Summary
The specificity of RNAi pathways is determined by several classes of small RNAs, which include siRNAs, piRNAs, endo-siRNAs, and microRNAs (miRNAs). These small RNAs are invariably incorporated into large Argonaute (Ago)-containing effector complexes known as RNA-induced silencing complexes (RISCs), which they guide to silencing targets. Both genetic and biochemical strategies have yielded conserved molecular components of small RNA biogenesis and effector machineries. However, given the complexity of these pathways, there are likely to be additional components and regulators that remain to be uncovered. We have undertaken a comparative and comprehensive RNAi screen to identify genes that impact three major Ago-dependent small RNA pathways that operate in Drosophila S2 cells. We identify subsets of candidates that act positively or negatively in siRNA, endo-siRNA and miRNA pathways. Our studies indicate that many components are shared among all three Argonaute-dependent silencing pathways, though each is also impacted by discrete sets of genes.
Introduction
Despite similarities in their form and overall function, small RNAs that bind Argonaute (Ago) proteins in Drosophila arise from compartmentalized biogenesis pathways and join effector complexes with specialized properties (Zamore and Haley, 2005). Drosophila small interfering RNAs (siRNAs) are most often generated from exogenously introduced double-stranded RNAs (dsRNAs), though the replication products of RNA viruses can enter this pathway (Wang et al., 2006). Double-stranded RNA can also be produced from the Drosophila genome itself, either from loci encoding extensively structured RNAs or by hybridization of convergently transcribed mRNAs (Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008). These bind Ago2 to form a complex that can efficiently cleave complementary targets. miRNAs are generated via a two-step processing pathway from endogenously transcribed primary miRNAs (primiRNAs). miRNAs guide Ago1 via a 5’ “seed” sequence to mRNA targets, which are primarily repressed at the translational level (Bartel, 2004).
A great deal of progress has been made in deciphering small RNA-based regulatory networks; however, it is clear that many additional components are pending identification and functional characterization. Genome-scale screens for components of siRNA or miRNA pathways have been carried out, with some overlap between components identified (Dorner et al., 2006; Eulalio et al., 2007; Kim et al., 2005; Parry et al., 2007; Saleh et al., 2006; Ulvila et al., 2006). However, none of these screens have addressed endo-siRNA pathways nor have they attempted comparative studies in the same experimental model. Here we report comparative and comprehensive RNAi screens that identify components of the Argonaute-dependent small RNA pathways (siRNA, miRNA and endo-siRNA) in cultured Drosophila cells.
Results
Assay Systems to Monitor the siRNA/miRNA Pathways
We constructed robust assay systems that allowed us to interrogate the miRNA and siRNA pathways individually. For probing the siRNA pathway, we created an S2 cell line (RZ-14) stably expressing both the Renilla luciferase and a 688-bp perfect inverted repeat that directs Renilla silencing (Fig. S1A). To identify components of the miRNA pathway, we embedded an artificial miRNA sequence (CXCR4) into the Bantam pri-miRNA (Fig. S1B). This construct was transiently introduced into S2 cells together with an expression construct for a Renilla luciferase gene with multiple imperfect CXCR4 complementary sites in its 3’ UTR (Doench et al., 2003). In both assays, an expression construct for the firefly luciferase gene served as a normalization control. To prevent the half-life of reporter proteins from confounding our analysis, all transgenes were expressed from the inducible metallothionein promoter.
Both assays systems performed as expected upon knockdown of known components of either pathway. Silencing of Dcr-2 or Ago2 caused significant de-repression of siRNA reporters, whereas dsRNAs against Drosha, Dcr-1 or Ago1 had no effect (Fig. S1C). Conversely, RNAi knockdown of Drosha or Ago1 led to a marked decrease in miRNA-mediated gene silencing while treatment with dsRNAs against LacZ, Dcr-2 or Ago2 had no effect (Fig. S1D).
Comprehensive Identification of siRNA/miRNA Pathway Components
We screened a collection of ~21,000 dsRNAs for those that impacted the siRNA and miRNA pathways. To assess reproducibility, dsRNAs targeting each positive emerging from the two primary screens were re-synthesized and tested multiple times using both assay systems. To minimize potential off-target effects, we also generated additional independent dsRNAs targeting each gene and assessed their impacts on the siRNA and miRNA pathways. Only genes represented by two or more independent consistently scoring dsRNAs were selected as final candidates. We found that Dcr-2 and Ago2 were among the siRNA pathway genes, whereas Drosha and Ago1 were among the miRNA pathway candidates (Fig. 1C and Table S1), providing an internal validation of each screen.
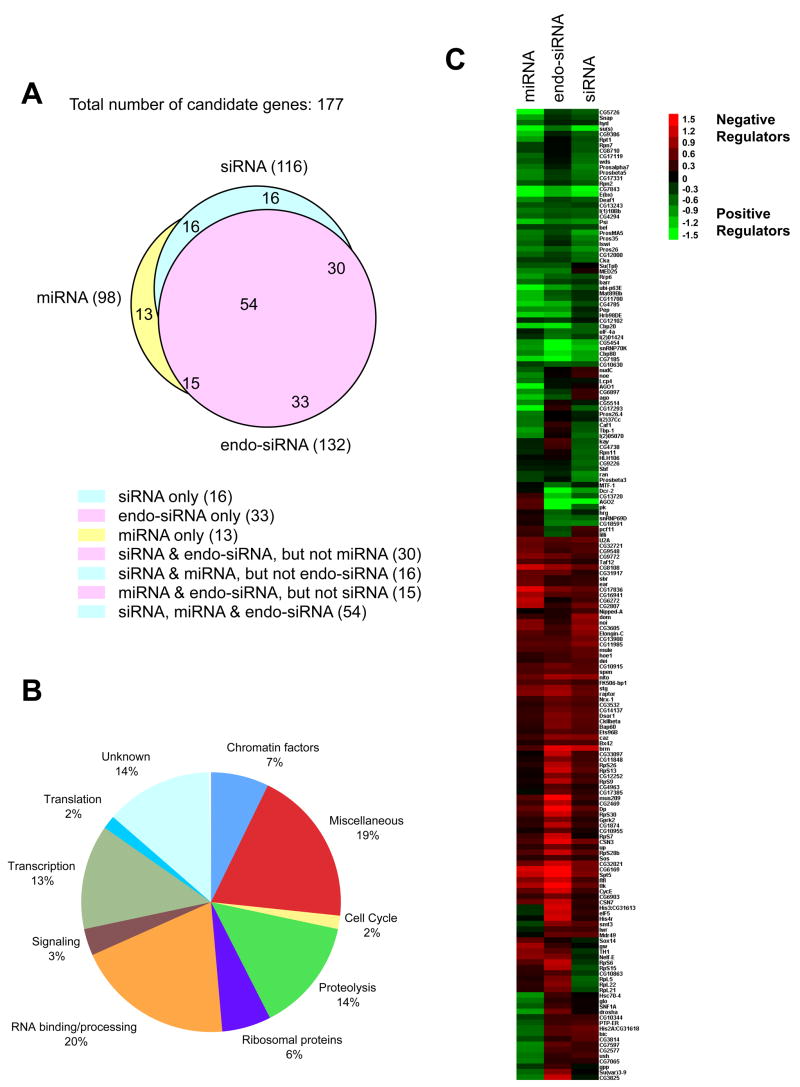
Fig. 1. Candidates identified from the screens.
(A) A Venn diagram showing the impact of the candidates on the siRNA, endo-siRNA and miRNA assays. (B) Candidates were sorted into various categories based on their annotated/verified function. (C) A heat map of the candidates and their scoring patterns in multiple assays. The scores assigned to individual genes in a given assay reflect the relative activity of the pathway upon knockdown of candidate gene expression. Red indicates an increase in silencing and green indicates a decrease. As each candidate is represented by multiple independent dsRNAs, presented are the average scores from all independent dsRNAs targeting a given gene. Scores for individual dsRNAs are shown in Table S1.
Recently, an extensive collection of endogenous siRNAs has been characterized in Drosophila (Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008). The biogenesis and function of these endo-siRNAs depend upon canonical RNAi pathway components, including Dcr-2 and Ago2. In some cases, endo-siRNA production depends much more heavily on Loquacious rather than on the canonical Dcr-2 partner, R2D2 (Czech et al., 2008; Okamura et al., 2008). We tested all candidates emerging from the miRNA and siRNA pathway screens for their impacts on a sensor for an abundant endo-siRNA, esi-2.1 (Fig. S1E). In summary, 177 candidates were identified that affected at least one of the three pathways when knocked down (Fig. 1A). Among the 116 candidates for the siRNA pathway, 84 also altered the endo-siRNA pathway, and 70 the miRNA pathway. Significant overlap (69) was also observed between the endo-siRNA (132) and miRNA pathway (98) candidates. Notably, 54 candidates affected all three pathways.
Candidate Genes Identified from the Screens
Based on their annotations, candidates that affect small RNA pathways could be assigned to several functional categories. Notably, genes encoding RNA-binding/processing factors and translation factors were enriched by ~10-fold (Fig. 1B), since only ~2% of all Drosophila genes belong to this class (Lasko, 2000). These included splicing factors, RNA helicases, and proteins involved in poly-adenylation. Splicing factors could have scored in the siRNA pathway screen because the artificial hairpin transcript, which triggers silencing, carries an intron. However, neither the miRNA expression construct nor the endo-siRNA triggers contain intronic sequences, yet most splicing factors still impacted their function. Moreover, in a screen for RNAi pathway components in C. elegans, a number of splicing factors as well as the ortholog of the U1-associated factor, PSI, were identified (Kim et al., 2005).
Many candidates emerging from the screens showed direct protein-protein interactions, fit into functional modules or joined known multi-protein complexes (Tables S1 and S2). For example, six U2 snRNP proteins displayed a similar scoring pattern in the screen. We also observed common behavior for two U1 snRNP proteins, snRNP70K and CG5454, and for PSI, which physically and functionally interacts with snRNP70K (Labourier et al., 2001; Salz et al., 2004).
Knockdown of several ribosomal proteins impacted small RNA pathways. While this effect could be indirect, some ribosomal proteins have been implicated in the siRNA pathway. For example, RpL11 and RpL5 have been shown to reside in a protein complex containing FMR, Dmp68 and Ago2 in S2 cells (Ishizuka et al., 2002).
Silencing a number of proteasomal components consistently impacted all three small RNA pathways. Indeed, a regulatory subunit, Pros45, has previously been implicated in RNAi (Ulvila et al., 2006). We find that knockdown of Tbp-1, CG12000, or l(2)05070 led to a decrease in miRNA-mediated silencing and a concomitant reduction in miRNA levels (Fig. 2A). It is not yet clear whether these effects are direct or indirect.
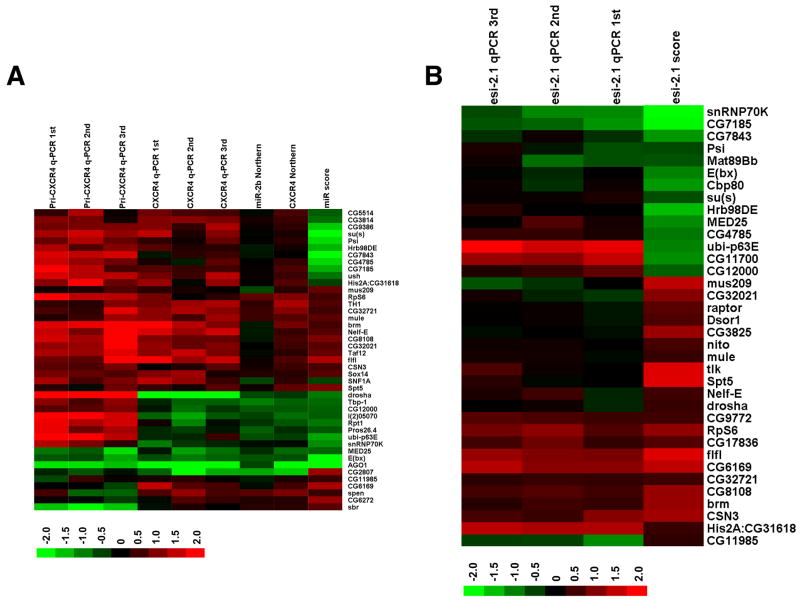
Fig. 2. Mapping candidates along the small RNA pathway.
(A) A heat map of steady-state pri-miRNA and miRNA levels upon knockdown of candidates. Steady state levels of CXCR4 and those of endogenous miR-2b upon knockdown of 43 selected candidates were examined by Northern blotting and semi-quantitative RT-PCR (q-PCR). miRNA levels were quantified and normalized first against U6 RNA levels, and then against the average of multiple controls (cells treated with dsRNA against LacZ). Also shown are steady-state levels of pri-CXCR4 measured by q-PCR, and the relative miRNA pathway activities associated with the representative dsRNA. Red indicates an increase in RNA levels, or an increase in the activity of the miRNA pathway, and green indicates a decrease. Presented are the average from 2 to 4 independent Northern blotting experiments and results from individual independent q-PCR assays. The scores associated with each candidate were from experiments involving one representative dsRNA targeting that gene (Table S3). (B) A heat map of endo-siRNA levels upon knockdown of candidates. Steady-state levels of esi-2.1 upon knockdown of 36 selected candidates were examined by q-PCR using multiple independent RNA samples. Quantification was performed as described in A. Also shown are the relative esi-2.1-mediated gene silencing activities. The scores associated with each candidate were from experiments involving one representative dsRNA (Table S4).
Placing Candidates within the siRNA/miRNA Pathway
Candidates affecting the miRNA pathway could impact miRNA biogenesis/stability or miRNA effector functions. To map candidates along the pathway, we examined the effects of their knockdown on steady state levels of the CXCR4 miRNA mimetic, on endogenous miR-2b, and on those of pri-CXCR4. Knockdown of several candidates led to consistent alterations in mature miRNA levels that paralleled effects on miRNA-mediated silencing (Fig. 2A and Table S3). For example, Drosha and Ago1 silencing reduced both miRNA levels and miRNA function, and Drosha silencing also led to a coincidental accumulation of pri-CXCR4. This class of positives likely affects miRNA processing and/or stability. Unexpectedly, silencing of Ago1 also had an impact on pri-miRNA levels. While this could represent a feedback mechanism, it is also possible that a miRNA indirectly regulates the metallothionein promoter used to express the pri-CXCR4 in this study. Another set of candidates (such as CG5514, CG3814 and CG2807) impacted miRNA-mediated repression without corresponding effects on mature miRNA levels. These positives most likely impact directly or indirectly effector steps within the pathway.
We also examined steady levels of esi-2.1 upon knockdown of candidates. As shown in Fig. 2B, knockdown of a group of candidates (e.g., CG7185 and snRNP70K) caused a decrease in esi-2.1-mediated gene silencing, which correlates with a reduction in esi-2.1 levels. This suggests involvement in siRNA production and/or stability. In contrast, knockdown of ubi-p63E, CG11700, or CG12000 caused a change in esi-2.1 activity that did not correlate with alterations in esi-2.1 RNA levels. Considered together, our analyses of the steady-state levels of small RNAs and their precursors following candidate knockdown provide clues on the placement of candidates within small RNA pathways.
Validation of Belle as a bona fide RNAi Pathway Component
bel emerged from our screen and from other studies as an RNAi pathway candidate (Kim et al., 2005; Ulvila et al., 2006). It encodes a DEAD-box RNA helicase, which is required for viability and in the germ line (Johnstone et al., 2005). We chose to validate bel both in animals and through biochemical approaches.
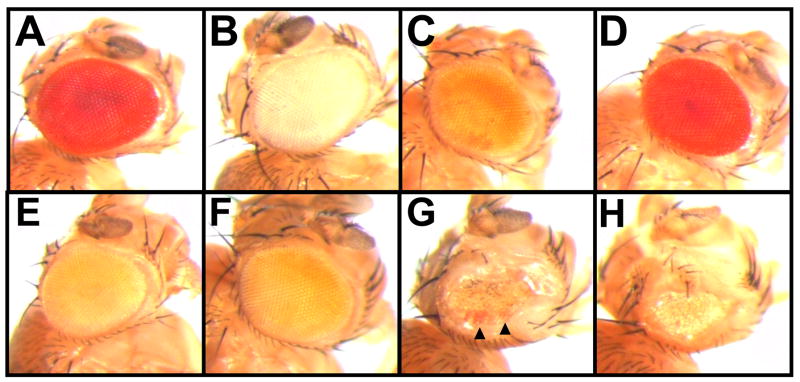
In order to assess RNAi efficiency in flies, we used transgenics carrying an inverted repeat of the white (w) gene under the control of the eye-specific GMR promoter (GMR-wIR; Fig. 3). These flies display a pale eye color due to strong suppression of white. Because bel is essential, we used an eye-specific mitotic recombination system to generate mosaics in which we could assay the impact of a bel allele (bel6) on w silencing in clones (Stowers and Schwarz, 1999). As previously described, eye cells homozygous for a Dcr-2 mutation, Dcr-2fsL811X, display a dark red color indicating loss of silencing (Fig. 3D and Lee et al., 2004). Eyes predominantly homozygous for bel6 are rough and small, suggesting that bel is required for cell viability. Importantly, patches of cells with increased pigmentation are observed in homozygous bel6 clones (Fig. 3G, arrow heads). We examined hsc70-4 mutant eyes, which are also small and rough, and found pigmentation to be unaffected, suggesting that the bel6 phenotype is specific (Fig. 3H). These observations suggest that bel6 mutant clones are defective in RNAi.
Fig. 3. bel6 flies are defective in RNAi.
Eye phenotypes of the corresponding genotypes are shown.
(A) OreR
(B) w1118
(C) w+,GMR-wIR/yw; dcr-2L811fsX,FRT42D/CyO; EGUF/+
(D) w+,GMR-wIR/yw; dcr-2L811fsX,FRT42D/FRT42D,GMR-hid,Cl; EGUF/+
(E) w+,GMR-wIR/yw; EGUF/+; FRT82B/FRT82B,Cl,GMR-hid
(F) w+,GMR-wIR/yw; EGUF/+; bel6,FRT82B/TM2
(G) w+,GMR-wIR/yw; EGUF/+; bel6,FRT82B/FRT82B,Cl,GMR-hid
(H) w+,GMR-wIR/yw; EGUF/+; hsc70-454.1,FRT82B/FRT82B,Cl,GMR-hid
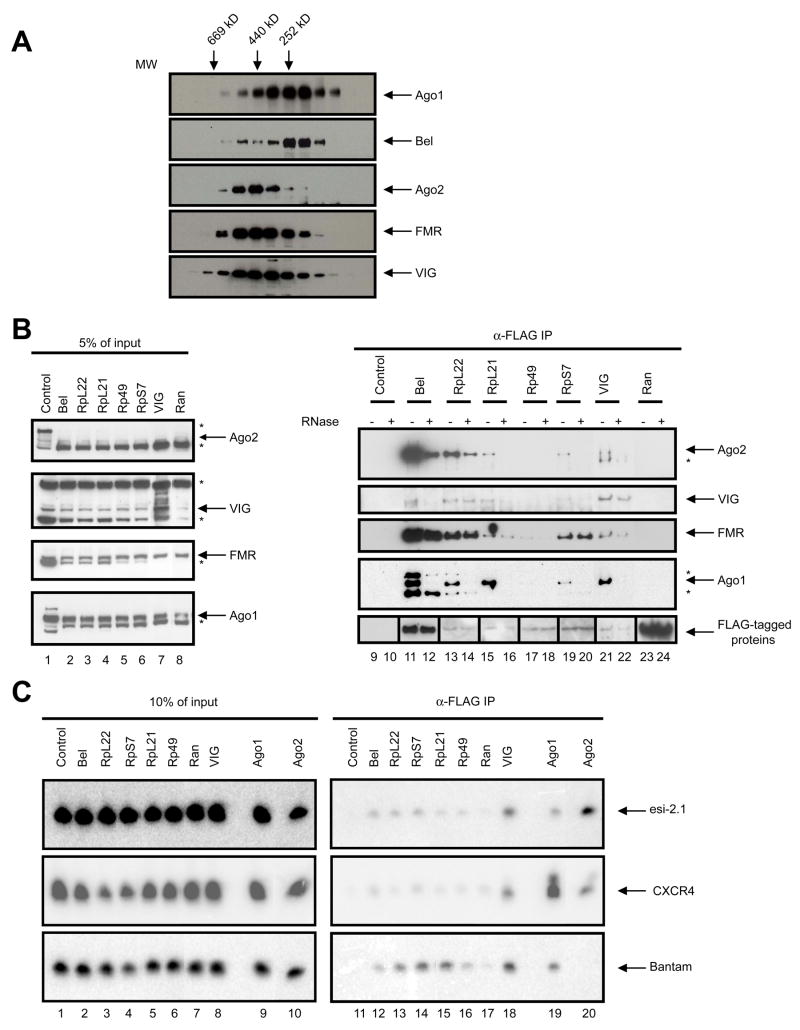
Next we tested whether Bel associates with RNAi components. Cytoplasmic extract was prepared from S2 cells and fractionated by gel-filtration. Individual fractions were immunoblotted using antibodies against Bel and components of RISC, including VIG, FMR, Ago2 or Ago1. The majority of Bel co-fractionates with Ago1, whereas a smaller fraction co-elutes with Ago2, FMR and VIG, components of the siRISC (Fig. 4A). We also immunoprecipitated FLAG-tagged Bel from S2 cells and found robust co-precipitation of Ago2, FMR or VIG (Fig. 4B). RNase treatment reduced interaction with Ago2 and VIG but not as substantially with FMR (Fig. 4B).
Fig. 4. Bel and RpL22 interact with components of the RISC.
(A) Cytoplasmic extract from S2-NP cells was fractionated by size exclusion chromatography, and the fractions were immunoblotted using antibodies against Bel and components of the RNAi pathway, as indicated. The elution profile of molecular markers is shown on top of the panel. (B) FLAG-tagged proteins (as labeled above the panel) were expressed in S2-NP cells and anti-FLAG immunoprecipitates were prepared. The samples were evenly split, and one set were treated with RNase while the other served as control. The samples were immunoblotted sequentially using antibodies against VIG, Ago2, FMR, Ago1 and FLAG. A small amount of cell extract was processed in parallel as input control. Non-specific bands are marked with a “*”. Note that the images shown in lanes 9 through 12 and 23 through 24 of the anti-FLAG panel were from films that were subjected to different exposure times from the rest due to differences in expression levels. (C) Small RNAs are enriched in the Bel and ribosomal protein immunoprecipitates. Expression constructs for FLAG-tagged proteins together with that for the CXCR4 siRNA mimetic were transfected into S2-NP cells. Total RNA was extracted from FLAG immunoprecipitates, and subjected to Northern blotting using probes against Bantam, esi-2.1 and CXCR4. To avoid signal saturation, approximately ~25% of the RNA recovered from the Ago1 and Ago2 samples was loaded compared to the rest of the samples. Total RNAs recovered from 10% of the cell extracts were processed in parallel as input control.
To test interactions of Bel with small RNAs, we expressed FLAG-tagged Bel or known components of the RISC together with an artificial siRNA (CXCR4) generated from a perfectly complementary hairpin expression construct. As expected, robust CXCR4 signals could be detected in the Ago2 or VIG complexes (Fig. 4C). Importantly, CXCR4 was also present in the Bel immunoprecipitate, as was esi-2.1, though they are probably bound directly to another protein in the complex. We conclude that Bel likely acts directly as part of the RNAi machinery, as it resides in a complex that also contains both protein and RNA components of RISC. Interestingly, more CXCR4 siRNA was present in the Ago1 immunoprecipitate than was detected in the Ago2 sample (Fig. 4C). While this could be attributed to some intrinsic characteristics of the CXCR4 siRNA mimetic, it is also possible that the coupling between miRNA processing and loading steps accounts for this observation.
Ribosomal Proteins Associate with the RNAi Machinery
We found that RpL22, a candidate from the screen, could co-immunoprecipitate components of the siRISC, including VIG, FMR, and Ago2, suggesting either a direct or indirect interaction. Similar behavior was observed for several other ribosomal proteins (Fig. 4B). These interactions seemed to display different degrees of dependence on RNA. For example, RNase treatment caused a moderate decrease in the levels of Ago2 in the RpL22 immunoprecipitate, whereas the levels of FMR and VIG remained unaffected (Fig. 4B). Similar observations were also made for the RpS7 and RpL21 samples. Moreover, both CXCR4 and esi-2.1 are present in the immunoprecipitates of a number of ribosomal proteins, including RpL22, RpS7, RpL21 and Rp49 (Fig. 4C). These observations indicate that these ribosomal proteins reside in large RNA-protein complex(es) that also contain core components of the RNAi machinery.
Discussion
In Drosophila, siRNA and miRNA pathways have been viewed as being biochemically compartmentalized. However, the boundary between these pathways has been blurred by recent observations that, depending on the configuration of their precursors, miRNAs (and possibly siRNAs) can be partitioned between Ago1 and Ago2 (Forstemann et al., 2007; Tomari et al., 2007). Moreover, Loquacious plays roles both in miRNA biogenesis and in the production of some endo-siRNAs (Czech et al., 2008; Okamura et al., 2008). Intimate connections between siRNA and miRNA pathways are also suggested by the observation that knockdown of Ago2 leads to more pronounced silencing by miRNAs (Fig. 1C), possibly by increasing the access of Ago1 to miRNAs or other limiting components that were previously bound by Ago2.
Analysis of miRNA and siRNA screens shows extensive overlap between genes that impact these pathways. Suppression of many candidates reduces the efficiency of the target small RNA pathways, indicating that those genes might be components of siRNA- or miRNA-mediated responses. Silencing of a roughly equal number of genes increases silencing, indicating that they encode negative regulators of small RNA pathways. These so-called “enhancer of RNAi” phenotypes might indicate attractive targets for genetic manipulation or small molecule inhibitors that could increase the activity of RNAi in either experimental or therapeutic settings. It is worth noting that our analysis of steady-state levels of small RNAs upon candidate knockdown revealed that for some, enhanced silencing is correlated with increased levels of the small RNA silencing trigger, as is the case for the miRNA pathway candidates such as CG32721, mule, TH1 or flfl (Fig. 2A). In contrast, while knocking down CG2807 led to markedly enhanced silencing by the miRNA mimetic CXCR4, the steady-state levels of both CXCR4 and miR-2b significantly decreased. While these effects on the small RNA pathways could be indirect, these observations suggest that some of these negative regulators of RNAi are primarily involved in the biogenesis and/or stability of the small RNA silencing trigger, while others are implicated in the downstream effector steps.
Each pathway was uniquely or differentially affected by a number of genes (Fig. 1C). For example, knockdown of one class of genes (caf1, CG17293 and genes encoding ribosomal proteins L5, L21, L22 and S15) led to decreased silencing by exogenous siRNAs, but enhanced silencing by endo-siRNAs. Suppression of such a class of genes might enhance the production or loading of endo-siRNAs into RISC, thereby depleting the pool available for products of exogenously introduced dsRNAs, a model that has been previously proposed for some loci in C. elegans (Duchaine et al., 2006). Knockdown of another group of genes (hsc70-4, CG3825 and CG2577) decreased silencing by miRNAs but enhanced silencing by endo-siRNAs. A number of possibilities, including effects on small RNA sorting might account for these observations.
We validated Bel as a bona fide component of the RNAi pathway. Bel most likely functions at step(s) downstream of siRNA processing and loading, as neither steady state levels of esi-2.1 nor the levels of Ago2-bound esi-2.1 are affected by Bel knockdown (Figs. S2 and S3). Interestingly, Ago1 and Bantam are also present in the Bel immunoprecipitate, consistent with the co-fractionation of Bel with miRISC (Figs. 4B, C). Thus, Bel may also participate in the miRNA pathway. While none of the bel dsRNAs met the scoring criteria in the miRNA assay, they did trend consistently (Table S1).
Ago1 and Bantam were present in a number of ribosomal protein immunoprecipitates, and the association between Ago1 and these ribosomal proteins was abolished by RNase treatment (Figs. 4B, C). These observations are consistent with the notion that miRISC associates with the translation machinery. Both protein and RNA components of the siRISC are also present in these immunoprecipitates and that knockdown of a number of ribosomal proteins consistently leads to enhanced silencing by endo-siRNAs (Table S1). Thus, the integrity and function of the translational machinery as a whole may be impacting the small RNA pathways.
In summary, our comparative genome-wide screens (Table S5) generate a rich resource for further study of the three Argonaute-dependent small RNA regulatory pathways in Drosophila. These studies not only point to extensive overlap and interplay among small RNA directed silencing machineries in flies but also highlight specific players in each of the three pathways.
Experimental Procedures
DNA constructs and cell culture
Detailed description of DNA constructs can be found in Supplementary Methods. S2-NP cells were maintained in Schneider’s medium (Invitrogen) supplemented with 10% FBS and 1% pen-strep (Invitrogen). To generate the RZ-14 stable cell line, S2-NP cells were transfected with pRmHa-3-Firefly-long, pRmHa-3-Renilla and pRmHa-3-Renilla-hairpin together with pHS-neo, using Effectene (Qiagen). Transfected cells were selected and maintained in growth medium supplemented with 400 μg/mL G418.
RNAi screening
Detailed description of the RNAi screening and bioinformatic analysis can be found in Supplementary Methods.
RNA isolation, Northern blotting and q-PCR assays
S2-NP cells were transiently transfected with DNA constructs for the miRNA assay (pMT-Renilla-CXCR4-6B, pMT-D05 and pRmHa-3-Firefly-long). Two days after transfection, ~3X106 cells were incubated in 1.5 mL serum-free Schneider’s medium containing 10 μg of candidate dsRNAs in 6-well plates, and 3 mL serum-containing medium was added 45 minutes later. After 3 days of dsRNA treatment, cells were induced with 200 mM CuSO4 for 24 hours. A small aliquot of cells were subjected to luciferase assays to examine the effect of dsRNA treatment on reporter activity. Total RNAs were extracted from the rest of the samples using Trizol (Invitrogen). Northern blotting was performed as previously described (Czech et al., 2008). miRNA signals were normalized against that of the U6 RNA. A score was assigned to each sample based on the average results from two or four independent experiments. To quantify steady-state levels of pri-and mature miRNAs, semi-quantitative RT-PCR (q-PCR) assays were performed (Applied Biosystems) using primers specific for pri-Bantam, CXCR4 and U6.
Immunoprecipitation of proteins and RNAs
Cells were transfected with expression constructs for epitope-tagged proteins, induced with 500 μM CuSO4 2 days after transfection, and harvested another 24 hours later. Immunoprecipitation was performed as described (Czech et al., 2008). For RNase treatment, the immunoprecipitates were split into two sets, and one was incubated in lysis buffer with 100 μg/mL RNase A at 4° C for 15 minutes prior to washing. For RNA immunoprecipitation, cells were transfected with protein expression constructs together with pMT-F12, an expression vector for CXCR4 derived from a perfectly base-paired precursor, and induced with 500 μM CuSO4 2 days after transfection. Cells were harvested and cell lysates prepared 24 hours later. About 80% of the cell lysates were subject to immunoprecipitation using M2 agarose beads. After washing, 10% of the immunoprecipitates were analyzed by Western blotting to verify the expression of the epitope-tagged proteins. Total RNAs were extracted from the remaining immunoprecipitates and cell lysates and analyzed by Northern blotting.
Size Exclusion Chromatography
S2-NP cells were lysed in hypotonic buffer [20 mM Hepes (pH 7.0), 2 mM MgCl2, 0.2 mM CaCl2, 1 mM DTT], and spun at 30,000g for 20 minutes. The supernatant was spun at 200,000g for 2.5 hours, and the resulting pellet was subsequently resuspended in buffer A [20 mM Hepes (pH 7.0), 2 mM MgCl2, 1 mM DTT, 0.5% octyl glucoside, 400 mM KCl], and spun at 200,000g for another 2.5 hours. The supernatant was fractionated on Superose-6 HR10/10 (Pharmacia).
Fly Stocks
bel mutant flies (bel6) were from the Bloomington stock center and from Dr. Paul Lasko; EGUF flies were from the Bloomington stock center and from Drs. Thomas Schwarz and Stephen Stowers; and the GMR-wIR and dcr-2L811fsX flies were from Drs. Richard Carthew and Sara Cherry.
Supplementary Material
Acknowledgments
We thank members of the Hannon and Perrimon labs, especially Sara Cherry for helpful discussion, and Julius Brennecke, Ben Czech and Colin Malone for sharing unpublished data and reagents. We also thank the DRSC staff for technical support, the Bloomington stock center, Drs. Paul Lasko, Thomas Schwarz, Stephen Stowers, Richard Carthew and Sara Cherry for fly stocks. We are grateful to Dr. Phillip Sharp for the CXCR4 constructs, and to Dr. Paul Lasko for the Bel antibody. We are indebted to Drs. Bernard Mathey-Prevot and Richard Binari for critically reading the manuscript. R.Z. is supported by a Special Fellowship from the Leukemia and Lymphoma Society. This work was supported by grants from the NIH (N.P. and G.J.H.), and by a kind gift from K. W. Davis (G.J.H.). N.P. and G.J.H. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S, Lum L, Kim M, Paro R, Beachy PA, Green R. A genomewide screen for components of the RNAi pathway in Drosophila cultured cells. Proc Natl Acad Sci U S A. 2006;103:11880–11885. doi: 10.1073/pnas.0605210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Labourier E, Adams MD, Rio DC. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol Cell. 2001;8:363–373. doi: 10.1016/s1097-2765(01)00311-2. [DOI] [PubMed] [Google Scholar]
- Lasko P. The drosophila melanogaster genome: translation factors and RNA binding proteins. J Cell Biol. 2000;150:F51–56. doi: 10.1083/jcb.150.2.f51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK, Mancebo RS, Nagengast AA, Speck O, Psotka M, Mount SM. The Drosophila U1-70K protein is required for viability, but its arginine-rich domain is dispensable. Genetics. 2004;168:2059–2065. doi: 10.1534/genetics.104.032532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.