SUMMARY
Cryo-electron microscopy (cryoEM) studies of the bacteriophage ϕ29 DNA packaging motor have delineated the relative positions and molecular boundaries of the 12-fold symmetric head-tail connector, the 5-fold symmetric prohead RNA (pRNA), the ATPase that provides the energy for packaging, and the procapsid. Reconstructions, assuming 5-fold symmetry, were determined for proheads with 174-base, 120-base, 71-base pRNA, proheads lacking pRNA; proheads with ATPase bound, and proheads in which the packaging motor was missing the connector. These structures are consistent with pRNA and ATPase forming a pentameric motor component around the unique vertex of proheads. They suggest an assembly pathway for the packaging motor, and a mechanism for DNA translocation into empty proheads.
INTRODUCTION
Linear dsDNA viruses, including tailed bacteriophages, adenoviruses, herpesviruses, and poxviruses, first package their genomes into preformed protein shells (Anderson and Reilly, 1993; Bazinet and King, 1985; Black, 1989; Casjens and Hendrix, 1988) and later inject their DNA, via elaborate tail structures in the case of bacteriophages, into the cytoplasm of the host cell (Kostyuchenko et al., 2005; Leiman et al., 2004; Molineux, 2001; Xiang et al., 2006). The process of genome encapsidation by dsDNA viruses is remarkable considering the entropic, electrostatic, and bending energies of DNA that must be overcome in order to package DNA to near crystalline densities (Chang et al., 2006; Fokine et al., 2004; Jiang et al., 2006; Lander et al., 2006; Xiang et al., 2006). Phage DNA packaging motors are among the most powerful biological motors known, capable of generating forces in excess of 57 picoNewtons (Smith et al., 2001). DNA packaging is driven by a molecular motor that converts the energy obtained from ATP hydrolysis to mechanical energy required for translocation of DNA into proheads (Jardine and Anderson, 2006). The viral ATPases that power genome encapsidation belong to a large family of ATPases whose members are involved in various DNA remodeling tasks including cell division, chromosome segregation, DNA recombination, strand separation, and conjugation (Mitchell et al., 2002; Sun et al., 2007).
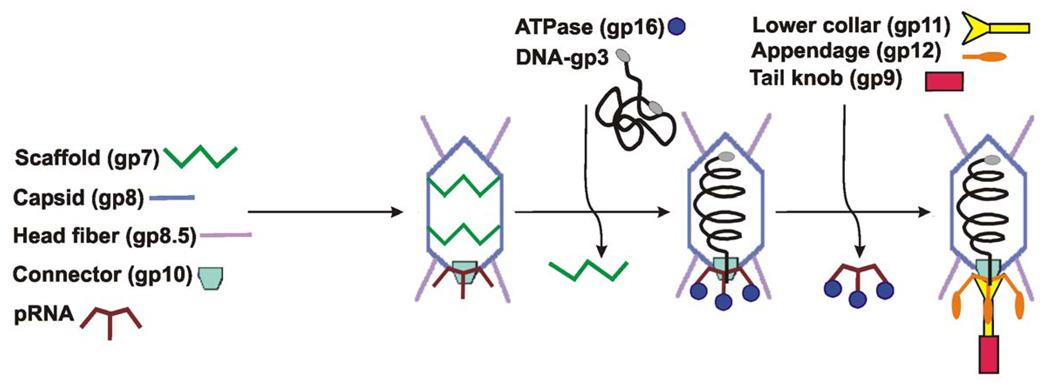
Bacteriophage ϕ29, a tailed dsDNA virus (family Podoviridae) that infects Bacillus subtilis, has been used for studying DNA packaging. An efficient in vitro packaging system has been developed for ϕ29 that has been adapted for both bulk (Grimes et al., 2002) and single-particle assays (Chemla et al., 2005; Fuller et al., 2007; Hugel et al., 2007; Smith et al., 2001). The ϕ29 genome is approximately 19.3 kb, codes for 20 proteins and is covalently linked at both 5′ ends to the phage protein gene product 3 (gp3). Proheads, the first particles assembled during morphogenesis (Figure 1), consist of the head-tail connector protein (gp10), scaffolding protein (gp7) (Morais et al., 2003), the major capsid protein (gp8) (Morais et al., 2005), the head fiber protein (gp8.5) (Morais et al., 2005), and a phage-encoded, 174-base RNA molecule (pRNA) (Tao et al., 1998). A virus-encoded ATPase (gp16) drives packaging of the phage genome into the prohead.
Figure 1. Assembly pathway of bacteriophage ϕ29.
Various phage proteins are shown schematically, with the names of the proteins written next to their schematic representations.
At present, the only component of the ϕ29 DNA packaging motor that has been determined to atomic resolution is the dodecameric connector (Simpson et al., 2000). The connector is a 75 Å-long cone-like structure measuring 138 Å across the wider, capsid-embedded end and 74 Å across the narrower, protruding end. The connector's central channel is composed of α-helices arranged with predominantly negatively-charged amino acid side chains facing the interior, thus facilitating the smooth passage of dsDNA through the connector into the head (Simpson et al., 2000). Based on sequence comparisons and common function, it is likely that the structure of the ϕ29 DNA packaging ATPase gp16 resembles the known structure of its counterpart in T4, gp17, which has a typical six-stranded nucleotide-binding fold with Walker A and B motifs positioned to bind the nucleotide (Draper and Rao, 2007; Mitchell et al., 2002; Sun et al., 2007). A cryoEM reconstruction of ϕ29 proheads flash-frozen in the midst of packaging suggests that the ATPase binds to the pRNA and interacts directly with the DNA (Simpson et al., 2000).
Here we report a series of cryoEM reconstructions which locate the different components of the packaging motor. CryoEM reconstructions were determined for various particles missing different components of the motor, such as the connector, the ATPase, full-length 174-base pRNA, as well as missing different domains within the pRNA. The resulting reconstructions confirmed that the pRNA is a pentamer on proheads, delineate the molecular boundaries of individual motor components, and suggest a mechanism for translocation of the genomic DNA.
RESULTS
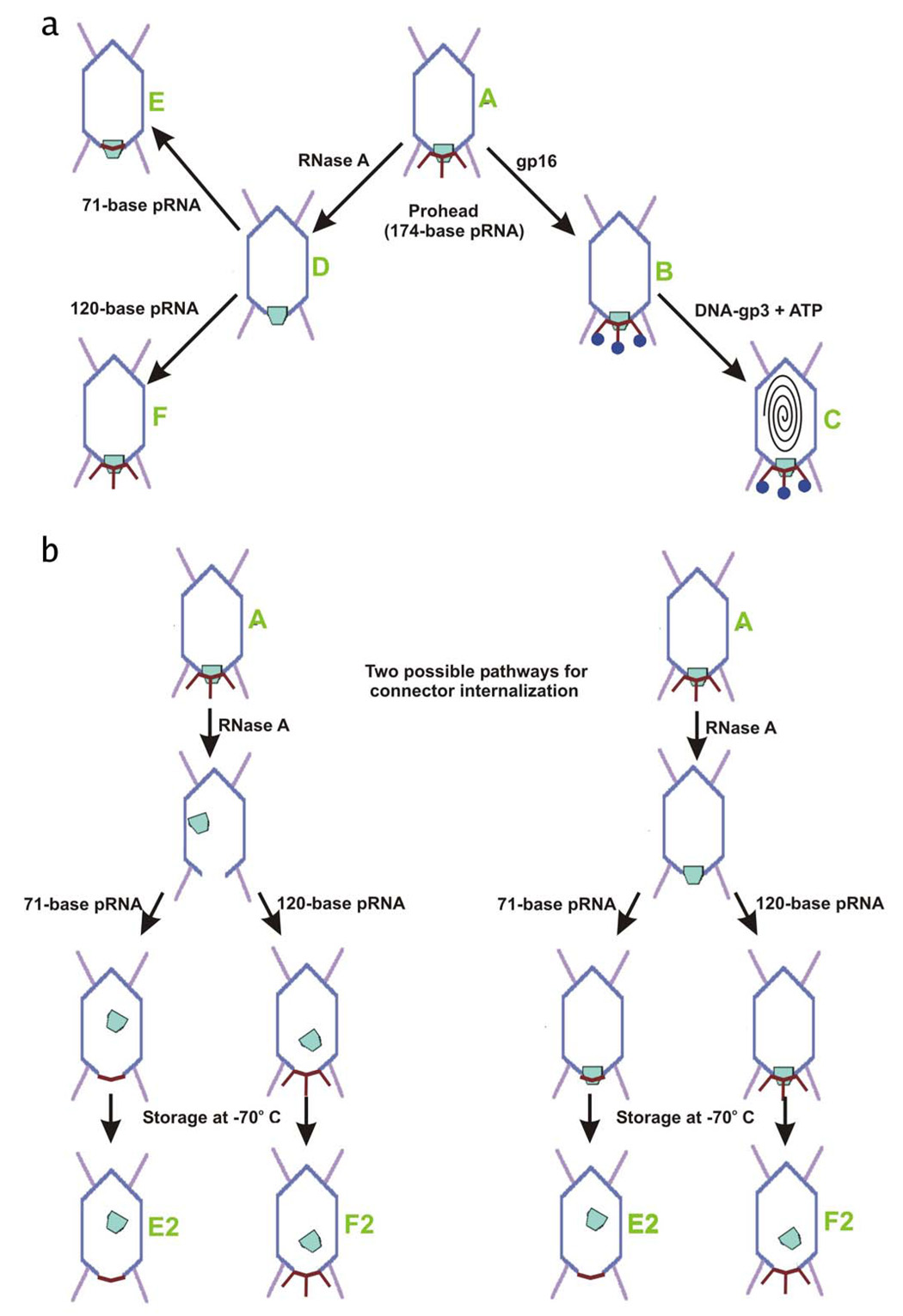
A three-step procedure was used to determine domain boundaries within the pRNA: First purified proheads were incubated with RNase to remove pRNA. Second these RNA-free proheads were repurified and then reconstituted with pRNA molecules of various lengths produced by in vitro transcription. In the third step the resulting capsid-pRNA complexes, some incubated with the ATPase gp16, were flash-frozen for cryoEM analyses (Figure 2A and Figure 2B, Table 1). Five-fold symmetry was assumed in calculating all the cryoEM reconstructions of the various particles described here
Figure 2. Diagram of particle production.
Phage proteins are colored as in Figure 1. Different particles are identified as A – F (green text).
(A) Production of all particles which retained the connector.
(B) Two possible pathways, both beginning with particle A, for the formation of connector-internalized particles (E2 and F2). The two pathways differ only in which step connectors are displaced.
Table 1.
EM Data
| Reconstruction* | pRNA (bp) | ATPase | Connector | Number of Particles | Defocus range, µm | Resolution, Å |
|---|---|---|---|---|---|---|
| A | 174 | no | yes | 1572 | 2.1–3.4 | 17.8 |
| B | 120 | yes | yes | 1611 | 1.8–3.5 | 18.0 |
| D | 0 | no | yes | 1497 | 2.0–4.1 | 21.6 |
| E | 71 | no | yes | 1234 | 2.1–4.1 | 20.2 |
| F | 120 | no | yes | 1353 | 2.1–4.1 | 21.0 |
| E2 | 71 | no | no | 1348 | 1.9–3.8 | 20.8 |
| F2 | 120 | no | no | 1461 | 1.9–4.1 | 21.4 |
The identity of each particle, A–F, is as described in Figure 2.
Proheads with or without Bound pRNA Connector or the gp16 ATPase
A reconstruction of proheads that had their pRNA removed by treatment with RNase (Figure 2A, particle D; Figure 3B, middle; and Table 1) confirmed (Simpson et al., 2000) the location of the pRNA by a comparison with the prohead (Figure 2A, particle A, Table 1). As expected, comparison of the two reconstructions indicates that the pRNA binds to proheads at the connector vertex, and that it interacts with both the narrow end of the connector and with the capsid proteins which surround the connector/pRNA complex.
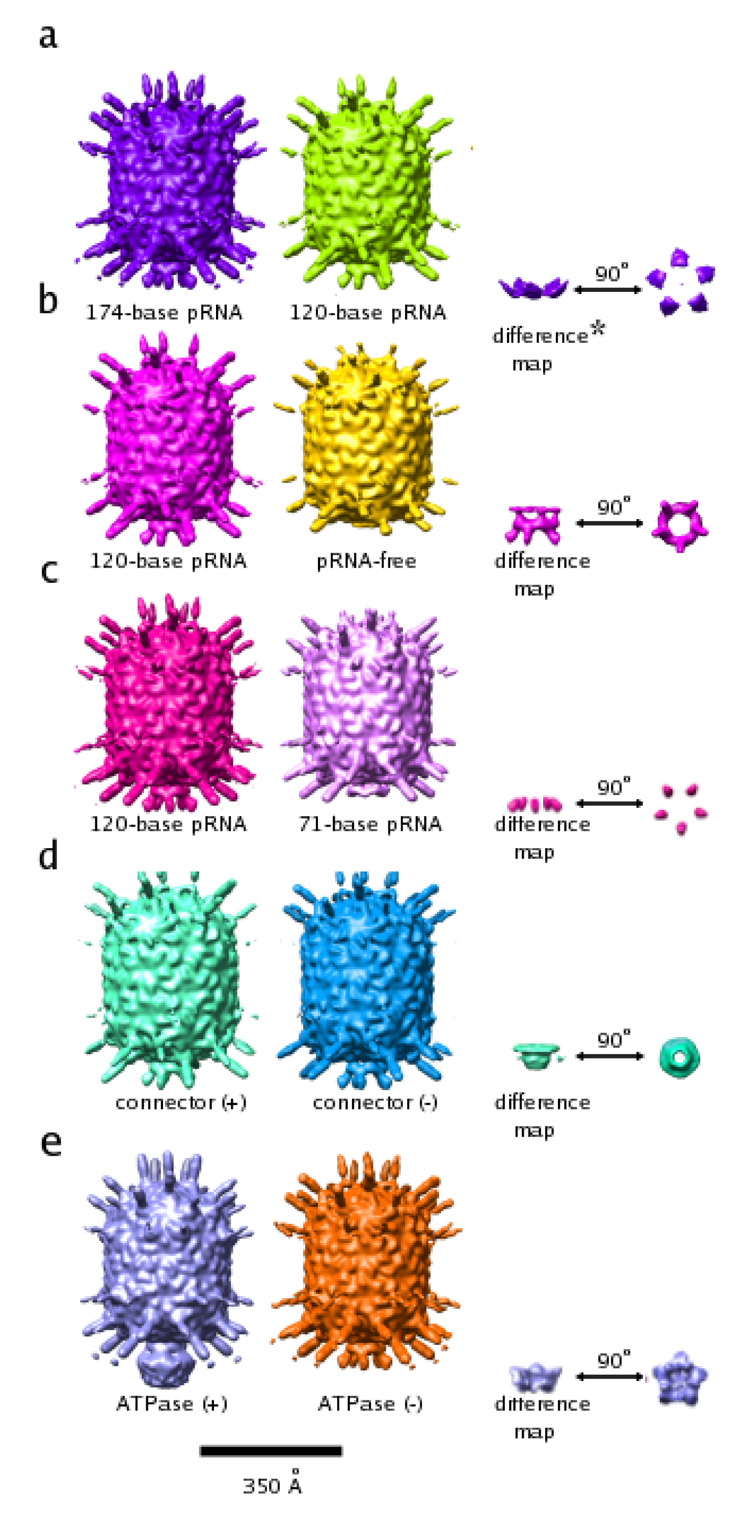
Figure 3. Reconstructions of ϕ29 proheads.
All reconstructions are shown as surface-shaded renderings contoured at 1.5 standard deviations above the mean of the map. A short description is written below each particle. Although Figures 3A (middle), B (left), C (left), and D (left) all show the same reconstruction, they are colored differently. As a consequence, the resultant structural features in the difference maps come out in different colors. The color of the difference densities are maintained in all figures shown in this paper.
(A) Prohead with 174-base pRNA (left), prohead with 120-base pRNA (middle), difference map between the two (right; side view (near right), end-on view (far right)).
(B) Prohead with 120-base pRNA plus connector (left), pRNA-free prohead (middle), difference map between the two (right; side view (near right), end-on view (far right)).
(C) Prohead with 120-base pRNA plus connector (left, same reconstruction as in a), shown again to clarify the difference map shown on the right), prohead with 71-base pRNA plus connector (middle), difference map between the two (right; side view (near right), end-on view (far right)).
(D) Prohead with 120-base pRNA plus connector (left same reconstruction as in a) and b), shown again to clarify difference map shown on the right), connector internalized prohead (middle), difference map between the two (right; side view (near right), end-on view (far right)).
(E) Prohead with 174-base pRNA plus connector (middle) and the same prohead with bound ATPase (left), difference between the two (right; side view (near right), end- on view (far right)).
* The asterisk indicates that the feature shown is contoured at less than 0.1 standard deviations above the mean electron density value of the map. Additional noise features also visible at this contour level were masked out manually so that the feature in question would be visible in the figure.
Due to the impostion of five-fold symmetry, it is not possible to identify the envelopes of individual monomers within the volume of density corresponding to the dodecameric connector. However, the overall envelope of the dodecamer will still be essentially correct (Simpson et al., 2000). Placement of the crystal structure of the ϕ29 connector (Protein Data Bank accession code 1IJG) into its cryoEM density shows that the central helical region of the connector has a larger diameter in the pRNA-free proheads than in the crystal structure (Figure 4A). Such a conformational change was not apparent when the atomic coordinates of the connector were placed into a cryoEM reconstruction of proheads with pRNA (Morais et al., 2005; Simpson et al., 2000). Thus, the observed conformational increase in diameter of the connector may be the result of removing the pRNA, possibly as a result of connector-capsid interactions which are otherwise blocked by the pRNA.
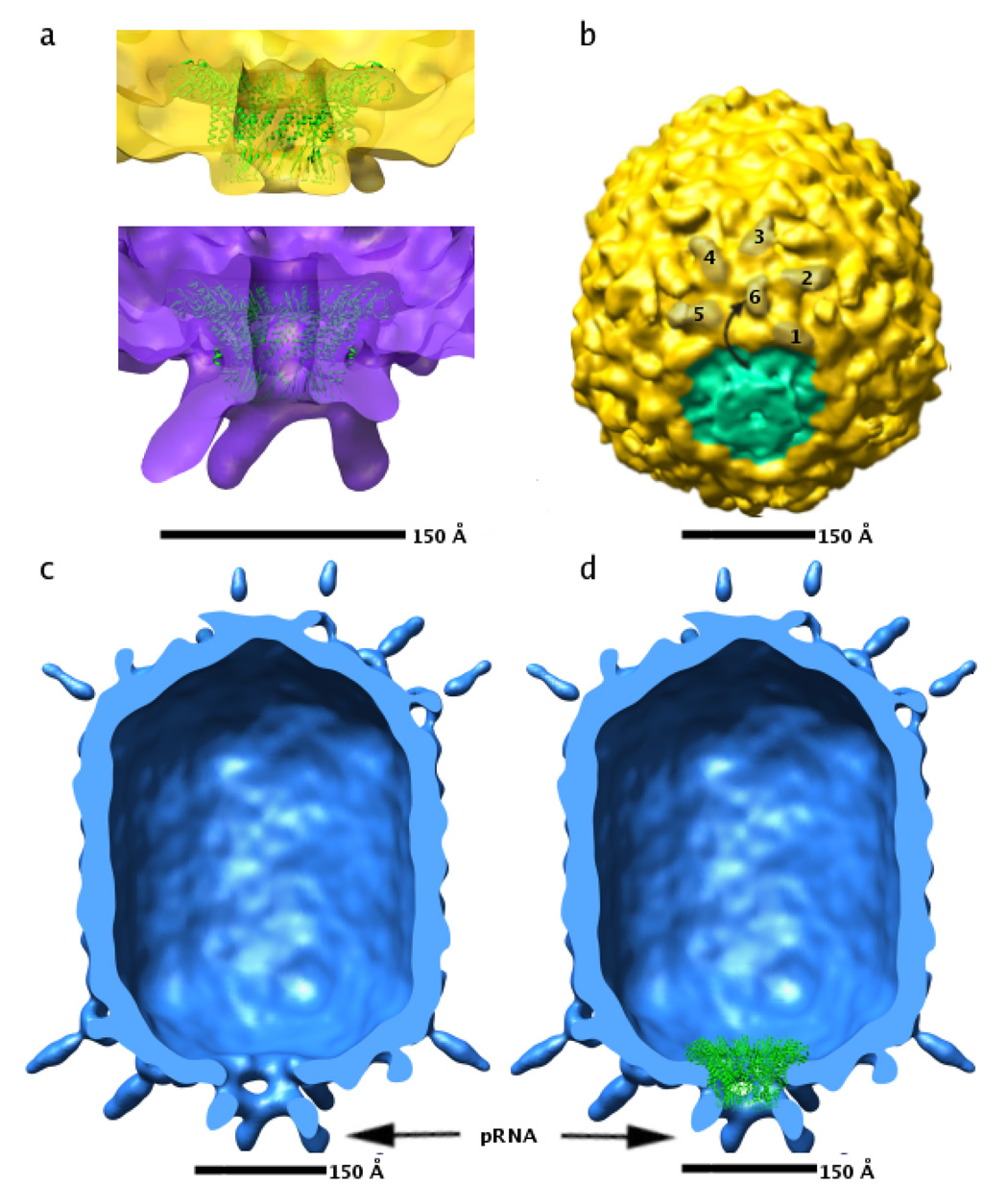
Figure 4. CryoEM reconstructions of pRNA-free and connector-free particles.
(A) Fit of the atomic coordinates of the connector (green ribbon, Protein Data Bank accession code 1IJG) to density corresponding to the connector in pRNA-free proheads (top, yellow density) and to 174-base proheads (bottom, purple density).
(B) Skewed view of pRNA-free particle showing rotation of the BIG II domain in capsid proteins abutting the connector. Six BIG II domains in one of the five hexamers surrounding the connector are shaded brown, and numbered 1–6. The ~180 degree rotation of the BIG2 domain labeled six is indicated by a curved arrow. Approximate density corresponding to the connector is shaded green.
(C) Cut-away view of connector internalized particle with the front half of the particle removed.
(D) Same as (C), but with the atomic coordinates of the connector (shown as green ribbon) placed according to their fit to wild-type cryoEM density maps. Density corresponding to the pRNA in panels (C) and (D) is indicated by a black arrow.
It had been previously shown that the bacterial immunoglobulin-like domains (BIG-II) in the capsid proteins that border the connector-pRNA complex are rotated approximately 180° from their position in the other capsid proteins, possibly in order to accommodate binding of the pRNA (Morais et al., 2005). This conformational change is maintained in particles whose pRNA has been removed (Figure 4B).
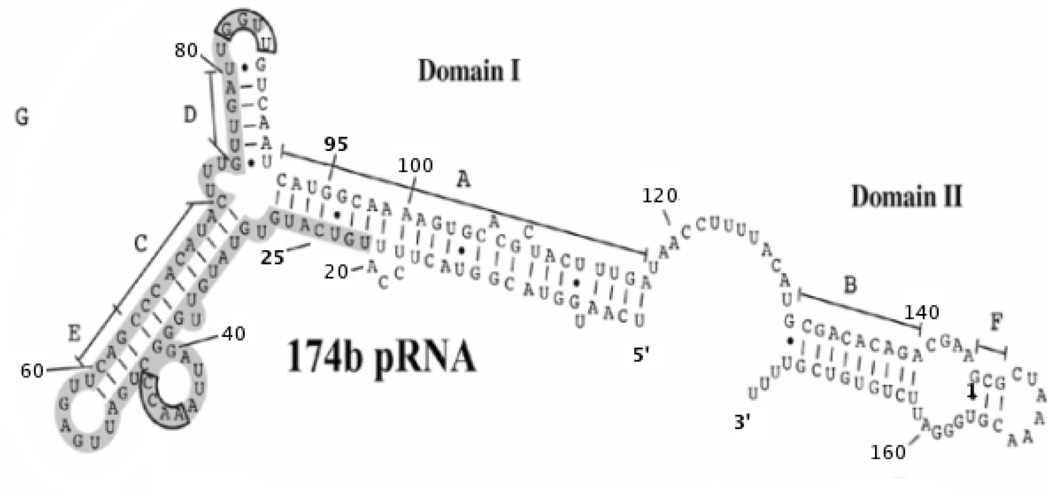
Secondary structure prediction suggests that the pRNA of ϕ29 has two domains (Bailey et al., 1990), with domain I consisting of the first 117 bases at the 5′-end, and domain II the last 44 bases at the 3′-end, with a 13-base single-stranded region connecting the two domains (Figure 5). Proheads with 120-base pRNA (domain I only) are fully capable of packaging DNA (Grimes et al., 2002; Wichitwechkarn et al., 1992). Thus, domain II is dispensable for in vitro packaging. However, the conservation of the domain II counterpart in the pRNAs of related phages suggests a biological function. In order to find the location of domain II in the prohead, a reconstruction of proheads reconstituted with 120-base pRNA, was determined (Figure 2A, particle F; Figure 3A, middle; and Table 1) and compared to a previous reconstruction of fiberless proheads with 174-base pRNA (Figure 2A, particle A; Figure 3A, left) (Morais et al., 2005). Additional density in the reconstruction of particles with 174-base pRNA, possibly corresponding to pRNA domain II, could be seen at the end of the pRNA arms. However, this density is weak and diffuse, indicating that domain II is either disordered or extremely flexible. Similarly a difference map between proheads reconstructed with 174base full length pRNA and proheads reconstructed with 120 base pRNA (Figure 3A, right) showed weak diffuse density at the end of the pRNA spokes, but the density height of this feature is less than 0.5 standard deviations above the mean density value of the difference map.
Figure 5. Secondary structure and domain prediction for pRNA.
Domains I and II are indicated. Base paired helices are labeled A through G. Bases believed to be involved in prohead binding are boxed in gray. Every 20th residue is labeled and residues 25 and 95, the residues that mark the ends of the 71 base pRNA are marked in bold font. The 3’ and 5’ ends of the 174-base pRNA are also marked in bold font.
Within domain I of the pRNA, bases 1 through 28 are predicted to form the A-helix with bases 92 through 117 (Figure 5). The A-helix is essential for DNA packaging (Reid et al., 1994; Zhang et al., 1995; Zhang et al., 1994). Simpson et al. (2000) proposed that the A-helices in the pRNA oligomer correspond to the five spokes observed in cryoEM reconstructions of the ϕ29 prohead. A cryoEM reconstruction of pRNA-free proheads reconstituted with a 71-base pRNA construct (residues 25 through 95, lacking the A-helix and domain II) (Figure 2A, particle E; Figure 3C, middle; and Table 1). is missing the pRNA spokes, confirming the identification of the spokes as corresponding to the predicted A helix.
Reconstructions of proheads subjected to RNase treatment, reconstituted with either 71-or 120-base pRNA, and then stored for 3 months at −70°C had no connector density (Figures 2A, particle E2, F2, 3D, middle; Figure 4C, 4D; and Table 1), that either initial RNase treatment or storage at −70°C and subsequent thawing, and/or flash-freezing, resulted in connectors being dislodged from the unique pentameric vertex (Figure 2B). Considering that the wide end of the connector is situated within the prohead, and that its diameter is much larger than the diameter of the pentameric vertex where the connector is located, it does not seem likely that intact connectors could exit proheads. Presumably, the above described prohead treatment caused the connector assembly to be released and drift into the interior of proheads. Because the connector would be in a different location within each individual prohead, they would not be visible after averaging over many particle images during the reconstruction procedure. This interpretation is consistent with SDS-PAGE analyses, which show a protein band with the same molecular weight and intensity relative to the capsid protein as the connector protein in wild-type prohead particles (data not shown). The strong density corresponding to the pRNA (Figure 4C, 4D; and Figure 3C, middle) shows that pRNA remains bound to the capsid in these particles, even in the absence of bound connector.
A cryoEM reconstruction of proheads with 174-base pRNA, incubated with the gp16 ATPase (Figure 2A, particle B, Figure 3E, left; and Table 1). had a significant volume of additional density at the distal end of the A-helix, when compared to the cryoEM reconstruction of proheads without gp16 (Figure 2A, particle A, Figure 3E, middle; and Table 1). The identification of this density as gp16 is consistent with the results obtained by Simpson et al. (2000).
Determining Molecular Envelopes for the Individual Components of the ϕ29 DNA Packaging Motor
Calculation of appropriate difference maps (Table 2) allowed the determination of the molecular envelopes of the various components of the packaging motor (Figure 3). In all difference maps, the density height of the anticipated structural features was always at least four times greater than that of the next highest noise peak. The strongest noise features were at the edge of the virus shell where artifacts due to small errors in the application of the contrast transfer function (CTF) tend to accumulate.
Table 2.
Difference Maps that Delineate the Envelops of the Various Components of the ϕ29 DNA Packaging Motor
| Object | Map1 | Map2 | Map1-Map2 |
|---|---|---|---|
| (Fig 2 particle) | (Fig 2 particle) | Fig 3 | |
| 120 pRNA | 120b pRNA | Prohead | 3a |
| Particle F | Particle D | ||
| A helix | 120b pRNA | 71b pRNA | 3b |
| Particle F | Particle E | ||
| Connector | 120b pRNA | 120b pRNA, | 3c |
| Internalized connector | |||
| Particle F | Particle F2 | ||
| ATPase gp16 | 174b pRNA + gp16 | 174b pRNA | 3d |
| Particle B | Particle A |
The difference maps described above can be combined to generate a picture of the ϕ29 DNA packaging motor (Figure 6). The pRNA forms a ring-like structure with ten arms emanating alternatively in up and down conformations (“down” means away from and “up” means toward the prohead). The five downward-pointing arms correspond to the A-helix (see above). The identity of the bases in the upward-pointing arms is not known and cannot be explained by the pRNA secondary structural predictions (Hoeprich and Guo, 2002; Simpson et al., 2000). The connector forms a cone-like structure, consistent with the X-ray structure (Simpson et al., 2000), that fits snugly into the pRNA ring such that the wide end of the connector is supported by the five upward-pointing pRNA arms and the narrow end of the connector is associated with the central ring of the pRNA. Thus, the upward-pointing arms of the pRNA are wedged between the prohead and the central section of the connector, in agreement with the previously reported structural data on proheads (Morais et al., 2005). The ATPase gp16 resembles a five-bladed propeller bound to the A-helix of the pRNA such that the five blades are inter-digitated with the five A-helices of the pRNA. There is some weakerdensity (less than 3 sigma) associated with the ATPase in the central channel of the motor that might be an artifact resulting from errors, CTF corrections, or from symmetry averaging close to the 5-fold axis of the capsid. However, it is likely that at least part of this density represents a true feature. Its proximity to the DNA suggests that it could play a role in translocation of the phage genome.
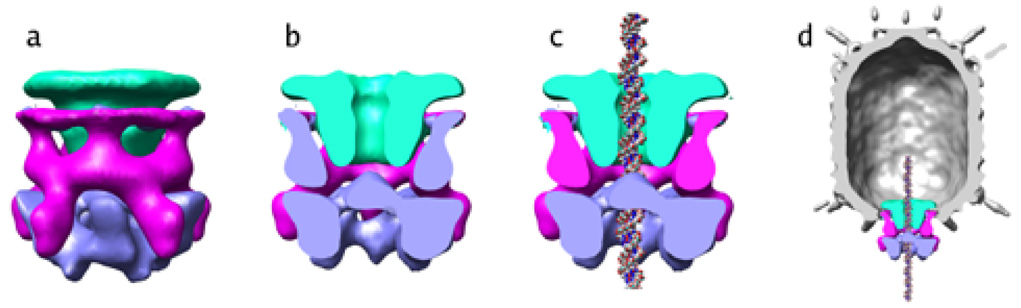
Figure 6. Combination of difference maps.
Density from difference maps (Figure 3) isolating the connector (green), pRNA (magenta) and ATPase (blue) were combined to visualize the DNA packaging motor in ϕ29.
(A) End-on view of the motor, looking from the pRNA down the long axis of the phage.
(B), Cut-away of side view of the motor, with the front half of the motor density removed.
(C) Cut-away of side view of the motor, with the front half of the motor density removed and the structure of DNA placed in the central channel of the packaging motor.
(D).The motor is shown in the context of the entire prohead. The front half of the prohead density has been removed so all the motor components can be seen.
DISCUSSION
Symmetry and Assembly of the ϕ29 Packaging Motor
There has been some controversy as to whether there are five or six pRNA molecules in the ϕ29 DNA packaging motor. Intermolecular base-pairing between adjacent monomers had suggested that the pRNA forms a hexamer in solution (Guo et al., 1998; Zhang et al., 1998). However, our cryoEM reconstructions of the ϕ29 prohead, comparing 5 and 6 fold averaging, showed that there were only five pRNA molecules in the packaging motor (Simpson et al., 2000; Tao et al., 1998). Subsequently Ibarra et al. (2000) also used cryoEM reconstruction studies that suggested 6-fold symmetry, The controversy appeared to have been settled in favor of five molecules in packaging-capable proheads by using an asymmetric reconstruction that made no symmetry assumptions (Morais et al., 2001), but, a recent publication (Shu et al., 2007) that utilized fluorescent labeled pRNA molecules inferred that there were six pRNA molecules in the motor, although their presence on the prohead was not confirmed. The cryoEM reconstructions presented here confirm that there are five pRNA molecules in the motor because the reconstructions (using 5-fold symmetry) show the difference in length of the pRNA A-helix when there are 71 bases in the pRNA fragment (no A-helix) and 120 bases (including the A-helix). Had 6-fold symmetry been assumed, pRNA density would have been blurred beyond recognition, as previously demonstrated by Morais et al. (2001). Furthermore, the presence of pRNA in the connector internalized particles shows that pRNA-prohead interactions occur not only via the connector, but also through the capsid. As the capsid is 5-fold symmetric, binding a pentameric pRNA would be favored over binding a hexameric pRNA.
The strong pRNA density in connector-internalized particles (Figure 3C; Figure 2B particles E2 and F2, Figure 4C, and 4D) was unexpected since previous biochemical studies had suggested that pRNA binds to the connector (Atz et al., 2007; Robinson et al., 2006; Xiao et al., 2005). However, extensive interactions of pRNA with the capsid had been noted in the 12.5 Å resolution cryoEM reconstruction of wild-type proheads (Morais et al., 2005) and had also been presumed by Simpson et al. (2000). An assembly scenario compatible with the seemingly conflicting data is one in which the pRNA initially recognizes and binds to the connector, followed by a conformational change in the capsid which creates a new, tight binding site on the capsid for the pRNA. This proposed conformational change may involve the previously described rotation of the BIG II domains in capsid proteins surrounding the pRNA (Morais et al., 2005), which would uncover the pRNA binding site on the capsid. As described above, this conformational change is present in prohead particles whose pRNA has been removed by RNase digestion.
Mechanism of DNA Translocation
Genome encapsidation by phage dsDNA packaging motors requires two valves that alternately grip and release the DNA during translocation. The opening, closing, and translation operations require energy, and must be appropriately synchronized. It is apparent from the data presented here that the ATPase would make extensive contacts with DNA, representing the first valve. Furthermore, the structure shows that the mechanical energy resulting from the ATP hydrolysis can only be transmitted directly to the DNA, not indirectly through the pRNA and connector as had been suggested by Simpson et al. (2000). The position of the second valve, whose function would be to restrain the already packaged DNA from exiting the prohead while the first valve is recovering, might be the connector. The central section of the connector consists of a series of parallel slanted á-helices (Simpson et al., 2000). The angle of slant is opposite to the right-handed DNA helix, causing the alpha helices of the connector to cross the ribose-phosphate backbone of the DNA. Although connector rotation during packaging is unlikely (Baumann et al., 2006; Hugel et al., 2007), the available data does not rule out small motions caused by conformational transitions. Thus, if the connector were to stretch slightly length wise, it would reduce the diameter of the connector, gripping the DNA in a non-specific binding action as would be required for a pump. As the pRNA binds both the ATPase and the connector, their activities might be coordinated through the ring of five pRNA molecules. The mechanism described above is similar to that proposed by Simpson et al., except that the connector is not required to rotate, and is also similar to the ATPase ratchet model of DNA translocation in phage T3 of Fujisawa and Morita (1997).
EXPERIMENTAL PROCEDURES
Production of ϕ29 Particles
Fibered proheads with 174-base pRNA were produced in B. subtilis SpoOA12(sup−) cells infected with the mutant sus16(300)-sus14(1241) that is defective for the DNA packaging ATPase gp16 as described previously (Morais et al., 2005). These proheads were treated with RNAse A (1mg/ml) to digest pRNA, purified by sucrose density gradient centrifugation, reconstituted with in vitro transcribed 71- or 120-base pRNA, and repurified on a sucrose density gradient. Proheads with 174-base pRNA were incubated with gp16 to produce the prohead-ATPase complex. Proheads without connectors were observed in RNaseA-treated proheads which were reconstituted with in vitro transcribed 120-base pRNA and stored at −70°C for 3 months.
Electron Microscopy
Prohead particles were flash-frozen on holey grids in liquid ethane. Images were recorded at 39,000 × magnification with a CM200 FEG microscope, with electron dose levels of approximately 20 e−/Å2. All micrographs were digitized at 3.59 Å pixel−1 using either a Zeiss SCAI scanner or a Nikon Super CoolScan 9000 scanner.
For all reconstructions, individual particle images were boxed, floated, and preprocessed to normalize mean intensities and variances and to remove linear background gradients. For each data set, reference projections of a previously published prohead reconstruction (Morais et al., 2005) were used to initially classify particles for three-dimensional reconstruction. The resulting model was used to recalculate reference projections for better particle classification. Several cycles of iterative particle classification and reconstruction were performed until convergence had been reached. Structure factor phases were modified as indicated by the parameters of the contrast transfer function. All steps of the reconstruction process, including determination of the contrast transfer function parameters, were performed with the program EMAN (Ludtke et al., 1999). Five-fold symmetry was assumed in all the reconstructions. The number of particles incorporated into each reconstruction, as well as the final resolution of each reconstruction, is summarized in Table 1. The resolutions of these reconstructions were determined by the Fourier shell correlation method using a correlation coefficient of 0.5 between independent half data sets as the cut-off criterion.
Difference maps between the reconstructions were calculated after adjusting radial and density scale factors using RobEM (http://cryoem.ucsd.edu/programs.shtm). In order to eliminate artifacts that might arise as a result of calculating difference maps between reconstructions of differing resolutions, the same low-pass filter with a Gaussian fall-off was applied to each of the two reconstructions in a given difference map prior to scaling and subsequent map subtraction. The cut-off value of this low-pass filter was chosen to match the poorer resolution of the two maps being compared.
ACKNOWLEDGEMENTS
We thank Ye Xiang, Kay Choi and Rockney Atz for many helpful discussions. We are grateful to Sheryl Kelly and Cheryl Towell for help in preparation of the manuscript. The work was supported by an NSF grant (MCB-0443899) to MGR and an NIH grant (DE-03606) to DLA and MGR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson D, Reilly B. Morphogenesis of bacteriophage ϕ29. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 859–867. [Google Scholar]
- Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine Scanning and Fe-BABE probing of the bacteriophage ϕ29 prohead RNA connector interaction. J. Mol. Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Wichitwechkarn J, Johnson D, Reilly BE, Anderson DL, Bodley JW. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J. Biol. Chem. 1990;265:22365–22370. [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. New York: Plenum Press; 1988. pp. 15–75. [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J. Mol. Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DA. Ionic effects on viral DNA packaging and portal motor function in bacteriophage ϕ29. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes S, Jardine PJ, Anderson DL. Bacteriophage phi29 DNA packaging. Adv. Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage φ29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Hoeprich S, Guo P. Computer modeling of three-dimensional structure of DNApackaging RNA (pRNA) monomer, dimer, and hexamer of ϕ29 DNA packaging motor. J. Biol. Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetheringron CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra B, Castón JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. Topology of the components of the DNA packaging machinery in the phage ϕ29 prohead. J. Mol. Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- Jardine PJ, Anderson DL.Calendar R.DNA packaging in double-stranded DNA phages The Bacteriophages 2006New York: Oxford Press; 49–65.1 [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30:4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of ϕ29. Mol. Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BAL, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage ϕ29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- Morais MC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage ϕ29. J. Struct. Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJ, Bodley JW, Anderson D. Identification of bacteriophage phi 29 prohead RNA domains necessary for in vitro DNA-gp3 packaging. J. Biol. Chem. 1994;269:9084–9089. [PubMed] [Google Scholar]
- Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, Stonehouse NJ. Affinity of molecular interactions in the bacteriophage phi 29 packaging motor. Nucleic Acids Res. 2006;34:2698–2709. doi: 10.1093/nar/gkl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Zhanq H, Jin J, Guo P. Counting of six pRNA-s of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL et al. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichitwechkarn J, Johnson D, Anderson D. Mutant prohead RNAs in the in vitro packaging of bacteriophage ϕ29 DNA-gp3. J. Mol. Biol. 1992;223:991–998. doi: 10.1016/0022-2836(92)90257-k. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage ϕ29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Moll WD, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacteriophage phi29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Garver K, Guo P. Inhibition of phage ϕ29 assembly by antisense oligonucleotides targeting viral pRNA essential for DNA packaging. Virology. 1995;211:568–676. doi: 10.1006/viro.1995.1439. [DOI] [PubMed] [Google Scholar]
- Zhang C, Lee CS, Guo P. The proximate 5' and 3' ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage ϕ29. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St.-Arnaud D, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage ϕ29 DNA in vitro. Mol. Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]