characteristic structural feature of kaps is a domain capable of binding RanGTP. Ran (Gsp1p in yeast) is primarily found in the nucleus, where the chromatin-bound Ran–guanine nucleotide exchange factor (GEF; Rcc1 in vertebrates and Prp20p in yeast) maintains it in a GTP-bound form. For importins, RanGTP binding induces conformational changes in the kaps that release cargoes in the nucleoplasm. Conversely, RanGTP binding to exportins increases their affinity for NES-containing cargoes. The resulting trimeric complex is stable until exported into the cytoplasm, where it encounters a second Ran effector, Ran–GTPase-activating protein (GAP) (RanGAP1 in vertebrates and Rna1p in yeast), which binds RanGTP and stimulates its GTPase activity. Conversion to RanGDP destabilizes the export complex and allows cargo release.
In addition to their roles in nucleocytoplasmic trafficking, various elements of the transport machinery have been implicated in mitotic events, with most of this work conducted in vertebrate model systems. For example, the vertebrate Nup107–160 subcomplex is recruited to kinetochores during mitosis (Loiodice et al., 2004), where it may function in kinetochore–microtubule attachment (Zuccolo et al., 2007). These Nups may also function as a binding site at kinetochores for the exportin Crm1 (Zuccolo et al., 2007). During mitosis, Crm1 associates with kinetochores and together with RanGTP targets RanGAP1 and Nup358/RanBP2 to kinetochores (Arnaoutov et al., 2005). Disrupting these events results in both microtubule–kinetochore attachment defects and errors in chromosome segregation (Arnaoutov et al., 2005).
The concentration of vertebrate RanGAP1 at mitotic kinetochores suggests that the conversion of RanGTP to RanGDP predominates at kinetochores. What role this plays in kinetochore function is unclear. Interestingly, adjacent to kinetochores, RanGEF is concentrated on condensed chromosomes, and RanGTP predominates near these structures. One function of this RanGTP cloud is to activate spindle assembly factors (for review see Kalab and Heald, 2008). These observations represent a growing number of results linking Ran and Ran-binding proteins, including kaps in spindle, centrosome, and kinetochore structure and function.
A less well-understood observation is the sequestration of two spindle assembly checkpoint (SAC) proteins, Mad1 and Mad2, at NPC (Campbell et al., 2001; Iouk et al., 2002). These proteins reside at NPC in interphase and are recruited to kinetochores before spindle attachment. Here, they function to transduce a metaphase arrest signal from unattached kinetochores by producing a soluble complex containing Mad2 that is capable of binding Cdc20 and inhibiting its activation of the anaphase-promoting complex until all kinetochores are engaged by spindle microtubules (for review see Musacchio and Salmon, 2007). Why Mad1 and Mad2 associate with NPC has remained an enigma. A study of yeast Mad1p and Mad2p has shown that in checkpoint-arrested cells, the majority of Mad1p (>60%) rapidly cycles between kinetochores and NPCs by a mechanism that is energy dependent (Scott et al., 2005). In this study, we show that the exportin Xpo1p together with RanGTP functions in an intranuclear-targeting event that directs Mad1p and Xpo1p to unattached kinetochores upon SAC activation. Moreover, Xpo1p, RanGTP, and the conversion of RanGTP to RanGDP are required for the cycling of Mad1p between NPC-binding sites and kinetochores in SAC-activated cells. These results have revealed a role for exportins in intranuclear targeting events and their regulation by the SAC machinery.
Results and discussion
Monitoring Mad1p movement between NPCs and kinetochores
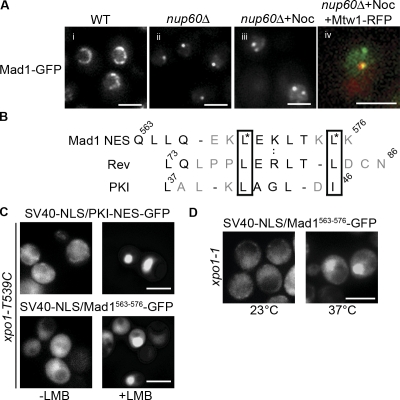
Yeast Mad1p is bound to NPCs throughout the cell cycle; however, after activation of SAC, a pool of Mad1p is visible at unattached kinetochores until termination of the checkpoint (Iouk et al., 2002; Gillett et al., 2004). The association of Mad1p with the NPC is dependent on Nup60p and two proteins, Mlp1p and Mlp2p, that form fibers extending from the nucleoplasmic face of NPC. Removal of Nup60p (nup60Δ) releases Mlp1p and Mlp2p from NPC, leading to their concentration in a single intranuclear focus that retains Mad1p binding (Fig. 1 A; Feuerbach et al., 2002; Scott et al., 2005). This focus can be clearly distinguished from kinetochores, allowing visualization of Mad1p movement from its NPC-binding sites to kinetochores after nocodazole-induced disruption of microtubules and activation of the SAC (Figs. 1 A and 2 A; Scott et al., 2005).
Figure 1.
Mad1p contains a functional Xpo1p-dependent NES. (A) Mad1-GFP was visualized by fluorescence microscopy in WT (Y3028, i) or nup60Δ cells (Y3040, ii and iii; Y3057, iv). Mad1-GFP intranuclear Mlp1p/Mlp2p foci in the absence of Nup60p (ii). Treatment of nup60Δ cells with nocodazole induces recruitment of Mad1-GFP to kinetochores (iii) as conferred by colocalization with Mtw1-RFP (iv). (B) An alignment of the predicted NES in Mad1p with the NESs of HIV Rev and rabbit PKI is shown. Residues required for NES function in HIV Rev and PKI are boxed together with leucine residues in the Mad1p NES (asterisks). (C) Plasmids encoding SV40-NLS/PKI-NES-GFP and SV40-NLS/MAD1563–576-GFP3 were introduced into a strain expressing xpo1-T539C (Y3105 + pKW711). The localization of these reporters was examined by fluorescence microscopy before or 30 min after the addition of LMB. (D) SV40-NLS/MAD1563–576-GFP3 was introduced into the xpo1-1 Ts strain (Y3105 + pKW457) and observed at the permissive (23°C) or restrictive temperature (30 min at 37°C). Bars: (A) 2 μm; (C and D) 5 μm.
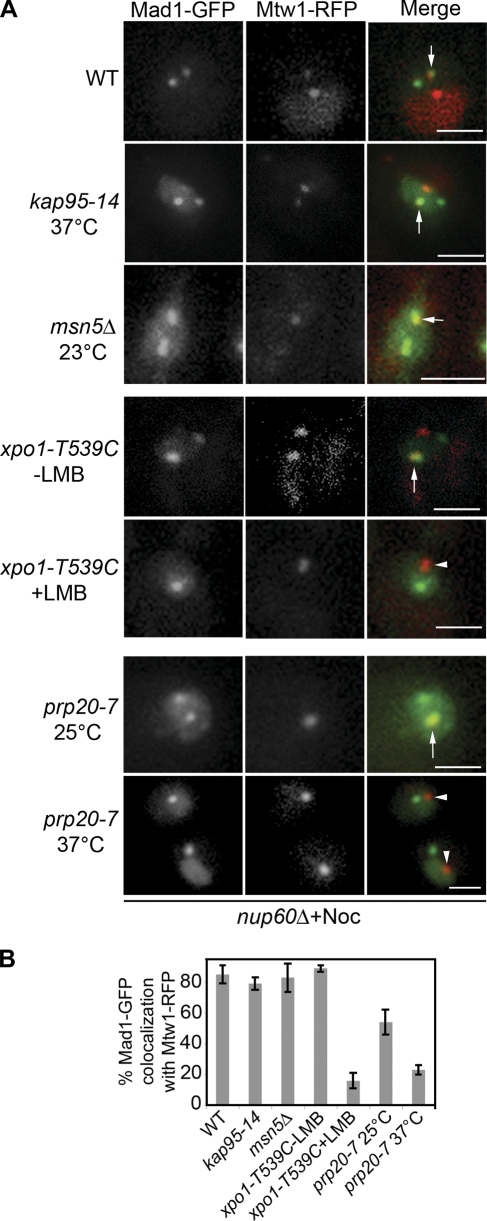
Figure 2.
Xpo1p-dependent targeting of Mad1p to kinetochores. (A) MAD1-GFP and MTW1-RFP in nup60Δ strains containing an otherwise WT genotype (Y3057) or the mutant alleles kap95-14 (Y3161), msn5Δ (Y3116), xpo1-T539C (Y3156), or prp20-7 (Y3091) were examined under the indicated conditions of temperature or LMB addition and after nocodazole addition. The localization of Mad1-GFP and the kinetochore protein Mtw1-RFP was examined by confocal microscopy. Note that as described previously (Gillett et al., 2004), some nocodazole-treated cells contain two kinetochore foci, with one lacking attached microtubules and being capable of binding Mad1p. Arrows point to overlapping Mad1-GFP and Mtw1-RFP foci. Arrowheads point to kinetochores devoid of Mad1-GFP. (B) Quantification of Mad1-GFP and Mtw1-RFP colocalization from the experiments presented in A. The results of three experiments were combined, and SD (error bars) is shown in each case. Bars, 2 μm.
Mad1p contains a functional NES
Mad1p contains a Kap60p/Kap95p-specific NLS that is required for its efficient association with NPCs (Scott et al., 2005). We also detected a potential NES within residues 563–576 that aligned with the NESs of HIV Rev and protein kinase inhibitor (PKI). The Mad1p NES contains critical leucine residues and adjacent charged residues, suggesting that this region lies at the surface of Mad1p (Fig. 1 B). To test the functionality of the putative NES, we used a competition assay in which NES and an NLS are fused to three tandemly repeated GFP molecules (Stade et al., 1997). If both signals are functional, the fusion equilibrates between the nuclear and cytoplasmic compartments. Inactivation of the cognate importin or exportin results in a redistribution of the reporter to the cytoplasm or nucleus as a result of the continued activity of the unaffected kap. To test whether Mad1563–576 functions as an NES, we constructed an SV40-NLS/Mad1563–576-GFP3 fusion and examined its localization in an xpo1-T539C mutant. The mutant protein functioned like wild-type (WT) Xpo1p but was sensitive to the fungal toxin leptomycin B (LMB; Neville and Rosbash, 1999). A control reporter, SV40-NLS/PKI-NES-GFP3, and the SV40-NLS/Mad1563–576-GFP3 chimera were visible throughout the cell (Fig. 1 C, top left). Upon addition of LMB to cells, both reporters accumulated in the nucleus, suggesting that Xpo1p recognizes Mad1563–576 as an NES (Fig. 1 C). Blocking Xpo1p function using an xpo1-1 temperature-sensitive (Ts) mutant yielded similar results (Fig. 1 D, 37°C). Of note, we also observed that the xpo1-1 mutation caused a decrease in NPC-associated Mad1p (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200804098/DC1). We conclude from these data that Mad1p contains an Xpo1p-specific NES that contributes to its NPC localization.
SAC-induced accumulation of Mad1p to kinetochores is dependent on Xpo1p
We further examined whether kaps play a role in the association of Mad1p with kinetochores during SAC arrest. Mutations in KAP95 (kap95-14; Leslie et al., 2002), MSN5 (msn5Δ), or XPO1 (xpo1-T539C) were introduced into a nup60Δ strain, and the localization of Mad1-GFP was examined after nocodazole-induced activation of the SAC. Mad1-GFP targeting to kinetochores was unaffected by the msn5Δ or kap95-14 mutation and appeared similar to that seen in WT cells. In contrast, LMB inactivation of xpo1-T539C prevented Mad1-GFP accumulation at kinetochores (Fig. 2). These results implicated Xpo1p in the targeting of Mad1p to kinetochores. However, LMB did not affect the localization of the kinetochore protein Mtw1p or the SAC protein Bub1p (Fig. 2 and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200804098/DC1), suggesting that Xpo1p is not playing a general role in kinetochore structure.
Because the efficient association of Xpo1p with its cognate NES occurs when Xpo1p is bound to RanGTP (Fornerod et al., 1997; Maurer et al., 2001), we examined kinetochore targeting of Mad1p in the presence of reduced nuclear levels of RanGTP, a condition that can be induced by a Ts mutation in the RanGEF Prp20p (prp20-7; Amberg et al., 1993; Dilworth et al., 2001). Although nocodazole-induced kinetochore binding of Mad1-GFP occurs normally at the permissive temperature in a prp20-7 nup60Δ strain, a shift to 37°C before nocodazole treatment inhibited Mad1-GFP accumulation at kinetochores in large-budded cells (Fig. 2 A).
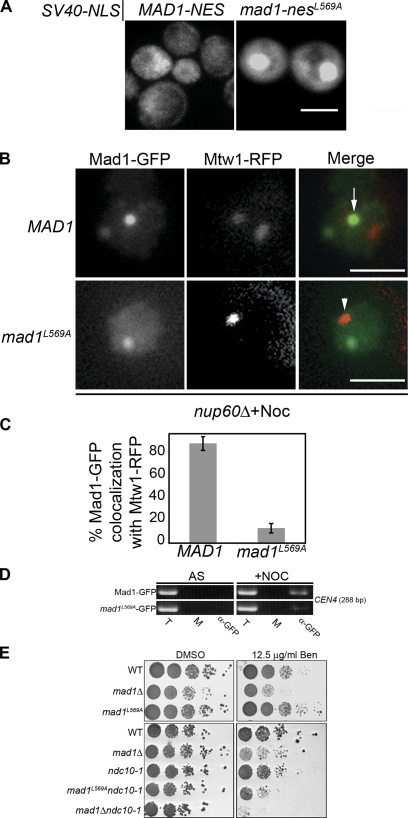
To further examine the link between Xpo1p function and kinetochore targeting of Mad1p, we constructed a point mutation in the Mad1p NES predicted to inhibit its interaction with Xpo1p. This mad1-nesL569A mutation, when incorporated into SV40-NLS/Mad1563–576-GFP3, inhibited export of the reporter, leading to its accumulation in the nucleus (Fig. 3 A). Importantly, when introduced into GFP-tagged endogenous MAD1, the resulting mad1L569A-GFP protein failed to accumulate at kinetochores after SAC arrest (Fig. 3, B and C). This result was further confirmed by chromatin immunoprecipitation (IP [ChIP]) analysis showing that levels of mad1L569A-GFP at centromeric DNA were greatly reduced in SAC-arrested cells (Fig. 3 D). These effects could not be attributed to a decrease in cellular levels of the mad1L569A mutant protein (unpublished data). Cumulatively, these findings support the conclusion that a Mad1p–Xpo1p–RanGTP complex functions in the efficient targeting of Mad1p to kinetochores during SAC arrest.
Figure 3.
Inactivation of the Mad1p NES dramatically reduces targeting of Mad1p to kinetochores. (A) The cellular distribution of SV40-NLS/MAD1563-576-GFP3 or SV40-NLS/mad1-nesL569A-GFP3 containing the Mad1p NES mutant in WT cells was examined by epifluorescence microscopy. (B) nup60Δ cells expressing MAD1-GFP (Y3151) or mad1L569A-GFP (Y3154) and MTW1-RFP were arrested with nocodazole and examined by epifluorescence microscopy. The arrow points to Mad1-GFP overlap with Mtw1-RFP. The arrowhead points to kinetochores devoid of Mad1-GFP. (C) Quantification of Mad1-GFP and Mtw1-RFP colocalization from the experiments presented in B. 100 cells were analyzed at random for colocalization between Mad1-GFP and Mtw1-RFP, and the results of three experiments were combined. SD (error bars) is shown in each case. (D) ChIP was performed on cells expressing MAD1-GFP (Y3028) or mad1L569A-GFP (Y3170) and growing logarithmically (asynchronous [AS]) or arrested in nocodazole (+Noc). (E) The indicated strains (WT [Y3162], mad1Δ [Y3165], mad1L569A [Y3164], ndc10-1 [3166], mad1L569Andc10-1 [Y3167], and mad1Δ ndc10-1 [Y3168]) were grown on YPD plates containing DMSO or DMSO plus benomyl for 2 or 4 d (bottom right). T, total chromatin used in each IP; M, mock minus antibody IP; α-GFP, anti-GFP antibody IP. Bars: (A) 5 μm; (B) 2 μm.
Despite the inability of the mad1L569A protein to accumulate at kinetochores, mad1L569A mutants exhibited characteristics suggesting they possess a functional SAC; they grew similar to WT cells on benomyl-containing plates (Fig. 3 E), and they arrested as large-budded cells with sustained, elevated Clb2p levels (not depicted). The mad1L569A mutant also rescues synthetic sick or lethal phenotypes of double mutants of mad1Δ and various null mutants tested, including ctf3Δ, ctf4Δ, ctf18Δ, ctf19Δ, erg3Δ, nup133Δ, sic1Δ, and cdc40Δ (unpublished data). We conclude that either the SAC function of Mad1p can be performed outside of kinetochores or that reduced levels of kinetochore-bound Mad1p (i.e., below that detectable by GFP tagging) are sufficient for SAC arrest. Such an interpretation would also be consistent with studies in HeLa cells, where depletion of Hec1 (Martin-Lluesma et al., 2002; DeLuca et al., 2003) or Nup358/RanBP2 (Salina et al., 2003; Joseph et al., 2004) does not inhibit SAC arrest but leads to greatly reduced or undetectable levels of Mad1 and Mad2 at kinetochores.
The mad1L569A mutant does have consequences. We have detected a synthetic fitness defect between the mad1L569A mutation and an ndc10-1 Ts mutation. The mad1L569Andc10-1 mutant exhibits an increased sensitivity to benomyl as compared with the ndc10-1 mutant (Fig. 3 D). Ndc10p is a component of the CBF3 complex (Lechner and Carbon, 1991). Mutations in Ndc10p disrupt kinetochore structure (Goh and Kilmartin, 1993) and recruitment of the SAC machinery (Gillett et al., 2004). The effects of the ndc10-1 mutation combined with the decreased efficiency of mad1L569A targeting to kinetochores may explain the apparent compromise in SAC function.
Xpo1p is targeted to kinetochores during SAC arrest
Our data raise the question of whether Xpo1p is recruited to kinetochores with Mad1p. Xpo1p shuttles between the nucleus and the cytoplasm but displays a predominantly nuclear localization (Fig. 4 A; Stade et al., 1997). In addition to its diffuse nuclear localization, we also detected Xpo1-GFP at one or two foci per nucleus. In asynchronous cultures, Xpo1-GFP foci did not appear to coincide with the kinetochore marker protein Mtw1-RFP (Fig. 4, A and G) but instead with the spindle pole body protein Spc42-RFP (Fig. 4, B and G). However, after incubation with nocodazole and activation of the SAC, Xpo1-GFP localization changed. Xpo1-GFP was now concentrated at kinetochores together with Mtw1-RFP and was no longer detected at spindle pole bodies (Fig. 4, C, D, and G). Consistent with these results, ChIP analysis detected Xpo1-GFP in association with centromeric DNA only in SAC-arrested cells (Fig. 4 H). Importantly, this association is dependent on Mad1p. Cells lacking Mad1p (mad1Δ) or Bub1p (bub1Δ), which is necessary for Mad1p binding to kinetochores (Gillett et al., 2004), failed to accumulate Xpo1p at kinetochores (Fig. S2). The relationship between Xpo1p kinetochore association and SAC activation was also examined in a cdc26-null mutant. At 37°C, cdc26Δ mutants arrest in metaphase without activating the SAC (Zachariae et al., 1996; Hwang and Murray, 1997), and Xpo1p remained associated with spindle pole bodies (Fig. 4 E). However, upon addition of nocodazole, the Xpo1-GFP signal at spindle pole bodies was lost, and the protein accumulated on kinetochores, suggesting that this event is dependent on SAC activation (Fig. 4 F).
Figure 4.
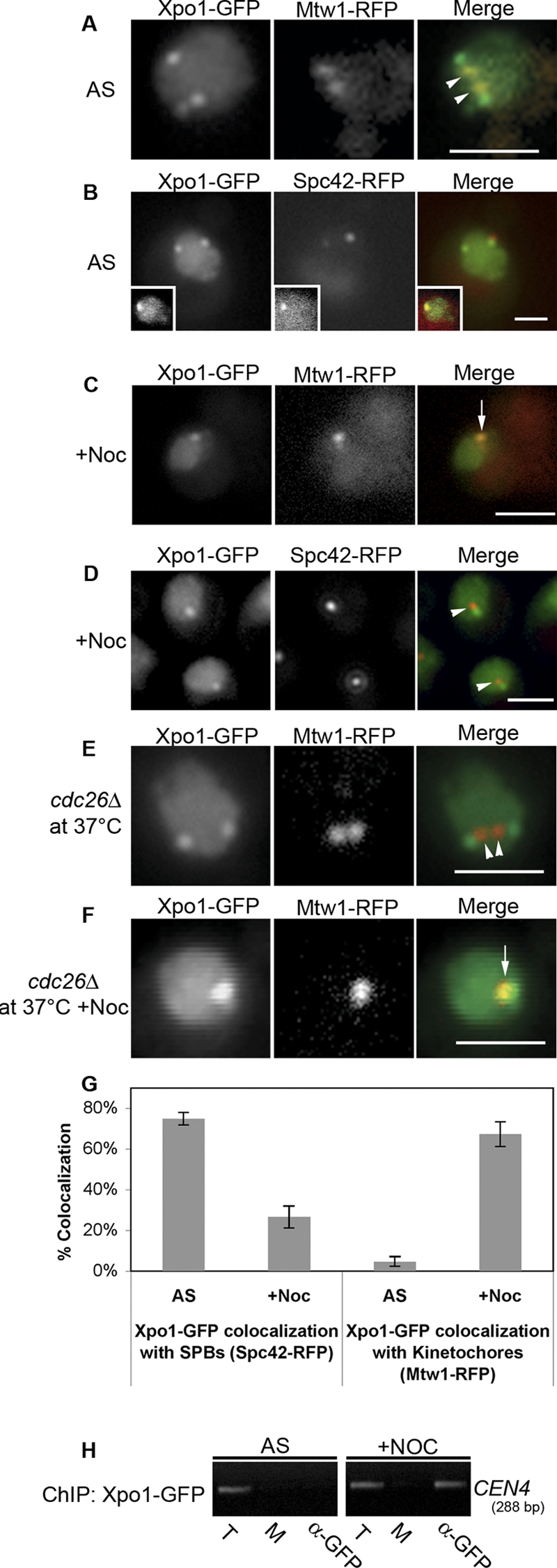
Xpo1p associates with spindle pole bodies in logarithmically growing cells and redistributes to kinetochores during SAC arrest. (A–D) Cells synthesizing Xpo1-GFP and Mtw1-RFP (Y3109) or Xpo1-GFP and Spc42-RFP (Y3101) were analyzed by epifluorescence microscopy after either logarithmic growth (A and B) or nocodazole-induced arrest (C and D). Note that the insets in B show a G1 cell. (E and F) A cdc26Δ mutant was introduced into Y3109 to produce Y3131. Cultures of these cells were shifted to 37°C, and the localization of Mtw1-RFP and Xpo1-GFP was assessed in the absence (E) and presence (F) of nocodazole. The arrows in C and F highlight colocalization of Xpo1-GFP and Mtw1-RFP, and the arrowheads in A and E point to kinetochores lacking Xpo1-GFP signal. The arrowheads in D indicate spindle poles devoid of Xpo1-GFP signal. (G) Quantification of Xpo1-GFP colocalization with either Spc42-RFP or Mtw1-RFP from the experiments presented in A–D. 30 cells were analyzed for each condition in each of three independent experiments, and results are presented as a mean with SD (error bars). (H) ChIP analysis was performed on cells producing Xpo1-GFP (Y3169) growing logarithmically (asynchronous [AS]) or arrested in nocodazole (+Noc). T, total chromatin used in each IP; M, mock minus antibody IP; α-GFP, anti-GFP antibody IP. Bars, 2 μm.
Although its kinetochore association is clearly linked to Mad1p, it remains possible that Xpo1p could target additional proteins to kinetochores. This is the case in vertebrate cells in which the Xpo1p counterpart, Crm1p, facilitates the binding of Nup358 (RanBP2) and RanGAP1 to kinetochores (Arnaoutov et al., 2005). These proteins are in turn required for Mad1 association with kinetochores (Joseph et al., 2004). However, whether Crm1 functions in directly targeting Mad1 to vertebrate kinetochores is unclear.
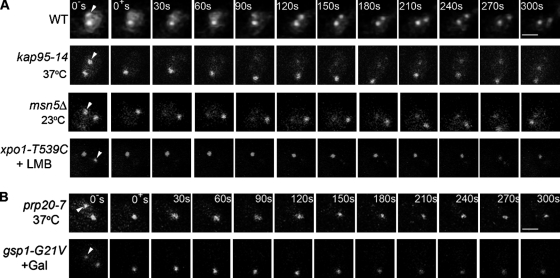
Mad1p turnover on kinetochores during SAC arrest requires Xpo1p, RanGTP, and RanGTP conversion to RanGDP
Mad1p cycles between NPCs and kinetochores during SAC arrest (Scott et al., 2005). As Xpo1p is required for Mad1p targeting to kinetochores (Fig. 2), we examined the role of kaps in the turnover of Mad1p at kinetochores. In these experiments, the SAC was activated in nup60Δ strains containing specific kap mutations, allowing Mad1-GFP to target to kinetochores. Arrested kap95-14 and xpo1-T539C mutants were shifted to their nonpermissive condition. Neither a shift to 37°C for 40 min (kap95-14) nor treatment of cells with LMB (xpo1-T539C) altered the kinetochore association of Mad1-GFP (Fig. 5 A, 0− s). This allowed us to monitor Mad1-GFP turnover at kinetochores by FRAP analysis. In the absence of kap mutations, Mad1-GFP signal at postphotobleached kinetochores recovered to ∼60% of the original signal with a t1/2 of recovery of ∼33 s (WT; Fig. 5 A and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200804098/DC1), which is consistent with previous experiments (Scott et al., 2005). Similar recovery rates were also detected in the kap95-14 strain at 37°C (t1/2 = ∼40 s) and the msn5Δ mutant (t1/2 = ∼38 s). In contrast, the kinetochore-associated Mad1-GFP signal failed to recover in the xpo1-T539C strain treated with LMB. We conclude from these results that once the SAC is activated and Mad1p is positioned at unattached kinetochores, Xpo1p binding is not required for Mad1p's stable kinetochore association, but it is necessary for its turnover.
Figure 5.
Mutations in Xpo1p, Ran, and Prp20p abrogate the turnover of Mad1p on kinetochores during SAC arrest. (A and B) MAD1-GFP and MTW1-RFP in nup60Δ strains containing an otherwise WT genotype (Y3057) or the mutant alleles kap95-14 (Y3161), msn5Δ (Y3116), or xpo1-T539C (Y3156; A) and prp20-7 (Y3091) or gsp1-G21V (Y3096; B) were arrested in nocodazole-containing media. The function of the mutant allele was inhibited by shifting to a nonpermissive growth condition (37°C or +LMB), or, in the case of gsp1-G21V, its expression was induced by the addition of galactose. WT and msn5Δ cells were examined directly after nocodazole arrest. Kinetochore-associated Mad1-GFP (prebleach; 0− s) was identified by colocalization with Mtw1-RFP (not depicted). Kinetochore-associated Mad1-GFP was photobleached, and recovery was monitored by acquiring images immediately after bleaching (postbleach; 0+ s) and at 15-s intervals. Images from 30-s intervals are shown. The arrowheads point to bleached kinetochores (0− s). Bars, 1 μm.
As shown in Fig. 2, depletion of nuclear RanGTP before activation of the SAC prevented the recruitment of Mad1p onto kinetochores (Fig. 2). We also examined whether RanGTP plays a role in maintaining Mad1p at kinetochores or assists in the turnover process once the checkpoint is activated. To test this, prp20-7 nup60Δ cells were treated with nocodazole at the permissive temperature, allowing Mad1-GFP to target onto unattached kinetochores (unpublished data). Cells were shifted to 37°C to inactivate prp20-7p. Under these conditions, Mad1-GFP remained bound to the Mlp focus and kinetochores (Fig. 5 B, 0− s). Thus, either RanGTP plays no role in the turnover of Mad1-GFP at kinetochores or it becomes immobilized there in the absence of RanGTP, similar to what was observed upon inactivation of Xpo1p. To distinguish between these possibilities, kinetochore-associated Mad1-GFP was photobleached, and fluorescent recovery was monitored after inactivation of prp20-7p. We observed that kinetochore-bound Mad1-GFP signal failed to recover under these conditions of reduced nuclear RanGTP (Fig. 5 B).
Using a similar methodology, we also investigated whether GTP hydrolysis by Ran plays a role in Mad1p turnover at kinetochores. To test this, we used a dominant-negative mutant of yeast Ran (gsp1-G21V) that stabilizes Ran in the GTP-bound form (Schlenstedt et al., 1995). A GAL1/10-gsp1-G21V gene cassette was introduced into a nup60Δ strain, producing Mad1-GFP. After nocodazole-induced SAC arrest, expression of gsp1-G21V was induced with galactose. As we observed after depletion of RanGTP, blocking its conversion to RanGDP did not alter the Mad1-GFP signal at kinetochores but prevented its recovery after photobleaching (Fig. 5 B and Fig. S1). This effect was specific for the gsp1-G21V allele, as overexpression of WT GSP1 had no effect on Mad1-GFP turnover (unpublished data). Importantly, the effects of the gsp1-G21V allele are unlikely linked to an inhibition of nuclear transport or loss of nuclear Mad1-GFP, as a pool of Mad1-GFP remained bound to the intranuclear Mlp foci (Fig. 5 B), and this provides Mad1-GFP for kinetochore turnover (Scott et al., 2005).
We conclude that although required for the initial binding of Mad1p to kinetochores after nocodazole–induced SAC activation, Xpo1p and RanGTP are not essential for maintaining this interaction during periods of arrest. However, turnover of Mad1p at kinetochores requires active Xpo1p, RanGTP, and hydrolysis of GTP by Ran. On the basis of our results, we have developed a working model in which a trimeric Mad1p–Xpo1p–RanGTP complex facilitates the initial binding of Mad1p to kinetochores after SAC activation. In this model, conversion of RanGTP to RanGDP is predicted to release Xpo1p from Mad1p at kinetochores. During SAC arrest, we envisage that Mad1p would remain attached to kinetochores until displaced by a Mad1p–Xpo1p–RanGTP complex. Subsequent conversion of RanGTP to RanGDP and the release of Xpo1p from Mad1p would complete the turnover cycle.
A key feature of this model is the conversion of Xpo1p-bound RanGTP to RanGDP at kinetochores. At present, the only known yeast RanGAP is Rna1p. This protein is found in the cytoplasm, and it is assumed to be excluded from the nucleoplasm to avoid depleting nuclear RanGTP. However, several observations suggest that RanGAP can enter the nucleus and potentially be targeted to specific locations. Both Schizosaccharomyces pombe and Saccharomyces cerevisiae Rna1p contain functional NLSs and NESs, suggesting that they shuttle between the nucleus and cytoplasm (Feng et al., 1999; Nishijima et al., 2006). Moreover, S. pombe Rna1 binds histone H3, and it was proposed that it functions in heterochromatin assembly (Nishijima et al., 2006). These results suggest that Rna1p may function in the nucleus but only within specific contexts and at certain locations. Kinetochores may represent one such location. As mentioned in the Introduction, in metazoan cells, RanGAP1 associates with kinetochores during M phase (Joseph et al., 2002; Arnaoutov and Dasso, 2003; Joseph et al., 2004), and its presence there has been linked to Crm1 function (Arnaoutov and Dasso, 2005; Arnaoutov et al., 2005). In addition, RanBP1, which is known to stimulate RanGTP hydrolysis mediated by RanGAP, is also required for Mad2 binding to kinetochores (Li et al., 2007) in HeLa cells. It remains to be determined whether these events are conserved in yeast. Our preliminary experiments have failed to detect Rna1-GFP at kinetochores (unpublished data). An alternative possibility is that another kinetochore-bound protein could activate the GTPase activity of Ran.
Our results reveal a previously undefined function for Xpo1p in enhancing the intranuclear targeting of Mad1p to kinetochores and facilitating the cycling of Mad1p between kinetochores and NPCs. This latter event may contribute to the depletion of Mad1p from attached kinetochores and its sequestration at the NPC after termination of the SAC. It also seems reasonable to assume that this interplay reflects a broader functional relationship between the nuclear transport machinery and kinetochores, which could include SAC-induced alterations in nuclear transport. This idea is particularly intriguing, as previous experiments have shown that specific nuclear import pathways are inhibited in nocodazole-arrested cells (Makhnevych et al., 2003). Future studies will undoubtedly reveal unanticipated links between these various pathways.
Materials and methods
Yeast strains and media
Strains are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200804098/DC1). All yeast strains were grown at 30°C in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose) supplemented with 40 μg/ml adenine or synthetic media (SM) supplemented with appropriate nutrients and 2% glucose. Benomyl plates were made as described previously (Scott et al., 2005). For growth on YPD + DMSO or YPD + 12.5 μg/ml of benomyl plates, 10-fold serial dilutions of cell cultures were placed onto plates and incubated for 2–4 d at 25°C. Transformations were performed by electroporation. Integrations of the GFP+ or RFP ORFs were performed as previously described (Scott et al., 2005) using the plasmids pGFP+/HIS5, pGFP+/NatR, pRFP/HIS5, or pRFP/NatR (described below). Synthesis of the GFP and RFP fusions was confirmed by immunoblotting and microscopy.
Plasmids
pSV40-NLS/Mad1-NES-GFP3 was constructed by removing Mad1-NLS from pMad1-NLS-GFP3 (Scott et al., 2005) and inserting the coding regions for SV40-NLS (PKKKRKV) and Mad1 residues 563–576. A point mutation in NES (L569A) was made using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies). The mutation was designed (la Cour et al., 2004) based on studies of the Rev and PKI NESs (Wen et al., 1995; Meyer et al., 1996). Plasmids for integrating MAD1 and mad1-L569A were assembled in pRS306 to produce pRS306-Mad1 and pRS306-mad1L569A. Plasmids encoding XPO1 (pKW440), xpo1-1 (pKW457), xpo1-T539C (pKW711), and the SV40-NLS/PKI-NES-GFP construct were provided by K. Weis (University of California, Berkeley, Berkeley, CA). The GSP1- and gsp1-G21V–integrating plasmids were assembled in pRS306 GAL1/CYC1 (pTM1012). Plasmids were cut with StuI to direct their integration to the URA3 locus.
Fluorescence microscopy
The fluorescent proteins used in this study were GFP (+) and monomeric RFP (1.5). All images of fusion proteins were acquired from live cells in SM supplemented with appropriate nutrients and the indicated reagents using either (a) an epifluorescence microscope (IX-81; Olympus), a 100× objective with a U Plan Apo oil immersion lens, and a digital camera (IX2-UCB; Olympus) or (b) a microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a confocal scanning system (LSM 510 META; Carl Zeiss, Inc.) and a 63× objective with a Plan Apochromat oil immersion lens. Epifluorescent images were acquired using In vivo 3 (Media Cybernetics) and were logged to and analyzed in ImagePro 3-D (Media Cybernetics). All confocal images were acquired using LSM 510 software and viewed using LSM Image Browser (Carl Zeiss, Inc.). Image processing was performed with ImageJ software (National Institutes of Health) and Photoshop (Adobe).
Mad1p kinetochore-targeting experiments
The strains Y3057 (WT), Y3116 (msn5D), Y3156 (xpo1-T539C; −LMB), and Y3091 (prp20-7 at 25°C) were grown to an OD600 of ∼0.5 at 25°C and were treated with 12.5 mg/ml nocodazole (Sigma-Aldrich) for 90 min. Alternatively, Y3161 (kap95-14) and Y3091 (prp20-7) were shifted to 37°C for 60 and 30 min followed by the addition of 12.5 μg/ml nocodazole for 90 min. To inactivate xpo1-T539C, 100 ng/ml LMB was added to Y3156 for 30 min before addition of 12.5 μg/ml nocodazole. In all instances, cells were rapidly harvested, resuspended in SM supplemented with appropriate nutrients and containing 12.5 μg/ml nocodazole, and immediately imaged. The Y3156 (xpo1-T539C; +LMB) cell suspension also contained 100 ng/ml LMB. To quantify the colocalization of Mad1-GFP or mad1L569A-GFP with Mtw1-RFP, 100 cells were examined in three independent experiments, results were averaged, and SD from the mean was determined. For the Y3154 and Y3057 strain, cells were arrested in G1 with α factor, released into media containing 12.5 μg/ml nocodazole for 75 min at 25°C, and imaged.
In vivo cross-linking and ChIP
ChIP experiments were performed as described previously (Meluh and Koshland, 1997). Cells expressing XPO1-GFP, Mad1-GFP, or mad1L569A were grown in 200 ml YPD to an OD600 of 0.6 at 30°C, split, and either left untreated or treated with 12.5 μg/ml nocodazole for 90 min. IPs were performed using a commercial anti-GFP antibody (Roche), and antibody-bound complexes were purified using protein A–Sepharose CL-4B beads (GE Healthcare). The amount of CEN4 DNA associated with GFP fusions was determined by PCR. CEN4 DNA from cells expressing XPO1-GFP, Mad1-GFP, or mad1L569A was amplified with 23, 38, or 38 cycles of PCR using 2% of the total immunoprecipitated samples or 0.2% of the total chromatin input. Reactions were analyzed on agarose gels using ethidium bromide staining.
Laser photobleaching of Mad1-GFP
All photobleaching experiments were performed on a microscope (LSM 510; Carl Zeiss, Inc.). Strains for photobleaching were grown to an OD600 of ∼0.3 and arrested in G1 phase using α factor for 2 h at 25°C. Cells were released into YPD containing 12.5 μg/ml nocodazole at room temperature. After 75 min, WT (Y3057) and msn5Δ (Y3116) cells were washed, resuspended in SM supplemented with appropriate nutrients and containing 12.5 μg/ml nocodazole, visualized, and photobleached. xpo1-T539C (Y3156) cells were further treated with 100 ng/ml LMB (Sigma-Aldrich) for 15 min, and kap95-14 (Y3161) and prp20-7 (Y3091) cells were shifted to 37°C for 60 and 30 min in the presence of nocodazole before photobleaching. gsp1-G21V (Y3096) cells were grown in YP raffinose, arrested in G1 phase with α factor, and released into media containing 12.5 μg/ml nocodazole for 60 min. Galactose was added to a final concentration of 3%, and the incubation continued for 90 min. In all cases, 488- and 543-nm light scans were used to identify Mad1-GFP at kinetochores (Mtw1-RFP). Kinetochore-associated Mad1-GFP was bleached with 15 iterations of full-intensity 488-nm light. Images were acquired every 15 s after bleaching. Photobleaching experiments were repeated four times. Plots of integrated fluorescence intensity were obtained as previously described, and half-times of recovery were derived from these plots (Scott et al., 2005).
Online supplemental material
Fig. S1 shows plots of integrated fluorescence intensity at kinetochores at various times after photobleaching. Fig. S2 shows that Mad1p and Bub1p are required for checkpoint-induced kinetochore localization of Xpo1p. Fig. S3 shows that Xpo1p is required for NPC association of Mad1p but not the kinetochore association of Bub1p or Mtw1p. Table S1 contains a list of the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200804098/DC1.
Supplementary Material
Acknowledgments
We would like to thank those listed in the text for providing reagents used for this work. We thank members of the Wozniak laboratory for thoughtful discussions during preparation of the manuscript.
This work is supported by funds provided by the Canadian Institutes of Health Research, the Alberta Heritage Foundation for Medical Research, and the Howard Hughes Medical Institute.
Abbreviations used in this paper: ChIP, chromatin IP; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; IP, immunoprecipitation; kap, karyopherin; LMB, leptomycin B; NES, nuclear export signal; NPC, nuclear pore complex; PKI, protein kinase inhibitor; SAC, spindle assembly checkpoint; SM, synthetic media; Ts, temperature sensitive; WT, wild type.
References
- Amberg, D.C., M. Fleischmann, I. Stagljar, C.N. Cole, and M. Aebi. 1993. Nuclear PRP20 protein is required for mRNA export. EMBO J. 12:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaoutov, A., and M. Dasso. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell. 5:99–111. [DOI] [PubMed] [Google Scholar]
- Arnaoutov, A., and M. Dasso. 2005. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 4:1161–1165. [DOI] [PubMed] [Google Scholar]
- Arnaoutov, A., Y. Azuma, K. Ribbeck, J. Joseph, Y. Boyarchuk, T. Karpova, J. McNally, and M. Dasso. 2005. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 7:626–632. [DOI] [PubMed] [Google Scholar]
- Campbell, M.S., G.K. Chan, and T.J. Yen. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953–963. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., B.J. Howell, J.C. Canman, J.M. Hickey, G. Fang, and E.D. Salmon. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103–2109. [DOI] [PubMed] [Google Scholar]
- Dilworth, D.J., A. Suprapto, J.C. Padovan, B.T. Chait, R.W. Wozniak, M.P. Rout, and J.D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W., A.L. Benko, J.H. Lee, D.R. Stanford, and A.K. Hopper. 1999. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J. Cell Sci. 112:339–347. [DOI] [PubMed] [Google Scholar]
- Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier, E. Gilson, J.C. Olivo-Marin, H. Scherthan, and U. Nehrbass. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4:214–221. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., M. Ohno, M. Yoshida, and I.W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 90:1051–1060. [DOI] [PubMed] [Google Scholar]
- Gillett, E.S., C.W. Espelin, and P.K. Sorger. 2004. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J. Cell Biol. 164:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, P.Y., and J.V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M.W., T.C. Walther, and I.W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347–380. [DOI] [PubMed] [Google Scholar]
- Hwang, L.H., and A.W. Murray. 1997. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol. Biol. Cell. 8:1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk, T., O. Kerscher, R.J. Scott, M.A. Basrai, and R.W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S.H. Tan, T.S. Karpova, J.G. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S.T. Liu, S.A. Jablonski, T.J. Yen, and M. Dasso. 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14:611–617. [DOI] [PubMed] [Google Scholar]
- Kalab, P., and R. Heald. 2008. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 121:1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17:527–536. [DOI] [PubMed] [Google Scholar]
- Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 64:717–725. [DOI] [PubMed] [Google Scholar]
- Leslie, D.M., B. Grill, M.P. Rout, R.W. Wozniak, and J.D. Aitchison. 2002. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22:2544–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.Y., W.P. Ng, C.H. Wong, P.A. Iglesias, and Y. Zheng. 2007. Coordination of chromosome alignment and mitotic progression by the chromosome-based Ran signal. Cell Cycle. 6:1886–1895. [DOI] [PubMed] [Google Scholar]
- Loiodice, I., A. Alves, G. Rabut, M. Van Overbeek, J. Ellenberg, J.B. Sibarita, and V. Doye. 2004. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 15:3333–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych, T., C.P. Lusk, A.M. Anderson, J.D. Aitchison, and R.W. Wozniak. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 115:813–823. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., V.M. Stucke, and E.A. Nigg. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270. [DOI] [PubMed] [Google Scholar]
- Maurer, P., M. Redd, J. Solsbacher, F.R. Bischoff, M. Greiner, A.V. Podtelejnikov, M. Mann, K. Stade, K. Weis, and G. Schlenstedt. 2001. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p). Mol. Biol. Cell. 12:539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B.E., J.L. Meinkoth, and M.H. Malim. 1996. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J. Virol. 70:2350–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and E.D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. [DOI] [PubMed] [Google Scholar]
- Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima, H., J. Nakayama, T. Yoshioka, A. Kusano, H. Nishitani, K. Shibahara, and T. Nishimoto. 2006. Nuclear RanGAP is required for the heterochromatin assembly and is reciprocally regulated by histone H3 and Clr4 histone methyltransferase in Schizosaccharomyces pombe. Mol. Biol. Cell. 17:2524–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, D., P. Enarson, J.B. Rattner, and B. Burke. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt, G., C. Saavedra, J.D. Loeb, C.N. Cole, and P.A. Silver. 1995. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+ RNA in the cytoplasm. Proc. Natl. Acad. Sci. USA. 92:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.J., C.P. Lusk, D.J. Dilworth, J.D. Aitchison, and R.W. Wozniak. 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 16:4362–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., C.S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 90:1041–1050. [DOI] [PubMed] [Google Scholar]
- Wen, W., J.L. Meinkoth, R.Y. Tsien, and S.S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell. 82:463–473. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., T.H. Shin, M. Galova, B. Obermaier, and K. Nasmyth. 1996. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 274:1201–1204. [DOI] [PubMed] [Google Scholar]
- Zuccolo, M., A. Alves, V. Galy, S. Bolhy, E. Formstecher, V. Racine, J.B. Sibarita, T. Fukagawa, R. Shiekhattar, T. Yen, and V. Doye. 2007. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 26:1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.