SUMMARY
MDM2 is the predominant negative regulator of p53 that functions to maintain the appropriate level of expression and activity of this central tumor suppressor. Mdm2-a is a commonly identified splice variant of Mdm2; however, its physiological function is unclear. To gain insight into the activity of MDM2-A and its potential impact on p53, an Mdm2-a transgenic mouse model was generated. Mdm2-a transgenic mice displayed a homozygous-lethal phenotype that could be rescued by a reduction in p53 expression, demonstrating a dependence upon p53. Mdm2-a hemizygous mice exhibited reduced longevity, and enhanced senescence was observed in their salivary glands. In addition, the transgenic mice lacked typical, accelerated aging phenotypes. Growth of transgenic mouse embryonic fibroblasts (MEFs) was inhibited relative to wild-type MEFs, and MDM2-A was shown to bind to full-length MDM2 in an interaction that could increase p53 activity via reduced MDM2 inhibition. Evidence of p53 activation was shown in the Mdm2-a transgenic MEFs, including p53-dependent growth inhibition and elevated expression of the p53 target protein p21. In addition, MDM2-A increased senescence in a p21-independent manner. In conclusion, unexpected roles for MDM2-A in longevity and senescence were identified in a transgenic mouse model, suggesting that Mdm2 splice variants might be determinants of these phenotypes in vivo.
INTRODUCTION
p53 is a tumor suppressor protein that plays a central role in controlling cell cycle progression (Kastan et al., 1991) and apoptosis (Yonish-Rouach et al., 1991). The expression and activity of p53 are precisely regulated in order to maintain appropriate levels of cellular proliferation and death (Momand et al., 1992). MDM2 is the best-characterized negative regulator of p53 and itself is classified as an oncogene (Fakharzadeh et al., 1991). Inhibition of p53 by MDM2 results in strict control of p53 protein levels through MDM2-mediated ubiquitylation and proteasomal degradation (Li et al., 2003). Alterations in MDM2 levels lead to shifts in p53 activity, which result in dramatic biological outcomes (Donehower, 2002). At one end of the spectrum, when MDM2 is highly overexpressed, p53 cannot suppress growth sufficiently and transformation results (Oliner et al., 1992; Ladanyi et al., 1993; Reifenberger et al., 1993; Corvi et al., 1995). At the other end of the spectrum, the deletion of Mdm2, as observed in Mdm2-null embryos, results in unchecked p53 activity, and the enhanced growth inhibition consequently hinders organ development (Jones et al., 1995; Montes de Oca Luna et al., 1995). Additionally, several mouse models have been developed that exhibit varying levels of p53 activity. As expected, reduced p53 activity leads to tumor susceptibility (Donehower et al., 1992; Jacks et al., 1994), whereas enhanced p53 activity confers tumor protection (Garcia-Cao et al., 2002; Tyner et al., 2002; Maier et al., 2004; Mendrysa et al., 2006), and surprisingly, in certain model systems, accelerates aging (Tyner et al., 2002; Maier et al., 2004).
Many splice variants of Mdm2 have been identified both in tumors (Sigalas et al., 1996; Bartel et al., 2002a) and in normal tissues (Bartel et al., 2004); however, the functional characterization of the MDM2 isoforms encoded by these variants is limited. Mdm2-a is one of the most common variants identified to date (Bartel et al., 2002a) and the most common variant detected in pediatric rhabdomyosarcoma tumors (Bartel et al., 2002b). The protein encoded by this particular variant has a deletion of a large portion of the amino terminus and, therefore, lacks a functional p53-binding domain (Sigalas et al., 1996; Bartel et al., 2002b). However, it maintains the central acidic domain and the carboxy-terminal RING finger domain, which contain important sequences for binding to other proteins and for the ubiquitylation of p53 (Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997; Bartel et al., 2002b). Previous data have suggested that, unlike several similar MDM2 isoforms, MDM2-A [or Δ28-220 as referred to by Fridman et al. (Fridman et al., 2003)] does not enhance tumorigenesis (Fridman et al., 2003). These data are consistent with a model proposed previously suggesting that splice variants with an intact C-terminal RING domain bind to full-length MDM2 protein, resulting in increased p53 activity and a growth-inhibitory, rather than growth-promoting, phenotype (Evans et al., 2001; Dang et al., 2002). However, these data are inconsistent with the frequent expression of MDM2 splice variants in tumors.
In order to assess the physiological function of MDM2-A in vivo, we generated an Mdm2-a transgenic mouse model. These mice, and mouse embryonic fibroblasts (MEFs) derived from them, provide evidence of a p53-dependent phenotype of MDM2-A and demonstrate the impact of this splice variant on growth inhibition, senescence and longevity.
RESULTS
Mdm2-a transgenic mice are homozygous lethal in a p53-dependent manner
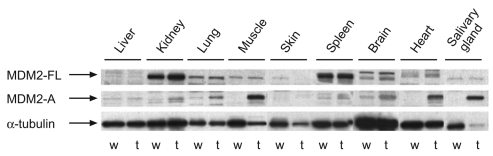
Transgenic mice expressing the Mdm2-a cDNA were generated by standard pronuclear injection methods, as detailed in the Methods. Despite multiple rounds of microinjection of the pCAGGS–Mdm2-a construct, only one founder line was obtained that integrated the Mdm2-a cDNA without any mutations (Schuster and Harris, 2007). In addition, despite the use of a ubiquitous promoter, expression of the transgene was restricted to only a few tissues (Fig. 1). Analysis of transgene expression in Mdm2-a mice revealed high levels of MDM2-A in muscle, the heart and salivary glands, moderate levels in the brain and lungs, and low levels of expression in the kidney and spleen (Fig. 1). To rule out any chromosomal integration positional effects of the transgene, Mdm2-a was localized to a region on chromosome 13 that was at least 150 kb, 5′ and 3′, from the nearest coding regions.
Fig. 1.
The Mdm2-a transgene has limited tissue expression. Western blot analysis of various tissues from either wild-type (w) or Mdm2-a transgenic (t) mice were probed with antibodies to detect either full-length MDM2 (MDM2-FL) and its splice variant, MDM2-A, or the loading control α-tubulin.
Interestingly, when Mdm2-a transgenic mice were bred, only hemizygous mice were viable (Table 1). Analysis of newborn litters revealed that Mdm2-a homozygous pups were born at the expected Mendelian ratio and were anatomically indistinguishable from hemizygous and wild-type littermates; however, the homozygous pups died of unknown causes several hours after birth. Earlier embryonic stages were evaluated to determine whether any abnormalities could be observed. At embryonic day 14.5 (E14.5), all Mdm2-a homozygotes begin to display marked edema, specifically in the subcutaneous tissue of the nuchal region, which is absent in both hemizygous and wild-type embryos (Fig. 2A-E). The fluid in this region contained foamy macrophages indicative of active clearance of the edema (Fig. 2E). The edematous phenotype suggested a possible lymphatic defect, but detailed pathological analyses of lymph and blood vessels, as well as complete blood counts from both Mdm2-a homozygous neonates and embryos, revealed neither gross anatomical malformation (Fig. 2F,G) nor hematopoietic defects (data not shown). In addition, all organ systems appeared normal in the homozygous embryos and neonates. Consequently, the actual cause of death of the Mdm2-a homozygous neonates remains unresolved.
Table 1.
Mdm2-a homozygous mice are viable only when p53 gene dosage is reduced
|
Mdm2-a genotype |
||||
|---|---|---|---|---|
| Cross | Wild-type | Mdm2-a+/– | Mdm2-a+/+ | Total |
| Mdm2-a+/–intercross | 90 | 115 | 0 | 205 |
| p53+/–/Mdm2-a+/– intercross | 9 | 13 | 7* | 29 |
| p53 genotype of the Mdm2-a+/+ mice
|
||||
|---|---|---|---|---|
| Mdm2-a genotype | Wild-type | p53+/– | p53–/– | Total |
| Mdm2-a+/+ | 0 | 5 | 2 | 7 |
The lower half of this table shows the p53 genotype of these seven mice.
Fig. 2.
Mdm2-a homozygous embryos exhibit edema. Wild-type (A) and Mdm2-a homozygous (B) embryos harvested at E14.5 show Mdm2-a-specific edema. Cross-sections through the nuchal regions of wild-type (C) and Mdm2-a homozygous (D) E14.5 embryos. Bar, 600 μm. (E) Edematous region of Mdm2-a homozygous embryo. Arrowheads designate foamy macrophages within the region of edema. Bar, 100 μm. Whole-body cross-sections of wild-type (F) and Mdm2-a transgenic (G) E18.5 neonates exemplify the apparent normal development of Mdm2-a neonates. Bar, 2.5 mm.
The only function currently assigned to Mdm2 splice variants is the aforementioned p53 activation that occurs as a consequence of splice variant binding to full-length MDM2 (Evans et al., 2001; Dang et al., 2002). To assess directly the role of p53 on the MDM2-A-mediated perinatal lethality, transgenic mice were crossed onto a p53-null background. Interestingly, Mdm2-a homozygous animals were viable and present at the expected Mendelian ratio, but only in the presence of a concomitant reduction in p53 gene dosage (Table 1). These data demonstrate that the perinatal lethality of the Mdm2-a homozygous mice was mediated by p53 and that MDM2-A activities in vivo are, at least in part, p53 dependent.
Mdm2-a transgenic mice exhibit reduced longevity
In contrast to their homozygous counterparts, Mdm2-a hemizygous mice were viable with no obvious phenotype compared with wild-type littermates. Therefore, to assess the in vivo role of this splice variant further, these animals were used in all subsequent experiments. Mdm2-a transgenic mice and their wild-type littermates were observed for any health problems for up to 30 months.
After reaching 1 year of age, many animals developed typical age-related health problems that required euthanasia, such as uterine hyperplasia, inguinal hernia and retinal degeneration. Too few tumors developed in either the wild-type or Mdm2-a transgenic animals to allow any statistical analysis; however, both the frequency and tumor type appeared to be similar between the two groups of mice (Table 2). Interestingly, although there were no differences in the causes of death between transgenic and wild-type mice, Mdm2-a mice had significantly shorter life spans as shown by the Kaplan-Meier survival curve (Fig. 3). The median survival of wild-type mice was 25 months, whereas the Mdm2-a mice had a median survival of only 20 months.
Table 2.
Tumors detected in Mdm2-a hemizygous transgenic mice and wild-type littermates
| Mouse line | Number of tumors/total (%) | Tumor types (number of animals) | Age at death (months) |
|---|---|---|---|
| Wild-type | 2/55 (4) | Squamous cell carcinoma (1) | 25 |
| Soft tissue sarcoma (1) | 24 | ||
| Mdm2-a transgenic | 4/64 (6) | Alveolar bronchiolar adenoma (1) | 12 |
| Soft tissue sarcoma (1) | 19 | ||
| Osteoma (1) | 8 | ||
| Soft tissue mass* leg (1) | 13 |
Diagnosis unclear.
Fig. 3.
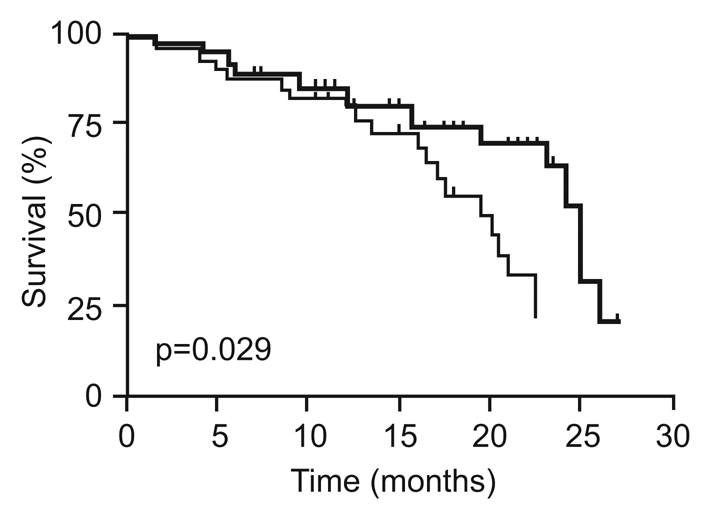
Mdm2-a transgenic mice display reduced longevity. Kaplan-Meier survival curve of wild-type (bold line) and Mdm2-a transgenic (thin line) mice (p=0.029). The survival curve is based on data from 55 wild-type and 64 Mdm2-a transgenic mice.
MDM2-A-mediated reduction in longevity was not the result of accelerated aging
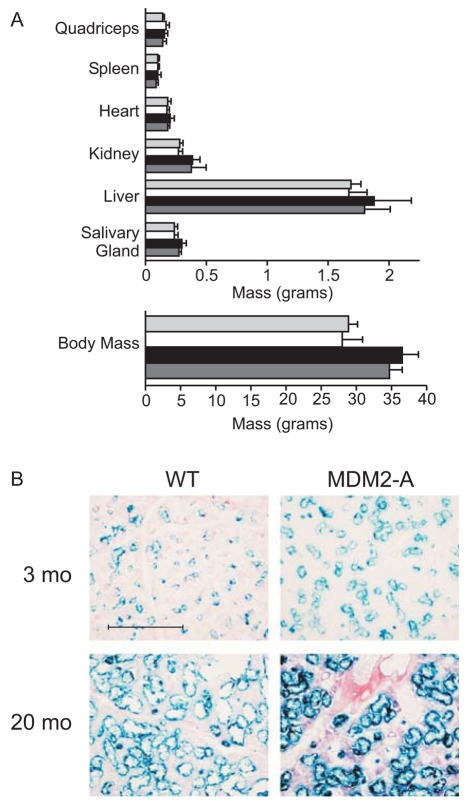
To determine the role of MDM2-A in the shortened life span of the transgenic animals, we assessed various aging parameters in cohorts of young (3 months) and aged (18–20 month) mice. Total body mass, various organ masses, bone density, dermal adipose thickness and shaved hair re-growth were measured and compared between transgenic and wild-type mice. As expected, total body mass, as well as that of most organs, increased as both wild-type and Mdm2-a transgenic mice progressed from 3 to 20 months of age (Fig. 4A). There was a trend towards reduced body and organ masses (Fig. 4A), as well as a reduced rate of hair growth (data not shown), in the Mdm2-a transgenic mice at both 3 and 20 months of age; however, the differences were not statistically significant. Additionally, there was no detectable difference in either bone density or dermal adipose thickness between transgenic and wild-type animals (data not shown). Thus, despite their shortened life span, there appeared to be only subtle changes in the aging of Mdm2-a mice.
Fig. 4.
Mdm2-a transgenic mice do not show accelerated aging, but display enhanced senescence in aged tissue. (A) Tissue and total body mass from 3-month-old wild-type (light gray bars) and Mdm2-a (white bars) male mice, and from 20-month-old wild-type (black bars) and Mdm2-a (dark gray bars) male mice. Female mice exhibited the same trend (data not shown). (B) SA-βgal staining of salivary gland tissue from young and aged wild-type and Mdm2-a transgenic mice. Bar, 50 μm.
Tissues from Mdm2-a transgenic mice display enhanced senescence
Although there were no significant differences in various physical parameters of aging, it remained possible that there were biochemical differences in tissues that might have contributed to the reduced life span of Mdm2-a transgenic animals. To this end, the role of MDM2-A in senescence was evaluated in vivo. The activity of β-galactosidase at pH 6.0 is the current standard marker of cellular senescence (Dimri et al., 1995). Therefore, senescence-associated β-galactosidase (SA-βgal) assays were performed on salivary gland, muscle, heart, liver and brain tissue samples from both transgenic and wild-type mice. Although SA-βgal-positive cells could be detected in several organs, no significant differences were found between wild-type and transgenic tissues, with one exception. In the salivary gland tissue from Mdm2-a mice, SA-βgal activity increased appropriately with age, and stained more robustly than that of wild-type littermates, particularly in aged tissue (Fig. 4B). Even though we observed enhanced senescence in only the salivary gland, these data suggest that there might have been other tissues in which senescence was enhanced by MDM2-A expression, potentially contributing to the reduction in longevity observed in the Mdm2-a mice.
MDM2-A causes growth inhibition in vitro and interacts with full-length MDM2
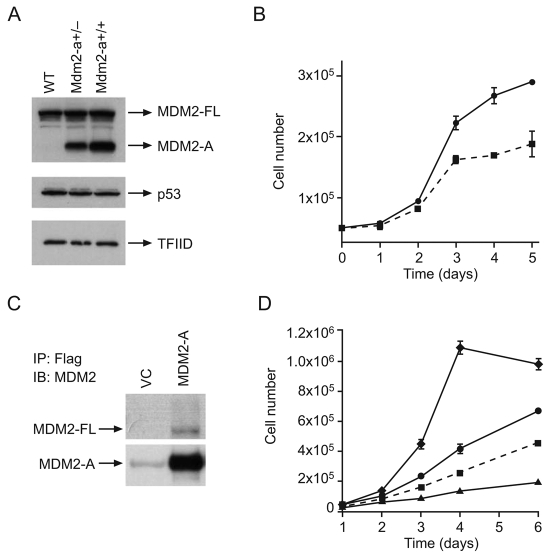
To gain further insight into the function of MDM2-A, MEFs were isolated from transgenic and wild-type mice for in vitro analyses of cellular growth and p53 activation. Western blot analyses showed that neither p53 nor full-length MDM2 protein levels were altered by the presence of MDM2-A (Fig. 5A). However, cellular growth assays demonstrated that MEFs expressing MDM2-A grew at a slower rate than wild-type MEFs (Fig. 5B), suggesting that MDM2-A confers a growth-inhibitory phenotype.
Fig. 5.
MDM2-A is growth-inhibitory and binds to full-length MDM2 in vitro. (A) Western blot analysis of MEFs to detect expression of full-length MDM2 (MDM2-FL), the MDM2-A transgene, p53 and the loading control TFIID. (B) Growth curves for wild-type (circles, solid line) and MDM2-A transgenic (squares, dotted line) MEFs isolated from embryos from the same litter. (C) Immunoprecipitation (IP) of FLAG-tagged MDM2-A that was retrovirally expressed in wild-type MEFs. Immunoblot (IB) analysis with anti-MDM2 antibody was used to visualize the full-length MDM2 (MDM2-FL) and MDM2-A proteins. The faint band visible in the vector control (VC) lane represents antibody heavy chain. (D) Transduction of wild-type MEFs with retroviral vectors containing cDNAs of different MDM2 isoforms. MDM2-A and MDM2-M3 inhibit the growth of wild-type MEFs, whereas full-length MDM2 results in enhanced cell growth. Symbols: MDM2-A (squares, dotted line); vector control (circles, solid line); full-length MDM2 (diamonds, solid line); truncated MDM2-M3 (triangles, solid line).
Other MDM2 splice variants have been shown to bind to full-length MDM2 and result in growth inhibition as a consequence of increased p53 activity. Technical difficulties precluded analysis of the interaction between full-length MDM2 and MDM2-A in the transgenic MEFs. Therefore, to evaluate their potential interaction, FLAG-tagged MDM2-A was retrovirally expressed in wild-type MEFs and immunoprecipitated with an anti-FLAG antibody. By probing the western blot with an anti-MDM2 antibody, full-length MDM2 was found to co-immunoprecipitate weakly with MDM2-A (Fig. 5C), demonstrating that the MDM2-A splice variant is capable of binding to full-length MDM2 protein. This interaction could potentially disrupt the endogenous protein interactions of full-length MDM2, particularly with p53, to mediate a growth-inhibitory phenotype.
Exogenous expression of MDM2-A in wild-type MEFs following retroviral transduction could also inhibit cell growth (Fig. 5D). MEFs transduced with retroviral vectors expressing either full-length MDM2 or MDM2-M3 (the RING domain only) were also evaluated. As expected, full-length MDM2 enhanced growth owing to the inhibition of p53 activity. MDM2-M3 suppressed growth in a similar manner to MDM2-A when compared with vector-control-transduced cells.
MDM2-A-mediated activation of p53
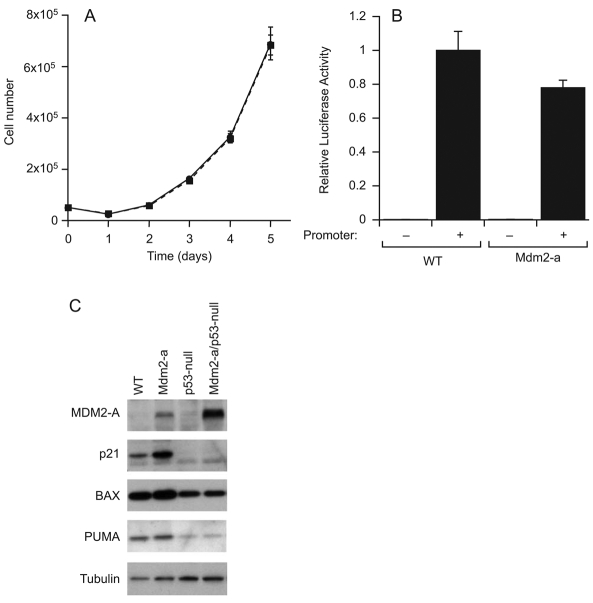
To assess the role of p53 in the Mdm2-a-mediated in vitro growth inhibition, p53-null MEFs were generated with, and without, Mdm2-a transgene expression, and assayed for cell growth. In contrast to the growth inhibition observed in MEFs containing Mdm2-a and expressing wild-type p53 (Fig. 5B), p53-null MEFs grew at the same rate in the presence or absence of MDM2-A (Fig. 6A). Thus, when p53 is deleted, the growth inhibition of Mdm2-a-expressing MEFs is abolished, demonstrating that this phenotype is p53 dependent. In addition, p53-null MEFs were insensitive to the growth-inhibitory effects of exogenous MDM2-A expression, again confirming the dependence on p53 (data not shown).
Fig. 6.
p53 activation in the presence of MDM2-A. (A) Growth-curve analysis of p53-null (circle, solid line) and Mdm2-a–p53-null (squares, dotted line) MEFs. (B) Relative luciferase activity of the p53-responsive p21WAF1/CIP1 promoter linked to the luciferase reporter gene in both wild-type and MDM2-A transgenic MEFs. (C) Western blot analysis of expression of the p53 target genes p21, bax and PUMA in both wild-type and p53-null MEFs with, and without, MDM2-A. Tubulin is shown as a loading control.
To assess directly p53 activation in Mdm2-a MEFs, luciferase reporter assays were performed using p53-responsive promoters. Surprisingly, when using either the p21WAF1/CIP1 promoter (Fig. 6B) or the bax promoter (data not shown), there was no significant difference in p53 transcriptional activity in MEFs that express MDM2-A relative to wild-type MEFs.
To explore further the potential p53 activation in the presence of MDM2-A, several p53 target genes were assessed for alterations in their expression in wild-type and transgenic MEFs. Levels of p21WAF1/CIP1, a cyclin-dependent kinase inhibitor (El-Deiry et al., 1994), were elevated in the presence of MDM2-A, whereas the expression of endogenous MDM2 (Fig. 5A) and the pro-apoptotic proteins Bax (Miyashita and Reed, 1995) and PUMA (Nakano and Vousden 2001; Yu et al., 2001) were unaltered by the transgene (Fig. 6C). The elevation of p21 was shown to be p53 dependent because MDM2-A transgenic MEFs that were p53-null showed no p21 activation (Fig. 6C). As expected, p53-null MEFs displayed reduced expression of the p53 target genes bax and PUMA. These data demonstrate that specific p53 targets are affected by MDM2-A, but there was no global increase in the expression of p53-responsive genes in the presence of MDM2-A.
Mdm2-a MEFs display enhanced senescence
In vivo data suggested that Mdm2-a plays a role in enhancing senescence in the tissues of Mdm2-a transgenic mice (Fig. 4B). To assess this phenotype in vitro, Mdm2-a and wild-type MEFs were stained for SA-βgal from passages 4–16. As expected, both wild-type and Mdm2-a MEFs contained low levels of SA-βgal-positive cells at both very early and late passages owing to their mitotically active or immortal states, respectively (data not shown). A peak in the proportion of senescent cells was observed at passage 6, and significant differences in the degree of senescence between Mdm2-a and wild-type MEFs were observed (Fig. 7A,B). Mdm2-a MEFs displayed a greater fraction of senescent cells compared with wild-type MEFs, which probably contributed to their phenotype of slower growth.
Fig. 7.
MDM2-A enhances senescence in a p21-independent manner. (A) SA-βgal staining in wild-type and Mdm2-a transgenic MEFs. (B) Histogram of passage 6, SA-βgal-positive cells in wild-type, p53-null and p21-null MEFs with, and without, MDM2-A. (C) Growth curve of p21-null (circles, solid line) and Mdm2-a–p21-null (squares, dotted line) MEFs.
Because p53 was shown to be responsible for the growth inhibition observed in the Mdm2-a MEFs, the role of p53 in MDM2-A-mediated senescence was also evaluated. When p53 was deleted, little or no senescence was observed in either wild-type or Mdm2-a MEFs (Fig. 7B), confirming the p53-dependence of this phenotype (Harvey et al., 1993). However, because senescence was suppressed in both wild-type and transgenic MEFs, it was difficult to assess the contribution of p53 to the increased senescence observed in the presence of Mdm2-a.
To determine whether p21 was the p53 target gene involved in the MDM2-A-mediated growth inhibition and senescence, both the wild-type and the Mdm2-a phenotypes were assessed in the absence of p21. In p21-null MEFs that were generated with and without the transgene, Mdm2-a-mediated growth inhibition persisted, suggesting that p21 was not involved (Fig. 7C). Senescence was also quantitated in p21-null MEFs with and without MDM2-A, and overall this phenotype was suppressed in the absence of p21. However, there was a slight increase in SA-βgal-positive staining in Mdm2-a–p21-null MEFs compared with Mdm2–p21-null MEFs – a trend similar to that observed in MEFs containing wild-type p21. Even though this difference was not significant (Fig. 7B), these data suggested that p21 was probably not responsible for the enhanced senescence observed in the presence of Mdm2-a.
DISCUSSION
Mdm2 splice variants have long been considered to be either tumor-specific isoforms that actively promote tumorigenesis or the passive by-products of deregulated splicing in transformed cells (Bartel et al., 2002a). Here, we provide evidence for a novel and apparently contrasting role for the Mdm2-a splice variant in normal tissues. We show that MDM2-A causes growth inhibition, resulting in either p53-dependent perinatal lethality of Mdm2-a transgenic homozygotes or reduced longevity of hemizygous mice in vivo. These phenotypes are a consequence of p53 activation, which probably leads to elevation of specific downstream targets. Additionally, and in accordance with growth inhibition and reduced life span, MDM2-A expression was shown to enhance senescence in MEFs and in certain aged tissues of the transgenic mice.
As has been described previously for other MDM2 splice variants, the presence of MDM2-A appears to reduce the inhibitory capacity of full-length MDM2 by disrupting its normal interaction with p53, which in turn results in increased p53 activity (Evans et al., 2001). The impact of MDM2-A in vivo mirrors other previously described mouse models in which the MDM2-p53 balance is altered. The MDM2-A-mediated perinatal lethality, which can be rescued by p53 reduction, is similar to, albeit less robust than, the embryonic lethality observed when Mdm2 is deleted. Mdm2-null embryos die at an early developmental stage shortly after implantation and can only be rescued by deletion of p53 (Jones et al., 1995; Montes de Oca Luna et al., 1995), whereas Mdm2-a homozygotes die shortly after birth and survive only when mice are concomitantly heterozygous or null for p53.
Reduced longevity has also been observed in models of p53 activation that have been described previously. Mutant p53 mice (p53+/m) that express a truncated form of p53, which is capable of activating the wild-type allele, display reduced longevity, enhanced aging and reduced tumor formation (Tyner et al., 2002). Similarly, p44 mice express a highly active p53 isoform and display a shortened life span accompanied by accelerated aging, activation of p53 target genes and enhanced senescence (Maier et al., 2004). The phenotype of Zmpste24-null mice, which lack a key metalloproteinase involved in nuclear architecture, was also proposed to be the result of p53 activation (Varela et al., 2005). These animals display reduced longevity, enhanced aging and upregulation of several p53 target genes (Varela et al., 2005). However, not all models in which p53 is elevated have shown the same phenotypes (Garcia-Cao et al., 2002; Mendrysa et al., 2006). The cause of phenotypic differences between the mice within each model system is unclear, but probably reflects variation in the degree of p53 activation in specific tissues. The modest changes observed in the Mdm2-a mice are probably a consequence of the limited tissue expression of the transgene in a hemizygous animal.
Interestingly, p53 activation in the presence of MDM2-A was not evident in a p53-responsive reporter assay in MEFs, and p21 was the only target gene observed to display elevated expression. However, this result is consistent with previous observations of enhanced p53 activity. For instance, the mutant p53 expressed in the p53+/m mice only showed increased p53 activity in a luciferase reporter assay when the mutant allele was ectopically expressed in Saos-2 cells (Tyner et al., 2002). Additionally, not all p53 target genes evaluated were found to be elevated in p44- and Zmpste24-null cells (Maier et al., 2004; Varela et al., 2005). Remarkably, despite multiple indirect indicators of p53 activation, these models, including that of Mdm2-a, fail to demonstrate any changes in the p53 protein itself, or a global activation of p53-responsive genes. However, these models do support the hypothesis that p53 target genes can be specifically and selectively upregulated depending on the cellular context and degree of p53 activation (Bouvard et al., 2000). Therefore, p21 upregulation together with reduced longevity, p53-dependent growth inhibition and perinatal lethality, which are all observed in the presence of MDM2-A, are all indicators of p53 activation.
Because p21 was elevated upon MDM2-A expression and is known to play a role in growth inhibition and senescence (Noda et al., 1994), we evaluated p21 as a candidate p53 target gene that could potentially mediate these phenotypes as a consequence of MDM2-A expression. The data demonstrate that MDM2-A remains growth-inhibitory in p21-null MEFs, indicating that p21 is not involved. In addition, although p21 is important for senescence of both wild-type and MDM2-A transgenic MEFs, it appears that p21 does not play a role in the MDM2-A-phenotype-enhanced senescence. Therefore, growth inhibition and potentially the senescence observed in the presence of MDM2-A are p53 mediated; however, the downstream effectors responsible for these phenotypes remain unknown.
The data described here indicate that MDM2-A acts through p53 to slow growth and increase senescence, resulting in phenotypes that are potentially tumor protective (Campisi, 2001). However, Mdm2-a mice were generally healthy and developed few tumors. The extremely low level of tumorigenesis in these mice probably obscured any tumor protection conferred by MDM2-A. It will be important in the future to investigate the impact of MDM2-A on both tumorigenesis and carcinogenesis to evaluate whether MDM2-A expression is tumor-protective when the incidence of tumor formation is increased.
In conclusion, to the best of our knowledge, we have identified a novel physiological role for the Mdm2 splice variant MDM2-A. This splice variant, and potentially other Mdm2 splice variants, might be important determinants of aging, senescence and longevity. However, the data presented here are in contrast to those of others describing an oncogenic phenotype for other, similar, MDM2 variants (Fridman et al., 2003; Steinman et al., 2004). In future work, the challenge will be to evaluate the expression levels of different variants in specific tissue types and to determine the consequences of their coordinated expression on aging, senescence and transformation when expressed at physiological levels.
METHODS
Generation of the Mdm2-a cDNA expression plasmid
The Mdm2-a cDNA was generated by the PCR-based method of splicing by overlapping extension. The murine full-length Mdm2 cDNA (provided by G. Lozano, MD Anderson, TX) was used as a template, and exons 4-9 (Δ28-220 amino acids) were deleted to generate a murine sequence equivalent to the human MDM2-A sequence (GenBank U33199). Primers flanking the N-terminal and C-terminal regions of Mdm2-a included EcoRI and XhoI restriction sites, respectively (underlined), and were as follows: N-terminal primer, 5′-CCGAATTCCGCCAATGTGCAATACCAACAT-3′ and C-terminal primer, 5′-CCGCTCGAGGCTAGTTGAAG-TAACTTAGCACA-3′. Mutually complementary super-primers that overlap the junction between exon 3 and exon 10 were generated: antisense, 5′-CTTACGCCATCGTCAAGATCCA-GAGTCTCTCTTGTTCCGAAG-3′ and sense, 5′-CTTCGGAA-CAAGAGACTCTGGATCTTGACGATGGCGTAAG-3′. The recombinant Mdm2-a cDNA was inserted into the pCAGGS/MCS vector (Niwa et al., 1991) (provided by J. Cunningham, St Jude Children’s Research Hospital, TN) to potentially allow constitutive ubiquitous expression of MDM2-A protein in transgenic mice. The plasmid was digested with HincII and PstI to remove unnecessary plasmid DNA. A 3.1 kb DNA fragment containing the chicken actin promoter, CMV enhancer, Mdm2-a cDNA and rabbit β-globin polyadenylation signal was generated for microinjection.
Generation of Mdm2-a transgenic mice
Transgenic mice were generated in the Transgenic Core Unit at St Jude by standard microinjection of DNA into the pronuclei of fertilized single-cell mouse embryos (FVB/NJ) (Hogan et al., 1994). The Institutional Animal Care and Use Committee (IACUC) approved all animal work, and all experiments conformed to the applicable regulatory standards. Transgenic mice and their wild-type littermates were monitored for tumor development and disease. Animals showing signs of pain or distress were euthanized according to IACUC guidelines. Euthanized mice were examined for lesions and tissue abnormalities.
Transgene genomic localization and genotyping
The Mdm2-a insertion site was mapped within the mouse genome using the BD GenomeWalker Universal Kit (BD Biosciences, Palo Alto, CA) according to the manufacturer’s protocol. Briefly, genomic DNA was prepared from tail tips using the DNEasy kit (Qiagen, Valencia, CA) and digested individually with DraI, EcoRV, PvuII and StuI blunt-cutting restriction enzymes to generate four DNA libraries. BD GenomeWalker adaptors were subsequently ligated to the purified DNA, and primary and secondary PCRs were performed. PCR reactions were prepared using the HotStar Taq Master Mix kit (Qiagen) and used the following gene-specific primers in combination with adaptor-specific primers: TG2, 5′-AGGTGGCTATAAAGAGGTCATCAGTA-3′; TG2N, 5′-AGATTTTTCCTCCTCTCCTGACTACT-3′. The resultant amplicons were isolated from gels using the QiaexII kit (Qiagen) and sequenced and mapped using BLAST searches of the mouse genome. Localization of the transgene to a region of chromosome 13 allowed for the construction of the following flanking and internal primers for determination of zygosity: Mdm2-aTg, 5′-TTCATTGCAATAGTGTGTTGGA-3′; Chr13A, 5′-TGCATCATTTTGAATCACAGC-3′; and Chr13B, 5′-TTAAGCAATCACCTGCCAAT-3′. A 311 bp product identifies wild-type animals, whereas a 587 bp product is present in Mdm2-a homozygous animals; hemizygous mice contain both products.
Embryo/neonate sectioning and staining
Both embryos and neonates were fixed in 10% formalin overnight, embedded in paraffin and sectioned in serial 4-micron intervals. Sections were stained with hematoxylin and eosin using standard methods and evaluated by the St Jude diagnostic laboratory.
Aging analysis
Several aging parameters were assessed in both young (3-month) and aged (18–20-month) wild-type and Mdm2-a transgenic male and female mice. For hair re-growth analysis, approximately 1 square inch of dorsal fur was shaved and, after 3 weeks growth, scored as either positive or negative. Mice were then euthanized and X-rayed (Faxitron X-ray, Wheeling, IL) at 35 kV for 10 seconds for bone density analysis. Whole-body and individual organ masses were determined and organs were either cryopreserved or fixed. Brain, heart, kidney and liver were cryopreserved for sectioning and subsequent SA-βgal staining, as described below. Skin samples were fixed in formalin, sectioned and stained with hematoxylin and eosin. Muscle, adipose and epidermal layers of the skin were measured and the average of at least 30 measurements of each was used for comparison.
MEF preparation and growth curves
MEFs were generated by standard methods (Hogan et al., 1994) from E14.5 embryos and cultured in DMEM containing 10% FBS, 2 mM glutamine, 0.1 mM β-mercaptoethanol, 100 U/ml penicillin/streptomycin and non-essential amino acids. Growth curves were generated using P3–P5 MEFs. Cells were plated in triplicate at 5×104 cells/well in 6-well dishes and counted daily for 5 days.
Protein extraction, immunoprecipitation and western blotting
To generate cell extracts, MEFs were pelleted, suspended in sonication buffer (50 mM Tris pH 8.0, 300 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 0.1% NP40, 1 mM sodium orthovanadate, 10 mM NaF, 200 μM PMSF and 10 μg/ml of each of the protease inhibitors leupeptin, aprotinin and antipain), freeze-thawed three times and centrifuged at 20,000 g at 4°C to remove the insoluble fraction. To obtain protein extracts from mouse tissues, samples were homogenized in pre-chilled sonication buffer and sonicated twice at 4°C for 10 seconds before centrifugation at 20,000 g at 4°C. Protein concentrations were determined by measuring the color intensity of protein assay dye reagent (Bio-Rad; Hercules, CA) at 595 nm. Protein samples were separated by electrophoresis on a Novex 4-20% gradient Tris-glycine polyacrylamide gel (Invitrogen, Carlsbad, CA) and transferred to a polyvinylidenedifluoride (PVDF) membrane (Immobilon, Millipore; Billerica, MA) by electroblotting. For immunoprecipitation (IP), pelleted MEFs were resuspended in cooled IP-lysis buffer [50 mM Tris (pH 8.0), 5 mM EDTA, 150 mM NaCl, 1% NP40 and protease inhibitors, as described for protein extract preparation], immunoprecipitated with an anti-FLAG-M2 agarose affinity gel (Sigma, St Louis, MO) and washed three times in cooled IP wash buffer [5% sucrose, 5 mM Tris (pH 7.4), 5 mM EDTA, 0.5 M NaCl, 1% NP40 and protease inhibitors] as described previously (McKenzie et al., 2002). Proteins were visualized by immunoblotting with the following antibodies: anti-hMDM2 (R&D Systems, Minneapolis, MN), anti-MDM2 (C-18, Santa Cruz Biotechnology, Santa Cruz, CA), anti-p53 (JA1308, Calbiochem, La Jolla, CA), anti-p21 (F-5, Santa Cruz Biotechnology), anti-bax (P-19, Santa Cruz Biotechnology), anti-PUMA (4976, Cell Signaling, Danvers, MA), anti-α-tubulin (B-7, Santa Cruz Biotechnology), anti-β-tubulin (clone 2.1, Sigma) or anti-TFIID (N-12, Santa Cruz Biotechnology). Secondary antibodies conjugated to horseradish peroxidase were obtained from Amersham (Piscataway, NY) and visualized by incubation of the membrane with either ECL reagents (Amersham) or SuperSignal (Pierce, Rockford, IL), and subsequent exposure to BioMax MR film (Eastman Kodak, Rochester, NY).
Senescence-associated β-galactosidase staining
Tissue cryosections and plated MEFs were stained using the β-galactosidase staining kit (US Biological, Swampscott, MA) according to the manufacturer’s protocol. Briefly, cells were washed in PBS, fixed in 2% formaldehyde and 0.2% glutaraldehyde in PBS for 5 minutes, and then washed again in PBS. Cells were incubated in staining solution [1 mg/ml X-gal, 40 mM citric acid/sodium phosphate (pH 6.0), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, 2 mM magnesium chloride] and staining was quantitated after 24 hours.
Retroviral expression
Subconfluent 293T cells were plated and co-transfected with the helper pEQECO packaging vector (provided by Dr E. Vanin) and MSCV-IRES-GFP retroviral vectors containing the Mdm2-a-FLAG cDNA. The medium was changed after 24 hours and virus-containing supernatant was harvested at 48–72 hours post-transfection. Early-passage (≤4) wild-type MEFs were transduced with filtered retroviral supernatant containing 1 μg/ml polybrene (Sigma). Two days after transduction, cells were sorted for GFP expression by flow cytometry under sterile conditions, and GFP-positive cells were used to prepare cell extracts.
Luciferase reporter assay
Early passage (≤4) MEFs were co-transfected with the p53-responsive p21WAF1/CIP1 promoter firefly luciferase reporter plasmid (a gift from Wafik El-Diery, University of Pennsylvania, Philadelphia PA) and the pRL-SV40 transfection control renilla plasmid (Promega, Madison, WI) using Fugene 6 (Roche, Indianapolis, IN) and standard methods. At 48-72 hours post-transfection, cell extracts were harvested and assayed according to the manufacturer’s protocol using the dual luciferase reporter assay system (Promega) and the Optocomp I luminometer (GEM Biomedical, Hamden, CT). After normalizing data for transfection efficiency based upon expression from the pRL-SV40 plasmid, data were presented relative to the activity measured in wild-type MEFs.
Acknowledgments
We thank John Raucci, George Heath and the Transgenic Core Facility at St Jude for the generation of the transgenic mice; Misty Cheney, June Bursi and the Animal Resource Center for their technical assistance; and Dr Kelli Boyd and the Diagnostic Lab for evaluation of mouse tissues. We also acknowledge the St Jude Hartwell Center for primer production and sequence analysis and the St Jude Flow Cytometry Lab for sorting of GFP-positive cells. We would also like to thank Julie Groff of the St Jude Biomedical Communication Department for preparing the figures and Dr Martine Roussel for the full-length MDM2 and MDM2-M3 retroviral constructs. This work was supported by NIH grants CA92401, CA21765 and the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Bartel F., Taubert H., Harris L. C. (2002a). Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2, 9–15 [DOI] [PubMed] [Google Scholar]
- Bartel F., Taylor A. C., Taubert H., Harris L. C. (2002b). Novel mdm2 splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines. Oncol. Res. 12, 451–457 [DOI] [PubMed] [Google Scholar]
- Bartel F., Pinkert C. A., Fiedler W., Wurl P., Schmidt H., Taubert H. (2004). Expression of alternatively and aberrantly spliced transcripts of the MDM2 mRNA is not tumor-specific. Int. J. Cancer 24, 143–151 [DOI] [PubMed] [Google Scholar]
- Bouvard V., Zaitchouk T., Vacher M., Duthu A., Canivet M., Choisy-Rossi C., Nieruchalski M., May E. (2000). Tissue and cell-specific expression of the p53-target genes: bax, fas, mdm2 and waf1/p21, before and following ionising irradiation in mice. Oncogene 19, 649–660 [DOI] [PubMed] [Google Scholar]
- Campisi J. (2001). Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 11, S27–S31 [DOI] [PubMed] [Google Scholar]
- Corvi R., Savelyeva L., Breit S., Wenzel A., Handgretinger R., Barak J., Oren M., Amler L., Schwab M. (1995). Non-syntenic amplification of MDM2 and MYCN in human neuroblastoma. Oncogene 10, 1081–1086 [PubMed] [Google Scholar]
- Dang J., Kuo M. L., Eischen C., Stepanova L., Sherr C., Roussel M. (2002). The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 62, 1222–1230 [PubMed] [Google Scholar]
- Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., et al. (1995). A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A. (2002). Does p53 affect organismal aging? J. Cell Physiol. 192, 23–33 [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr, Butel J. S., Bradley A. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- El-Deiry W. S., Harper J. W., O’Conner P. M., Velculescu V. E., Canman C. E., Jackman J., Pietenpol J. A., Burrell M., Hill D. E., Wang Y., et al. (1994). WAF1/CIP1 is induced upon p53-mediated growth arrest and apoptosis. Cancer Res. 54, 1169–1174 [PubMed] [Google Scholar]
- Evans S. C., Viswanathan M., Grier J. D., Narayana M., El-Naggar A. K., Lozano G. (2001). An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 20, 4041–4049 [DOI] [PubMed] [Google Scholar]
- Fakharzadeh S. S., Trusko S. P., George D. L. (1991). Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 10, 1565–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman J. S., Hernando E., Hemann M. T., de Stanchina E., Cordon-Cardo C., Lowe S. W. (2003). Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res. 63, 5703–5706 [PubMed] [Google Scholar]
- Garcia-Cao I., Garcia-Cao M., Martin-Caballero J., Criado L. M., Klatt P., Flores J. M., Weill J. C., Blasco M. A., Serrano M. (2002). “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21, 6225–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M., Sands A. T., Weiss R. S., Hegi M. E., Wiseman R. W., Pantazis P., Giovanella B. C., Tainsky M. A., Bradley A., Donehower L. A. (1993). In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8, 2457–2467 [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington R., Costnatini F., Lacy E. (1994). Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Honda R., Tanaka H., Yasuda H. (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B. O., Schmitt E. M., Halachmi S., Bronson R. T., Weinberg R. A. (1994). Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 [DOI] [PubMed] [Google Scholar]
- Jones S. N., Roe A. E., Donehower L. A., Bradley A. (1995). Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208 [DOI] [PubMed] [Google Scholar]
- Kastan M. B., Onyekwere O., Sidransky D., Vogelstein B., Craig R. W. (1991). Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51, 6304–6311 [PubMed] [Google Scholar]
- Kubbutat M. H., Jones S. N., Vousden K. H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- Ladanyi M., Cha C., Lewis R., Jhanwar S. C., Huvos A. G., Healey J. H. (1993). MDM2 gene amplification in metastatic osteosarcoma. Cancer Res. 53, 16–18 [PubMed] [Google Scholar]
- Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003). Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- Maier B., Gluba W., Bernier B., Turner T., Mohammad K., Guise T., Sutherland A., Thorner M., Scrable H. (2004). Modulation of mammalian life span by the short isoform of p53. Genes Dev. 18, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie P. P., McPake C. R., Ashford A. A., Vanin E. F., Harris L. C. (2002). MDM2 does not influence p53-mediated sensitivity to DNA-damaging drugs. Mol. Cancer Ther. 1, 1097–1104 [PubMed] [Google Scholar]
- Mendrysa S. M., O’Leary K. A., McElwee M. K., Michalowski J., Eisenman R. N., Powell D. A., Perry M. E. (2006). Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 20, 16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T., Reed J. C. (1995). Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80, 293–299 [DOI] [PubMed] [Google Scholar]
- Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992). The MDM2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R., Wagner D. S., Lozano G. (1995). Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206 [DOI] [PubMed] [Google Scholar]
- Nakano K., Vousden K. H. (2001). PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- Noda A., Ning Y., Venable S. F., Pereira-Smith O. M., Smith J. R. (1994). Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res. 211, 90–98 [DOI] [PubMed] [Google Scholar]
- Oliner J. D., Kinzler K. W., Meltzer P. S., George D. L., Vogelstein B. (1992). Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83 [DOI] [PubMed] [Google Scholar]
- Reifenberger G., Liu L., Ichimura K., Schmidt E. E., Collins V. P. (1993). Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 53, 2736–2739 [PubMed] [Google Scholar]
- Schuster K., Harris L. C. (2007). Selection for mutations in the cDNAs of transgenic mice upon expression of an embryonic lethal protein. Transgenic Res. 16, 527–530 [DOI] [PubMed] [Google Scholar]
- Sigalas I., Calvert A. H., Anderson J. J., Neal D. E., Lunec J. (1996). Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat. Med. 2, 912–917 [DOI] [PubMed] [Google Scholar]
- Steinman H. A., Burstein E., Lengner C., Gosselin J., Pihan G., Duckett C. S., Jones S. N. (2004). An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J. Biol. Chem. 279, 4877–4886 [DOI] [PubMed] [Google Scholar]
- Tyner S. D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., Brayton C., et al. (2002). p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53 [DOI] [PubMed] [Google Scholar]
- Varela I., Cadinanos J., Pendas A. M., Gutierrez-Fernandez A., Folgueras A. R., Sanchez L. M., Zhou Z., Rodriguez F. J., Stewart C. L., Vega J. A., et al. (2005). Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 437, 564–568 [DOI] [PubMed] [Google Scholar]
- Yonish-Rouach E., Resnitzky D., Lotem J., Sachs L., Kimchi A., Oren M. (1991). Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352, 345–347 [DOI] [PubMed] [Google Scholar]
- Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001). PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]