Abstract
The antitumor drug 5-fluoro-2’-deoxyuridine (FdUrd) also sensitizes tumor cells to ionizing radiation in vitro and in vivo. While radiosensitization with FdUrd requires dTTP depletion and S-phase arrest, the exact mechanism by which these events produce radiosensitization remains unknown. We hypothesized that the depletion of dTTP produces DNA mismatches which, if not repaired prior to irradiation, would result in radiosensitization. We evaluated this hypothesis in mismatch repair (MMR)-deficient HCT116 0–1 cells which lack the expression of the required MMR protein MLH1 (inactive MLH1), and MMR-proficient (wildtype MLH1) HCT116 1–2 cells. Although HCT116 0–1 cells were less sensitive to FdUrd (IC50= 3.5 µM) versus HCT116 1–2 cells (IC50 = 0.75 µM), when irradiation followed FdUrd (IC50) the MLH1-inactivated cells exhibited greater radiosensitization compared to MMR- wildtype cells (radiation enhancement ratio (RER) = 1.8 ± 0.28 vs. 1.1 ± 0.1, respectively) and an increase (≥ 8-fold) in nucleotide misincorporations. In SW620 cells and HCT116 1–2 MLH1-wildtype cells, FdUrd (IC50) did not produce radiosensitization nor did it increase the mutation frequency, but following shRNA-directed suppression of MLH1 this concentration produced excellent radiosensitization (RER = 1.6 ± 0.10, and 1.5 ± 0.06) and an increase in nucleotide misincorporations (8-fold and 6-fold respectively). Incubation with higher concentrations of FdUrd (IC90) after suppression of MLH1 produced a further increase in IR sensitivity in both SW620 and HCT116 1–2 cells (RER = 1.8 ± 0.03, and RER = 1.7 ± 0.13, respectively) and nucleotide misincorporations (>10-fold in both cell lines). These results demonstrate an important role for MLH1 and implicate mismatches in radiosensitization by FdUrd.
The fluoropyrimidines (FPs) 5-fluorouracil (FUra) and 5-fluoro-2’-deoxyuridine (FdUrd) form the mainstay of treatment of gastrointestinal cancers. In addition to their chemotherapeutic effects, they can sensitize tumor cells to ionizing radiation (IR) resulting in synergistic tumor cell killing, and are some of the most widely used radiation sensitizers in patients (Edward et al., 2003; Sobrero et al., 1997; van Laar et al., 1998). Although these antimetabolites have been used commonly in conjunction with IR, the mechanism(s) underlying the radiosensitization effect remains to be fully elucidated.
The FPs exert their cytotoxic effects primarily through activation to 5-fluoro-2’-deoxyuridine 5’-monophosphate (FdUMP), which is a potent inhibitor of thymidylate synthase (TS) resulting in depletion of dTTP and subsequently inhibition of DNA synthesis. Previous studies have determined that the radiosensitizing effect of FdUrd correlates with dTTP depletion but is not dependent on cytotoxicity (Davis et al., 1995). The ability of FdUrd to radiosensitize under minimally or noncytotoxic conditions was demonstrated to be sequence dependent, occurring only when cells were exposed to drug prior to radiation (Bruso et al., 1990) and to correlate with the accumulation of cells in early- to mid-S-phase (McGinn et al., 1994; Miller and Kinsella, 1992). However, cell cycle redistribution alone or killing of radioresistant S-phase cells by FdUrd can not sufficiently explain the effectiveness of drug and radiation since, compared with mid-S phase control cells, mid-S phase FdUrd treated cells were markedly radiosensitized, and to the same degree as unsorted cells treated with the same concentration of FdUrd (Lawrence et al., 1996). In addition, aphidicolin blocked FdUrd radiosensitization when given to FdUrd treated cells prior to radiation, while the late G1 p53-mediated checkpoint was determined not to be crucial to radiosensitization by FdUrd (Lawrence et al., 2000). Therefore, studies suggest that the ability of cells to traverse the G1/S checkpoint followed by progression of DNA replication on a damaged template may be key elements of radiosensitization by FdUrd (Lawrence et al., 1996) .
Ionizing radiation produces many types of DNA damage, and it is thought that the ineffective repair of DNA double strand breaks (dsbs) contributes most strongly to cytotoxicity (Ward, 1990). Until recently the most commonly proposed models for radiosensitization by antimetabolites included either an increase in DNA dsbs prior to or with radiation compared to radiation alone, or the inhibition of the repair of DNA dsbs after irradiation. While FdUrd has been demonstrated to enhance cytotoxicity when administered with radiation by inhibiting the repair of radiation-induced DNA damage, radiosensitization can also occur in the absence of detectable DNA dsbs, thus suggesting another mechanism exists to explain the radiosensitizing effect of these drugs (Bruso et al., 1990; Davis et al., 1995). Our recent studies with another antimetabolite radiosensitizer, gemcitabine (dFdCyd), demonstrated that, at radiosensitizing concentrations, the dFdCyd-mediated depletion in dATP pools via inhibition of ribonucleotide reductase (RR) produced DNA mismatches that, if left unrepaired, resulted in radiosensitization (Flanagan et al., 2007). These studies also demonstrated a role for the mismatch repair (MMR) protein MLH1 whereby cells deficient in MLH1 were unable to repair drug induced DNA mismatches and were more easily radiosensitized than MLH1-expressing, presumably MMR-proficient cells.
The FdUMP-mediated inhibition of TS and subsequent depletion in dTTP results in the inhibition of DNA synthesis (Martomo and Mathews, 2002; Meyers et al., 2003), and induces perturbations in the levels of the other deoxynucleotides (dATP, dGTP, and dCTP) through feedback mechanisms (Longley et al., 2003). dNTP imbalances (in particular dATP/dTTP ratio) are thought to severely disrupt DNA synthesis and repair (Houghton et al., 1995) and can produce errors in DNA replication such as single base substitutions, and insertions or deletions, resulting in frameshift mutagenesis and a damaged template (Bebenek et al., 1992; Martomo and Mathews, 2002). The depletion of dTTP pools in the presence of FdUrd may contribute to the decreased ability to perform DNA repair (Lawrence et al., 1993). The MMR system plays a role in correcting DNA mismatches during replication (Kunkel and Erie, 2005) and has been demonstrated to play a role in cytotoxicity of FdUrd whereby MMR-deficient cells are significantly more resistant to the drug than their MMR-proficient counterparts (Carethers et al., 1999; Meyers et al., 2001; Meyers et al., 2005). However, a role for MMR in the radiosensitizing property of FdUrd has not been explored.
We hypothesize that MMR deficiency will enhance radiosensitization by FdUrd by preventing correction of misincorporated nucleotides in DNA produced by depleted dTTP. We have evaluated this hypothesis in two colorectal carcinoma cell lines, HCT116 and SW620 cells, that differ in their expression of MLH1, a required MMR protein. While we postulate that dTTP depletion leads to nucleotide misincorporation in DNA, it has not been demonstrated that FdUrd can produce these lesions. The present study directly tests the hypothesis that the dNTP pool imbalances produced by FdUrd can produce mismatches in DNA, and that these are the lesions that result in radiosensitization.
Materials and Methods
Cell Culture, Plasmid, and Drug Preparation
HCT116 colon carcinoma cells are MMR-deficient due to inactivation of MLH1. The HCT116 1–2 cell line was produced from the parental HCT116 colon carcinoma cell line and contains wild-type MLH1 cDNA while the HCT116 0–1 cell line contains the vector without the MLH1 insert (Jacob et al., 2001). SW620 colon carcinoma cells are considered MMR- proficient since they express the two major MMR proteins, MLH1 and MSH2 (Taverna et al., 2000). All cells were maintained in Dulbecco’s modified essential medium (DMEM) (Invitrogen), supplemented with 10% fetal calf serum (Invitrogen), and 2 mM L-glutamine (Fisher Scientific). FdUrd (Sigma Chemical Co., St. Louis, MO) was dissolved in PBS. Cell cycle distribution was determined by dual parameter (propidium iodide(PI)/5-bromo-2’-deoxyuridine (BrdUrd)) flow cytometric analysis as described (Ostruszka and Shewach, 2000), and DNA synthesis was measured by BrdUrd incorporation (Ostruszka and Shewach, 2003).
Cell Survival and Radiosensitization Assay
Cells were left untreated or treated with FdUrd at various concentrations for 24 h prior to irradiation [Co60 (AECL Theratron 80) at 1–2 Gy/min]. Following FdUrd and/or IR (0, 2, 5, 7.5, and 10 Gy), cells were assessed for clonogenic survival as described previously (Shewach et al., 1994). Radiation sensitivity is expressed in terms of the mean inactivation dose (D-bar), which represents the area under the cell survival curve (Fertil et al., 1984). Radiosensitization is expressed as an enhancement ratio, which is defined as the mean inactivation dose (control)/mean inactivation dose (drug).
Determination of nucleotide pools
Nucleotides were extracted from cells using 0.4 N perchloric acid, neutralized, and ribonucleotides were removed using a boronate affinity column (Shewach et al., 1994). Cellular dNTPs were separated and quantified using a strong anion exchange column (Whatman, Hillshore, OR) with a high pressure liquid chromatography (HPLC) system (Waters Milford, MA) equipped with a photodiode array detector and controlled by Millennium 2010 software. Nucleotides were eluted at 2 ml/min with a linear gradient of ammonium phosphate buffer (0.15 M, pH 2.8 to 0.60 M, pH 2.9 or 3.4). Nucleotides were identified based on their UV absorbance spectrum and quantified at 254, 281, or 292 nm by comparison to the absorbance of a known amount of authentic standard.
pSP189 Plasmid Mutation Assay
The pSP189 plasmid can replicate in either bacterial or mammalian cells, and contains supF suppressor tRNA that corrects an amber mutation in MBM7070 E. coli. A single mutation at nearly any site in the coding sequence for the supF gene sequence prevents the expression of beta-galactosidase (Seidman et al., 1985). The assay was performed as previously described (Flanagan et al., 2007). Briefly, cells were transfected with the pSP189 plasmid overnight, incubated with FdUrd for 24 hr and plasmid extracted 24 hr later. Replicated plasmid DNA was electroporated into MBM7070 E. coli, and transformants were grown on agar plates with ampicillin and X-gal. White and blue colonies were enumerated, and mutation frequencies were calculated as # white colonies / # (white + blue) colonies. DNA from some control and all mutant clones was isolated and sequenced at the University of Michigan DNA Sequencing Core using the 20-mer primer (5’-GGCGACACGGAAATGTTGAA).
Transfection with shRNA
Small interfering RNAs (siRNAs) were expressed from short hairpin RNA (shRNA) lentiviral plasmids (pLKO.1-purp) containing MLH1 (GenBank accession number NM_000249) target sequences (targeting nucleotides 2186–2206 of human MLH1; Sigma MISSION™, SHGLY-NM_000249; Sigma Chemical Co., St. Louis, MO). The control shRNA contains a hairpin insert that will generate siRNAs but does not target any known human gene. HEK293FT cells in 6-well plates were cotransfected with lentiviral plasmid, shRNA to MLH1 or control shRNA (1.0 µg), lentiviral packaging vector (pCMVΔdR8.91, 1.0 µg), and the vesticular stomatitis virus G glycoprotein expression vector pVSV-G (0.5 µg) using Superfect Transfection Reagent (Qiagen, Inc., Chatsworth, CA). Viral supernatants were collected 48 hr post-transfection, isolated by centrifugation then purified by filter sterilization (0.45 µM). One µg/μL of polybrene (Fisher Scientific, Tustin, CA) was added to viral samples and SW620 cells were transduced at 37°C overnight. Virus containing media was removed; stably expressing cells were selected with puromycin (2 µg/ml) and harvested at the appropriate time for determination of MLH1 protein expression. Five shRNAs to MLH1 were tested for knockdown efficiency and we chose to use the individual construct MISSION™ TRNCN0000040053-249.2.2358 shRNA to MLH1 containing the sequence CCGGGCTTCGCCAGAGCATCAGCTTCTCGAGAAGCTGATG CTCTGGCGAAGCTTTTT since it produced the strongest and longest suppression of MLH1.
Western Blot Analysis
Cell lysates were prepared in RIPA (radio-immunoprecipitation assay) lysis buffer (0.5 M Tris-HCL, 1.5 M NaCl, 2.5% deoxycholic acid, 10% NP-40, 10 mM EDTA, pH 7.4), with the addition of protease and phosphatase inhibitors [complete mini protease inhibitor cocktail tablet (Roche), 1 mM sodiumorthovanadate, and 1 mM sodium fluoride]. Proteins were separated by SDS-PAGE on 10% acrylamide gels and transferred onto Immobilon-P transfer membrane (Millipore Corp., Bedford, MA). Membranes were probed with MLH1 polyclonal rabbit IgG antibodies (Santa Cruz Biotechnology) at 1:100 and anti-rabbit IgG horseradish peroxidase linked antibodies at 1:20 000 dilutions. Proteins were detected and visualized using an enhanced chemiluminescence detection system (Pierce, Rockford, IL).
Results
Cytotoxicity and radiosensitization with FdUrd in matched HCT116 cell lines
The HCT116 MLH1-inactivated cell line was less sensitive to FdUrd than the matched HCT116 MLH1- wildtype cell line (Table 1; IC50 = 3.5 µM and 0.75 µM for HCT116 0–1 and HCT116 1–2 cells, respectively). The ability of FdUrd to radiosensitize HCT116 cell lines was examined by irradiating cells after a 24 hr incubation with FdUrd at equitoxic concentrations (IC10 or IC50). As illustrated in Table 1, MLH1-inactivated HCT116 cells were radiosensitized at a non-cytotoxic concentration, whereby ≥IC10 FdUrd produced an excellent radiation enhancement ratio (RER = 1.8). In contrast, incubation with ≤IC50 of FdUrd did not significantly enhance the sensitivity of the HCT116 MLH1-wildtype cells to radiation-induced cytotoxicity (RER ≤ 1.2), however significant radiosensitization was observed at the IC90 (RER = 1.4 ± 0.1).
Table 1.
Effect of FdUrd on the sensitivity of HCT 116 0–1, HCT116 1–2 and SW620 cells to ionizing radiation.
| MLH1 expression | Cell line | [FdUrd] | Radiation Enhancement Ratio | D-bar (no drug) |
|---|---|---|---|---|
| Inactivated | HCT116 0–1 | 0.25µM (IC10) | 1.8 ± 0.33† | |
| 3.5 µM (IC50) | 1.8 ± 0.28† | 3.1 ± 0.19 | ||
| Wildtype | HCT116 1–2 | 0.125 µM (IC10) | 1.2 ± 0.03 | |
| 0.75 µM (IC50) | 1.1 ± 0.10 | 2.9 ± 0.14 | ||
| 3.5 µM (IC90) | 1.4 ± 0.10*† | |||
| Suppressed | HCT116 1–2 + | 0.75 µM | 1.5 ± 0.06*† | 2.8 ± 0.21* |
| shRNA-MLH1 | 3.5 µM | 1.7 ± 0.13*† | ||
| 2.8 ± 0.13* | ||||
| Wildtype | HCT116 1–2 + shRNA-non- specific |
0.75 µM | 1.1 ± 0.10* | |
| wild-type | SW620 | 0.35 µM (IC50) | 1.2 ± 0.12 | |
| 3.5 µM (IC90) | 1.4 ± 0.04† | 2.2 ± 0.14 | ||
| Suppressed | SW620 + shRNA-MLH1 | 0.35 µM | 1.6 ± 0.10† | 2.3 ± 0.24 |
| 3.5 µM | 1.8 ± 0.03*† | |||
| Wildtype | SW620 + shRNA-non-specific | 0.35 µM | 1.2 ± 0.08 | 2.3 ± 0.17 |
Radiation enhancement ratios (mean ± SE) are shown for all cell lines after a 24 hr drug incubation followed by irradiation. Sensitivity to radiation in control cells (no drug) is shown as the D-bar.
Each value is an average of at least three separate (mean ± SE), or two separate (mean ± SD*) experiments.
significantly >1 (p < 0.05)
Effect of FdUrd on dNTP Pools and Cell Cycle Distribution
Since dTTP depletion is necessary for radiosensitization with FdUrd (Davis et al., 1995), we wished to determine whether the inability of the MLH1 proficient HCT116 1–2 cells to undergo radiosensitization by FdUrd was due to lesser depletion of dTTP compared to the MLH1 inactivated HCT116 0–1 cells. Equitoxic concentrations (IC50) of FdUrd produced similar changes in dTTP and other dNTPs in each cell line at 4 hr, with ~ 40% reduction in dTTP and > 50% reduction in dGTP with a concomitant ≥35% increase in dATP (Table 2). This pattern of dNTP effects is typical following FP administration (Wadler et al., 1996). At 24 hr the HCT116 MLH1-inactivated cells displayed an increase in dTTP, dATP and dGTP while all four dNTPs were depressed in the MLH1-wildtype cells.
Table 2.
Endogenous dNTP pools in HCT116 and SW620 cells following incubation FdUrd.
| % control value | ||||||
|---|---|---|---|---|---|---|
| Cell Line | MLH1 Status | Time (h) | dCTP | dTTP | dATP | dGTP |
| HCT116 0–1 | inactivated | 4 | 101.5 ± 17.3 | 62.0 ± 9.5 | 136.1 ± 18.5 | 42.2 ± 6.6 |
| 24 | 95.6 ± 46.2 | 233.7 ± 49.9 | 338.5 ± 91.6 | 122.0 ± 48.3 | ||
| HCT116 1–2 | wildtype | 4 | 85.0 ± 7.1 | 56.5 ± 12.6 | 174.4 ± 14.9 | 41.6 ± 0.7 |
| 24 | 33.4 ± 12.6 | 51.5 ± 0.5 | 71.2 ± 2.0 | 18.5 ± 0.4 | ||
| SW620 | wildtype | 4 | 91.0 ± 1.9 | 10.14 ± 0.5 | 143.1 ± 9.6 | 18.2 ± 2.5 |
| 24 | 283.4 ± 96.3 | 17.06 ± 1.5 | 524.6 ± 92.6 | 41.8 ± 7.5 | ||
| SW620 + non-specific shRNA | Suppressed | 4 | 91.4 ± 12.1 | 8.9 ± 0.5 | 161.3 ± 7.0 | 18.2 ± 4.2 |
| 24 | 560.2 ± 77.1 | 24.9 ± 7.5 | 790.5 ± 38.9 | 89.1 ± 27.7 | ||
| SW620 + MLH1 targeted shRNA | wildtype | 4 | 87.1 ± 0.5 | 11.3 ± 0.8 | 152.1 ± 8.6 | 26.62 ± 9.2 |
| 24 | 410.0 ± 17.0 | 22.27 ± 0.07 | 781.9 ± 76.0 | 46.75 ± 22.0 | ||
Exponentially growing HCT116 MLH1-wildtype and MLH1-inactivated cells were incubated with an IC50 for FdUrd or left untreated (control). SW620 cells were left untreated or transduced with shRNA targeted to MLH1 mRNA or non-specific (NS) shRNA. Four days following shRNA treatment, exponentially growing cells were incubated with IC50 for FdUrd or left untreated (control). dNTP pools were extracted and analyzed as described in “Materials and Methods”. The data are presented as a % of the corresponding control value and represent the mean ± SD from duplicate determinations. Control values (pmol dNTP/106 cells): HCT116 0-1 cells, dCTP: 3.43 ± 1.0, dTTP: 29.7 ± 3.0, dATP: 6.1 ± 1.1, dGTP: 2.05 ± 0.6. HCT116 1-2 cells, dCTP: 4.0 ± 1.7, dTTP: 32.9 ± 2.8, dATP: 6.7 ± 0.8, dGTP: 1.97 ± 0.4. SW620, dCTP: 3.67 ± 0.5, dTTP: 20.05 ± 1.3, dATP: 3.55 ± 0.2, dGTP: 0.85 ± 0.04. SW620 +NSshRNA, dCTP: 7.06 ± 0.6, dTTP: 37.6 ± 3.1, dATP: 8.1 ± 0.2, dGTP: 1.84 ± 0.1. SW620 +MLH1shRNA, dCTP: 5.73 ± 1.3, dTTP: 21.42 ± 0.8, dATP: 4.93 ± 0.4, dGTP: 1.54 ± 0.5.
Previous studies have demonstrated that radiosensitization with FdUrd depends upon the ability of cells to enter S-phase during drug exposure (McGinn et al., 1994; Miller and Kinsella, 1992). To determine whether MLH1 inactivation altered the cell cycle progression of HCT116 cells treated with FdUrd, cell cycle distribution was measured for 72 hr following the end of drug exposure by dual parameter (BrdU and PI) flow cytometry. Both cell lines exhibited similar S-phase accumulation (>66%) following incubation for 24 h with FdUrd (IC50) (Table 3). Thus, although IC50 FdUrd resulted in similar cell cycle distributions plus depressed dNTP pools to at least the same degree and for a longer period of time in the MLH1-wildtype compared to MLH1 inactivated cells, MLH1-expressing cells were not radiosensitized at this drug concentration.
Table 3.
Effect of FdUrd on cell cycle distribution of HCT116 and SW620 cells.
| Cell Line | MLH1 Status | FdUrd (IC50) | Time (h) | G1 | S | G2/M | Cell # x106 |
|---|---|---|---|---|---|---|---|
| HCT116 0–1 | inactivated | Control | 0 | 71.8 | 21.7 | 6.5 | 4.7 |
| FdUrd | 0 | 11.7 | 84.1 | 4.2 | 3.6 | ||
| 24 | 33.3 | 28.7 | 37.9 | 2.9 | |||
| 48 | 72.6 | 14.0 | 13.4 | 5.7 | |||
| 72 | 42.4 | 39.4 | 18.1 | 6.7 | |||
| HCT116 1–2 | wildtype | Control | 0 | 75.1 | 18.8 | 6.1 | 2.7 |
| FdUrd | 0 | 18.9 | 66.8 | 14.2 | 1.9 | ||
| 24 | 59.8 | 18.5 | 21.7 | 3.8 | |||
| 48 | 78.6 | 14.3 | 7.0 | 5.7 | |||
| 72 | 78.4 | 15.2 | 6.4 | 5.9 | |||
| SW620 | wildtype | Control | 0 | 55.6 | 44.4 | 9.1 | 2.6 |
| FdUrd | 0 | 8.9 | 89.4 | 1.7 | 1.8 | ||
| 24 | 6.1 | 79.2 | 14.8 | 2 | |||
| 48 | 40.8 | 35.8 | 23.4 | 2.2 | |||
| 72 | 41.7 | 39.3 | 19.0 | 2.3 | |||
| SW620 + NS-shRNA | suppressed | Control | 0 | 72.1 | 20.3 | 7.6 | 2.6 |
| FdUrd | 0 | 67.8 | 24.1 | 8.1 | 2.0 | ||
| 24 | 5.1 | 90.5 | 4.4 | 1.8 | |||
| 48 | 49.8 | 33.8 | 16.4 | 2.0 | |||
| 72 | 53.0 | 28.1 | 18.9 | 2.1 | |||
| SW620 + MLH1-shRNA | wildtype | Control | 0 | 68.0 | 24.3 | 7.7 | 2.5 |
| FdUrd | 0 | 43.3 | 51.7 | 5.1 | 2.1 | ||
| 24 | 6.7 | 88.8 | 4.5 | 1.8 | |||
| 48 | 41.3 | 38.1 | 20.6 | 2.1 | |||
| 72 | 57.6 | 23.1 | 19.3 | 2.2 | |||
Zero hour represents the time at drug washout after a 24-hr incubation with IC50 FdUrd. Cells were harvested at the time indicated and cell cycle distribution was analyzed by dual flow cytometry as described in “Materials and Methods”.
Effect of FdUrd on Mutation Frequency in HCT116 cell lines
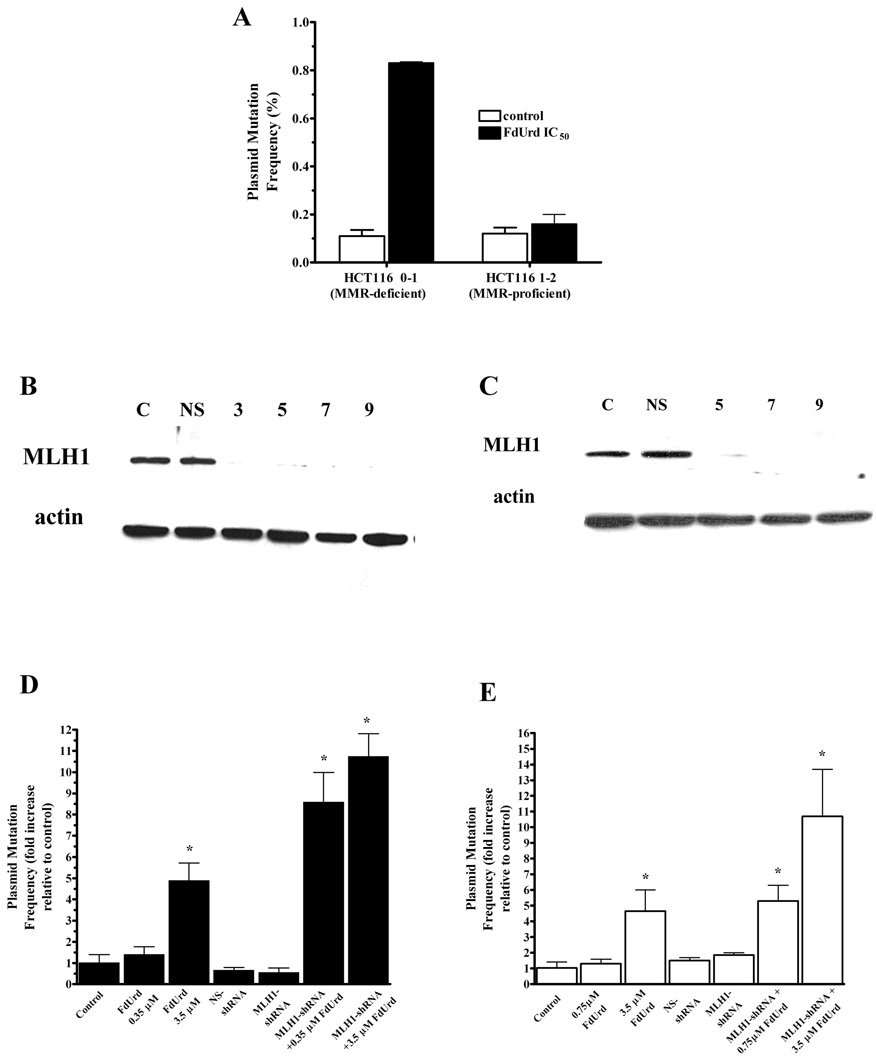
Since differential effects on dNTP pools or cell cycle distribution could not explain the inability of FdUrd to radiosensitize the MLH1-wildtype HCT116 cells, we evaluated the functional effect of dNTP pool imbalances on producing errors in DNA replication using a plasmid mutation assay. Cells were transfected with pSP189 plasmid, and FdUrd or no drug (control) was added for 24 hr. The mutation frequencies in plasmids replicated in the untreated control group of both cell lines were similar (0.11 % ± 0.025, and 0.12 % ± 0.025, in MLH1-inactivated or MLH1-wildtype cells, respectively; p < 0.05), as reported by others (Jeong et al., 1999; Tobi et al., 1999). Although FdUrd (IC50) decreased dTTP by >40% for at least 24 hr in MLH1-expressing HCT116 1–2 cells, there was no significant increase in the plasmid mutation frequency at this drug concentration. In contrast, in HCT116 0–1 MLH1-inactivated cells, FdUrd (IC50) decreased dTTP by ~40% for <24 hr, yet this concentration resulted in a significantly (nearly 8-fold) increased plasmid mutation frequency compared to control (0.83% ± 0.005, and 0.11% ± 0.025, respectively; p<0.0001; Fig. 1A).
Fig. 1. MLH1 deficiency promotes an increase in FdUrd-mediated DNA mismatches and potentiates radiosensitization by FdUrd.
(A) pSP189 plasmid mutation frequency in HCT116 MLH1-inactivated and HCT116 MLH1-wildtype cells. Cells were transfected with pSP189 plasmid overnight, washed and incubated with FUrd (IC50) for 24 h. Plasmids were harvested 24 h after drug washout. DNA was harvested from replicated plasmids, electroporated into E. coli and mutation frequency calculated as # white colonies / # (white + blue) colonies. The supF sequence of a portion of the control plasmids and all of the mutant plasmids were confirmed by DNA sequencing. The data presented are the means ± SD. *significantly > control (P < 0.05), within each cell line. (B and C) Whole cell lysates were analyzed by Western blotting for MLH1. Expression of β-actin is shown as a loading control. A representative blot from a minimum of three separate experiments is shown. (D and E) pSP189 plasmid mutation frequency in SW620 cells (D) and HCT116 1–2 cells (E). Both cell lines were left untreated or transduced with MLH1-shRNA or NS-shRNA. Five days following the absence or presence of shRNA treatment, cells were incubated with IC50 or IC90 for FdUrd or left untreated (control). Results are expressed as the –fold increase relative to the untreated control cells within each treatment group. The data presented are the means ± SE of at least three separate experiments. * significantly > control (P < 0.05).
Radiosensitization by FdUrd in MMR-proficient cells following shRNA-mediated suppression of MLH1 expression
After demonstrating that MLH1-inactivation was associated with radiosensitization by FdUrd and an increased plasmid mutation frequency, we used shRNA technology to decrease expression of wildtype MLH1 in the HCT116 1–2 cells and examined the ability of FdUrd to enhance IR sensitivity. In addition, we examined another cell line, SW620, to determine whether the effect of MLH1 on radiosensitization and cytotoxicity by FdUrd was unique to the HCT116 cell lines or could also be reproduced in other cells. Previous studies demonstrated that SW620 cells were only minimally radiosensitized by FdUrd at concentrations up to IC50 (Davis et al., 1995). In our studies, SW620 cells did not exhibit significant radiosensitization until a highly toxic concentration of FdUrd (3.5 µM, IC90) was used (RER = 1.4 ± 0.04), similar to the HCT116 MLH1-wildtype cells (Table 1). After transduction of SW620 and HCT116 1–2 cells with lentivirus-delivered MLH1 shRNA and selection for transduced cells, nearly complete depletion of MLH1 protein was observed in both cell lines by 5 days and it remained depressed through at least 9 days post-transduction (Fig. 1B and C). The ability of FdUrd to radiosensitize SW620 and HCT116 1–2 cells was examined by irradiating cells after a 24 hr incubation with FdUrd. As illustrated in Table 1, SW620 MLH1-expressing cells did not exhibit significant radiosensitization at 0.35 µM (IC50), but when MLH1 expression was suppressed with shRNA, the same concentration of FdUrd produced excellent radiosensitization (RER = 1.6 ± 0.10 in SW620+shRNA-MLH1 cells). Similarily, incubation with 0.75 µM FdUrd (IC50) did not significantly enhance the sensitivity of HCT116 1–2 MLH1-wildtype cells, but when MLH1 expression was suppressed with shRNA the same concentration of FdUrd produced enhanced IR sensitivity (RER = 1.5 ± 0.06 in HCT116 1–2+shRNA-MLH1 cells). Incubation with higher concentrations of FdUrd (IC90) after suppression of MLH1 produced a further increase in IR sensitivity in both SW620 and HCT116 1–2 cells (RER = 1.8 ± 0.03, and RER = 1.7 ± 0.13, respectively). Control SW620 and HCT116 cells treated with non-specific shRNA and then incubated with FdUrd (IC50) and radiation did not exhibit radiosensitization (RER = 1.2 ± 0.08, and RER = 1.1 ± 0.1, respectively) and treatment of either cell line with shRNA for MLH1 alone or a non-specific shRNA alone did not increase radiation sensitivity according to D-bar values (p > 0.05) . Further examination of the SW620 cell line revealed that these differences in radiosensitization were not mediated by differences in dNTP pool effects, since an equimolar concentration of FdUrd (0.35 µM) produced similar depletion of dTTP (≥ 90% reduction in dTTP) and comparable changes in the other dNTP pools in SW620 control cells or after treatment with shRNA (Table 2). In addition, similar S-phase accumulation and subsequent cell cycle progression was observed following 24 hr FdUrd (0.35 µM) in all SW620 cell lines, regardless of MLH1 status (Table 3). Thus, despite similar effects on cytotoxicity, dNTP pools and cell cycle distribution, 0.35 µM FdUrd produced radiosensitization in the SW620 + shRNA-MLH1 cells but not in the SW620 control or SW620 + NS-shRNA cells (Table 1) suggesting an important role for MLH1 in radiosensitization with this drug.
Effect of shRNA-mediated suppression of MLH1 on plasmid mutation frequency with FdUrd in HCT116 MLH1-wildtype and SW620 cells
Since shRNA-mediated suppression of MLH1 in HCT116 MLH1-wildtype cells and SW620 cells resulted in increased sensitivity of these cells to FdUrd-mediated radiosensitization, we wished to determine whether this corresponded to an increase in the frequency of misincorporation in DNA using the plasmid mutation assay. Compared to control HCT116 MLH1-wildtype cells and control SW620 cells expressing MLH1 (mutation frequency = 0.12 ± 0.02 % and 0.09 ± 0.02% respectively), FdUrd did not induce a significant increase in plasmid mutation frequency at IC50 (0.16 ± 0.04 % and 0.12 ± 0.05 %, respectively, p > 0.05) but, at a highly toxic (IC90) radiosensitizing concentration, a significant increase (> 5-fold in HCT116 1–2 MLH1-wildtype cells, and >4.5-fold in SW620 cells, respectively) was observed (Fig. 1D and E). Following shRNA-induced suppression of MLH1 and incubation with FdUrd (IC50), conditions which induced radiosensitization, the mutation frequency increased approximately 6- fold in HCT116 1–2 MLH1-wildtype cells and 8-fold in SW620 cells compared to control cells treated only with shRNA. Incubation with higher concentrations of FdUrd (IC90) after suppression of MLH1 produced a further increase(>10-fold) in mutation frequency in both cell lines. Compared to untreated control cells, plasmid mutation frequency was not significantly different with addition of MLH1 shRNA alone, non-specific shRNA alone or with FdUrd in either cell line. Thus, only a radiosensitizing concentration of FdUrd produced an increase in plasmid mutation frequency.
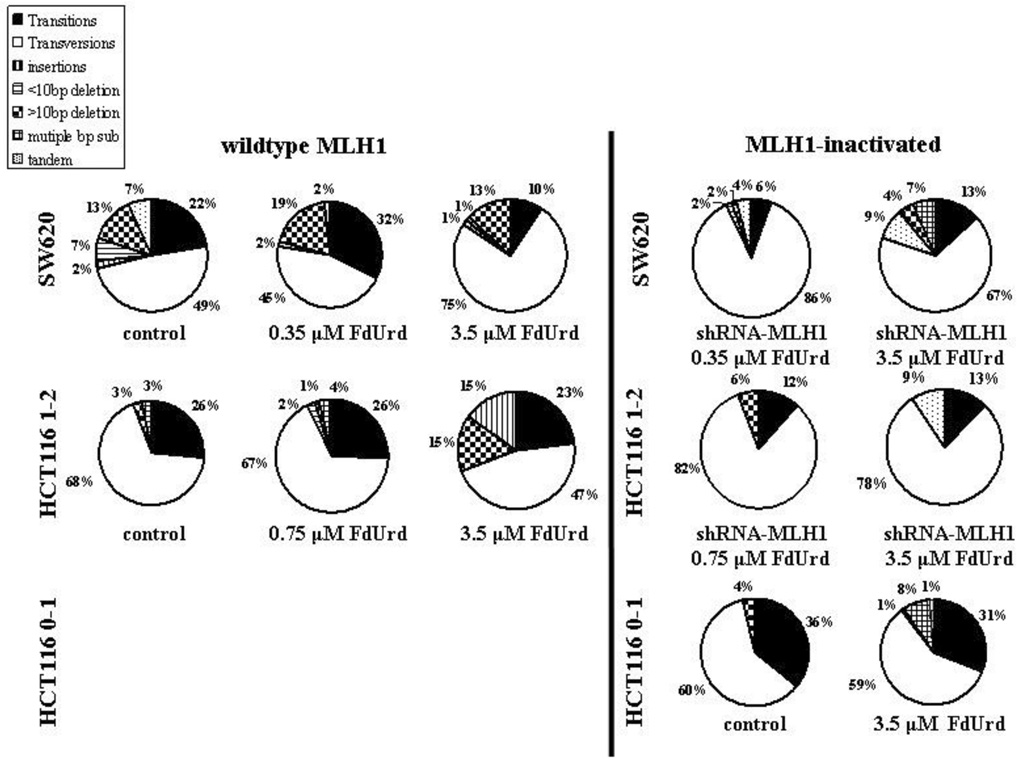
Type and frequency of mutations in pSP189
DNA sequencing results from muta006Et colonies demonstrated that the majority of plasmid mutations generated in MMR-proficient and MMR-deficient cells were single base substitutions in the control (no drug) or FdUrd treated cells (≥ 90% and ≥ 70% of total plasmid mutations HCT116 and SW620 cells, respectively) (Figure 2). Insertions and deletions accounted for the remainder of mutations observed within each group. The most common mutations observed with FdUrd in both HCT116 MLH1-wildtype and MLH1-inactivated cell lines were transversions with the largest increase in base substitution events in all cells observed at A:T sites, as expected for a drug that elicits a decrease in dTTP. Following a moderate FdUrd (IC50) concentration in HCT116 or SW620 cells, the relative contribution of base substitutions to total replication errors did not change from control (no drug). However, plasmids replicated in HCT116 1–2 MLH1-wildtype cells and SW620 cells following a high FdUrd (IC90) or following the suppression of MLH1 protein revealed small changes in the relative contribution of base substitutions to total replication errors compared to control, but transversions remained largely dominant regardless of treatment.
Fig. 2. Type and frequency of mutations in the supF sequence in pSP189 plasmids replicated in HCT116 cells and SW620 cells.
HCT116 0–1 MLH1-inactivated, HCT116 1–2 MLH1-wildtype and HCT116 MLH1-inactivated, and SW620 MLH1-wildtype and SW620 MLH1-inactivated cells were transfected with pSP189 plasmid overnight and exposed to no drug (control), or to 3.5 µM (IC50) (HCT116 0–1 cells) or 0.75 µM (IC50) and 3.5 µM (IC90) (HCT116 1–2 cells), or 0.35 µM (IC50) and 3.5 µM (IC90) (SW620 cells) FdUrd for 24 h. Mutant colonies were picked and grown in LB broth followed by plasmid extraction and DNA sequencing. Mutation frequency was calculated as # white colonies / # (white + blue) colonies. n, the total number of mutant colonies, all of which were submitted for DNA sequencing. At least 26 mutant colonies were counted within each group.
Discussion
It is generally accepted that depletion of dTTP, due to FdUMP-mediated inhibition of TS, is the primary effect that produces radiosensitization with FPs. Although it has been suggested that dTTP depletion may slow the rate of repair of radiation-induced dsbs, that radiosensitization can occur in the absence of detectable DNA dsbs suggests other mechanism(s) exist to explain the radiosensitizing effect of these drugs (Bruso et al., 1990). Here we provide evidence that the FdUrd-mediated decrease in dTTP produces mismatches in DNA which, while not required for cytotoxicity, are associated with radiosensitization. This is the first demonstration of a lesion in DNA that leads to radiosensitization with FdUrd. Furthermore, demonstration that MLH1 deficiency induced both mismatches in DNA and radiosensitization suggests that errors of replication play an integral role in radiosensitization with FdUrd.
To evaluate the role of MLH1 in radiosensitization with FdUrd, we initially measured the cytotoxicity of FdUrd ± ionizing radiation in the HCT116 0–1 (inactivated MLH1) and HCT116 1–2 (wildtype MLH1) cells. As reported by others, the HCT116 1–2 cells were more sensitive to FdUrd (Meyers et al., 2003; Meyers et al., 2005); however, innate sensitivity to ionizing radiation was similar. Therefore, to evaluate both a non-cytotoxic (IC10) and a cytotoxic (IC50) concentration of FdUrd on radiosensitization in the cell lines, we used equitoxic rather than equimolar concentrations of FdUrd. Under these conditions only the MLH1-inactivated HCT116 0–1 cells exhibited radiosensitization. Both cell lines exhibited similar effects on the two parameters required for radiosensitization with FPs, dTTP depletion and accumulation in S-phase. Indeed, dTTP depletion was more prolonged in the HCT116 1–2 cells, yet no radiosensitization was observed at ≤IC50.
To further support our hypothesis that MLH1 plays a role in FdUrd-mediated radio-sensitization, we used shRNA technology to return the HCT116 1–2 MLH1-wildtype cells to the MLH1 deficiency status of the parental cell line. Following MLH1 suppression we were able to radiosensitize these cells at a concentration of FdUrd shown here to be unable to increase cytotoxicity by ionizing radiation (Table 1). Similarly, the suppression of MLH1 expression in the SW620 cell line induced radiosensitization and, as in the HCT116 matched cell lines, insufficient dTTP depletion or lack of S-phase accumulation could not explain the lack of radiosensitization in parental SW620 cells. Since it has been demonstrated previously that radiosensitization requires the addition of FdUrd at least 8 hr prior to irradiation (Bruso et al., 1990), we reasoned that radiosensitization requires an effect of dTTP depletion on DNA replication and not simply dNTP pool imbalance. Therefore, we have hypothesized that radiosensitization with FdUrd is due to the ability of dTTP depletion to produce mismatches in DNA which, if not repaired prior to irradiation, will result in radiosensitization. Our results demonstrate that, in the HCT116 and SW620 cells, radiosensitization with FdUrd occurred only under conditions which produced mismatches in DNA. Thus, only cells with MLH1 inactivated (HCT116 0–1 cells) or suppressed with shRNA (HCT116 1–2 and SW620 cells) exhibited mismatches and radiosensitization at concentrations ≤IC50. Radiosensitization was observed in the wildtype MLH1 expressing HCT116 1–2 and SW620 cells only at the corresponding IC90 for FdUrd, a concentration that also significantly increased mismatches.
Compared to the matched HCT116 cell lines, cultured separately over many years, suppression of MLH1 expression with lentivirus-delivered shRNA in the SW620 cells should allow a more accurate determination of the effect of MLH1 on radiosensitization and plasmid mutation frequency with FdUrd. Indeed, the demonstration that MLH1 suppression induced both mismatches and radiosensitization with FdUrd demonstrates a causal role for MLH1 deficiency in these processes.
Both HCT116 1–2 and SW620 cells express high levels of at least one MMR protein (Taverna et al., 2000) and are resistant to radiosensitization at moderate concentrations of FdUrd. However, MMR proficient cells expressing reduced levels of MMR proteins can be radiosensitized at lower concentrations of FdUrd. For example, HT29 colon carcinoma cells have about 2-fold less MLH1 protein and 40-fold less MSH2 than SW620 cells (Taverna et al., 2000), but are more susceptible to the combination of FdUrd and ionizing radiation (Davis et al., 1995). In our studies only a toxic concentration of FdUrd (IC90) in SW620 cells and HCT116 1–2 MLH1-wildtype cells produced radiosensitization and an increase in plasmid mutations. We hypothesize that, at low levels of FdUrd, the existing MMR capability is sufficient to correct errors of replication resulting from an imbalance in dNTP pools, but can be overcome at sufficiently high concentrations of FdUrd that induce an increase in misincorporated nucleotides.
Since FdUrd can exert its cytotoxic effects through incorporation into DNA as well as through the inhibition of TS (Mader et al., 1998), it is difficult to eliminate the contribution of DNA incorporation to radiosensitization. However, since FdUMP incorporation into DNA is associated with cytotoxicity (Ingraham et al., 1982), the decreased cytotoxicity of FdUrd in MLH1-deficient cells compared to MLH1-wildtype cells under radiosensitizing conditions suggests that radiosensitization can not be attributed to an increase in FdUMP incorporation into DNA. Furthermore, previous studies have determined that the radiosensitizing effect of FdUrd is not dependent on cytotoxicity (Davis et al., 1995). TS inhibition also produces an accumulation of dUMP, which may lead to increased levels of dUTP (Biserka et al., 1994), and a possible increase in dUTP incorporation into DNA (Ingraham et al., 1982), thus supplying another possible contributor to radiosensitization. We did not observe appreciable levels of dUTP in the HCT116 cell lines following FdUrd exposure. SW620 cells displayed a small amount of dUTP (data not shown), although the amount was similar regardless of MLH1 status. Therefore, this metabolite does not provide an explanation for the radiosensitization that occurred when MLH1 was suppressed or inactivated.
Following FdUrd exposure, all of the cells used in our studies, regardless of MLH1 status, exhibited S-phase cell cycle arrest, a response strongly correlated with radiosensitization by FdUrd (Lawrence et al., 1996; McGinn et al., 1994; Miller and Kinsella, 1992). Some studies have described a shorter G2 arrest after FP treatment in HCT116 MMR-deficient versus their MMR-proficient counterparts, (Meyers et al., 2001) whereas others did not (Carethers et al., 1999), although drug concentration and exposure time varied. We used a moderate and equitoxic concentration of FdUrd for a moderate exposure time and did not observe a difference in G2 response between MMR-proficient and MMR-deficient cells. Thus, differential effects on cell cycle progression cannot explain FdUrd radiosensitization observed in MLH1-inactivated but not MLH1-wildtype cells. Although the HCT116 MLH1-wildtype and MLH1-inactivated cells continued to progress through the cell cycle after FdUrd washout whereas the SW620 cells progressed little (Table 3), within a cell line progression was similar regardless of MLH1 status. Furthermore, since a similar rate of DNA synthesis (as determined by the incorporation of BrdUrd, data not shown) was observed after drug washout within each cell line, regardless of MLH1 status, differences in mutation frequency cannot be attributed to DNA synthesis inhibition.
Despite similar effects on cytotoxicity, dNTP pools, and cell cycle distribution, only a radiosensitizing concentration of FdUrd produced an increase in plasmid mutation frequency in two different cell lines. These studies support our previous findings with dFdCyd and hydroxyurea, radiosensitizers that produce an imbalance in dNTP pools (primarily a decrease in dATP) due to inhibition of RR, where mismatches in DNA occurred only under radiosensitizing conditions, and MLH1 deficiency enhanced radiosensitization (Flanagan et al., 2007). Together these studies strongly support errors of replication as a general mechanism of radiosensitization for drugs that produce imbalances in dNTPs. Importantly, we have demonstrated that a decrease in dTTP produces different replication errors than drugs that produce decreases primarily in dATP (Flanagan et al., 2007), yet a strong relationship between DNA errors and radiosensitization still exists. These results demonstrate an important role for MLH1 and further implicate insufficient MMR in radiosensitization with drugs that produce dNTP imbalances.
Finally, these data suggest that tumors with innate or acquired deficiency in MLH1 would be most sensitive to radiosensitization with FdUrd, but that higher doses of drug could be used in MLH1-expressing tumors to increase their radiosensitivity. Furthermore, the dependence of radiosensitization on DNA mismatches and not cytotoxicity suggests that, if clinical treatment with FdUrd and IR could be titrated to maximize DNA mismatches in tumors rather than cytotoxicity, normal tissue toxicity may be lessened. An understanding of the lesions and repair pathways leading to radiosensitization will aid us in optimizing chemoradiotherapy with the clinically important fluoropyrimidines.
Acknowledgments
Grant Support: This work was supported in part by grants CA83081, CA76581 and CA120244 from the NIH.
Abbreviations used
- FdUrd
5-Fluoro-2’-deoxyuridine
- FPs
fluoropyrimidines
- FUra
5-fluorouracil
- FdUMP
5’-monophosphate
- RR
5-fluoro-2’-deoxyuridine; ribonucleotide reductase
- dFCyd
gemcitabine
- TS
thymidylate synthase
- HRR
homologous recombination repair
- dsbs
DNA double strand breaks
- MMR
mismatch repair
Reference List
- Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem. 1992;267:3589–3596. [PubMed] [Google Scholar]
- Biserka M, Josie P, Saul M, Charles E. Biochemical effects of folate-based inhibitors of thymidylate synthase in MGH-U1 cells. Cancer Chemother Pharmacol. 1994;35:109–114. doi: 10.1007/BF00686631. [DOI] [PubMed] [Google Scholar]
- Bruso CE, Shewach DS, Lawrence TS. Fluorodeoxyuridine-induced radiosensitization and inhibition of DNA double strand break repair in human colon cancer cells. International Journal of Radiation Oncology Biol Phys. 1990;19:1411–1417. doi: 10.1016/0360-3016(90)90352-k. [DOI] [PubMed] [Google Scholar]
- Carethers J, Chauhan D, Fink D. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Tang HY, Maybaum J, Lawrence TS. Dependence of fluorodeoxyuridine-mediated radiosensitization on S phase progression. International Journal of Radiation Biology. 1995;67:509–517. doi: 10.1080/09553009514550621. [DOI] [PubMed] [Google Scholar]
- Edward C, Marc AC, Michael PF, John CS. Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother Pharmacol. 2003;V52:80–89. doi: 10.1007/s00280-003-0625-9. [DOI] [PubMed] [Google Scholar]
- Fertil B, Dertinger H, Courdi A, Malaise EP. Mean inactivation dose: a useful concept for intercomparison of human cell survival curves. Radiat Res. 1984;99:73–84. [PubMed] [Google Scholar]
- Flanagan SA, Robinson BW, Krokosky CM, Shewach DS. Mismatched nucleotides as the lesions responsible for radiosensitization with gemcitabine: a new paradigm for antimetabolite radiosensitizers. Mol Cancer Ther. 2007;6:1858–1868. doi: 10.1158/1535-7163.MCT-07-0068. [DOI] [PubMed] [Google Scholar]
- Houghton JA, Tillman DM, Harwood FG. Ratio of 2'-deosyadenosine-5'-triphosphate/thymidine-5'-triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin Cancer Res. 1995;1:723-720. [PubMed] [Google Scholar]
- Ingraham HA, Tseng BY, Goulian M. Nucleotide levels and incorporation of 5-fluorouracil and uracil into DNA of cells treated with 5-fluorodeoxyuridine. Mol Pharmacol. 1982;21:211–216. [PubMed] [Google Scholar]
- Jacob S, Aguado M, Fallik D, Praz F. The Role of the DNA Mismatch Repair System in the Cytotoxicity of the Topoisomerase Inhibitors Camptothecin and Etoposide to Human Colorectal Cancer Cells. Cancer Res. 2001;61:6555–6562. [PubMed] [Google Scholar]
- Jeong JK, Wogan GN, Lau SS, Monks TJ. Quinol-Glutathione Conjugate-induced Mutation Spectra in the supF Gene Replicated in Human AD293 Cells and Bacterial MBL50 Cells. Cancer Res. 1999;59:3641–3645. [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lawrence TS, Burke R, Davis MA. Lack of effect of T53 status on fluorodeoxyuridne-mediated radiosensitization. Radiat Res. 2000;154:140–144. doi: 10.1667/0033-7587(2000)154[0140:loeots]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lawrence TS, Davis MA, Normolle DP, Maybaum J. The use of biphasic linear ramped pulsed field gel electrophoresis to quantify DNA damage based on fragment size distribution. Int J Radiat Oncol Biol Phys. 1993;27:659–663. doi: 10.1016/0360-3016(93)90393-a. [DOI] [PubMed] [Google Scholar]
- Lawrence TS, Davis MA, Tang HY, Maybaum J. Fluorodeoxyuridine-mediated cytotoxicity and radiosensitization require S phase progression. Int J Radiat Oncol Biol Phys. 1996;70:272–280. doi: 10.1080/095530096145003. [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical straegies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- Mader RM, Muller M, Steger GG. Resistance to 5-Fluorouracil. General Pharmacology: The Vascular System. 1998;31:661–666. doi: 10.1016/s0306-3623(98)00191-8. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Mathews CK. Effects of biological DNA precursor pool asymmetry upon accuracy of DNA replication in vitro. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2002;499:197–211. doi: 10.1016/s0027-5107(01)00283-4. [DOI] [PubMed] [Google Scholar]
- McGinn CJ, Miller EM, Lindstrom MJ, Kunugi KA, Johnston PG, Kinsella TJ. The role of cell cycle redistribution in radiosensitization: Implications regarding the mechansim of fluorodeoxyuriidne radiosensitization. Int J Radiat Oncol Biol Phys. 1994;30:851–859. doi: 10.1016/0360-3016(94)90360-3. [DOI] [PubMed] [Google Scholar]
- Meyers M, Hwang A, Wagner MW, Bruening AJ, Veigl ML, Sedwick WD, Booothman DA. A role for DNA mismatch repair in sensing and responding to fluoropyrimidine damage. Oncogene. 2003;22:7376–7388. doi: 10.1038/sj.onc.1206941. [DOI] [PubMed] [Google Scholar]
- Meyers M, Wagner MW, Hwang Hs, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA Mismatch Repair Protein in Fluoropyrimidine-mediated Cell Death and Cell Cycle Responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA Mismatch Repair-dependent Response to Fluoropyrimidine-generated Damage. J Biol Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- Miller EM, Kinsella TJ. Radiosensitization by Fluorodeoxyuridine: Effects of Thymidylate Synthase Inhibition and Cell Synchronization. Cancer Res. 1992;52:1687–1694. [PubMed] [Google Scholar]
- Ostruszka LJ, Shewach DS. The Role of Cell Cycle Progression in Radiosensitization by 2',2'-Difluoro-2'-deoxycytidine. Cancer Res. 2000;60:6080–6088. [PubMed] [Google Scholar]
- Ostruszka L, Shewach D. The role of DNA synthesis inhibition in cytotoxicity of 2',2'-difluoro-2'-deoxycytidine. Cancer Chemother Pharmacol. 2003;52:325–332. doi: 10.1007/s00280-003-0661-5. [DOI] [PubMed] [Google Scholar]
- Seidman MM, Dixon K, Razzaque A, Zagursky RJ, Berman ML. A shuttle vector plasmid for studying carcinogen-induced point mutations in mammalian cells. Gene. 1985;38:233–237. doi: 10.1016/0378-1119(85)90222-7. [DOI] [PubMed] [Google Scholar]
- Shewach DS, Hahn TM, Chang E, Hertel LW, Lawrence TS. Metabolism of 2',2'-difluorodeoxycytidine and radiation sensitization of human colon carcinoma cells. Cancer Res. 1994;54:3218–3223. [PubMed] [Google Scholar]
- Sobrero AF, Aschele C, Bertino JR. Fluorouracil in colorectal cancer--a tale of two drugs: implications for biochemical modulation. J Clin Oncol. 1997;15:368–381. doi: 10.1200/JCO.1997.15.1.368. [DOI] [PubMed] [Google Scholar]
- Taverna P, Liu L, Hanson AJ, Monks A, Gerson SL. Characterization of MLH1 and MSH2 DNA mismatch repair proteins in cell lines of the NCI anticancer drug screen. Cancer Chemother Pharmacol. 2000;46:507–516. doi: 10.1007/s002800000186. [DOI] [PubMed] [Google Scholar]
- Tobi SE, Levy DD, Seidman MM, Kramer K. Sequence-dependent mutations in a shuttle vector plasmid replicated in a mismatch repair deficient human cell line. Carcinogenesis. 1999;20:1293–1301. doi: 10.1093/carcin/20.7.1293. [DOI] [PubMed] [Google Scholar]
- van Laar JAM, Rustum YM, Ackland SP, van Groeningen CJ, Peters GJ. Comparison of 5-fluoro-2'-deoxyuridine with 5-fluorouracil and their role in the treatment of colorectal cancer. European Journal of Cancer. 1998;34:296–306. doi: 10.1016/s0959-8049(97)00366-3. [DOI] [PubMed] [Google Scholar]
- Wadler S, Horowitz R, Mao X, Schwartz EL. Effect of interferon on 5-fluorouracil-induced perturbations in pools of deoxynucleotide triphosphates and DNA strand breaks. Cancer Chemother Pharmacol. 1996;38:529–535. doi: 10.1007/s002800050522. [DOI] [PubMed] [Google Scholar]
- Ward JF. The Yield of DNA Double-strand Breaks Produced Intracellularly by Ionizing Radiation: A Review. International Journal of Radiation Biology. 1990;57:1141–1150. doi: 10.1080/09553009014551251. [DOI] [PubMed] [Google Scholar]