Abstract
Malaria parasite sporozoites prepare for transmission to a mammalian host by upregulation of UIS (Upregulated in Infectious Sporozoites) genes. A number of UIS gene products are essential for the establishment of the intrahepatocytic niche. However, the factors that regulate the expression of genes involved in gain of infectivity for the liver are unknown. Herein, we show that a conserved Plasmodium sporozoite low-complexity asparagine-rich protein, SAP1 (Sporozoite Asparagine-rich Protein 1), has an essential role in malaria parasite liver infection. Targeted deletion of SAP1 in the rodent malaria parasite Plasmodium yoelii generated mutant parasites that traverse and invade hepatocytes normally but cannot initiate liver-stage development in vitro and in vivo. Moreover, immunizations with Pysap1(−) sporozoites confer long-lasting sterile protection against wild-type sporozoite infection. Strikingly, lack of SAP1 abolished expression of essential UIS genes including UIS3, UIS4 and P52 but not the constitutively expressed genes encoding, among others, sporozoite proteins CSP and TRAP. SAP1 localization to the cell interior but not the nucleus of sporozoites suggests its involvement in a post-transcriptional mechanism of gene expression control. These findings demonstrate that SAP1 is essential for liver infection possibly by functioning as a selective regulator controlling the expression of infectivity-associated parasite effector genes.
Introduction
The first step of malaria transmission is the injection of sporozoites into a mammalian host by an anopheline mosquito bite (Vanderberg and Frevert, 2004; Amino et al., 2006). Initially, sporozoites form in mosquito midgut oocysts and subsequently invade and reside inside the salivary glands (reviewed in Matuschewski, 2006). In the mosquito salivary glands, sporozoites gain infectivity that is critical to support their transmission and life cycle progression in the mammalian liver (Vanderberg, 1974; 1975). Previous work has demonstrated that gain of infectivity is accompanied by extensive differential upregulation of unique gene products called UIS (Upregulated in Infectious Sporozoites) (Matuschewski et al., 2002). Indeed, UIS genes were shown to be essential for malaria parasite liver infection. UIS3 and UIS4 (Mueller et al., 2005a,b; Tarun et al., 2007) are proteins of the parasitophorous vacuole membrane (PVM), the principal host–parasite interface during cell infection (Mueller et al., 2005b; Mikolajczak et al., 2007a). Deletion of UIS3 and UIS4 leads to complete early arrest of liver-stage development inside the PVM (Mueller et al., 2005a,b; Tarun et al., 2007). Recently, it was shown that UIS3 interacts with liver fatty acid-binding protein (L-FABP) indicating a potential role of this protein in fatty acid uptake from the host hepatocyte (Mikolajczak et al., 2007a). Simultaneous deletion of the UIS gene P52 (also called P36p), a putative GPI-anchored protein, and a non-UIS gene, P36, a putative secreted protein, renders sporozoites unable to form a PVM during infection and leads to complete developmental arrest at the early stage of hepatocyte infection (van Dijk et al., 2005; Ishino et al., 2005a; Labaied et al., 2007). Therefore, a number of UIS proteins critically contribute to establishing the intracellular parasitic niche either by the formation or modification of the host– parasite interface (reviewed in Mikolajczak and Kappe, 2006). However, it remains unknown what factors regulate the expression of UIS genes and consequently liver infectivity of sporozoites. Herein, we have identified a cytoplasmic low-complexity asparagine-rich protein, SAP1 (Sporozoite Asparagine-rich Protein 1) that is essential for liver infection possibly by means of regulating the expression of effector proteins such as P52, UIS3 and UIS4. Targeted deletion of PySAP1 generated mutant parasites that traverse host cells, invade hepatocytes and form a PVM but cannot initiate liver-stage development and consequently completely lose mammalian infectivity in vivo. Drastically reduced transcript levels of liver infection-associated UIS genes in SAP1-deficient sporozoites in combination with SAP1's putative cytoplasmic localization suggest a post-transcriptional regulation of gene expression function of SAP1 in malaria parasite liver infection.
Results
SAP1 is a conserved Plasmodium sporozoite protein with an asparagine-rich low-complexity domain
We searched for putative cytoplasmic proteins that are highly expressed in sporozoites but not in blood stages because they might uniquely contribute to regulation of sporozoite infectivity. SAP1 was first identified as a sporozoite-expressed gene in a suppression subtractive hybridization (SSH) screen of Plasmodium yoelii salivary gland sporozoites versus blood-stage merozoites (designated S22, sporozoite-specific gene 22) (Kaiser et al., 2004). PySAP1 (gene identifier PY03269) has orthologues in other Plasmodium species including the human malaria parasite P. falciparum (gene identifier PF11_0480) (Carlton et al., 2002; Gardner et al., 2002). Alignment of PfSAP1 and PySAP1 revealed that the PySAP1 open reading frame (ORF) was incomplete. Bioinformatics analysis and direct sequencing revealed that the PySAP1 coding sequence was dispersed over two unassembled sequence contigs (described in Text S1 in Supplementary material). We confirmed the overlap between the two contigs encoding PySAP1 by genomic PCR and reverse transcription polymerase chain reaction (RT-PCR) analysis. The correct start and stop codons as well as the correct exon–intron organization were also confirmed by RT-PCR analysis of salivary gland sporozoite RNA (Fig. 1A). The corrected PySAP1 ORF nucleotide sequence (9723 nucleotides) and the predicted protein sequence (3240 amino acids) were deposited in NCBI GenBank (Accession No.: EU652769, Text S1). PySAP1 encodes a large putative protein with a predicted 370 kDa molecular mass. PySAP1 has one large exon followed by two small exons (Fig. 1A). Signal sequences, transmembrane domain(s), enzymatic or structural motifs were not identifiable in any of the predicted PlasmodiumSAP1 protein sequences examined. SAP1 proteins are characterized by the presence of an extended internal asparagine-rich low-complexity domain, with an asparagine content of 27% in P. yoelii flanked by predicted globular domains with low asparagine content (Fig. 1B). Interestingly, these (N)- and (C)-terminal regions are highly conserved among Plasmodium species. The PySAP1 N-terminus shares 70% amino acid sequence identity with the N-terminus of the P. falciparum orthologue, and the PySAP1 C-terminus shares 89% amino acid sequence identity with C-terminus of PfSAP1 (Fig. 1B). However, the overall amino acid sequence identity of SAP1 between P. yoelii and P. falciparum is only 26% due to the sequence divergence in the asparagine-rich domain (Fig. 1B).
Fig. 1.
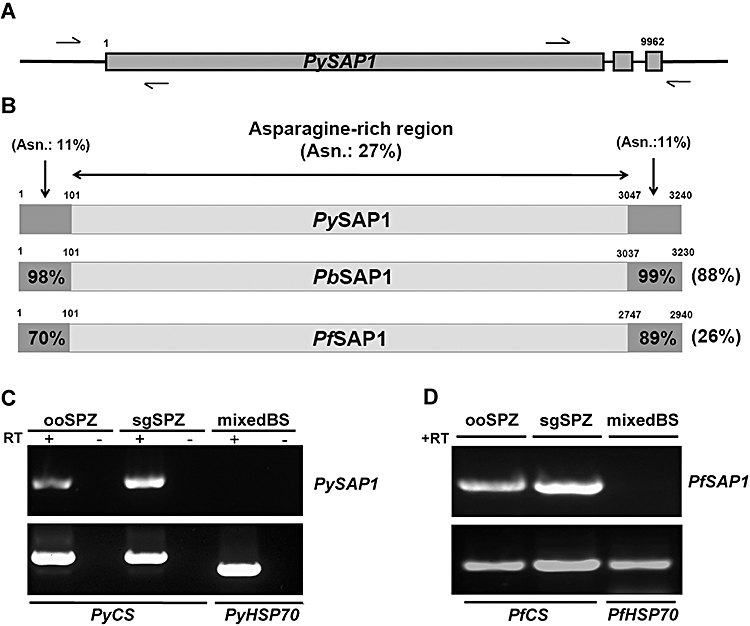
SAP1 gene structure, protein structure, conservation among Plasmodium species and transcriptional profiling.
A. A schematic representation of the SAP1 gene organization: arrows show the locations of primers used for RT-PCR to identify the start and stop codons as well as exon 2 and exon 3.
B. Alignment of the putative PySAP1 with PbSAP1 and PfSAP1. The asparagine-rich regions are shown as light grey boxes bordered by non-asparagine-rich N- and C- termini shown as dark grey boxes. Total amino acid sequence identities to PySAP1 are shown to the right and amino acid sequence identities between the N-termini and the C-termini are shown inside their respective boxes. Per cent (%) asparagine content is shown only for PySAP1.
C. RT-PCR analysis of RNA isolated from P. yoelii sporozoites shows the expression of PySAP1 in ooSPZ (oocyst sporozoites) and sgSPZ (salivary gland sporozoites) but not in mixed blood stages (mixedBS). PyCS (circumsporozoite protein) is a positive RT-PCR control for sporozoite expression and PyHSP70 is a positive RT-PCR control for mixed blood stages.
D. RT-PCR analysis of different P. falciparum life cycle stages shows expression of PfSAP1 in ooSPZ and sgSPZ but a lack of expression in blood stages (mixedBS).
Sporozoite-specific expression profile of PySAP1 and PfSAP1
RT-PCR analysis revealed that PySAP1 is transcribed in oocyst and salivary gland sporozoites (Fig. 1C). As expected from the results of the previous SSH screen (Kaiser et al., 2004), no transcripts were detected in unsynchronized mixed blood stages (Fig. 1C). A similar expression pattern of SAP1 was observed in P. falciparum oocyst and salivary gland sporozoites. No transcripts were detected in mixed blood stages. (Fig. 1D). Therefore, the sporozoite-specific expression profile of PfSAP1 is similar to the expression profile of PySAP1.
Localization of PySAP1
To determine the cellular localization of SAP1, we generated rabbit polyclonal antisera against a peptide in the C-terminus of PySAP1 and tested the antisera in immunoflourescence assays (IFAs) using P. yoelii sporozoites. A specific sporozoite-internal staining that excluded the nucleus and was distinct from circumsporozoite (CS) protein staining was observed (Fig. 2). SAP1 localization appeared uneven and clustered within the sporozoite cytoplasm. Its localization appeared also distinct when compared with cytoplasmic heat shock protein (HSP70) staining, which was induced by incubation of the sporozoites at 37°C for 1 h (Fig. 2). SAP1 staining was only observed in sporozoites after membrane permeabilization, indicating that SAP1 localizes exclusively to the interior of the sporozoite. Together, the data suggest that SAP1 localizes to the cytoplasm, intracellular organelles or other structures within the cytoplasm as predicted by the lack of a secretory signals in the SAP1 sequence. Pre-immune sera did not show reactivity with sporozoites (data not shown).
Fig. 2.
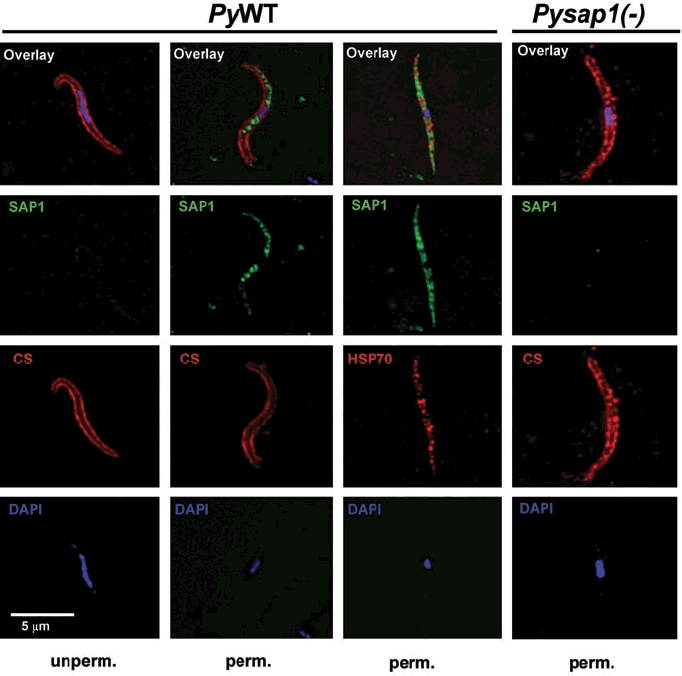
SAP1 is localized to the cell interior of Plasmodium sporozoites. Immunofluorescence assays on air-dried PyWT or Pysap1(−) salivary gland sporozoites show the predicted internal localization of SAP1 in PyWT, which is detected only after sporozoite permeabilization with saponin (perm.). SAP1 staining was not detected in Pysap1(−) sporozoites after sporozoite permeabilization. Note the absence of SAP1 staining in the nucleus of PyWT sporozoites. Scale bar is 5 μm.
Targeted deletion of PySAP1 and knockout phenotype in blood and mosquito stages
Targeted gene deletion of PySAP1 was conducted by double-cross-over homologous recombination to replace the majority of the coding sequence with the TgDHFR/TS selection marker cassette (Fig. 3A) (Menard and Janse, 1997). Deletion-specific genomic PCR analysis confirmed the successful double-cross-over recombination event and the successful isolation of a Pysap1(−) parasite clone with pure gene deletion background (Fig. 3B). Therefore, PySAP1 was successfully deleted in the erythrocytic stages with no observed deficiency of blood-stage development (data not shown). In addition, the morphology of male and female gametocytes in thin infected-blood smears and male gamete exflagellation in wet mounts of infected blood were indistinguishable from P. yoelii wild-type (WT) parasites (data not shown). Transmission of Pysap1(−) parasites to mosquitoes resulted in normal midgut infection and oocyst development. Pysap1(−) oocyst sporozoites developed in a similar manner as PyWT oocyst sporozoites (Table S1 in Supplementary material). Importantly, Pysap1(−) sporozoites accumulated in the salivary glands in numbers comparable to WT, indicating normal salivary gland infection (Table S1). RT-PCR analysis confirmed the absence of PySAP1 transcripts in Pysap1(−) sporozoites (Fig. 3C). IFAs with the anti-SAP1 antisera were negative, confirming that Pysap1(−) sporozoites do not express SAP1 (Fig. 2). The data give further support to the specificity of the SAP1 antisera. We conducted all initial experiments with two independent clones of Pysap1(−) that were identical in their phenotypes (data not shown). Thereafter, experiments were conducted with a single Pysap1(−) clone.
Fig. 3.

Targeted deletion of PySAP1.
A. Schematic representation of the replacement strategy to generate Pysap1(−) parasites. The endogenous PySAP1 genomic locus is targeted with a replacement fragment containing the 5′ and 3′ sequence within exon 1 of PySAP1 flanking the Toxoplasma gondii DHFR/TS-positive selection marker. Diagnostic wild type-specific or integration-specific test amplicons are indicated by lines.
B. PCR genotyping shows the gene replacement using oligonucleotide primer combinations that can only amplify from the recombinant locus (Test 1 and Test 2). The wild type-specific PCR reaction (WT) confirms the absence of wild-type parasites in the clonal Pysap1(−) parasites.
C. RT-PCR analysis (35 cycles) shows the loss of PySAP1 transcripts in RNA isolated from Pysap1(−) sgSPZs. The PySAP1-specific amplicon used for the analysis is shown above in (A) as the wild type (WT) test. PyCSP was used as a positive control.
Pysap1(−) sporozoites fail to induce blood-stage infection and elicit sterile protection against PyWT sporozoite challenge
We tested the infectivity of PySAP1-deficient salivary gland sporozoites in susceptible BALB/c mice. Mosquito bite experiments with more than 50 Pysap1(−)-infected mosquitoes/mouse did not result in blood-stage infection (data not shown). Strikingly, intravenous (iv) injection of escalating doses of Pysap1(−) salivary gland sporozoites did not lead to blood-stage parasitaemia, tested daily by blood smears until day 14 post infection (Table 1). Even with extremely high doses of more than 2 million sporozoites no subsequent blood-stage parasitaemia was observed. This highest dose corresponded to a ∼200 000-fold increase over the minimal infectious dose of PyWT sporozoites administered to BALB/c mice by iv injection (Belmonte et al., 2003). Hence, we conclude that PySAP1 is essential for parasite pre-erythrocytic stage functions after transmission from the mosquito to the mammalian host.
Table 1.
PySAP1-deficient sporozoites are completely attenuated and do not cause blood-stage infection in BALB/c mice.
| Pysap1(−) | PyWT | |||
|---|---|---|---|---|
| No. of injected sporozoites | Infected | Pre-patent perioda | Infected | Pre-patent perioda |
| 20 | ND | ND | 2/2 | 4 days |
| 100 | ND | ND | 6/6 | 4 days |
| 10 000 | 0/30 | – | 8/8 | 3 days |
| 100 000 | 0/15 | – | 3/3 | 2.5 days |
| 500 000 | 0/8 | – | ND | ND |
| 1 000 000 | 0/4 | – | ND | ND |
| > 2 000 000 | 0/3 | – | ND | ND |
The period (in days) between sporozoite infection and the detection of erythrocytic stages in blood smears.
ND, not done.
We next tested whether Pysap1(−) salivary gland sporozoite immunization of mice can induce sterile protection against PyWT sporozoite challenge. Four groups of BALB/c mice were immunized iv with three doses of 10 000 Pysap1(−) salivary gland sporozoites, in 2-week intervals (Table 2). The first immunization group (group I) was challenged by iv injection of 10 000 PyWT sporozoites at day 7 after the last immunization dose. Two of the immunization groups (groups II and III) were challenged by iv injection of 10 000 PyWT sporozoites 30 and 210 days after the last immunization dose. The mice of group III were then challenged by PyWT erythrocytic stages 2 weeks after the last challenge with either 103 or 106 asexual blood stages injected iv or intraperitoneally (ip) into five mice each respectively (data not shown). The fourth group (group IV) was challenged by infectious mosquito bite 45 and 210 days after the last immunization dose. All mice were protected when challenged with PyWT sporozoites and did not develop any blood-stage infection (Table 2). However, mice challenged with blood-stage parasites developed blood-stage parasitaemia after 2 days (data not shown). The data demonstrate that Pysap1(−) salivary gland sporozoite immunizations induce stage-specific sterile immunity against subsequent PyWT sporozoite infection but not against asexual blood-stage infection.
Table 2.
Immunization with Pysap1(−) sporozoites confers sterile protection against wild-type sporozoite challenge.
| Group | Primary dose (days of booster dose) | Challenge dose/days after last boost | No. protected/No. challengeda | Mean pre-patent period (days) |
|---|---|---|---|---|
| I | 10 000 (14, 28) | 10 000/7 | 9/9 | − |
| II | 10 000 (14, 28) | 10 000/(30)/(210) | 15/15/15 | −/− |
| III | 10 000 (14, 28) | 10 000/(30)/(210) | 10/10/10 | −/− |
| IV | 10 000 (14, 28) | MBb/(45)/(210) | 5/5/5 | −/− |
Each immunization group had an age-matched naïve control group (minimum three mice) that all became patent at day 3 after each PyWT sporozoite challenge.
Infection through mosquito bite (MB) by allowing a minimum of 10 PyWT female infected mosquitoes, with midgut oocyst infectivity higher than 90%, to bite one mouse for at least 10 min.
Pysap1(−) sporozoites traverse and invade hepatocytes normally but suffer an early liver-stage developmental arrest in vitro
Failure of mutant salivary gland sporozoites to induce blood-stage infection in mice can be due to distinct knockout phenotypes (reviewed in Mikolajczak and Kappe, 2006). Pysap1(−) salivary gland sporozoites displayed continuous gliding motility that was undistinguishable from PyWT, tested on glass slides by direct microscopic examination (Vanderberg, 1974) (data not shown). Thereafter, we tested the cell-traversal capacity of Pysap1(−) salivary gland sporozoites using a cell-wounding assay (Vanderberg et al., 1990; Mota et al., 2001). Pysap1(−) sporozoites traversed hepatocytes and wounded cells at a rate comparable to PyWT sporozoites (Fig. 4A). In order to identify and characterize the deficiency of Pysap1(−) sporozoites in completing pre-erythrocytic infection, we conducted in vitro assays with the hepatoma cell line HepG2-CD81 which sustains productive P. yoelii sporozoite infection and liver-stage development (Silvie et al., 2006). Intrahepatocytic parasites were quantified by differential permeabilization at 1, 6, 12, 18 and 24 h post in vitro infection (pi) using fluorescence microscopy. Interestingly, at 1 h pi the number of Pysap1(−) intracellular parasites was similar to PyWT infections, but Pysap1(−) intracellular liver-stage numbers gradually decreased in comparison with PyWT infections at 6 h and 12 h pi (Fig. 4B). Pysap1(−) intracellular parasite numbers then sharply decreased at 18 h pi and at 24 h pi there were almost no intracellular parasites, when compared with PyWT (Fig. 4B). Most importantly, intrahepatocytic Pysap1(−) parasites failed to grow and develop. They appeared smaller and deficient in their transformation to liver-stage trophozoites when compared with PyWT liver stages (Fig. 4C). Surprisingly, immunostaining of the PVM-resident protein UIS4 showed that it was not detected in Pysap1(−) parasites at all time points tested, whereas in PyWT liver stages a UIS4 positive PVM staining pattern was observed at all time points later than 1 h pi (Fig. 4C). Evidently, Pysap1(−) sporozoites infect host hepatocytes but suffer growth arrest early during liver-stage development in vitro, suggesting that an early growth defect causes the lack of Pysap1(−) sporozoite infectivity to mice.
Fig. 4.
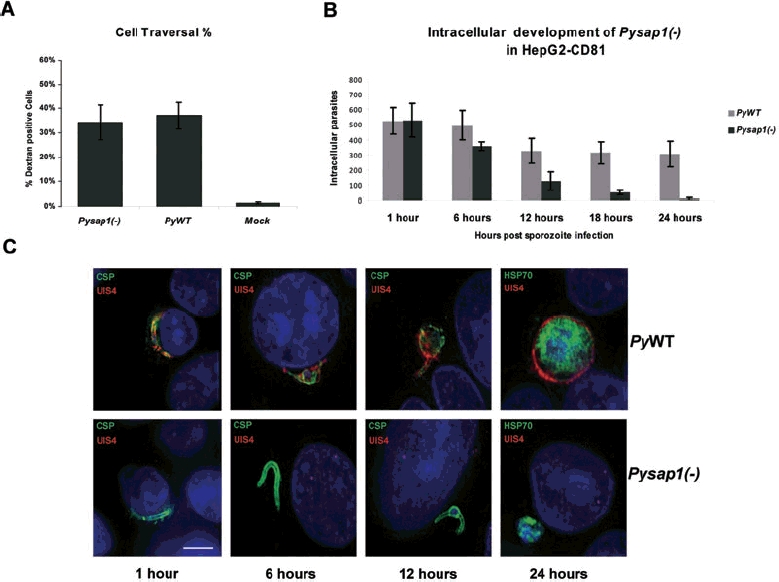
Pysap1(−) parasites traverse and invade hepatocytes but fail to complete liver-stage development in vitro.
A. Graph shows the per cent of dextran-positive hepatoma cells as an indication of sporozoites cell-traversal activity. No significant difference can be detected between Pysap1(−) and wild type (PyWT).
B. Graph shows the number of intracellular liver stages, determined by differential permeabilization, at different time points post sporozoite infection. The number of Pysap1(−) intracellular parasites is similar to PyWT at 1 and 6 h post infection but drastically decreases at 12 and 18 h. At 24 h virtually no Pysap1(−) liver stages survived. Note that the number of WT liver stages in this assay initially decreases but stabilizes after 12 h.
C. Immunofluorescence assays show liver-stage development at different time points after sporozoite infection. Pysap1(−) intracellular parasites (CSP and HSP70 staining in green) do not express the PVM marker UIS4 (red) and arrest in development. Growth-arrested Pysap1(−) liver stages shown at 24 h post infection were detected rarely in this assay. Scale bar is 5 μm.
Electron microscopic analysis of intrahepatocytic Pysap1(−) sporozoites
Intracellular malaria parasites need a PVM for development (reviewed in Mikolajczak and Kappe, 2006). Therefore, we examined whether the observed lack of UIS4 in intracellular Pysap1(−) parasites indicated a possible deficiency in PVM formation. We performed an electron microscopic analysis of intracellular WT and Pysap1(−) parasites 1 h after infection of HepG2-CD81 cells. Intracellular Pysap1(−) parasites were able to form a PVM (Fig. 5). Out of 15 intrahepatocytic Pysap1(−) parasites evaluated by EM, 4 exhibited a PVM and 11 appeared free in the cytoplasm. The latter may represent sporozoites in the process of cell traversal. However, it is also possible that Pysap1(−) sporozoites form a PVM but less efficiently than PyWT.
Fig. 5.
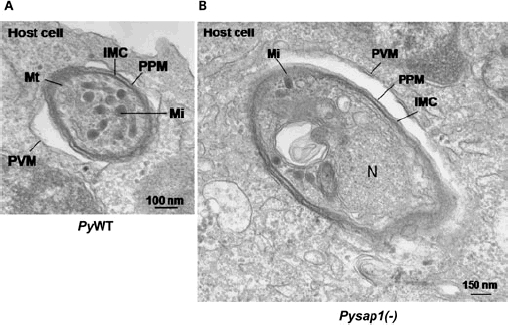
Pysap1(−) liver stages form a parasitophorous vacuole membrane (PVM). Electron microscopic analysis establishes that intrahepatocytic Pysap1(−) parasites form a PVM.
A. Transversal section of a PyWT parasite within a HepG2-CD81 cell 1 h post infection. The PVM is indicated.
B. Transversal section of Pysap1(−) sporozoite 1 h post infection in HepG2-CD81. The PVM is indicated. IMC, inner membrane complex; Mi, microneme; N, nucleus; PPM, parasite plasma membrane; PVM, parasitophorous vacuole membrane; Mt, mitochondrion.
UIS gene products are depleted in Pysap1(−) sporozoites
Despite of the presence of a PVM we noted the lack of PyUIS4 in Pysap1(−) liver stages (Fig. 4C). PyUIS4 is normally expressed in sporozoite secretory organelles as well as the liver-stage PVM and is essential for malaria parasite liver-stage development (Mueller et al., 2005b). To test whether PyUIS4, as well as other proteins, is expressed in Pysap1(−) sporozoites prior to hepatocyte invasion, we performed IFAs to test UIS4, UIS3 and MTIP (Bergman et al., 2003) expression in Pysap1(−) and PyWT salivary gland sporozoites. In PyWT sporozoites we detected expression of each protein (Fig. 6A). In contrast, we did not detect expression of UIS4 or UIS3 in Pysap1(−) sporozoites but did detect MTIP staining (Fig. 6A). To test whether this expression pattern is due to a reduction in UIS4 and UIS3 transcript abundance, which would potentially indicate transcript degradation (Parker and Sheth, 2007), we performed RT-PCR analysis on Pysap1(−) salivary gland sporozoite cDNA. Strikingly, we observed a severe reduction of PyUIS4 and PyUIS3 (Mueller et al., 2005a,b) transcript abundance (Fig. 6B and C). Furthermore, we saw a decrease in P52 (van Dijk et al., 2005; Ishino et al., 2005a,b; Labaied et al., 2007) transcript abundance as well as the transcripts of two uncharacterized UIS genes, UIS2 (putative secreted phosphatase) and UIS28 (putative secreted lipase) (Matuschewski et al., 2002) (Fig. 6B and C). Conversely, transcript abundance for genes that are involved in sporozoite functions prior to PVM formation and liver-stage development appeared not significantly reduced in Pysap1(−) sporozoites (Fig. 6B and C). These genes included CSP, TRAP (reviewed in Kappe et al., 2004; Baldacci and Menard, 2004), SPECT1 (Ishino et al., 2004), SPECT2 (Ishino et al., 2005b) and S4/CELTOS (Kaiser et al., 2004, Kariu et al., 2006). Therefore, lack of PySAP1 in sporozoites has a selective negative impact on UIS gene expression in P. yoelii.
Fig. 6.
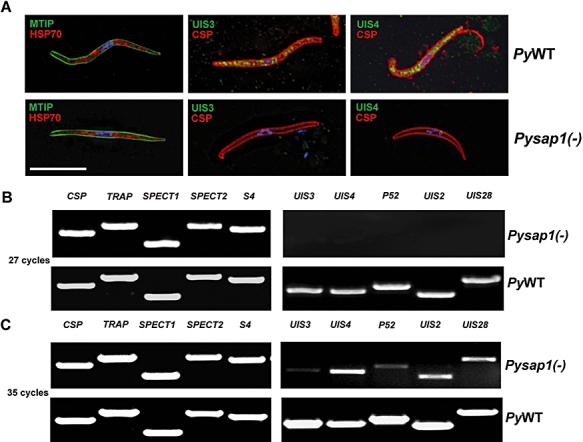
Essential UIS proteins are depleted in Pysap1(−) sporozoites.
A. Immunofluorescence assays on air-dried PyWT salivary gland sporozoites show expression of CSP and HSP70 (red), MTIP, UIS3 and UIS4 (green). Pysap1(−) sporozoites exhibit MTIP, HSP70 and CSP staining but lack UIS3 and UIS4. Scale bar is 5 μm.
B. Twenty-seven-cycle non-quantitative RT-PCR analysis with RNA isolated from PyWT and Pysap1(−) salivary gland sporozoites showing expression of UIS transcripts in WT sporozoites and the depletion of UIS transcripts in Pysap1(−) sporozoites (right). Transcripts of non-UIS genes that are essential for sporozoites prior to or in hepatocyte infection appear unaffected (left).
C. Thirty-five-cycle non-quantitative RT-PCR analysis, with the same RNA used in (B). At higher PCR cycle number UIS transcripts are detected.
Discussion
Successful hepatocyte infection and liver-stage development by the malaria parasite is dependent on establishment of a PVM as the functional host–parasite interface. Some UIS gene products uniquely expressed in salivary gland sporozoites and localized to the PVM in liver stages play essential roles in intrahepatocytic parasite survival (Mueller et al., 2005a,b; Mikolajczak et al., 2007a; Tarun et al., 2007). However, factors that allow salivary gland sporozoites lying-in-wait in the mosquito salivary glands to initiate a co-ordinated switch to mammalian host infection by differential expression of UIS have not been identified. Our work identifies SAP1 as such a potential factor. PySAP1 is the first identified cytoplasmic Plasmodium protein with an essential function for pre-erythrocytic stages. It has a large internal asparagine-rich low-complexity domain. Low-complexity domains are frequently found in Plasmodium proteins (Aravind et al., 2003). This is partially the consequence of the high A/T nucleotide content in the Plasmodium genome (Pizzi and Frontali, 2001; Singh et al., 2004). Low-complexity domains in Plasmodium proteins were hypothesized to be an evolutionary by-product with no significant function in the biology of the malaria parasite (Xue and Forsdyke, 2003). In contrast, low-complexity proteins have been proposed as virulence inducing factors in some pathogenic bacterial strains (Nandi et al., 2003). The low-complexity domain of SAP1 is flanked by two highly conserved non-asparagine-rich N- and C-terminal domains. The level of conservation in these domains is high among SAP1 proteins from distinct Plasmodium species, indicating that they represent functionally important regions. Pysap1(−) salivary gland sporozoites show extremely reduced transcript abundance for UIS3, UIS4 and P52 but not SPECTs, TRAP and CSP, indicating a selective mechanism of UIS transcript depletion. It will be interesting to determine whether transcript abundance for additional genes is affected in the Pysap1(−) sporozoites as we have shown here for the uncharacterized UIS2 and UIS28. Clearly, lack of UIS genes expression cannot be attributed to a defect in salivary gland invasion and residence as Pysap1(−) sporozoites infected the salivary glands with efficiencies that are comparable to WT sporozoites. Therefore, it appears likely that the reduction of UIS transcript expression in Pysap1(−) sporozoites is a direct effect of the lack of SAP1 and not an indirect effect of an altered biological behaviour of the Pysap1(−) mutants. Transcript abundance in eukaryotes is mainly regulated by transcriptional and post-transcriptional mechanisms. PySAP1 localization to the sporozoite cytoplasm and absence from the sporozoite nucleus suggests that PySAP1 is involved in as-yet-to-be-defined post-transcriptional mechanisms of UIS transcript regulation, as post-transcriptional regulation is expected to be executed in the cytoplasm of the cell (Elemento et al., 2007; Parker and Sheth, 2007). However, the C-terminus of PySAP1, which is recognized by the antisera, might be processed and left in the cytoplasm. In this scenario, the remaining part of the protein might translocate to the nucleus, where it might exert its effect by contributing to the regulation of transcription of UIS genes. Interestingly, post-transcriptional, but not transcriptional, regulation has been hypothesized to be the main pathway for controlling the expression levels of proteins in Plasmodium (Coulson et al., 2004; Hakimi and Deitsch, 2007). Indeed, it has been suggested that Plasmodium must rely on this mechanism for controlling the extensive differential gene expression required during the complex malaria parasite life cycle (Hall et al., 2005). Recently, it has been shown that a RNA helicase termed DOZI (Development of Zygote Inhibited) is expressed in the female gametocyte where it localizes to cytoplasmic protein complexes and is involved in translational repression of transcripts (Mair et al., 2006). DOZI knockouts showed a severe reduction in the levels of many sexual stage-specific transcripts, presumably because they were subject to rapid degradation when not protected in ribonucleoprotein (RNP) complexes. We speculate that SAP1 might be an essential component of such an RNP complex in sporozoites and protects UIS transcripts specifically from degradation. This scenario however requires further investigation. Interestingly, proteins with glutamine and asparagine-rich domains have recently been shown to be part of RNP complexes in yeast where they act as scaffolding proteins (Decker et al., 2007).
Pysap1(−) sporozoites showed complete attenuation of liver infection. This is clearly attributable to the lack of the essential proteins UIS3, UIS4 and P52 and possibly additional UIS in the knockout parasite and likely not to a lack of a direct effector function of PySAP1. Single- and double-gene-deletion sporozoites are effective live attenuated vaccines in mouse models (reviewed in Mikolajczak et al., 2007b) and we have shown herein that Pysap1(−) sporozoites also confer sterile long-lasting protection against PyWT sporozoite challenge. P. falciparum gene deletion mutants might go forward for testing as human malaria vaccines (Renia et al., 2006). We suggest that a putative P. falciparum sap1(−) sporozoite may be an attractive live attenuated vaccine candidate due to its quasi-multilocus attenuation. Together, our data give initial insights into the regulation of malaria parasite infectivity after mosquito transmission and these findings might advance efforts to develop measures for prevention of malaria infection.
Experimental procedures
Experimental animals, parasites and cell lines
Six- to 8-week-old female BALB/cJ (for in vivo infection studies and immunizations) or Swiss Webster (SW) mice (for parasite cycle maintenance) were purchased from the Jackson Laboratory (Bar Harbor, ME) or Harlan (Indianpolis, IN). Animal handling was conducted according to Institutional Animal Care and Use Committee-approved protocols. Wild-type P. yoelii 17XNL (non-lethal strain) clone 1.1 (Weiss et al., 1989) and Pysap1(−) parasites were cycled between SW mice and Anopheles stephensi mosquitoes. Infected mosquitoes were maintained on sugar water at 24°C and 70% humidity. Salivary gland sporozoites were extracted from infected mosquitoes between days 13 and 15 post blood meal infection as described before (Labaied et al., 2007; Tarun et al., 2007). The human hepatoma cell line HepG2-CD81 (Silvie et al., 2006) was used for all in vitro assays and was maintained in DMEM-F12 medium supplemented with antibiotics and 10% fetal calf serum (FCS).
Generation of Pysap1(−) parasites
Targeted deletion of PySAP1 by double-cross-over homologous recombination was achieved constructing a replacement plasmid in the b3D.DT.H Db targeting vector (Janse et al., 2006; Jongco et al., 2006). P. yoelii 17XNL genomic DNA (gDNA) was used as a template to amplify a 1.5 kb fragment of the 5′UTR of PySAP1 using oligonucleotide primers PySAP1rep1 forward (F) and PySAP1rep2 reverse (R) (primers sequences provided in Table S2). The amplified fragment was inserted into the transfection plasmid between KpnI and HindIII restriction enzymes sites. Similarly, a 1 kb DNA fragment from the 3′ORF sequence of PySAP1 was amplified using PySAP1rep3F and PySAP1rep4R primers and inserted between the SpeI and SacII restriction sites in the transfection vector. The resulting plasmids were digested with KpnI and SacII to release the replacement fragment used for the transfection. Transfection of P. yoelii 17XNL parasites using the Amaxa nucleofector device (Amaxa GmbH, Germany), resistant parasites selection and recombinant parasite cloning by serial limiting dilutions were all conducted as described elsewhere (Janse et al., 2006; Jongco et al., 2006). We obtained two independent Pysap1(−) clonal parasite populations that were phenotypically identical. Detailed analysis was performed with one representative clone. To confirm the targeted deletion and the new genetic recombination, integration-specific gDNA PCR amplification of the Pysap1(−) locus was generated using the specific primers combinations Test1 (TgF and PySAP1TestR) and Test2 (PySAP1TestF and TgR) (primers sequences provided in Table S2). Whereas PySAP1 locus-specific gDNA-PCR amplification was generated using the primers combination WT (PySAP1orfF and PySAP1orfR), the primers combination test for the WT PySAP1 locus was also used in RT-PCR to confirm the absence of the PySAP1 transcript from Pysap1(−) recombinant parasite clone.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen) and treated with TURBO DNase (Ambion) from P. yoelii salivary gland sporozoites (2 × 106), oocyst sporozoites (2 × 106) or mixed blood stages (1 × 107 infected red blood cells were isolated from an infected mouse by heart puncture bleeding and serial limiting dilution of infected blood). P. falciparum sporozoites and mixed blood-stage RNA materials were kindly provided by Dr Urszula Krzych and Dr Jack Williams (WRAIR, Silver Spring, MD). cDNA synthesis was performed using the Super Script III Platinum two-step qRT-PCR kit (Invitrogen). The sequences of specific primers used for amplification from cDNA are listed in Table S2. All PCR amplification cycles were performed at 95°C for 30 s for DNA denaturation, 55°C for 30 s for primer annealing, 60°C for 4 min for DNA strands extension.
Immunofluorescence assays (IFAs)
We generated rabbit polyclonal antiserum against a PySAP13020−3034 synthetic peptide (LRGRQVQQSFNHSAS). The antiserum was further affinity-purified against the synthetic peptide and the specific IgGs were concentrated. Pysap1(−) or PyWT sporozoites were air dried on poly-lysine-treated glass slides after incubation for 1 h at 37°C. Sporozoites were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature. This was followed by permeabilization with 0.2% saponin for 15 min at room temperature. After blocking in 10% FCS/PBS overnight (ON) at 4°C, primary antibodies were diluted 1:300 in 10%FCS/PBS and incubated with the sporozoites for 1 h at 37°C. Mouse monoclonal antibodies 2F6 (Potocnjak et al., 1980; Yoshida et al., 1980) and 2E6 (Tsuji et al., 1994) were used against PyCSP and PyHSP70, respectively, and rabbit polyclonal antisera against PySAP1, PyUIS4, PyUIS3, PyMTIP were used in sporozoite IFAs. Alexa Fluor (Molecular Probes, Eugene, OR) conjugated secondary antibodies were also diluted in 10%FCS/PBS and incubated with the sporozoites for 1 h at 37°C. Conjugated anti-rabbit Alexa Fluor 488 (green) and anti-mouse Alexa Fluor 594 (red) were used to visualize the bound primary antibodies. Nuclear staining with 4′,6′-diamidino-2-phenylindole (DAPI) was conducted within the last washing step with PBS (1:2000) and before mounting the slides with an antifade reagent (Fluoroguard; Bio-Rad, Hercules, CA). Preparations were analysed using fluorescence confocal microscopy (Olympus 1 × 70 Delta Vision).
Mouse infections and immunizations
For sporozoite immunizations and challenges, BALB/cJ mice were injected iv with sporozoites re-suspended in incomplete DMEM-F12 medium. Blood-stage patency was monitored daily by evaluation of Giemsa-stained blood smears from day 2 to day 14 post sporozoite infection. For blood-stage challenge experiments, immunized mice that had been challenged with sporozoites twice were injected either ip with 106 or iv with 103PyWT erythrocytic asexual stages in RPMI. PyWT blood stages were isolated from an infected mouse by heart puncture followed by serial limited dilution.
Cell-traversal assay
Hepatoma HepG2-CD81 cells were inoculated in eight-well chamber slides at a density of 60 000 cells well−1 2 days before the assay. A total of 20 000 sporozoites per well of PyWT or Pysap1(−), in addition to uninfected mosquitoes salivary gland extracts (mock), were re-suspended in incomplete DMEM-F12 medium with 3% bovine albumin serum (BSA) and 2 mg ml−1 fluorescein isothiocyanate (FITC)-dextran (Invitrogen-Molecular Probes, Eugene, OR). Sporozoites and mock suspensions were added to the cells, centrifuged for 2 min at 1000 r.p.m. and incubated for 1 h at 37°C. Thereafter, the cells were washed thoroughly with PBS and complete DMEM medium twice to remove any extracellular dextran, and the cells were allowed to grow for a further 3 h in complete DMEM medium. Flow cytometric quantitative analysis of dextran-positive cells was conducted using a flow cytometer (Cytopia, Seattle, WA) and the flow cytometry analysis program FlowJo version 7.0.3 (TreeStar, Ashland, OR) (Labaied et al., 2007).
In vitro infection assays
We standardized a differential permeabilization hepatoma infection assay to specifically quantify liver-stages parasites at different time points of infection by fluorescence microscopy. Hepatoma HepG2-CD81 cells were seeded in eight-well chamber slides at a density of 40 000–50 000 cells well−1 2 days before the assay. A total of 50 000 sporozoites of Pysap1(−) or PyWT re-suspended in incomplete DMEM-F12 medium were added per well. The sporozoites were incubated for 1 h with the cells at 37°C. The cells were washed with PBS and with complete DMEM F12 media twice to remove all non-invading and unbound sporozoites and mosquito debris. One hour post infection assays were fixed with 4% PFA (which does not permeabilize hepatocytes) for 10 min at room temperature, followed by permeabilization (or not) with ice-cold methanol for 5 min at room temperature and then blocking in 10%FCS/PBS (ON) at 4°C. The cells for other post-infection time points assays were further grown in complete DMEM medium until fixed, permeabilized (or not) and blocked at 6 h, 12 h, 18 h and 24 h. Primary antibodies against PyCSP, PyHSP70 and PyUIS4 were diluted to 1:300 in 10%FCS/PBS and incubated with the cells for 1 h at 37°C. Conjugated secondary anti-mouse Alexa Fluor 488 (green) and anti-rabbit Alexa Fluor 594 (red) were used to visualize the bound primary antibodies. Nuclear staining with DAPI (1:3000 diluted) was performed with the last wash and before mounting the slides with an antifade reagent. Preparations were analysed using fluorescence confocal microscopy (Olympus 1 × 70 Delta Vision). Intracellular parasites were determined as the total number of parasites counted in each permeabilized sample well as a fraction of the total number of parasites counted in the control unpermeabilized sample well.
Transmission electron microscopy
For thin-section transmission electron microscopy, 106PyWT and Pysap1(−) sporozoites were used to infect 106 subconfluent HepG2-CD81 cells. One hour post infection, cells were fixed with 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h at room temperature and processed as described previously (Quittnat et al., 2004), before examination with a Philips 410 electron microscope (Eindhoven, the Netherlands) under 80 kV.
Acknowledgments
We thank Dr Urszula Krzych and Dr Jack Williams for providing us with P. falciparum sporozoites and blood-stages materials. We also thank Xinxia Peng, Sasha DeLeon and Ronald Dumpit for excellent technical assistance and Dr Ashley Vaughan for critically reading this manuscript. This work was funded by a Grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative.
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/
j.1365-2958.2008.06271.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Amino R, Thiberge S, Martin B, Celli S, Shorte S, Frischknecht F, Menard R. Quantitative imaging of Plasmodium transmission from mosquito to mammal. Nat Med. 2006;12:220–224. doi: 10.1038/nm1350. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Baldacci P, Menard R. The elusive malaria sporozoite in the mammalian host. Mol Microbiol. 2004;54:298–306. doi: 10.1111/j.1365-2958.2004.04275.x. [DOI] [PubMed] [Google Scholar]
- Belmonte M, Jones TR, Lu M, Arcilla R, Smalls T, Belmonte A, et al. The infectivity of Plasmodium yoelii in different strains of mice. J Parasitol. 2003;89:602–603. doi: 10.1645/0022-3395(2003)089[0602:TIOPYI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bergman LW, Kaiser K, Fujioka H, Coppens I, Daly TM, Fox S, et al. Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. J Cell Sci. 2003;116:39–49. doi: 10.1242/jcs.00194. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MR, Douradinha B, Franke-Fayard B, Heussler V, van Dooren MW, van Schaijk B, et al. Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci USA. 2005;102:12194–12199. doi: 10.1073/pnas.0500925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Deitsch KW. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr Opin Microbiol. 2007;10:357–362. doi: 10.1016/j.mib.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- Ishino T, Yano K, Chinzei Y, Yuda M. Cell-passage activity is required for the malarial parasite to cross the liver sinusoidal cell layer. PLoS Biol. 2004;2:E4. doi: 10.1371/journal.pbio.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino T, Chinzei Y, Yuda M. Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol Microbiol. 2005a;58:1264–1275. doi: 10.1111/j.1365-2958.2005.04801.x. [DOI] [PubMed] [Google Scholar]
- Ishino T, Chinzei Y, Yuda M. A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell Microbiol. 2005b;7:199–208. doi: 10.1111/j.1462-5822.2004.00447.x. [DOI] [PubMed] [Google Scholar]
- Janse CJ, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Jongco AM, Ting LM, Thathy V, Mota MM, Kim K. Improved transfection and new selectable markers for the rodent malaria parasite Plasmodium yoelii. Mol Biochem Parasitol. 2006;146:242–250. doi: 10.1016/j.molbiopara.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kaiser K, Matuschewski K, Camargo N, Ross J, Kappe SH. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol Microbiol. 2004;51:1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x. [DOI] [PubMed] [Google Scholar]
- Kappe SH, Buscaglia CA, Nussenzweig V. Plasmodium sporozoite molecular cell biology. Annu Rev Cell Dev Biol. 2004;20:29–59. doi: 10.1146/annurev.cellbio.20.011603.150935. [DOI] [PubMed] [Google Scholar]
- Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol Microbiol. 2006;59:1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- Labaied M, Harupa A, Dumpit RF, Coppens I, Mikolajczak SA, Kappe SH. Plasmodium yoelii sporozoites with simultaneous deletion of P52 and P36 are completely attenuated and confer sterile immunity against infection. Infect Immun. 2007;75:3758–3768. doi: 10.1128/IAI.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, et al. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschewski K. Getting infectious: formation and maturation of Plasmodium sporozoites in the Anopheles vector. Cell Microbiol. 2006;8:1547–1556. doi: 10.1111/j.1462-5822.2006.00778.x. [DOI] [PubMed] [Google Scholar]
- Matuschewski K, Ross J, Brown SM, Kaiser K, Nussenzweig V, Kappe SH. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J Biol Chem. 2002;277:41948–41953. doi: 10.1074/jbc.M207315200. [DOI] [PubMed] [Google Scholar]
- Menard R, Janse C. Gene targeting in malaria parasites. Methods. 1997;13:148–157. doi: 10.1006/meth.1997.0507. [DOI] [PubMed] [Google Scholar]
- Mikolajczak SA, Kappe SH. A clash to conquer: the malaria parasite liver infection. Mol Microbiol. 2006;62:1499–1506. doi: 10.1111/j.1365-2958.2006.05470.x. [DOI] [PubMed] [Google Scholar]
- Mikolajczak SA, Jacobs-Lorena V, MacKellar DC, Camargo N, Kappe SH. L-FABP is a critical host factor for successful malaria liver stage development. Int J Parasitol. 2007a;37:483–489. doi: 10.1016/j.ijpara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Mikolajczak SA, Aly AS, Kappe SH. Preerythrocytic malaria vaccine development. Curr Opin Infect Dis. 2007b;20:461–466. doi: 10.1097/QCO.0b013e3282ef6172. [DOI] [PubMed] [Google Scholar]
- Mota MM, Pradel G, Vanderberg JP, Hafalla JC, Frevert U, Nussenzweig RS, et al. Migration of Plasmodium sporozoites through cells before infection. Science. 2001;291:141–144. doi: 10.1126/science.291.5501.141. [DOI] [PubMed] [Google Scholar]
- Mueller AK, Labaied M, Kappe SH, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005a;433:164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]
- Mueller AK, Camargo N, Kaiser K, Andorfer C, Frevert U, Matuschewski K, Kappe SH. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite–host interface. Proc Natl Acad Sci USA. 2005b;102:3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi T, Kannan K, Ramachandran S. The low complexity proteins from enteric pathogenic bacteria: taxonomic parallels embedded in diversity. In Silico Biol. 2003;3:277–285. [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pizzi E, Frontali C. Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 2001;11:218–229. doi: 10.1101/gr.152201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocnjak P, Yoshida N, Nussenzweig RS, Nussenzweig V. Monovalent fragments (Fab) of monoclonal antibodies to a sporozoite surface antigen (Pb44) protect mice against malarial infection. J Exp Med. 1980;151:1504–1513. doi: 10.1084/jem.151.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quittnat F, Nishikawa Y, Stedman TT, Voelker DR, Choi JY, Zahn MM, et al. On the biogenesis of lipid bodies in ancient eukaryotes: synthesis of triacylglycerols by a Toxoplasma DGAT1-related enzyme. Mol Biochem Parasitol. 2004;138:107–122. doi: 10.1016/j.molbiopara.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Renia L, Gruner AC, Mauduit M, Snounou G. Vaccination against malaria with live parasites. Expert Rev Vaccines. 2006;5:473–481. doi: 10.1586/14760584.5.4.473. [DOI] [PubMed] [Google Scholar]
- Silvie O, Greco C, Franetich JF, Dubart-Kupperschmitt A, Hannoun L, van Gemert GJ, et al. Expression of human CD81 differently affects host cell susceptibility to malaria sporozoites depending on the Plasmodium species. Cell Microbiol. 2006;8:1134–1146. doi: 10.1111/j.1462-5822.2006.00697.x. [DOI] [PubMed] [Google Scholar]
- Singh GP, Chandra BR, Bhattacharya A, Akhouri RR, Singh SK, Sharma A. Hyper-expansion of asparagines correlates with an abundance of proteins with prion-like domains in Plasmodium falciparum. Mol Biochem Parasitol. 2004;137:307–319. doi: 10.1016/j.molbiopara.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Tarun AS, Dumpit RF, Camargo N, Labaied M, Liu P, Takagi A, et al. Protracted sterile protection with Plasmodium yoelii pre-erythrocytic genetically attenuated parasite malaria vaccines is independent of significant liver-stage persistence and is mediated by CD8+ T cells. J Infect Dis. 2007;196:608–616. doi: 10.1086/519742. [DOI] [PubMed] [Google Scholar]
- Tsuji M, Mattei D, Nussenzweig RS, Eichinger D, Zavala F. Demonstration of heat-shock protein 70 in the sporozoite stage of malaria parasites. Parasitol Res. 1994;80:16–21. doi: 10.1007/BF00932618. [DOI] [PubMed] [Google Scholar]
- Vanderberg JP. Studies on the motility of Plasmodium sporozoites. J Protozool. 1974;21:527–537. doi: 10.1111/j.1550-7408.1974.tb03693.x. [DOI] [PubMed] [Google Scholar]
- Vanderberg JP. Development of infectivity by the Plasmodium berghei sporozoite. J Parasitol. 1975;61:43–50. [PubMed] [Google Scholar]
- Vanderberg JP, Frevert U. Intravital microscopy demonstrating antibody-mediated immobilisation of Plasmodium berghei sporozoites injected into skin by mosquitoes. Int J Parasitol. 2004;34:991–996. doi: 10.1016/j.ijpara.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Vanderberg JP, Chew S, Stewart MJ. Plasmodium sporozoite interactions with macrophages in vitro: a videomicroscopic analysis. J Protozool. 1990;37:528–536. doi: 10.1111/j.1550-7408.1990.tb01260.x. [DOI] [PubMed] [Google Scholar]
- Weiss WR, Good MF, Hollingdale MR, Miller LH, Berzofsky JA. Genetic control of immunity to Plasmodium yoelii sporozoites. J Immunol. 1989;143:4263–4266. [PubMed] [Google Scholar]
- Xue HY, Forsdyke DR. Low-complexity segments in Plasmodium falciparum proteins are primarily nucleic acid level adaptations. Mol Biochem Parasitol. 2003;128:21–32. doi: 10.1016/s0166-6851(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 1980;207:71–73. doi: 10.1126/science.6985745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


