Abstract
RNA polymerase is an essential enzyme for cellular gene expression. In an effort to further understand the enzyme’s importance in the cell’s response to temperature, we have probed the kinetic mechanism of E. coli RNA polymerase (RNAP) by studying the force-velocity behavior of individual RNAP complexes at temperatures between 7°C and 45°C using optical tweezers. Within this temperature range and at saturating nucleotide concentrations, the pause-free transcription velocity of RNAP was independent of force and increased monotonically with temperature with an elongation activation energy of 9.7 ± 0.7 kcal/mole. Interestingly, the pause density at cold temperatures (7 °C to 21 °C) was five times higher than that measured above room temperature. A simple kinetic model revealed a value of 1.29 ± 0.05 kcal/mol for the activation energy of pause entry, suggesting that pause entry is indeed a thermally accessible process. The dwell time distribution of all observable pauses was independent of temperature, directly confirming a prediction of the model recently proposed for Pol II in which pauses are diffusive backtracks along the DNA. Additionally, we find that the force at which the polymerase arrests (the arrest force) presents a maximum at 21 °C, an unexpected result as this is not the optimum temperature for bacterial growth. This observation suggests that arrest could play a regulatory role in vivo, possibly through interactions with specific elongation factors.
Keywords: RNAP, transcription, temperature, single molecule, optical tweezers
Introduction
RNA polymerase (RNAP) converts the genetic information encoded in DNA into RNA, which then serves as the template for protein synthesis. E. coli RNAP is a six-subunit polymerase possessing high processivity and fidelity. It has high sequence and structural homology with eukaryotic RNAPs such as Pol II. Compared to Pol II, however, the relatively simple initiation process of E. coli RNAP renders it an ideal model to study transcription. E. coli RNAP has been studied extensively using in vitro and in vivo biochemical assays 1; 2; 3, cryo-EM 4 and X-ray crystallographic methods 5; 6; 7, and single molecule assays using both optical tweezers 8; 9; 10; 11; 12; 13; 14 and fluorescence 15; 16. Single molecule methods are well-suited to study the dynamics of transcription as the details of transient, unsynchronized behavior such as pausing cannot be easily probed by bulk biochemical or structural methods.
Extensive studies have shown that transcription elongation is a dynamically complex process 8; 9; 10; 11; 12; 13. Elongation is often interrupted by pauses whose durations vary over several orders of magnitude. Different models have been proposed to explain the origin of these pauses. For example, biochemical and single molecule studies have suggested that at least some of the pauses are caused by the polymerase backtracking along the DNA 11; 12; 13; 17; 18. There may also exist short, non-backtracked pauses corresponding to catalytically inactive states of the polymerase [8 and references therein, 13; 19; 20]. However, recent work on Pol II has suggested that most, if not all, observed pauses in single molecule experiments are due to diffusive backtracking 21. To further study the process of transcription, more specifically the relationship between pausing and active elongation, including their activation energies and mechanisms, we investigated the temperature dependence of RNAP elongation using single-molecule experiments. A variable temperature assay will not only allow us to gain more information about the rates involved in transcription, but will also shed more light on the role that RNA polymerase plays in the cell’s ability to adapt to changes in temperature.
Our study used a dual-trap optical tweezers equipped with a temperature control module 22; 23; 24 to measure the rate of transcription elongation and describe the pausing behavior of individual E. coli RNAPs at temperatures ranging from 7 to 45°C. The present assay addresses a varying force regime and a much wider temperature range than that of a previous study that used constant force and a laser heating method with which only a limited temperature range could be studied (21 to 37 °C) 22. Through this study we find that temperatures below room temperature (RT) greatly increase the probability of pausing; feature which has allowed us to calculate precise values for the energetic barrier for pause entry as well as for the temperature dependence of elongation and pause entry rates. We have also verified that the pause distributions at every temperature tested are well fit by a single pause mechanism (diffusive backtracking of the enzyme along the DNA template) and are temperature independent. These new findings have allowed us to better understand the pausing mechanisms involved in transcription and even provided some insights on the role that pauses may play in vivo.
Results
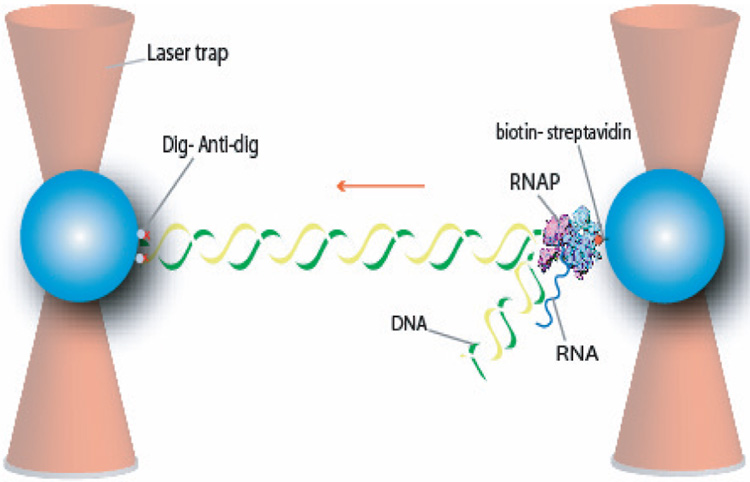
The experimental setup was composed of a single ternary complex consisting of RNAP, DNA template and nascent RNA tethered between two polystyrene beads in two optical traps (Figure 1). Stalled complexes were held at position 70 downstream of the λ-PR promoter of the DNA template by withholding CTP from the NTP cocktail (see Materials and Methods section for details). The biotinylated polymerase was bound to an optically-trapped, streptavidin-coated bead while the downstream end of the template DNA (11528 bp) was labeled with digoxigenin (dig) and attached to an anti-dig antibody-coated bead in a second optical trap. To re-initiate elongation, a complete set of NTPs was introduced into the flow cell at a saturating concentration of 1 mM. The two optical traps remained fixed during the passive mode experiment and, therefore, the force loading the polymerase increased and was recorded in real time (10 kHz). Changes in bead-to-bead distance during transcription were converted into DNA contour length using the worm-like chain model of DNA elasticity. The experiment was performed at different temperatures between 7 and 45 °C using a previously described temperature control module 23. Figure 2 shows representative traces at four of the six experimental temperatures (14°C, 21°C, 37°C, 45°C are shown; 7 °C, 29 °C are not). At all temperatures, the polymerase undergoes transcription periods (black) interrupted by pauses (red). It is interesting to note that at any given 25 temperature the instantaneous velocity of the enzyme remains constant throughout the run until the enzyme arrests 8; 9. The RNAP was assumed to be in an arrested state if the enzyme ceased to transcribe and did not resume active elongation after having paused for 10 minutes or more.
Fig. 1.
Experimental setup. The beads were held in two optical traps and a link was made between the polymerase and the downstream end of the DNA. As the polymerase transcribes, the load of the enzyme increases. The starting force for all traces was 2 pN (5 pN for 21°C). All runs go up to their arrest force.
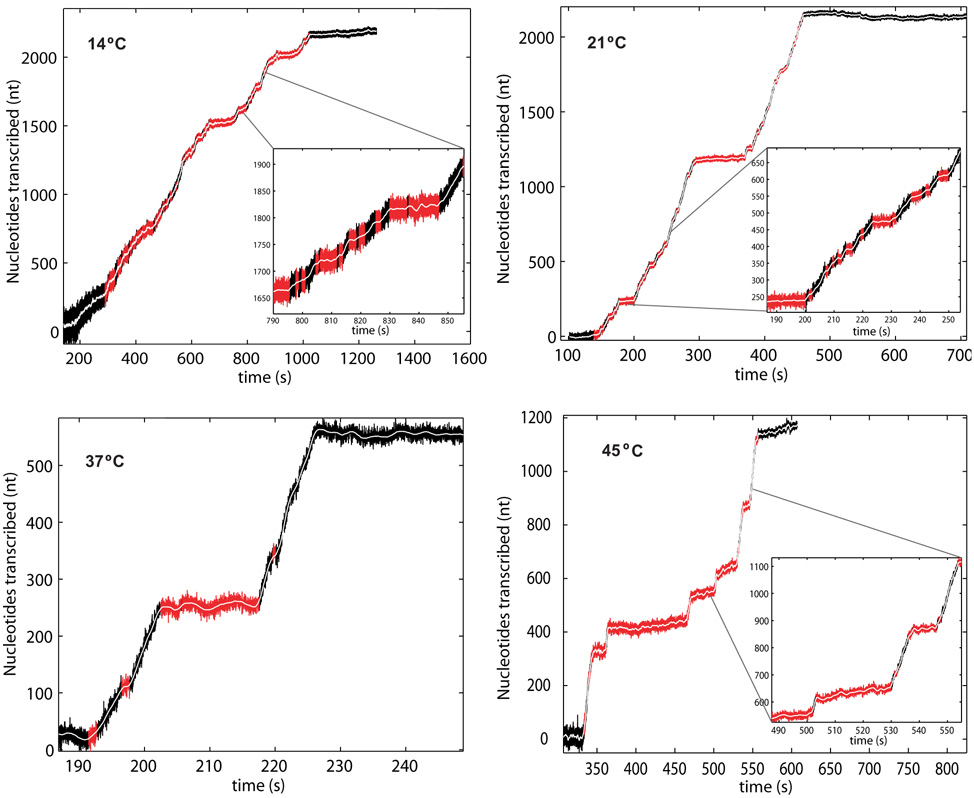
Fig. 2.
Transcription traces at 14, 21, 37 and 45°C. The data was filtered and pauses longer than 1 second were selected and are denoted (in red). Note that the slope (pause-less velocity, in black) becomes visibly steeper as temperature increases and the number of pauses increases dramatically for lower temperatures. Also note that the position noise decreases as transcription proceeds and the force increases. Insets: The insets (14°C, 21°C and 45°C) show an equal x-axis scale as the trace at 37°C, highlighting the changes in velocity and pause density with temperature. The starting force for all traces was 2 pN (5 pN for 21°C). All runs go up to their arrest force.
For all of the temperatures investigated, elongation velocity was nearly constant and independent of force throughout the range investigated as shown in plots of the normalized pause-free velocities as a function of force (Supplementary Data Fig. 1). This observation suggests that, in the temperature range from 7 °C to 45 °C and under NTP saturating conditions, the rate limiting process is not the actual enzyme translocation but some other biochemical process (such as NTP incorporation). As temperature changes, the rates of both biochemical and translocation processes vary respectively, however the biochemical rate remains the limiting one.
The non-normalized F-V curves (Supplementary Data Fig. 2) show an apparent gradual reduction in the velocity at high forces, a characteristic that has been observed in other single molecule studies at room temperature 8; 9. However, this feature is a reflection of the probabilistic spread in arrest forces since the velocities of individual enzymes remain constant until their arrest. In turn, the observed force-induced arrest is consistent with the idea that under increased opposing forces RNAP is likely to enter an off pathway state 11; 21 (see Figure 4b for the paused pathway). Thus, what we have termed an arrest force is strictly speaking not a thermodynamic stall force, but a kinetic arrest or an operational force limit 21.
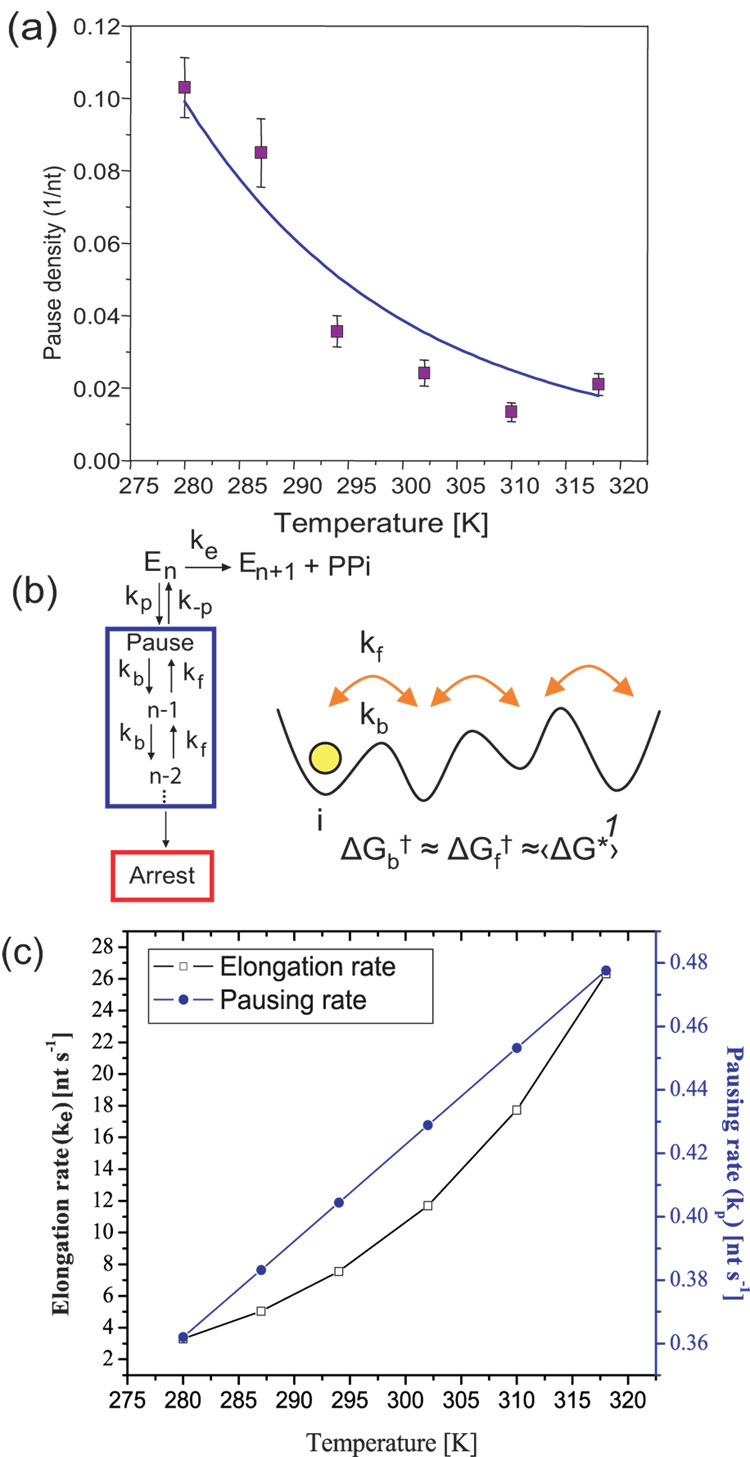
Fig. 4.
Kinetics of elongation and pausing. (a) The pause density (pauses/nt, for pauses >1 second) as a function of temperature. The fit to eqn 2 (solid line, R2= 0.9) suggests a kinetic competition between the elongation and pausing pathways and gives an activation energy of 1.29 ± 0.05 kcal/mol for entry into a pause. Error bars represent the standard error of the mean. (b) Simplest kinetic In this model ke is a convolution of the rate constants of all the processes that take the enzyme from n to n+1, including all reversible and irreversible steps, making ke an effective rate of transcription. The same way, kp is the effective rate for pause entry. Once in a pause, pauses are well described by diffusion in a 1D energy potential backwards along the DNA. The inset shows this diffusion schematically, where kf and kb are the forward and backwards rates, respectively; ΔGf† and ΔGb† are the corresponding activation energies. The active site is represented by 1. (c) Elongation (ke) and pausing rates (kp) vs. temperature plotted using the Arrhenius equation and the values obtained for ΔGe†, keo,ΔGp† and kpo, as described in the text. The elongation rate increases more rapidly than the pausing rate due to its increased activation energy. The starting force for all traces was 2 pN (5 pN for 21°C). All runs go up to their arrest force.
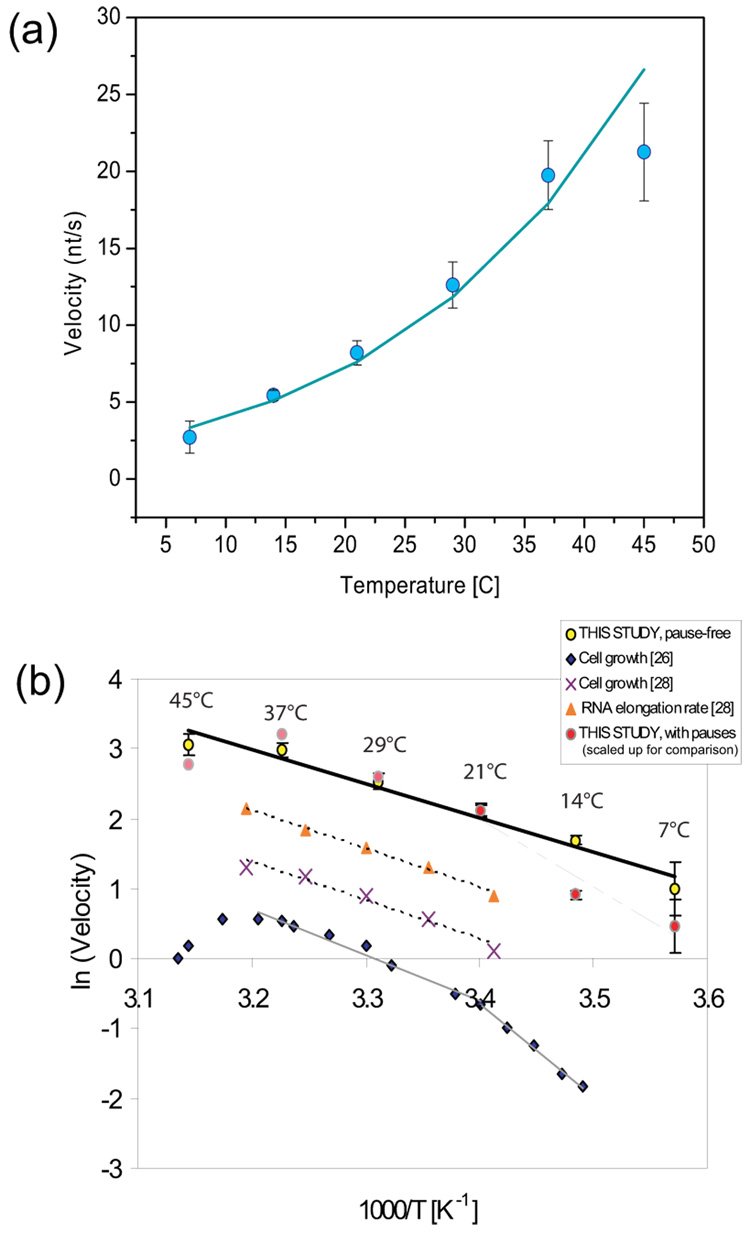
The maximum transcription rate at a given temperature corresponds to the velocity values at the plateaus of the F-V plots (values for 7 °C and 45°C are approximate, see Materials and Methods for details). Such pause-free velocities increase monotonically from 3 nt/s at 7°C to 22 nt/s at 45 °C. (Figure3a). Based on this information, an activation energy of transcription can be estimated by the Arrhenius equation:
| [eqn 1] |
where k is the rate constant, k0 is the pre-factor, ΔGe† is the elongation activation energy, kB is the Boltzmann constant and T is temperature. The plot of ln(k) vs 1/T is shown in Figure 3b. This plot yields a constant activation energy (ΔGe†) of 9.7 ± 0.7 kcal/mol for RNAP pause-free transcription from 7 to 45 °C. This value is slightly smaller than the one obtained in previous single molecule measurements (13±2 kcal/mol, 22) and is similar to values obtained from in vivo studies of the growth of E. coli (13.5 kcal/mol 26; 27; 11 kcal/mol 28) and the rates of RNA and peptide elongation within the range from 21 to 37 °C (11 kcal/mol 28; 29). Taken together, these results suggest that between 21 and 37 °C, transcription and translation are correlated processes in E. coli and that both are rate-limiting for cell growth. Below room temperature, however, the activation energy for bacterial growth (26.5 kcal/mol 26, Figure 3b; gray line, diamonds) is significantly higher than the value obtained here with the analysis of pause-free velocities. Reasons for this apparent discrepancy will be discussed later. Similarly, for temperatures higher than 37°C, the correlation between cell growth and transcription is lost indicating that another process likely becomes rate limiting in this temperature range (Figure 3b) 30.
Fig. 3.
Effect of temperature on RNAP’s pause-free velocity. (a) Transcriptional velocity increases with temperature and can be well fit by the Arrhenius equation. Error bars represent the standard error of the mean. (b) Arrhenius plot for the measured transcription velocity. Its slope gives an activation energy of approximately 9.67 ± 0.67 kcal/mol (yellow circles). Arrhenius-type plot of values published by Ryals et al.28 for cell growth (crosses) and RNA elongation rate (triangles) gives an activation energy of ~10.9 kcal/mol. Cell growth as measured by Herendeen et al. 26 (diamonds) gives a activation energy of 13.5 kcal/mol between 21 to 37°C, below room temperature the value increases to 26.5 kcal/mol. By including pauses in the single molecule velocities for temperatures below 21 °C (red circles) the activation energy measured for cell growth in this temperature range can be recovered. This highlights the relevance of transcriptional pausing at the cellular level. For temperatures above RT, including pauses in the velocities does not change the calculated activation energy ΔGe†. The average velocities shown in this plot have been scaled up to overlay them with the pause-free velocities for comparison, and comprise the data over all forces for each temperature. The starting force for all traces was 2 pN (5 pN for 21°C). All runs go up to their arrest force.
Off-pathway mechanisms
The pausing of RNAP at different temperatures was characterized by two parameters: the pause density and the pause duration. The pause density (Pd), defined as the number of pauses (longer than 1 sec) per nucleotide transcribed (pauses/nt), decreased towards a constant level as temperature increased (Figure 4a, 0.10 ± 0.008 bp−1 at 7 °C to 0.02 ± 0.003 bp−1 at 45 °C). Previously, Abbondanzieri et al. 22 observed no significant change in the pausing pattern for individual RNAP molecules within their studied range (21 to 37 °C). Our results reveal instead that pause density has a strong temperature dependence below 21°C and that the conclusion arrived at by the previous report reflects only the limited range of temperatures investigated in that study. This dependence has allowed us to obtain important kinetic parameters for pause entry by means of a simple kinetic model that is able to reproduce the observed experimental pause density values, as will be now described.
Past studies have found that pausing is a state off the main elongation pathway in kinetic competition with the elongating mechanism (Figure 4b) [11 and references therein]. Following this model, the probability of being in the paused state (Pp) can be written as:
| [eqn 2] |
where kpo and keo are the pre-factors for the paused and elongation rates, respectively, and ΔGp† and ΔGe† are the activation energies corresponding to an Arrhenius process. The pausing probability Pp is directly proportional to the pause density (Pd) as described by Pp = Pd *d, where d is the step size of the enzyme, which is 1 nt in this case. As a result, Pp = Pd. Using the measured value of ΔGe†, a two parameter fit (ke0/kp0 and ΔGp†) of the pause density using eqn 2 gives an activation energy of 1.29 ± 0.05 kcal/mol for the entry into the paused pathway (blue line, Figure 4a, R2 = 0.90). This value is much smaller than the activation energy of elongation, indicating that RNAP is an enzyme intrinsically prone to pause. Moreover, the magnitude of the activation energy to enter a pause is also within the range of thermal fluctuations, suggesting that the entry into the paused state is indeed spontaneous and thermally induced.
Using the measured values of ΔGe† (9.7 ± 0.7 kcal/mole) and keo (1.18*108 nt/s, from the y-intercept of Fig. 3b as described by the Arrhenius equation [eqn 1]), and the calculated values of ΔGp† (1.29 ± 0.05 kcal/mol) and kpo (3.69 nt/s) from the previous fit, the rate constants of elongation (ke) and pausing (kp) at different temperatures can be plotted as shown in Figure 4c. The plot reveals that the higher activation energy for elongation relative to that of pausing leads to a more rapid increase of the elongation reaction rate as temperature is raised. This prediction is consistent with the trend in pause density observed as a function of temperature (Fig 4a). At low temperatures, the RNA polymerase spends more time in the pausing pathway; however, as temperature increases, the enzyme is able to take more elongation steps before entering a pause. In fact, a rearrangement of eqn 2 shows that and so this simple model predicts that for ke≫ kp pause density would asymptotically approach a value of kp/ ke, and will tend to zero in the limit of infinite temperature. For the quantities measured here, the pause density above room temperature (RT) reaches an average value of 0.02 ± 0.003, which, as expected, is slightly higher than the asymptote value of 0.018 for the ratio kp/ ke (taken from the rates at the highest temperature in this study, which is the best way to fulfill the condition ke≫ kp) (Figure 4 a,c).
To investigate the pause durations, the time spent by the enzyme at every position along the template (‘dwell time’) for the whole length of the run, including both active translocation and pausing, was determined (see Materials and Methods). The processes with short dwell times (< 1 sec per 2nt) correspond to locations of active translocation (not shown), whereas those with longer dwell times (>1 sec per 2nt) correspond mostly to pauses. Figure 5 shows histograms representing dwell time probability distributions at different temperatures for dwell times greater than 1 sec. Pause dwell times can be described by a t(−3/2) power law (dotted red line) consistent with a random walk during diffusive backtracking21. As characterized for the eukaryotic RNA polymerase II21, this behavior implies that the pausing mechanism for RNAP is diffusion along the DNA template, initially backwards, with the pause ending (and elongation resuming) when the polymerase is again realigned with the 3’ end of the transcript. We note that the sequence studied here contains no well defined pause sequences, which might be expected to deviate from this mechanism. Deviations from power law behavior for pauses longer than 10 seconds can be attributed to the effect of an intrinsic forwards force, whose origin is currently being investigated (S.W. Grill and E.A. Galburt, personal communication). Nonetheless, the t(−3/2) power law better describes long pauses than a double exponential fit 8 (see 45 °C graph, purple dashed line Figure 5).
Fig. 5.
Dwell time distributions for different temperatures. Dwell times longer than 1 s correspond mostly to pauses. The single-parameter backtracking fit of the form A*t(−3/2) is shown by the (dotted) red line. The diffusive backtracking model can describe long pauses better than a double exponential fit (dashed purple line in 45°C plot) with time constants 1.2 and 6 seconds 6.
As predicted by the diffusive backtracking model21 the t(−3/2) scaling of dwell time distributions should be independent of temperature. Here we show that the distributions in Figure 5 do in fact verify this prediction. According to this model, the paused polymerase can be considered to diffuse among many intermediate states along the DNA. Once backtracked at a specific nucleotide position, the polymerase can either move forward towards the 3’ end of the RNA or backtrack even further (Figure 4b, inset), following a one-dimensional diffusion along the DNA template 12; 31; 32; 33. The equilibrium state of the enzyme at a particular nucleotide position n can be expressed as:
| [eqn 3] |
where kf0 and kb0 are the prefactors of the forward and backwards diffusional motions respectively, and ΔGf†n and ΔGb†n are the corresponding activation energies for RNAP at nucleotide position n (Figure 4b, inset). As RNAP diffuses while in the backtracked state, the energetic differences between specific base pair configurations and their interaction with the polymerase could in principle generate local energy minima 12. However, a pseudo-random sequence like the one used in this experiment would not bias the average movement of the enzyme. Hence, the movement of the polymerase is dominated by an average barrier height with ΔGb†n ≈ ΔGf†n≈‹ΔG†›. Using this assumption, the exponent in eqn 3 vanishes and, since the temperature dependence of the prefactors cancels out, leads to a temperature independent equilibrium constant . The probability that RNAP moves forward towards the active site from an arbitrary nucleotide position n is: . Since is temperature independent, this probability of return is not affected by temperature either. The dwell time distributions of Figure 5 show that the t−3/2 distribution is independent of temperature in accordance with the prediction of the backtracking model. This seemingly counter-intuitive result can be understood qualitatively by imagining that a change in the temperature is equivalent to changing the polymerase’s diffusion coefficient and so, in fact, this would only shift the entire dwell time distribution to shorter or longer times, but would not change its shape. In fact, on the logarithmic scale of these plots and within the limited temperature range, this shift is not significant. In this simplified analysis we have left out the effect of force, but it can be shown that, for a particular average external force, temperature variations of this magnitude do not result in significant changes in
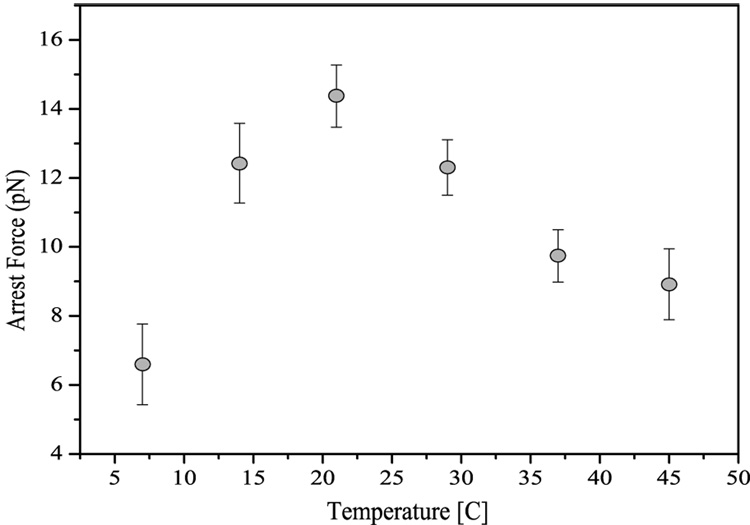
As described above, continuous transcription in these experiments results in an increase of tension on the DNA tether. The force at which the polymerase ceases translocation for more than 10 minutes is defined as the arrest force. In the case of RNA polymerase, this is not a thermodynamic arrest force, but is most likely a result of entry into the paused pathway and the subsequent inability of the enzyme to return to the elongation competent state which leads to enzyme arrest 11; 13; 17. It follows that an increase in the probability of pausing (the pause density), will offer more opportunities for arrest to occur and, as a consequence, will result in lower arrest forces. Since, as we have observed, pause durations are temperature independent, we would expect the temperature dependence of the observed arrest force to be determined by the temperature dependence of the pausing probability. Specifically, we would foresee the arrest force to increase asymptotically with increasing temperature. In accordance to this prediction, our experimental data shows an increase in the arrest force for temperatures below 21 °C (Figure 6). Nevertheless, to our surprise, the arrest force does not reach an asymptote, but decreases for higher temperatures, presenting a maximum of 14 ± 0.9 pN at room temperature. This result indicates that while the temperature dependence of the arrest forces below RT can be explained by the pause density decrease with temperature, another process dominates at higher temperatures. It is also interesting to note the asymmetry of this plot for temperatures above and below RT.
Fig. 6.
Temperature dependence of the arrest force. Contrary to expectations, the arrest force is not determined by the temperature dependence of the pausing probability, but it is largest at room temperature, a temperature which in addition is not the optimal temperature for bacterial growth. Further discussion is given in the text. Error bars represent the standard error of the mean.
Discussion
In this study we have found that the activation energy for transcription at the single molecule level correlates very closely to the activation energy of cell growth, transcription and translation as measured in vivo within the so-called ‘normal’ temperature range (21 to 37 °C). This result suggests that within this range transcription and translation are processes coupled in the cell and that together these processes are rate limiting for cell growth. Furthermore, this assay has for the first time explored temperatures below room temperature, where in contrast, the activation energy for cell growth is much bigger than the one calculated from the pause-free transcription rates at the single molecule level. However, at the cellular level the overall transcription rate (including pauses) is perhaps more important to the cell than the pause-free rates. In fact, for low temperatures (7 °C to 21 °C), the estimation of the activation energy using the average enzyme velocities (obtained including pauses), approximately doubles ΔGe† to 19.4 ± 2.2 kcal/mol. This is a consequence of the dramatic change in pause density for temperatures below RT (from 0.10 ± 0.008 bp−1 at 7 °C to 0.035 ± 0.004 bp−1 at 21 °C); a trend not observed for temperatures above RT, where pause density remains approximately constant and where the calculation of the activation energy is independent of the chosen velocity (average or pause-free). This new value of ΔGe † is much closer to the activation energy for bacterial growth at low temperature (26.5 kcal/mol, 13.5°C to 21°C 26). It is remarkable that by the inclusion of pauses in the in vitro transcription velocities we are able to recover the trends observed in vivo (Figure 3b, red dots). This observation suggests that pausing may play an important role in vivo in the cell’s response to temperature change. We further speculate that bacteria could use pausing as an added level of regulation and perhaps as a way of synchronizing processes in the cell as it adapts to changes in its environment [8 and references therein].
This study has also found the arrest force to be unexpectedly peaked at 21 °C, an observation that raises several questions. First, why doesn’t the arrest force continue to follow the trend posed by the pause density for temperatures above RT, and most importantly, what other mechanism is now responsible for the arresting of the enzyme? We hypothesize that at temperatures higher than RT the thermal stability of the enzyme might decrease and lead to force-assisted partial “melting” of the enzyme. Another possibility is that the enzyme itself is most stable at room temperature as has been found for other proteins such as RNAseH (Mao, unpublished results). Finally, it is also possible that as the polymerase transcribes faster the probability of hypertranslocated states increases which could then lead to arresting 34; 35. Nevertheless, further investigation is needed in order to determine the process or processes that contribute to the decrease of the arrest force at high temperatures. Secondly, why is the peak of the arrest force located at 21 °C instead of at 37 °C, the optimum temperature for bacterial growth? From this result we speculate that perhaps bacterial cells have not been evolutionally optimized to withstand high forces at their optimum temperature. The magnitude of the forces against which RNAP has to transcribe in vivo is not known, but perhaps forces inside the cell remain smaller than 12 pN, the enzyme’s arrest force at 37 °C, and therefore such a lower arrest force would not present a problem. Alternatively, the low arrest force observed at 37°C could be used by the cell as an additional regulatory mechanism: such marginal mechanical properties of the enzyme would allow for regulation of function via the participation of elongation factors such as GreA and GreB.
Finally, the pausing histograms have shown that most, if not all, pauses of the prokaryotic RNA polymerase are diffusive backtracks along the DNA as was originally proposed for the eukaryotic one. This further underscores the functional homology between these two systems and suggests the genetic conservation of the pausing mechanisms.
Materials and Methods
DNA construct
The DNA template was prepared from pPIA2–6 10, a plasmid which contains a λ PR promoter. The plasmid was double digested with EagI (New England Biolab (NEB), Ipswich, MA) and NdeI (NEB, Ipswich, MA) and later dephosphorylated using Antarctic phosphatase (NEB, Ipswich, MA). A 966 bp digoxigenin handle was made by amplifying (PCR) a pEIB plasmid 23 using dig-labeled nucleotides and then purified using a Qiagen PCR purification kit (Qiagen Inc.-USA, Valencia, CA). This fragment was then digested with EagI and ligated to the previously digested pPIA2–6. Finally, to purify the DNA construct the ligated products were run on a 0.7% agarose gel. The band corresponding to the desired length (11528 bp) was cut and loaded into an Elutrap to recover the DNA.
Stall complex preparation and transcription reaction
The stall complex was prepared by first incubating 3.78 nM of DNA prepared above with 40 nM of biotinylated RNAP (a gift from Irina Artsimovitch, Ohio State University) for 15 min at 37 °C in a buffer containing 20 mM Tris HCl, 20 mM NaCl, 10 mM MgCl2, 20 mM DTT and 0.2% NaN3 The transcription buffer had a pH of 7.9 adjusted at individual experimental temperatures ranging from 7 °C to 45 °C. After incubation, heparin (0.4 mg/ml) (Sigma-Aldrich, St. Louis, MO.) was added to eliminate polymerases non-specifically bound to the DNA. This was followed by a supply of 150 µM ApU (Sigma-Aldrich, St. Louis, MO), 2.5 µM ATP, 2.5 µM GTP and 2.5 µM UTP (Fermentas, Hanover, MD). The lack of CTP stalled the polymerase at the +70 position from the promoter. This stall complex was then mixed with anti-digoxigenin coated polystyrene beads (Spherotech, Libertyville, IL) in a transcription buffer that contained AseI restriction enzyme (NEB, Ipswich, MA) which cuts at a site between the promoter and the stall site and therefore eliminates complexes that did not start elongation. PPi (1 mM) (Sigma-Aldrich, St. Louis, MO) was then added to encourage pyrophosphorolysis as a method to reduce the number of backtracked complexes. After incubating for an hour and a half at room temperature, the beads were suspended in a transcription buffer, flowed into a glass chamber and trapped using a dual trap laser tweezers 24. Streptavidin-coated beads (Spherotech, Libertyville, IL) were flowed in and trapped separately. Both beads were then rubbed against each other until the biotinylated polymerase was bound to the streptavidin-coated bead and a tether was formed. The beads were kept at constant distance at a starting force of 5 pN for 21°C and 2 pN for the other temperatures. Transcription was re-initiated by flowing in transcription buffer containing all four NTPs at a saturating concentration of 1 mM and 1µM PPi.
Temperature control
The temperature control method used was recently developed and described by Mao et al. 23. It consists of two copper jackets placed over the objectives on both sides of the flow cell and through which an antifreeze solution of a determined temperature (7°C, 14°C, 21°C, 29°C, 37°C and 45°C ) was circulated, maintaining a stable and uniform temperature with little fluid convection in the sample chamber. The temperature inside the flow cell was monitored by a buried thermocouple and was maintained constant up to +/− 1 °C.
Data collection and analysis
As the polymerase translocated along the DNA, the beads were pulled closer to each other, increasing the force on the tether, which was measured by the displacement of the beads from the trap center. Thus, the forces for each run increase from 2 pN (5 pN for 21°C) to the arrest force. Using the initial contour length of the DNA template and the initial force, the initial extension is calculated using the extensible Worm-Like Chain (WLC) model 25. Changes in bead position reflect the changes in DNA extension. The updated extension and the measured force were used to calculate the new contour length by means of the WLC. The values used for the persistence length and the stretch modulus varied with temperature as measured by Mao and Arias-Gonzalez et al. (unpublished results).
Raw records were filtered with both a Gaussian filter with a standard deviation of 700 ms and a Savitzky-Golay filter with a time constant of 2.5 ms. Velocities were calculated by taking the derivatives of the time-dependent position of the enzyme along the template. The measurement of the velocity at 7 °C was limited by the signal-to-noise ratio which decreased significantly as the enzyme slowed down. At 45 °C the spread in the velocity distribution increased the error in the measurement. Pauses were removed both by using a threshold velocity and a threshold dwell time (time spent at a given nucleotide), with a value of 2 or 3 standard deviations from the average velocity and dwell time, adjusted based on the signal to noise ratio. Traces were visually inspected to verify that all observable pauses were correctly marked. The algorithms used for pause selection were shown to correctly identify pauses longer than 1 second. Once the pauses were identified, pause density was calculated by dividing the number of pauses in each trace by the total number of nucleotides transcribed. These values were then averaged together to get an average pause density for each temperature. The pooling of the data within the experimental force range is justified by the fact that both velocity and pause density (pauses/nt) are independent and/or very weakly dependent on force9; 11; 21.
Dwell time histograms and fits
Dwell times, defined as the time the polymerase remains at a particular nucleotide (s/nt), were calculated from the time required for the polymerase to advance by 2 nts along the template (the estimated spatial resolution of the filtered data). Dwell times shorter than 1s (not shown) correspond to stepping of the enzyme (equivalent to 1/Vmax), whereas those longer than 1s were attributed to periods of pausing. Dwell time histograms were calculated with variable bin sizes, such that each point corresponds to three events (measured dwell times); the bin size is given by the range of these three dwell times. The probability density for each bin (s−1) was calculated as the number of counts (3 in this case) divided by (total number of counts)*(binsize). These probability histograms were fit for all dwelltimes greater than 2 seconds with the unbiased diffusive backtracking model: P(t) = A*t(−3/2) 21 (Figure 5, dotted red line). Fits were taken from 2 seconds instead of 1 second because of undercounting in the histograms due to the 700 ms filter used for the filtering of the data.
Supplementary Material
Acknowledgements
We thank Irinia Artsimovich for the biotinylated polymerase and acknowledge Eric Galburt and Stephan Grill for the data analysis algorithm and many helpful discussions, in particular to Eric Galburt for the critical reading of this manuscript. This work was funded by NIH grant GM32543 and by U.S. Department of Energy grants DE-AC03-76SF00098 (C.B.), "Microscopies of Molecular Machines" (C.B.), and "Advanced Instrumentations for Microscopies of Molecular Machines" (C.B.)
The abbreviations used are
- DTT
Dithiothreitol
- PPi
Sodium Phyrophosphate
- nt
nucleotide
- bp
base-pair
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 2.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 3.Yager TD, von Hippel PH, Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2 Edition. Washington, D.C: ASM Press; 1987. [Google Scholar]
- 4.Finn RD, Orlova EV, Gowen B, Buck M, van Heel M. Escherichia coli RNA polymerase core and holoenzyme structures. Embo J. 2000;19:6833–6844. doi: 10.1093/emboj/19.24.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 6.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 7.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 8.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 10.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 11.Forde NR, Izhaky D, Woodcock GR, Wuite GJ, Bustamante C. Using mechanical force to probe the mechanism of pausing and arrest during continuous elongation by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2002;99:11682–11687. doi: 10.1073/pnas.142417799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert KM, Greenleaf WJ, Block SM. Single-Molecule Studies of RNA Polymerase: Motoring Along. Annu Rev Biochem. 2008 doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyduk T, Niedziela-Majka A. Fluorescence resonance energy transfer analysis of escherichia coli RNA polymerase and polymerase-DNA complexes. Biopolymers. 2001;61:201–213. doi: 10.1002/bip.10139. [DOI] [PubMed] [Google Scholar]
- 16.Andrecka J, Lewis R, Bruckner F, Lehmann E, Cramer P, Michaelis J. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci U S A. 2008;105:135–140. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng GH, Lee DN, Wang D, Chan CL, Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 19.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007 doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 22.Abbondanzieri EA, Shaevitz JW, Block SM. Picocalorimetry of transcription by RNA polymerase. Biophys J. 2005;89:L61–L63. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao H, Arias-Gonzalez JR, Smith SB, Tinoco I, Jr, Bustamante C. Temperature control methods in a laser tweezers system. Biophys J. 2005;89:1308–1316. doi: 10.1529/biophysj.104.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffitt JR, YR C, D I, C B. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci U S A. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 26.Herendeen SL, VanBogelen RA, Neidhardt FC. Levels of major proteins of Escherichia coli during growth at different temperatures. J Bacteriol. 1979;139:185–194. doi: 10.1128/jb.139.1.185-194.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingraham J, Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd Edition. Washington, D.C: ASM Press; 1987. [Google Scholar]
- 28.Ryals J, Little R, Bremer H. Temperature dependence of RNA synthesis parameters in Escherichia coli. J Bacteriol. 1982;151:879–887. doi: 10.1128/jb.151.2.879-887.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremer H, Dennis PP, Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2 Edition. Washington, D.C: ASM Press; 1987. [Google Scholar]
- 30.Ron EZ, Shani M. Growth Rate of Escherichia coli at Elevated Temperatures: Reversible Inhibition of Homoserine Trans-Succinylase. J Bacteriol. 1971;107:397–400. doi: 10.1128/jb.107.2.397-400.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 32.Sakata-Sogawa K, Shimamoto N. RNA polymerase can track a DNA groove during promoter search. Proc Natl Acad Sci U S A. 2004;101:14731–14735. doi: 10.1073/pnas.0406441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 34.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, Navaroli DM, Enuameh MS, Martin CT. Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism. Proc Natl Acad Sci U S A. 2007;104:10352–10357. doi: 10.1073/pnas.0606306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.