Abstract
Characterizing combinations of coding polymorphisms (cSNPs), alternative splicing and post-translational modifications (PTMs) on a single protein by standard peptide-based proteomics is challenging owing to <100% sequence coverage and the uncoupling effect of proteolysis on such variations >10–20 residues apart. Because top down MS measures the whole protein, combinations of all the variations affecting primary sequence can be detected as they occur in combination. The protein form generated by all types of variation is here termed the “proteotype”, akin to a haplotype at the DNA level. Analysis of proteins from human primary leukocytes harvested from leukoreduction filters using a dual on-line/off-line top down MS strategy produced >600 unique intact masses, 133 of which were identified from 67 unique genes. Utilizing a two-dimensional platform, termed multidimensional protein characterization by automated top down (MudCAT), 108 of the above protein forms were subsequently identified in the absence of MS/MS in 4 days. Additionally, MudCAT enables the quantitation of allele ratios for heterozygotes and PTM occupancies for phosphorylated species. The diversity of the human proteome is embodied in the fact that 32 of the identified proteins harbored cSNPs, PTMs, or were detected as proteolysis products. Among the information were three partially phosphorylated proteins and three proteins heterozygous at known cSNP loci, with evidence for non-1:1 expression ratios obtained for different alleles.
Mass spectrometry-based proteomics is now a crucial component of modern structural biology.1 The next phase of maturation for interrogation of proteomes involves more than a description of events, but how these diverse events occur in combination, thereby requiring a portion of the proteomic discussion to move from its “descriptive” beginnings (e.g., cataloging expressed proteins and site-specific modifications, etc.) into an integrative, combinatorial mode. Within the human proteome, much discussion is focused on modifications, splice variants, and polymorphisms/mutations; all the combinations of which serve to cloud the biology behind their presence.2,3 Integration of all such complexity into an array of protein forms from a single gene, “the proteotype”, produces molecular information akin to genetecists’ haplotypes. Categorizing the proteotype of a given gene product requires sophisticated mass spectrometric techniques to streamline detection of combinations of coding polymorphisms (cSNPs) and post-translational modifications (PTMs).
Application of established technology to clinically significant groups of samples (clinical proteomics) has ushered into existence the notion of “population proteomics”,4 wherein biomarkers at the protein level are detected and identified for further focus in drug target discovery and clinical diagnostics (e.g., ELISA-based screening).5 The majority of population proteomics studies to date employ standardized proteomic techniques including 2D PAGE-peptide mapping, gel electrophoresis-LC-MS, and MudPIT.4 Additionally, previous studies have focused primarily on individual proteins,6 utilizing immunoaffinity techniques, paying little attention to the “background noise” present in populations. With growing advances in proteomic analysis, which enable enrichment and localization of PTMs,7,8 improved sequence coverage, and the detection of cSNPs,9 the growing biomarker population proteomics field will benefit from implementing these techniques particularly in concert with the above established methods.
Top down MS/MS, the mass spectral analysis of intact molecular protein ions represents a promising method for complete characterization of proteins and large proteolytic peptides (<60 kDa).10 To date, top down has been applied to microorganisms11–13 and cancer cell lines8 for discovery proteomics and further to various tissues for hypothesis-driven protein characterization.7,14 In the above illustrations, top down was shown to precisely characterize unknown PTMs in the presence of genetic and alternative splicing variation, with relative quantitation of related protein forms.8
In order to improve top down proteomics, improved methods for sample handling and intact protein fractionation are required. Various two-dimensional (2D) platforms have been employed previously with moderate success. Recent utilization of anion exchange (AX) shows promise as a first dimension of separation of intact proteins with reversed-phase liquid chromatography (RPLC) as the second dimension of separation for top down MS analysis of the yeast proteome.15 Extension of a top down 2D-LC proteomics platform in discovery mode to populations from primary human material remains challenging; this is the first such report.
Developing the top down platform for clinical populations requires a source of viable and pure cells from a wide variety of subjects. With the increased implementation of leukoreduction (LR) in blood donation centers,16–18 the removal of leukocytes from donated blood components by use of specialized LR filters (LRFs),19 large amounts of leukocytes become available with adequate harvesting protocols. Recent publications report diverse methods for purification of specific cell types from LRFs,20–22 wherein LRFs are “back flushed” at moderate flow rates to free the entrapped leukocytes from the polymer mesh of the LRF.
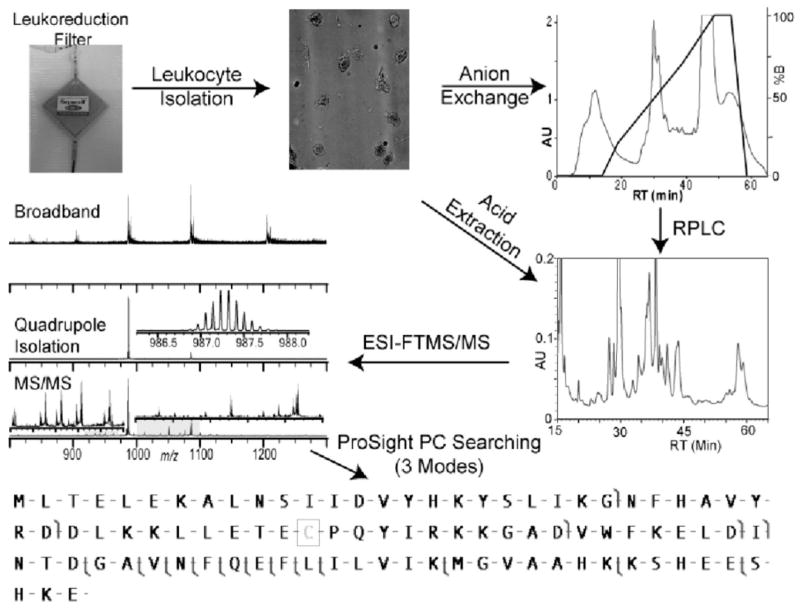
As a first example of top down population proteomics, we utilize leukocyte harvesting from LRFs with subsequent protein separations and MS analysis to enable proteotyping of wild-type proteins from multiple individuals (Figure 1). Application of the 2D top down proteomics platform to leukocytes harvested from LRFs establishes a basis for implementation of top down MS for population proteomics in clinically relevant studies.
Figure 1.
Schematic of the top down MudCAT platform applied to human leukocytes harvested from leukoreduction filters.
EXPERIMENTAL SECTION
Reagents and Leukoreduction Filters
All reagents were purchased from Sigma or Fisher with no modification. Genotyping primers were purchased from Integrated DNA Technologies (Coralville, IA). Prestorage LR of fresh red blood cells was performed by staff at Community Blood Services of Illinois, and only those containing no infectious diseases (e.g., only those that meet donor eligibility requirements) were transferred to the authors (in accord with UIUC IRB project # 06183).
Preparation of Erythrocyte-Free Leukocytes
Fresh LRFs (SepaCell R500, Asahi-Kasei, Tokyo, Japan) were maintained at 4 °C until leukocytes were harvested, always within 24 h of blood donation. Leukocytes were obtained by back-flushing LRFs with 1000 mL of erythrocyte lysis buffer (ELB; 165 mM NH4Cl, 1 mM EDTA, 7 mM K2CO3, pH 7.3) at ~200 mL/min. using a peristaltic pump. Eluate was centrifuged at 800 RCF for 12 min, and the supernatant discarded. Erythrocyte contamination was eliminated by 2 further treatments with 50 mL ELB with centrifugation at 600 RCF for 10 min after each wash. The resulting pellet was composed of 108–109 leukocytes, ~5% of which was stored for follow-up genotyping. Cell pellets were snap-frozen with liquid N2 and stored at −80 °C until lysis.
Genotyping cSNPs
Genotyping pellets were resuspended in ~1 mL of PBS and divided into 5–6 200-μL aliquots. One of these samples was used for DNA extraction following Qiagen’s DNeasy Tissue Handbook 03/2004: Purification of Total DNA from Cultured Animal Cells. For genotyping, the following primers were used: glucose-6-phosphate isomerase (G6PI) exon 6 (SNP at aa 208), 293 bp product: forward, 5′-ACC CCT CAT GGT GAC TGA AG -3′; reverse, 5′-AGG GCA GCT GTA CTG ACC TG -3′. G6PI exon 12 (SNP at aa 308), 936-bp product; forward, 5′-TGG GAG ACA GTG TTG CAG TC-3′; reverse, 5′-TCC CAT GGT GAT CAA ACT CA-3′; acyl CoA binding protein exon 5 (SNPs at aa 88, 92, 103), 223-bp product; forward, 5′-CCC ACC ATC CAC GGT ATT AG -3′, reverse: 5′-CTC TGG AGG CTG CTT GTT TC-3′; Charcot-Leyden crystal protein (CLC) exon 2 (SNP at aa 28), 836-bp product; forward, 5′-AGC TGG GTG TGG ACC AAT AG-3′; reverse, 5′-TTC TCC ATG GGT GGA AAG AG-3′.
Primers were designed using OligoPerfect Designer (http://www.invitrogen.com/). The polymerase chain reaction (PCR) mixture consisted of 200 ng of genomic DNA, 0.2 mM dNTP’s, 1 U Phusion High-Fidelity DNA polymerase (Finnzymes, Espoo, Finland), 0.5 μM each forward and reverse primer, and 10.0 μL of HF buffer (Finnzymes) suspended in a total volume of 50.0 μL. PCR was performed using a Px2 thermal cycler (Thermo Electron Corp., Waltham, MA) using the following conditions: 98.0 °C for 3 min, 35 cycles of 64.4 °C for 30 s followed by 72 °C for 30 s, with a final hold at 72 °C for 5 min. PCR products were purified using QIAquick’s PCR purification and the products sequenced using an ABI 3730XL capillary sequencer (Applied Biosystems, Foster City, CA) at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana–Champaign.
Cell Lysis and 2D-LC
For 2D-LC and 1D-RPLC processing, ~2 × 108 were lysed into 5 mL of lysis buffer (50 mM Tris-HCl, 1 mM AEBSF, 10 mM DTT, pH 8) with 7 cycles of sonication on ice (30 s on, 30 s off), followed by centrifugation at 2000 RCF to pellet debris. Total protein yields were typically 50–100 mg (by Bradford assay) with 10–20 mg loaded for each AX run. For AX runs, a 4.6-mm SynChropak AX300 column (Eichrom Technologies, Darien, IL) was used at a constant flow rate of 0.75 mL/min. at pH 8 with the following gradient: solvent A, 50 mM Tris-HCl, pH 8.0; solvent B, 50 mM Tris-HCl, 750 mM NaCl, pH 8.0; 0–15 min, 0% B; 25 min, 20% B; 55 min, 70% B; 80 min, 100% B. For RPLC analysis, 500 μL of each AX fraction was loaded onto a 4.6-mm Vydac C4 column (Grace Vydac, Columbia, MD) maintained at 50 °C and run at 1 mL/min. with the following gradient conditions: solvent A, 0.1% TFA in H2O; solvent B, 0.08% TFA in acetonitrile; 0–10 min, 5% B; 15 min, 25% B; 63 min, 55% B; 68 min, 95% B. Fractions were collected at 1-min intervals and those absorbing >40 mAu were dried on a SpeedVac for Fourier transform mass spectrometry (FTMS) analysis. Alternatively, lysate (prepared above) or by whole cell acid extraction (AE) with 0.4 N H2SO4 was loaded onto a 4.6-mm Vydac C4 column (Grace Vydac) at 1 mL/min. and fractions were collected at 1-min intervals with the above solvents and the following gradient conditions: 0–10 min, 5% B; 15 min, 30% B; 85 min, 55% B; 95 min, 95% B.
FTMS Analysis
Selected fractions were resuspended in 30–70 μL of electrospray solution (49.5% acetonitrile, 49.5% water, 1% formic acid) and nanoelectrosprayed using a NanoMate 100 (Advion Biosciences, Ithica, NY). The instrument used here was a custom quadrupole-FTMS of the Marshall design23,24 fitted with an ion funnel of the Jarrold design.25 Fractions were subjected to a quadrupole-marching script for precursor ion selection.11 For fragmentation, precursor ions were mass selected using the notch-filtering quadrupole with window sizes 1–4 m/z wide. Fragment ions were produced by collisionally activated dissociation (CAD) in the external octopole,10 infrared multiphoton dissociation (IRMPD),26 and electron capture dissociation27 within the penning trap, with typical spectra comprising 25–100 scans.
Off-Line Data Processing and Protein Identification
Tandem MS data were automatically processed using thorough high-resolation analysis of spectra by Horn (THRASH),27 with resulting peak lists and manually acquired intact masses used for database searching. ProSightPC was used to query a custom version of the UniProt human database that was Shotgun Annotated28 to contain combinations of known protein modifications, cSNPs, and alternative splice variants.8 Typical searches queried the database with an intact mass tolerance of 6000 Da and a fragment ion tolerance of 25 ppm. For those precursor ions not identified using intact mass searching, iterative searching was performed manually using sequence tag and then biomarker searches. A biomarker search within ProSight searches all protein subsequences within the human database with a defined intact mass tolerance (±1 Da); these forms are then queried and scored as described previously29 at 25 ppm. Proteins and peptides were considered identified with an expectation value of <0.01 and considered fully characterized if the intact mass and fragment ions contained no mass discrepancies (Δm’s). For characterization of Δm’s not consistent with PTMs or cSNPs housed in the database, manual characterization was required in single protein mode. All protein-level mass accuracy values given are for the intact mass of the protein unless otherwise specified.
“Middle Down” Analysis of Glucose-6-phosphate Iso-merase
For middle down processing of G6PI, 2D fractions were treated with 1 μg of endoproteinase LysC (Wako Chemicals, Richmond, VA) for 12 h at 37 °C in 50 mM Tris-HCl (pH 9.2). Peptides were analyzed by LC-MS on a 7-T LTQ FT to generate peptide maps. Peptide masses from high mass accuracy FTMS were queried against the Mascot human database (matrixscience-.com) for matches within 8 ppm. This example of middle down MS utilized the 7-T LTQ FT MS, although the 12-T system provides similar data for such an experiment.
On-Line MS on a 12-T LTQ FT
All top down on-line spectra were acquired on a custom 12-T LTQ FT Ultra built in collaboration with Thermo Scientific (San Jose, CA) and fitted with a TriVersa NanoMate in LC coupling mode from Advion Biosciences (Ithaca, NY). Anion-exchange fractions or acid extract from single individuals were loaded onto a 1-mm-i.d. C5 column from Phenomenex (Torrence, CA) thermostated at 55 °C and operating at a flow rate of 80 μL/min. with the following gradient conditions: solvent A, 0.2% formic acid/0.1% TFA in H2O; solvent B, 0.2% formic acid/0.1% TFA in 90% acetonitrile/10% 2-propanol; 0–10 min, 5% B; 90 min, 95% B. A five-segment “ion trap marching” script collected a single full ms scan in the ion trap followed by four consecutive 25 m/z ion trap isolation windows detected in the FT cell (4 microscans, 175 000 resolution). For each ion trap window, the most intense species was targeted for CID MS/MS in the LTQ. These on-line data were processed by THRASH modified for use on LTQ FT LC-MS files. The resulting THRASH files were filtered for redundancy and masses matching within 25 ppm of proteins identified by off-line experiments (performed on the 8.5 T) were identified by intact mass tags (IMTs).30,31
On-Line Quantitation of Heterozygote Expression Ratios from Charcot-Leyden Crystal Protein
Anion-exchange chromatography was performed on proteins from three human beings with collection of the early fractions containing CLC. Anion-exchange fractions containing CLC were combined for on-line LC-MS as described above. A three-segment MS method acquired a full MS ion trap scan (30 microscans), a broadband FTMS scan (4 microscans), and a 10 m/z wide mass selection scan (4 microscans) at 910 m/z (18+ charge state of CLC). Allele ratios were calculated as the quantitative weighted average of summed intensity (QWASI) for the spectrum averaged across the entire region of elution for CLC. Briefly, high-resolution MS absolute intensities of each allele were summed across all charge states of the signal averaged LC-MS scans. Next the allele ratios for each charge state were calculated (Figure S–2b, Supporting Information, SI). The QWASI absolute values were calculated by taking the sum of each ratio weighted for intensity across all charge states for each allele (Figure S–2c, SI), with simple calculations required to generate ratios.
On-Line Quantitation of Calgranulin B Phosphorylation
On-line LC-MS analysis of 400 μL of acid extract from three human subjects was performed as above for each subject. Intact MS scans in the area surrounding the retention time of calgranulin B were summed across the chromatogram. Phosphorylation ratios were generated using the QWASI method described above.
RESULTS AND DISCUSSION
Off-Line MS of a 2D-LC Fraction
Both the anion exchange RPLC and acid extraction RPLC platforms (Figure 1) produce a significant number of fractions containing multiple protein forms. Figure S–1 (SI) illustrates the spectral complexity of the intact MS of a single 2D–LC fraction, with no isotopic clusters observed clearly in the intact MS and two protein forms identified by deconvolution of a low-resolution spectrum. Through selective accumulation of ~25 m/z windows, four intact masses were observed with at least two charge states each, illustrating the enhancement in dynamic range by using this “quadrupole marching” approach.10 All four of these forms were identified by top down MS/MS (Figure S–1, bottom).
Characterization of Heterozygotes
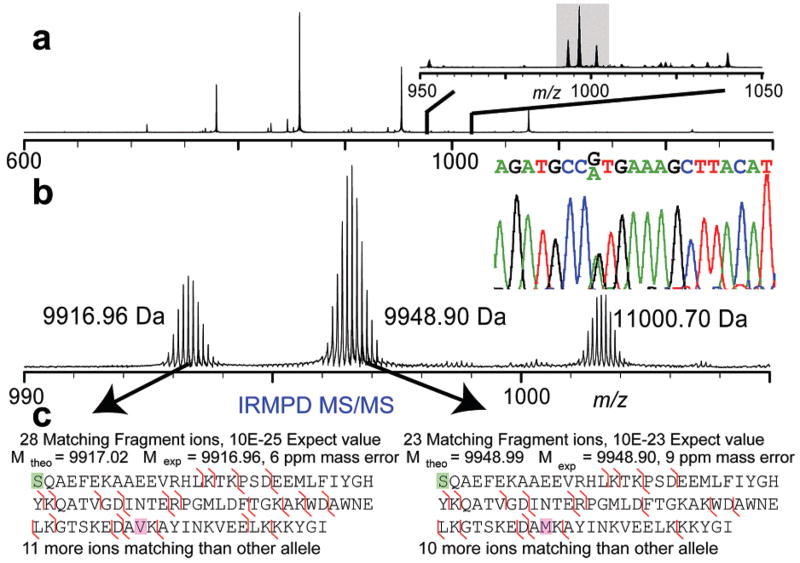
We present here top down characterization of two forms of acyl-CoA binding protein generated from 2D–LC of lysate produced from a single-subject leukocyte pellet (Figure 2). These forms are produced from the expression of heterozygous alleles at a single cSNP locus. Each allele is fully characterized by IRMPD MS/MS, with each individual allele yielding >10 matching ions more than the alternate allele. As illustrated, these alleles are not present at equivalent ratios; instead they are expressed at a ~2:3 ratio across all charge states and in all 2D–LC fractions in which they were observed. Given the similar nature of the amino acids involved (small, nonpolar, similar pKa), it is unlikely that this ratio arises as a result of ionization efficiency differences for this protein. Genotyping this subject at this locus indicates heterozygosity at the position of interest (Figure 2b, inset). To minimize bias, quantitation of these related protein forms was performed using low-resolution spectra in lieu of isotopically resolved spectra.30 Top down MS has previously been shown to fully characterize proteins from HeLa cells containing known coding polymorphisms;8 however, only in limited examples has MS-based analysis enabled protein-level mutation analysis6 and always on single protein targets. This first example of proteotyping heterozygotes by top down MS illustrates the utility of this technique in complex mixtures such as the human leukocyte proteome.
Figure 2.
Characterization of heterozygotes of acyl-CoA binding protein. (a) Intact MS spectrum illustrating enrichment by quadrupole isolation yielding ~85-fold S/N improvement (inset). (b) Expanded view of isolation window with corresponding low-resolution spectrum (inset b, left), illustrating the ~2:3 ratio of alleles, consistent across charge states and fractions containing this protein; DNA microsequencing results at loci in question illustrating both alleles present (inset b, right). (c) IRMPD MS/MS details of each individually fragmented allele localizing the (−31 Da) Δm to three residues containing the cSNP known to have a ~5% population frequency for the minor allele.
Intact Protein Identification with Improved Mass Range
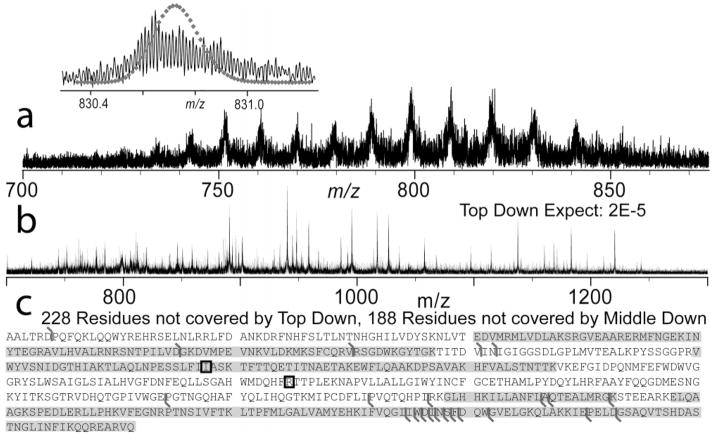
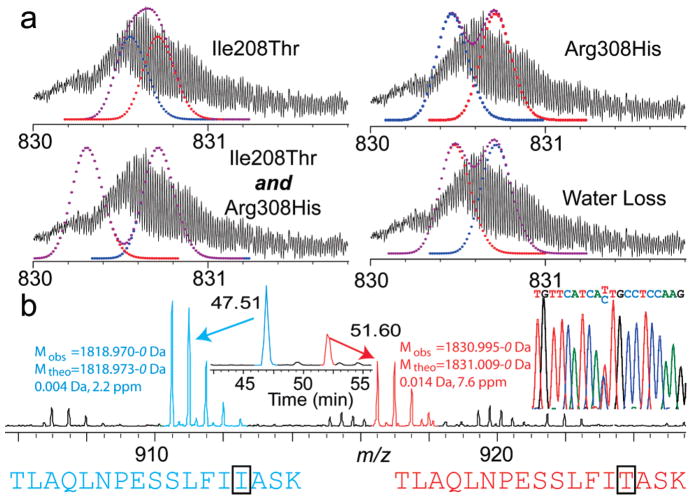
Previous top down experiments in discovery mode (i.e., not using highly purified samples) has generally been limited to proteins of <50 kDa. In this application of top down to human leukocytes from healthy donors, we identified endogenous G6PI, a 63-kDa protein, using CAD with an expectation value of 2 × 10−5 (Figure 3). With high-resolution MS, the intact isotopic distribution appears broadened toward the low mass side. Further analysis of these data indicates overlapping distributions with the existence of a second, lower mass form (Δm ~10 Da). Both cSNPs known on this protein yield lower mass products (Ile208Thr, Δm = −12.04 Da; Arg308His, Δm = −19.05 Da); calculation of possible distributions (Figure 4a) indicated the likely presence of both Ile208 and Thr208 alleles. Digestion of the 2D–LC purified protein with LysC for peptide mapping achieved 63% sequence coverage. The SNP at position 208 was covered, and the data indicated a heterozygous genotype at this locus with peptides matching within 2 and 8 ppm, respectively (Figure 4b). Genotyping of the locus containing each SNP confirmed the heterozygous genotype at position 208 (Figure 4b, inset) and the homozygous allele at position 308.
Figure 3.
Top down identification of G6PI, the largest protein as yet identified in top down for proteome-wide analysis. (a) Broadband MS illustrating minimal contamination and isotopic resolution using quad-SWIFT isolation with overlaid theoretical isotopic distribution for unmodified G6PI (inset). (b) CAD MS/MS of the intact species of a) inset. (c) Details of matching MS/MS ions from the intact protein (black flags) and peptides identified by “middle down” peptide analysis (highlighted in gray) for further protein characterization with known, validated cSNPs sites have a box around the amino acid position.
Figure 4.
Confirming a heterozygous genotype of G6PI. (a) Possible isotopic combinations of anticipated forms of G6PI. (b) LysC peptides of G6PI bearing the SNP at aa position 208. These two peptides indicate expression of both alleles within the accepted error tolerances of the LTQ FT; DNA microsequencing data are also shown for the locus containing the cSNP site for G6PI at aa position 208 (inset).
On-Line MS for Rapid Protein Characterization
It has been shown that proteins characterized by top down MS/MS may be identified using accurate intact mass alone (e.g., the intact mass tag or IMT approach).30,31 This approach entailed first identifying a protein form by either on- or off-line MS/MS; these identified species were then entered into a leukocyte-specific database. Upon subsequent observation by on-line MS within a tolerance of 25 ppm, these forms are rapidly identified by the IMT approach. Note that this IMT mass tolerance will shrink to ~5 ppm when both instruments used have automatic gain control. Off-line MS/MS analysis of human leukocytes provided a list of 133 protein forms from 67 unique genes, the intact masses from which are entered into a proteome-specific exclusion list (Table 1). Utilizing multi-dimensional protein characterization by automated top down (MudCAT), 32 anion-exchange fractions and the acid extract provided identification of 108 protein forms from 53 genes by online MS/MS in 4 days of instrument time. This method enables improved throughput for profiling of cSNPs, PTMs, and expression ratios on an LC time scale without the requirement for MS/MS.
Table 1.
Summary Table of All Proteins and Peptides Identified
| ID | uniprot # | Δm | M ob (kDa) | expect | function | % coverage | method used | on-line IMT |
|---|---|---|---|---|---|---|---|---|
| 1 | P63257 | 0 | 4.4 | 2.4 × 10−17 | actin, γ, residues 1–42, N-term acetyl | 11 | AE | |
| 2 | Q96Q14 | 0 | 4.8 | 8.0 × 10−5 | signal recognition particle 14kD, residues 2- 42 | 30 | 2D | |
| 3 | P63313 | 0 | 4.9 | 9.3 × 10−5 | thymosin β-10 | 1D, AE | ||
| 4 | P62328 | 0 | 5.0 | 2.9 × 10−7 | thymosin β-4 | 1D, AE | ||
| 5 | Q15417 | 0 | 5.4 | 7.3 × 10−7 | calponin-3, acidic isoform, residues 150–198 | 15 | AE | |
| 6 | Q2YD73 | 0 | 6.2 | 2.0 × 10−4 | coronin, actin binding protein 1A, residues 407461 | 12 | 1D | |
| 7 | P02788 | 0 | 7.5 | 6.0 × 10−6 | lactotransferrin, growth-inhibiting protein 12 residues 269–336 | 10 | 2D | X |
| 8 | Q15843 | 0 | 8.6 | 1.0 × 10−14 | none NEDD8, (ubiquitin-like protein Nedd8) (neddylin) | AE | X | |
| 9 | P62988 | 0 | 8.6 | 1.0 × 10−12 | ubiquitin | 1D, 2D, AE | X | |
| 10 | P35579 | 0 | 9.0 | 1.0 × 10−11 | myosin-9, residues 1650−1728 | 4 | 2D | X |
| 11 | Q0VGD5 | 0 | 9.3 | 4.4 × 10−6 | high-mobility group nucleosomal binding domain 2, HMG-17 | AE | X | |
| 12 | P15502 | 0 | 9.5 | 1.4 × 10−5 | elastin, tropoelastin, residues 603–708 | 13 | AE | |
| 13 | P84243 | 0 | 9.6 | 6.5 × 10−24 | histone H3.3, residues 53–135 | 41 | AE | X |
| 14 | P07108 | 0 | 9.9 | 7.0 × 10−25 | acyl-CoA-binding protein (ACBP), variant 2 | 1D, 2D, AE | ||
| 15 | P07108 | 0 | 9.9 | 8.0 × 10−23 | acyl-CoA-binding protein (ACBP), variant 1 | 1D, 2D, AE | X | |
| 16 | Q5RHS4 | 0 | 10.1 | 3.6 × 10−5 | S100-A6, calcyclin | AE | X | |
| 17 | Q3LUA8 | 0 | 10.2 | 2.0 × 10−3 | phospholipase C-eta2, residues 1209→1304 | 7 | 1D | |
| 18 | P80511 | 2 | 10.4 | 1.1 × 10−16 | S100-A12, calgranulin C CAGC CGRP | 1D, 2D, AE | X | |
| 19 | P07910 | 20 | 10.4 | 2.6 × 10−4 | heterogeneous nuclear ribonucleoproteins C1/C2, residues 11–107 | 32 | AE | X |
| 20 | P05109 | 0 | 10.8 | 1.2 × 10−18 | S100-A8, calgranulin A, MRP8 | 1D, 2D, AE | X | |
| 21 | P61604 | 0 | 10.8 | 7.0 × 10−03 | 10-kDa heat shock protein | 1D, 2D, AE | X | |
| 22 | P62805 | 0 | 11.3 | 2.7 × 10−8 | histone H4 + 28 form | 1D, AE | X | |
| 23 | Q5Y190 | 0 | 11.3 | 2.5 × 10−6 | anchor protein, residues 3486→3593 | 3 | 1D, AE | X |
| 24 | P62805 | 0 | 11.3 | 6.3 × 10−8 | histone H4, +70 Da | 1D, AE | X | |
| 25 | P11021 | 42 | 11.4 | 7.2 × 10−11 | 78 kDa glucose-regulated protein precursor, heat shock 70-kDa protein 5, residues 549- | 15 | AE | X |
| 26 | P26447 | 0 | 11.6 | 7.8 × 10−10 | S100-A4, metastasin, calvasculin | AE | X | |
| 27 | P31949 | 0 | 11.6 | 3.3 × 10−22 | S100-A11, calgizzarin | AE | X | |
| 28 | P62942 | 0 | 11.8 | 5.3 × 10−3 | FK506-binding protein 1A (peptidylprolyl cis-trans isomerase) | AE | X | |
| 29 | P16403 | 0 | 12.0 | 6.4 × 10−13 | histone–1.2, residues 1–120 | 57 | AE | X |
| 30 | P10412 | 0 | 12.0 | 2.1 × 10−11 | histone H1.4, residues 1–120 | 55 | AE | X |
| 31 | P11678 | 2 | 12.2 | 8.8 × 10−10 | eosinophil peroxidase precursor, residues 140–244 | 15 | AE | X |
| 32 | P99999 | 615 | 12.3 | 6.5 × 10−11 | cytochrome C | 2D | X | |
| 33 | P16401 | 0 | 12.3 | 2.9 × 10−5 | histone H1.5, residues 1–123 | 55 | AE | X |
| 34 | Q71DI3 | 0 | 12.4 | 2.5 × 10−16 | histone H3.2, residues 28–135 | 79 | AE | X |
| 35 | O75368 | 0 | 12.6 | 2.0 × 10−5 | SH3 domain-binding glutamic acid-rich-like protein | 1D, AE | X | |
| 36 | P06702 | 0 | 12.7 | 1.0 × 10−7 | S100-A9, calgraulin B, MRP14, alternative start Met | 1D, 2D, AE | X | |
| 37 | P68431 | 0 | 13.1 | 2.9 × 10−13 | histone H3.1, residues 21–135 | 85 | AE | X |
| P06702 | 0 | 13.1 | 1.0 × 10−6 | S100-A9, calgraulin B, MRP14 (full length) | 1D, 2D, AE | X | ||
| 38 | P62316 | 0 | 13.4 | 4.8 × 10−4 | small nuclear ribonucleoprotein Sm D2 | AE | X | |
| 39 | P49773 | 0 | 13.7 | 1.1 × 10−12 | histidine triad nucleotide-binding protein 1 | AE | X | |
| 40 | Q93079 | 0 | 13.8 | 1.0 × 10−4 | histone H2B type 1-H (H2B.j) | AE | X | |
| 41 | P23527 | 0 | 13.8 | 1.4 × 10−11 | histone H2B type 1-O (H2B.n) | AE | X | |
| 42 | Q96KK5 | 0 | 13.8 | 1.8 × 10−16 | histone H2A type 1-H | AE | X | |
| 43 | Q99878 | 0 | 13.8 | 3.5 × 10−12 | histone H2A type 1-J | AE | X | |
| 44 | Q99877 | 0 | 13.9 | 1.0 × 1−19 | histone H2B type 1-N (H2B.d) (H2B/d). | AE | X | |
| 45 | Q16777 | 0 | 13.9 | 5.5 × 10−17 | histone H2A type 2-C (H2A-GL101) (H2A/r) | AE | X | |
| 46 | P0C0S8 | 0 | 14.0 | 1.0 × 10−9 | histone H2A type 1 (H2A.1). | AE | X | |
| 47 | Q6FI13 | 0 | 14.0 | 6.8 × 10−8 | histone H2A type 2-A (H2A.2) | AE | X | |
| 48 | Q93077 | 0 | 14.0 | 4.2 × 10−7 | histone H2A type 1-C | AE | X | |
| 49 | P00338 | 2 | 14.4 | 2.0 × 10−4 | L-lactate dehydrogenase A chain, residues 70–199 | 39 | 2D | |
| 50 | Q5T0I0 | 0 | 14.8 | 6.7 × 10−18 | gelsolin (amyloidosis, Finnish type), residues 2–133 | 51 | 2D | |
| 51 | P07737 | 0 | 15.0 | 1.0 × 10−8 | profilin-1 | 1D, AE | X | |
| 52 | Q59EJ3 | 2 | 15.1 | 1.0 × 10−7 | heat shock 70kDa protein 1A variant, residues 571–709 | 20 | 2D | |
| 53 | P01922 | 0 | 15.1 | 1.0 × 10−25 | hemoglobin, α chain | 1D, 2D, AE | X | |
| 54 | P02023 | 0 | 15.9 | 1.0 × 10−33 | hemoglobin β chain | 1D, 2D, AE | X | |
| 55 | P00441 | 0 | 15.9 | 9.5 × 10−4 | superoxide dismutase [Cu-Zn] | AE | X | |
| 56 | Q05315 | 0 | 16.4 | 1.0 × 10−7 | Charcot-Leyden crystal protein | 2D | X | |
| 57 | Q6IB37 | 0 | 16.7 | 6.9 × 10−3 | glia maturation factor, γ (GMFG protein) | AE | ||
| 58 | P62937 | 0 | 17.9 | 3.0 × 10−4 | peptidylprolyl isomerase A (cyclophilin A) | 2D, AE | X | |
| 59 | P23528 | 0 | 18.4 | 3.0 × 10−4 | cofilin-1 (cofilin, nonmuscle isoform) | AE | X | |
| 60 | P59998 | 0 | 19.6 | 3.0 × 10−2 | actin-related protein 2/3 complex subunit 4 | 2D | ||
| 61 | Q99497 | 0 | 19.8 | 2.0 × 10−6 | protein DJ-1 (oncogene DJ1). | 2D | ||
| 62 | P30086 | 0 | 20.9 | 1.1 × 10−12 | phosphatidylethanolamine-binding protein 1 (PEBP-1) | 2D | X | |
| 63 | P04179 | 0 | 22.2 | 6.8 × 10−17 | superoxide dismutase [Mn], SOD2 | AE | ||
| 64 | P52566 | 0 | 22.9 | 9.0 × 10−6 | ρ GDP-dissociation inhibitor 2 (ρ GDI 2), (ρ-GDI beta) | AE | X | |
| 65 | Q5U071 | 0 | 23.6 | 8.7 × 10−10 | high-mobility group box 2 | AE | X | |
| 66 | Q6FHP9 | 1 | 26.5 | 3.0 × 10−4 | triosephosphate isomerase | 2D | X | |
| 67 | Q6FHU2 | 202 | 28.9 | 1.4 × 10−4 | phosphoglycerate mutase 1, PGAM | 2D | ||
| 68 | P04406 | 0 | 35.9 | 5.0 × 10−4 | glyceraldehyde-3-phosphate dehydrogenase | 2D | X | |
| 69 | P06744 | 0 | 63.0 | 1.4 × 10−6 | glucose-6-phosphate isomerase | 2D | X |
Quantitation of Heterozygotes in a Population
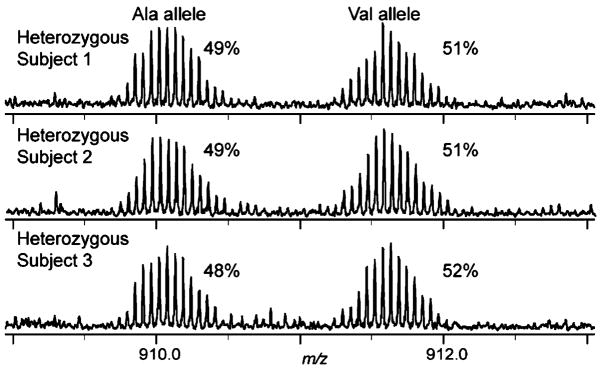
The ability to obtain ratios of modified protein forms by top down MS has been demonstrated previously.7 Quantitation of alleles by top down MS has been described above although not for multiple individuals with the identical genotype (i.e., multiple individuals that are heterozygous at a single position). We present here the quantitative weighted average of summed intensities (i.e., QWASI; see Experimental Section) of three distinct human subjects with heterozygous genotype at the locus encoding a cSNP on the sequence of Charcot-Leyden crystal protein by on-line MS (Figure 5). These ratios were obtained for species identified utilizing the IMT approach with accurate mass alone (no MS/MS). The QWASI ratios of the three heterozygotes indicate a 1:1 ratio of alleles expressed with no significant variation among the individuals, substantiating the applicability of top down MS for allele quantitation.
Figure 5.
Ratios of heterozygotes determined from multiple individuals. On-line 12-T intact MS from MudCAT of three unique individuals with heterozygous genotype at aa position 28 producing both Val28 and Ala28-containing protein forms. Percentage values were calculated using the QWASI method from Figure S-2 (SI).
Quantitation of Phosphorylation Occupancy
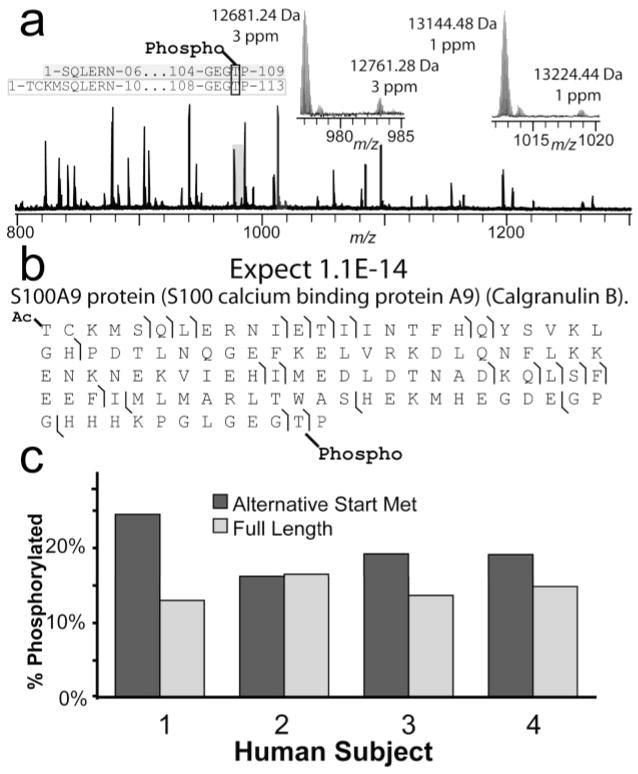
Utilizing MudCAT, the percent occupancy of a partially phosphorlyated species was determined. Within a complex region of the LC-MS chromatogram, two protein forms with +80-Da satellite peaks were observed, with the unmodified forms differing by 463 Da (Figure 6a). Top down MS/MS identifies these four forms as stemming from the gene encoding calgranulin B, a member of the s100 calcium-binding family. The two unmodified protein forms stem from alternate start Met codons, generating two protein forms differing by four residues at the N-terminus (Figure 6a). Further, the MS/MS data precisely localize the phosphorylation to the penultimate residue of the protein (Figure 6b). MudCAT was utilized to obtain this proteotype from four different human subjects. Figure 6c illustrates the phosphorylation levels for all individuals for each protein form, with phosphorylation levels constant at ~15% for the full-length form, with ~25% occupancy detected for the truncated form (n = 1).
Figure 6.
Phosphorylation occupancy levels determined by MudCAT. (a) Broadband MS of a single human subject illustrating two protein forms with +80-Da satellite peaks. (b) MS/MS details of the +80-Da satellite peak at 13224.44 Da (a, right inset) showing precise localization of the phosphorylation on a Thr residue near the C-terminus. (c) Graphical depiction of phosphorylation occupancies for 4 individuals for both calgranulin B protein forms.
Given that relatively few phosphorylated peptides coelute with the unmodified peptide, quantitation of phosphorylation occupancy using bottom up on specific sites is not often achieved. Further, phosphopeptide analysis typically takes advantage of phosphorylation-specific chromatography, often eliminating the unmodified peptide from the mixture. In the case of the calgranulin B proteotyping example above, any proteolysis-based approach would disconnect the phosphorylation events from the N-terminal differences, collapsing all phosphorylation information into a single peptide at the C-terminus. The ability to differentiate these two protein forms while obtaining phosphorylation levels highlights the characterization power unique to top down MS/MS.
Summary of Proteins Identified
In the first application of top down MudCAT analysis to primary human leukocytes, 621 unique protein forms were observed (≥3 times, nonredundant). Identification of 133 of these forms from 67 unique genes was accomplished by MS/MS, 70% requiring no manual validation. Using the above identifications, 108 protein forms from 53 genes were reidentified in just 4 days, the highest throughput to date for top down MS. The use of diverse methods for protein preparation prior to RPLC improves proteome coverage, increasing the odds of observing proteins of interest. Table 1 shows the methods employed to purify each identified protein form, illustrating the complementarity of 2D-LC, 1D-LC, and AE-LC, and denotes the proteins automatically identified by IMTs in an on-line fashion. The overlap between the 1D-RPLC, AE-RPLC, and 2D-LC platforms indicates the significant contamination of highly abundant proteins in any 2D run, but highlights the additional proteins not observed by single-dimension processing. Furthermore, the additional forms observed in the AE-RPLC may indicate protein loss as a result of 2D processing, again illustrating the complimentarity of protein handling methods. Among the proteins identified, 21% contain known, validated cSNPs (Table 2). Additionally, modifications including phosphorylation, acetylation (not N-terminal), heme, and biological truncations were observed in 17% of the proteins identified (Table 2).
Table 2.
Protein Forms Harboring cSNPs and PTMs
| ID UniProt # | Δm | Mobs | Mtheo | expect | function | notes | validated SNPs | %coverage | method used | |
|---|---|---|---|---|---|---|---|---|---|---|
| 70 | P63313 | 0 | 4933.5 | 4933.5 | 9.3E-53 | thymosin β-10 | M7R (0.32) | 1D, AE | ||
| 71 | Q2YD73 | 0 | 6176.1 | 6176.2 | 2.0E-04 | coronin, actin binding protein 1A, residues 407461 | proteolysis product | T443P (0.04) | 12 | 1D |
| 72 | Q15843 | 0 | 8554.6 | 8554.7 | 1.0E-14 | none NEDD8, (ubiquitin-like protein Nedd8) (Neddylin) | loss of C-term 5 residues | AE | ||
| 73 | P15502 | 0 | 9529.1 | 9529.1 | 1.4E-05 | elastin, tropoelastin, residues 603–708 | proteolysis product | G684R (0.026) | 13 | AE |
| 74 | P07108 | 0 | 9917.0 | 9917.0 | 7.0E-25 | acyl-CoA-binding protein (ACBP), variant 2 | heterozygote | M88V (0.04), G103R (0.02) | 1D, 2D, AE | |
| 75 | Q5RHS4 | 0 | 10084.3 | 10084.3 | 3.6E-05 | S100-A6, calcyclin | G90D (0.04) | AE | ||
| 76 | P62805 | 0 | 11299.5 | 11299.4 | 2.7E-08 | histone H4 +28 form | K22 dimethylation | 1D, AE | ||
| P62805 | 0 | 11341.2 | 11341.4 | 6.3E-08 | histone H4, +70 Da | acetylated+dimethylated | 1D, AE | |||
| 77 | P16403 | 0 | 11979.7 | 11979.8 | 6.4E-13 | histone H1.2, residues 1–120 | proteolysis product | A18V (0.32) | 57 | AE |
| 78 | P11678 | 2 | 12155.3 | 12157.2 | 8.8E-10 | eosinophil peroxidase precursor, residues 140–244 | proteolysis product, disulfide | 15 | AE | |
| 79 | P99999 | 615 | 12267.5 | 11652.1 | 6.5E-11 | cytochrome c | heme group | 2D | ||
| 80 | Q71DI3 | 0 | 12356.9 | 12356.7 | 2.5E-16 | histone H3.2, residues 28–135 | proteolysis product | D78E (0.46), S97R (0.5) | 79 | AE |
| 81 | P06702 | 0 | 12681.2 | 12681.3 | 1.0E-07 | S100-A9, calgraulin B, MRP14, alternative start Met | alternative start site, partial phosphorylation | 1D, 2D, AE | ||
| P06702 | 0 | 13144.3 | 13144.5 | 1.0E-06 | S100-A9, calgraulin B, MRP14(full length) | partial phosphorylation | 1D, 2D, AE | |||
| 82 | P00338 | 2 | 14398.6 | 14400.7 | 2.0E-04 | L-lactate dehydrogenase A chain, residues 70–199 | proteolysis product | S161R (0.49) | 39 | 2D |
| 83 | Q5T0I0 | 0 | 14819.5 | 14819.5 | 6.7E-18 | gelsolin (amyloidosis, Finnish type), residues 2–133 | A129T (0.118) | |||
| 84 | Q59EJ3 | 2 | 15092.3 | 15094.5 | 1.0E-07 | heat shock 70-kDa protein1A variant, residues 571–709 | proteolysis product, disulfide | 20 | 2D | |
| 85 | Q05315 | 0 | 16381.1 | 16381.2 | 1.0E-07 | Charcot-Leyden crystal protein | heterozygote | A28V (0.49) | 2D | |
| 86 | Q6IB37 | 0 | 16701.4 | 16701.4 | 6.9E-03 | glia maturation factor, γ (GMFG protein) | T15K (0.05) | AE | ||
| 87 | P23528 | 0 | 18401.5 | 18401.6 | 3.0E-04 | cofilin-1 (cofilin, non muscle isoform) | partial phosphorylation | AE | ||
| 88 | P04179 | 0 | 22190.3 | 22190.2 | 6.8E-17 | superoxide dismutase [Mn], SOD2 | signal peptide | R156W 0(0.01), G76R (0.01), E66V (0.01) | AE | |
| 89 | Q5U071 | 0 | 23629.4 | 23629.6 | 8.7E-10 | high-mobility group box 2 | loss of C-term EE | AE | ||
| 90 | P06744 | 0 | 63017.0 | 63017.0 | 1.4E-06 | glucose-6-phosphate isomerase | Heterozygote | I208T (0.03); R308H (0.1) | 2D |
Conclusions
The MudCAT platform presented here is the highest throughput protein characterization engine for top down reported to date and represents the first major step toward fully automated on-line top down MS analysis. Several challenges for top down methodology yet remain and have been recently reviewed.32 A few are addressed below including the “front end” problem of how to fractionate and effectively ionize intact proteins in complex mixtures. Top down MS has seen significant advances since the initial reports aimed at characterization of the primary sequence of individual proteins. For example, the 12-T LTQ FT system presented here has driven a large increase in throughput for top down LC/MS/MS and used alone could generate the data required for all protein identifications and characterizations shown here. Commercially available high-performance mass spectrometers continue to evolve with improving magnetic field strength and increasingly sophisticated software engines for smart acquisition of high-resolution MS/MS data on-the-fly. Software for high-throughput analysis of such top down and “middle down” data is keeping pace with these automated engines capable of interpreting “high res/high res” MS/MS data sets. ProSightHT is now capable of batch-processing large volumes of data to generate top down identification lists with no manual intervention or interpretation. This is a key step toward improving the top down methodology for general usage among the proteomic community. Given rich MS/MS spectra (i.e., cleavage at many backbone positions), the strategy of housing known protein information in a database28 allows automated interpretation of MS/MS spectra from proteins harboring multiple modifications or polymorphisms.8 Thus, the MudCAT approach is well positioned for implementation by protein analysts to generate highly informative, molecular-level characterization of alleles and modifications to enable tighter phenotypic correlations.
Supplementary Material
Acknowledgments
The authors are grateful to Patrick Kovar, Bobbie Burr, and particularly the donors with Community Blood Services of Illinois, Steve Horning, Mike Senko, Chris Hendrickson, Paul Thomas, and Craig Wenger for assistance with constructing the 12-T LTQ FT. This work was supported by the National Institutes of Health (GM 067193), the Henry and Lucille Packard Foundation, the Sloan Foundation, the Dreyfus Teacher-Scholar Award, and the Center for Cell-Cell Signaling and NeuroProteomics at the University of Illinois supported by the National Institutes of Health (P30 DA018310).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 2.Hondermarck H. Mol Cell Proteomics. 2003;2:281–291. doi: 10.1074/mcp.R300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Mann KG, Brummel-Ziedins K, Undas A, Butenas S. J Thromb Haemost. 2004;2:1727–1734. doi: 10.1111/j.1538-7836.2004.00958.x. [DOI] [PubMed] [Google Scholar]
- 4.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Mol Cell Proteomics. 2006;5:1811–1818. doi: 10.1074/mcp.R600006-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 6.Nepomuceno AI, Mason CJ, Muddiman DC, Bergen HR, 3rd, Zeldenrust SR. Clin Chem. 2004;50:1535–1543. doi: 10.1373/clinchem.2004.033274. [DOI] [PubMed] [Google Scholar]
- 7.Pesavento JJ, Mizzen CA, Kelleher NL. Anal Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 8.Roth MJ, Forbes AJ, Boyne MT, 2nd, Kim YB, Robinson DE, Kelleher NL. Mol Cell Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunger MK, Cargile BJ, Sevinsky JR, Deyanova E, Yates NA, Hendrickson RC, Stephenson JL., Jr J Protein Res. 2007;6:2331–2340. doi: 10.1021/pr0700908. [DOI] [PubMed] [Google Scholar]
- 10.Patrie SM, Ferguson JT, Robinson DE, Whipple D, Rother M, Metcalf WW, Kelleher NL. Mol Cell Proteomics. 2006;5:14–25. doi: 10.1074/mcp.M500219-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Forbes AJ, Patrie SM, Taylor GK, Kim YB, Jiang L, Kelleher NL. Proc Natl Acad Sci USA. 2004;101:2678–2683. doi: 10.1073/pnas.0306575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reid GE, Shang H, Hogan JM, Lee GU, McLuckey SA. J Am Chem Soc. 2002;124:7353–7362. doi: 10.1021/ja025966k. [DOI] [PubMed] [Google Scholar]
- 13.Sharma S, Simpson DC, Tolic N, Jaitly N. J Prot Res. 2007;6:602–610. doi: 10.1021/pr060354a. [DOI] [PubMed] [Google Scholar]; Analytical Chemistry vol 80. 2008 April;15(8):2857. doi: 10.1021/ac800141g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romanova EV, Roth MJ, Rubakhin SS, Jakubowski JA, Kelley WP, Kirk MD, Kelleher NL, Sweedler JV. J Mass Spectrom. 2006;41:1030–1040. doi: 10.1002/jms.1060. [DOI] [PubMed] [Google Scholar]
- 15.Parks BA, Jiang L, Thomas PM, Wenger CD, Roth MJ, Boyne MT, II, Burke PV, Kwast KE, Kelleher NL. Anal Chem. 2007;79:7984–7991. doi: 10.1021/ac070553t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackall DP. Curr Hematol Rep. 2003;2:493–494. [PubMed] [Google Scholar]
- 17.Pietersz RN. Transfusion Apheresis Sci. 2001;25:209–210. doi: 10.1016/s1473-0502(01)00128-8. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro MJ. Crit Care. 2004;8:S27–30. doi: 10.1186/cc2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dzik WH. Curr Opin Hematol. 2002;9:521–526. doi: 10.1097/00062752-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 20.Dietz AB, Bulur PA, Emery RL, Winters JL, Epps DE, Zubair AC, Vuk-Pavlovic S. Transfusion. 2006;46:2083–2089. doi: 10.1111/j.1537-2995.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 21.Meyer TP, Zehnter I, Hofmann B, Zaisserer J, Burkhart J, Rapp S, Weinauer F, Schmitz J, Illert WE. J Immunol Methods. 2005;307:150–166. doi: 10.1016/j.jim.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Teleron AA, Carlson B, Young PP. Transfusion. 2005;45:21–25. doi: 10.1111/j.1537-2995.2005.04191.x. [DOI] [PubMed] [Google Scholar]
- 23.Patrie SM, Charlebois JP, Whipple D, Kelleher NL, Hendrickson CL, Quinn JP, Marshall AG, Mukhopadhyay B. J Am Soc Mass Spectrom. 2004;15:1099–1108. doi: 10.1016/j.jasms.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Senko MW, Hendrickson CL, Pasa-Tolic L, Marto JA, White FM, Guan S, Marshall AG. Rapid Commun Mass Spectrom. 1996;10:1824–1828. doi: 10.1002/(SICI)1097-0231(199611)10:14<1824::AID-RCM695>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 25.Julian RR, Mabbett SR, Jarrold MF. J Am Soc Mass Spectrom. 2005;16:1708–1712. doi: 10.1016/j.jasms.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 27.Horn DM, Zubarev RA, McLafferty FW. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 28.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. J Am Chem Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor GK, Kim YB, Forbes AJ, Meng F, McCarthy R, Kelleher NL. Anal Chem. 2003;75:4081–4086. doi: 10.1021/ac0341721. [DOI] [PubMed] [Google Scholar]
- 30.Du Y, Parks BA, Sohn S, Kwast KE, Kelleher NL. Anal Chem. 2006;78:686–694. doi: 10.1021/ac050993p. [DOI] [PubMed] [Google Scholar]
- 31.Gomez SM, Nishio JN, Faull KF, Whitelegge JP. Mol Cell Proteomics. 2002;1:46–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Siuti NS, Kelleher NL. Nat Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.