Abstract
Activity-dependent gene expression plays an important role in mediating the effects of sensory experience on nervous system development and function. While several activity-dependent transcription factors have been identified, the mechanism by which calcium signaling converts a promoter from a silenced to an active state is not well understood. Here we show that a CREST-BRG1 complex plays a critical role in regulating promoter activation by orchestrating a calcium-dependent release of a repressor complex, and a recruitment of an activator complex. In resting neurons, transcription of the c-fos promoter is inhibited by BRG1-dependent recruitment of a phospho-Rb-HDAC repressor complex. Upon calcium influx, Rb becomes dephosphorylated at Serine 795 by Calcineurin, which leads to release of the repressor complex. At the same time there is increased recruitment of CBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 also binds to the NR2B promoter and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CBP, suggesting that this mechanism may be generally involved in regulating calcium-dependent transcription of neuronal genes.
Introduction
One of the most remarkable features of the nervous system is that its structure and function can be modified by sensory input. For example the pioneering work of Hubel and Wiesel showed that changes in postnatal visual experience can lead to lasting changes in cortical connectivity in cats (reviewed in Hubel and Wiesel, 1998). It is now well established now that neuronal activity plays a critical role in controlling many aspects of neural development and function, including neuronal viability, migration, morphogenesis and plasticity (reviewed in Katz and Shatz, 1996).
The effects of activity in the nervous system are primarily mediated by calcium signaling. Calcium influx leads to post-translational modification of synaptic proteins as well as induction of new gene expression. The lasting effects of neuronal activity, such as activity-dependent dendritic growth, long-term plasticity in sensory systems, and memory consolidation, require calcium-dependent transcription (Ghosh and Greenberg, 1995, West et al., 2001, Redmond et al., 2002). Much of our understanding of calcium-dependent transcription has come from studies on the regulation of Immediate Early Genes (IEGs), such as c-fos, which are rapidly induced upon calcium influx (reviewed in Ghosh and Greenberg, 1995). Among the major insights from these studies has been the recognition that a CREB-CBP complex plays a central role in regulating calcium-dependent transcription (reviewed in Mayr and Montminy, 2001; Chrivia et al., 1993; Chawla et al., 1998; Hu et al., 1999). Recent progress in chromatin biology and epigenetics suggests that covalent modification of histones, such as acetylation and methylation, also serve a critical role in regulating gene expression (Rosenfeld et al., 2006), but the role of these modifications in calcium-dependent transcription has not been extensively explored.
To gain additional insight into the mechanisms that mediate activity-dependent transcription we previously carried out a screen for calcium-dependent transactivators, and cloned a novel factor called CREST (Calcium RESponsive Transactivator; Aizawa et al., 2004). Here we report on our investigation of the mechanisms by which calcium signaling regulates transcription via the CREST complex. We show that CREST binds to CBP and BRG1via distinct domains. Whereas the association with CBP facilitates transcription, the association with BRG1 suppresses CREST-mediated transcription in resting neurons. Transcriptional repression by BRG1 is mediated by the Retinoblastoma protein (Rb), which recruits a Histone deacetylase (HDAC) complex to the promoter. Calcium-influx leads to release of the HDAC complex from Rb via Calciuneurin-dependent dephosphorylation of Rb. These findings reveal a novel regulatory mechanism for calcium-dependent transcription that is likely to play a critical role in mediating adaptive responses in the nervous system.
Results
Characterization of the CREST-BRG1 complex and bidirectional regulation of calcium-dependent c-fos transcription by CREST and BRG1
To identify components of the CREST complex, we carried out a yeast 2-hybrid screen and identified a homolog of BAF250b as a CREST-interacting protein (Shu-Ching Hu and Anirvan Ghosh, unpublished data). BAF250b is a component of the BRG1 chromatin-remodeling complex (Nie et al., 2000, Olave et al., 2002), which suggested that CREST and BRG1 might be part of the same complex. Co-immunoprecipitation experiments indicated that endogenous CREST associated with BRG1 in cortical neurons (Fig. 1A). To determine if this was due to a direct interaction between BRG1 and CREST, we purified GST fusion proteins of N- and C-terminus of BRG1, and tested the ability of BRG1 to bind to purified His-CREST using a GST pull-down assay. As shown in Fig. 1B, the N terminus of BRG1 (amino acid 1-282) directly interacts with CREST. To identify the domain of CREST that interacts with BRG1, we cotransfected CREST deletion constructs and the N-terminus fragment of BRG1 in 293 cells, and examined association by co-immunoprecipitation. These experiments revealed that the N terminus of CREST is required for its interaction with BRG1 (Fig. 1C). We have previously shown that the C-terminal domain of CREST interacts with CBP (Aizawa et al., 2004). Thus CREST can directly interact with both CBP and BRG1, raising the possibility that the BRG1-CREST-CBP protein complex regulates calcium-dependent gene expression.
Figure 1. CREST and BRG1 are present in a complex and exert opposite effects on calcium-dependent c-fos expression.
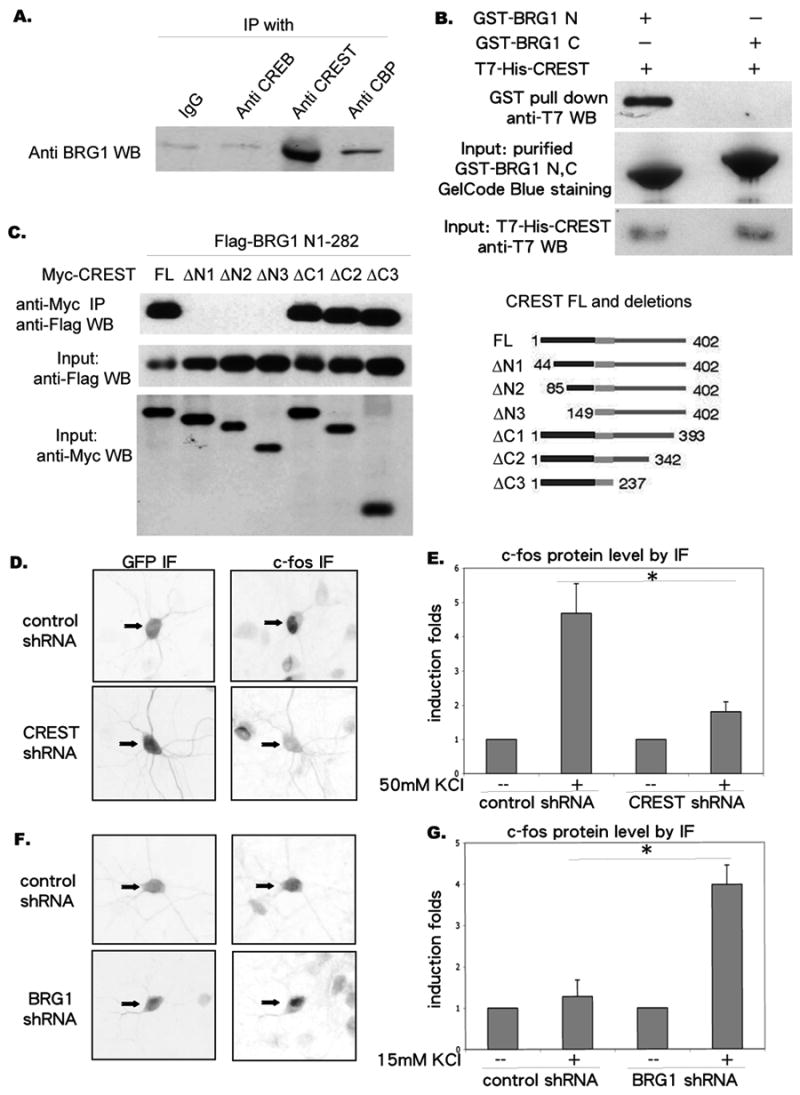
A. Immunoprecipitation of CREST with BRG1. Cortices from postnatal 4 days rat were dissected, homogenized and immunoprecipitated with antibodies as indicated. The immunoprecipitates were resolved by SDS-PAGE and the Western blot was probed with an anti-BRG1 antibody.
B. Interaction between N-terminus of BRG1 and CREST. His-T7-CREST, GST-BRG1 N and GST-BRG1 C were expressed in E.coli and purified. His-T7-CREST was combined with GST-BRG1 N and GST-BRG1 C respectively. The mixture was incubated for 30 minutes at room temp. After immobilizing GST-BRG1 N and GST-BRG1 C with GST affinity beads, mixtures were washed extensively, resolved by SDS-PAGE and the Western Blot was probed with anti-T7 antibody.
C. Interaction between N-terminus of CREST and BRG1. HEK 293 cells were transfected with Flag-tagged BRG1 N terminal a.a. 1-282 along with myc-tagged CREST deletion constructs (shown on the right), and immunoprecipitated with an anti-myc antibody. The myc immunoprecipitates were separated by SDS-PAGE and probed with an anti-Flag antibody (upper blot). Similar amounts of input Flag-tagged proteins in each sample were confirmed by probing with an anti-Flag antibody (middle blot). Similar amounts of input myc-tagged proteins in each sample were confirmed by probing with an anti-myc antibody (lower blot).
D. Examples of cortical neurons transfected with pSuper-control shRNA or pSuper-CREST shRNA and immunostained with antibodies indicated, after 50mM KCl stimulation for 4 hours.
E. Quantifications of c-fos protein levels measured by immunofluorescence intensity. (N=12 cells)
F. Examples of cortical neurons transfected with pSuper-control shRNA and pSuperBRG1 shRNA and immunostained with antibodies indicated, after 15mM KCl stimulation for 4 hours.
G. Quantifications of c-fos protein levels measured by immunofluorescence intensity. (N=12)
Asterisks indicate significance at p<0.05. Error bars represent +SD.
To determine if CREST and BRG1 contribute to calcium-dependent gene expression, we examined the consequence of shRNA-mediated knock-down of CREST and BRG1 on depolarization-induced c-fos expression (Fig. 2A, Supplemental Fig. 1). c-fos is the prototypical calcium-regulated gene and has been widely used to study mechanisms of calcium-dependent transcription (Sheng et al., 1990). The pSuper shRNA vector that was used in these experiments contains the PGK-GFP cassette, which allows transfected neurons to be identified based on GFP expression. After transfecting pSuper vector based short hairpin RNA against CREST and BRG1 into primary cortical neurons, respectively, we measured depolarization-induced c-fos protein level by immunofluorescence. Reduction of CREST expression strongly inhibited depolarization-induced c-fos expression (Fig. 1 D, E). In contrast, c-fos immunofluorescence was significantly increased following knock-down of BRG1 (Fig. 1 F, G). These observations suggest that CREST and BRG1 exert opposite regulatory influences on calcium-activated c-fos transcription – whereas CREST appears to facilitate c-fos transcription, BRG1 appears to suppress calcium activation of c-fos.
Figure 2. CREST regulates calcium-dependent c-fos expression via CBP recruitment.
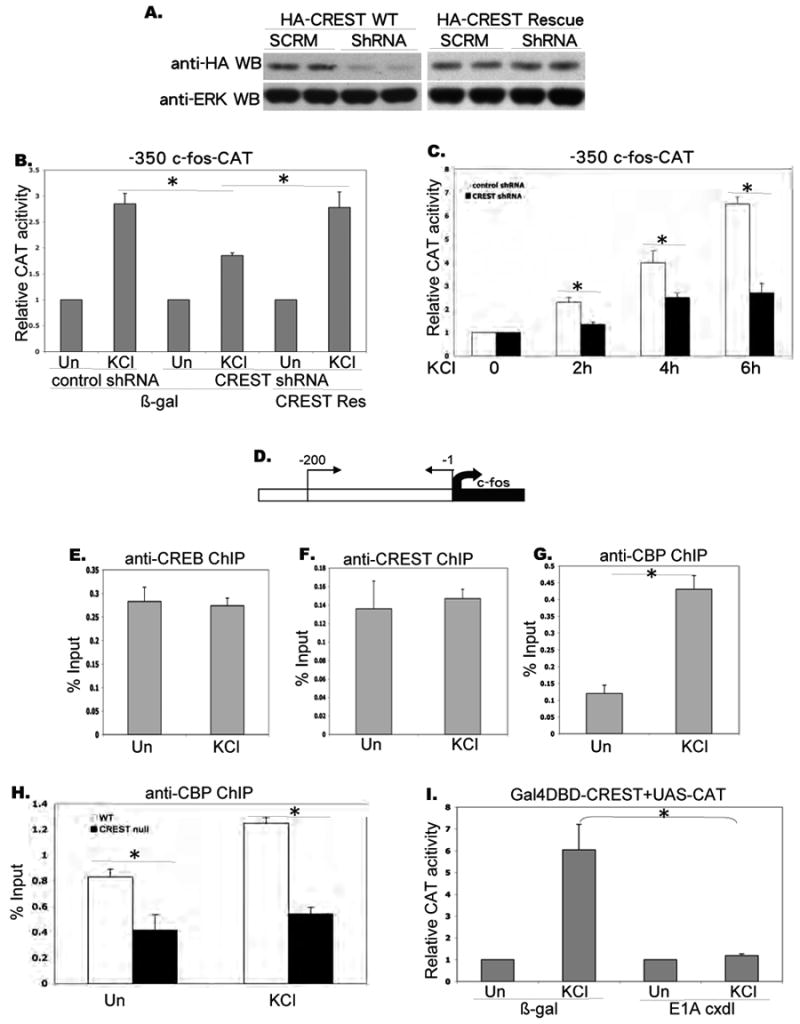
A. Inhibition of CREST expression by CREST Short hairpin RNA. HEK 293 cells were transfected with HA-CREST and HA-CREST rescue constructs along with pSuper vector harboring scrambled shRNA and shRNA against rat CREST. The cell lysates were separated by SDS-PAGE and probed with an anti-HA antibody (upper blot). Similar amounts of input proteins in each sample were confirmed by probing with an anti-ERK1 antibody (lower blot).
B. Relative CAT activity in E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with pSuper vector harboring control shRNA and CREST-specific shRNA constructs at 3DIV. The same amounts of b-galactosidase and HA-CREST rescue constructs were co-transfected to perform rescue experiments. Transfected cells were stimulated for 4 hours with 50mM KCl as indicated at 5 DIV.
C. Relative CAT activity in E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with pSuper vector harboring control shRNA and CREST-specific shRNA constructs at 3DIV. Transfected cells were stimulated for the number of hours indicated, at 5 DIV (50 mM KCl).
D. Diagram for c-fos promoter analyzed by chromatin immunoprecipitation assay.
E. F. G. Chromatin immunoprecipitation on c-fos promoter. Rat E18 cortical neurons were stimulated with KCl (50mM) for 10 minutes at 5DIV, and immunoprecipitated with antibodies as indicated. PCR reactions with endogenous c-fos primers (upper panel) were used to amplify endogenous c-fos promoter-specific DNA segments from -200 to -1.
Real time PCR was performed and signals were normalized as percentage of input. ChIP with IgG was always less than 0.05% of Input (Fig. 7).
H. Chromatin immunoprecipitation in CREST null and wild type neurons. E15 cortical neurons from CREST KO and wild type mice were cultured and stimulated for 10 minutes with KCl (50mM) at 5DIV. After immunoprecipitation with anti-CBP antibody, PCR reactions with endogenous c-fos promoter primers were used to amplify c-fos promoter-specific segments. Real time PCR was performed and signals were normalized as percentage of input.
I. Relative CAT activity in E18 cortical neurons transfected with Gal4DBD-CREST and UAS-CAT reporter along with β galactosidase and E1A cxdl constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
Asterisks indicate significance at p<0.05. Error bars represent SD.
To determine if BRG1 negatively regulates CREST-mediated transcription, we examined the consequences of suppressing BRG1 expression in cortical neurons using the BRG1 shRNA construct. Expression of BRG1 shRNA led to a marked increase in KCl-induced activation of Gal4-CREST, indicating that endogenous BRG1 negatively regulates CREST-mediated transcription (Supplemental Fig. 2A). The effects of BRG1 shRNA were reversed by overexpression of human BRG1 that is resistant to the shRNA against rat BRG1 (Supplemental Fig. 2A). Expression of BRG1 shRNA did not affect calcium activation of Gal4-CREB, indicating that suppression of BRG1 expression does not lead to a general increase in calcium-dependent transcription (Supplemental Fig. 2B). Consistent with a repressive role for BRG1 on CREST-mediated transcription, we found that expression of WT hBRG1 strongly suppressed calcium activation of Gal4-CREST (Supplemental Fig. 2C). Mutation of the N-terminal domain of CREST, which disrupts the association of CREST with BRG1, abrogated the ability of BRG1 to inhibit activation of Gal4-CREST (Supplemental Fig. 2D). Expression of WT BRG1 did not suppress calcium activation of Gal4-NeuroD1, indicating that BRG1 overexpression does not lead to a general suppression of calcium-dependent transcription (Supplemental Fig 2E). Thus, BRG1 directly binds to N-terminus of CREST and exhibits a negative effect to CREST-mediated transcription.
Regulation of the c-fos promoter by the CREST-CBP complex
To gain further insight into the role of CREST in calcium activation of the c-fos promoter, we examined the effects of expressing a CREST shRNA construct on depolarization-induced activation of a c-fos reporter. As shown in Fig. 2A, the CREST shRNA sharply reduced expression of HA-CREST in transfected 293 cells. This suppression could be reversed by expression of a HA-CREST rescue construct that contains same sense mutations in shRNA targeting sequences (Fig. 2A). Consistent with the effects on endogenous c-fos expression, the expression of CREST shRNA significantly decreased KCl induced activation of -350 c-fos-CAT, a construct that includes all major transcriptional regulatory sites (Fig. 2B). More importantly, this effect was reversed by co-expressing a shRNA resistant HA-CREST construct, indicating that this effect is specifically due to the loss of CREST. We found that knocking down CREST significantly reduces calcium-mediated c-fos reporter expression at various time points up to 6 hours (Fig. 2C).
To examine the effect of depolarization on recruitment of CREB, CREST, and CBP to the c-fos promoter, we performed chromatin immunoprecipitation (ChIP) experiments on extracts of control and KCl-treated neurons using CREB, CREST and CBP antibodies (Fig. 2D, E, F, G). In this and all subsequent experiments, the ChIP experiments were quantified using real time quantitative PCR. All three proteins were found to be bound to the endogenous c-fos promoter both before and after KCl stimulation. Whereas the binding of CREB and CREST to the promoter was largely unaffected by KCl stimulation, there was a significant increase in the association of CBP with the promoter (Fig. 2G). Since CREST binds to CBP, we next asked whether CREST contributes to the recruitment of CBP to the c-fos promoter by comparing CBP association with the promoter in cortical cultures from wild type and CREST knockout mice (Aizawa et al., 2004). As shown in Fig. 2H recruitment of CBP to the c-fos promoter was significantly decreased in CREST null neurons in both unstimulated and KCl stimulated cultures. This observation, together with the fact that CBP can mediate calcium-dependent transcription suggested that the transactivation activity of CREST may rely on CBP (Chawla et al., 1998; Hu et al., 1999; Impey et al., 2002). Indeed, Gal4-CREST mediated reporter gene expression was totally abolished when CBP function was inhibited by the viral protein E1A (cxdl) (Hu et al., 1999) (Fig. 2I). These data indicate that CREST mediates calcium activation of the c-fos promoter via recruitment of CBP.
Regulation of the c-fos promoter by the BRG1-Rb-HDAC complex
We next investigated the mechanism by which BRG1 regulates the c-fos promoter. BRG1 has extensively been studied as a component of a chromatin-remodeling complex, but its role in calcium-dependent transcription has not been explored. Consistent with the observation that BRG1 shRNA leads to an increase in calcium activation of the c-fos expression (Fig. 1G), expression of BRG1 shRNA increased expression of the c-fos-CAT reporter, whereas overexpression wild type BRG1 led to a suppression of KCl activation of c-fos CAT (Fig. 3A, B). To determine if transcriptional repression of the c-fos promoter by BRG1 might involve recruitment of an HDAC complex (Fass et al., 2003, Kumar et al., 2005), we examined the effect of BRG1 in the presence of trichostatin A (TSA), an HDAC inhibitor. Not only did BRG1 expression not inhibit KCl-induced transactivation in the presence of TSA, it actually enhanced transactivation, indicating that the effect of BRG1 on c-fos activation requires HDAC activity (Fig. 3C). The fact that BRG1 expression increases transcription in the presence of TSA suggests that BRG1 might also play a positive role in calcium-dependent transcription that is occluded by HDAC recruitment. In contrast to its effect on BRG1-transfected neurons, TSA did not reverse the inhibitory effect of acidic CREB on calcium activation of c-fos-CAT (Supplemental Fig. 3 A, B), suggesting that HDACs might specifically inhibit transcription of the c-fos promoter via the BRG1 complex. Consistent with this possibility, co-immunoprecipitation experiments from cortical lysates showed that HDAC1, HDAC2 and to some extent HDAC3 and mSin3A associate with BRG1 (Fig. 3D).
Figure 3. BRG1 recruits HDAC and Rb to inhibit c-fos activation.
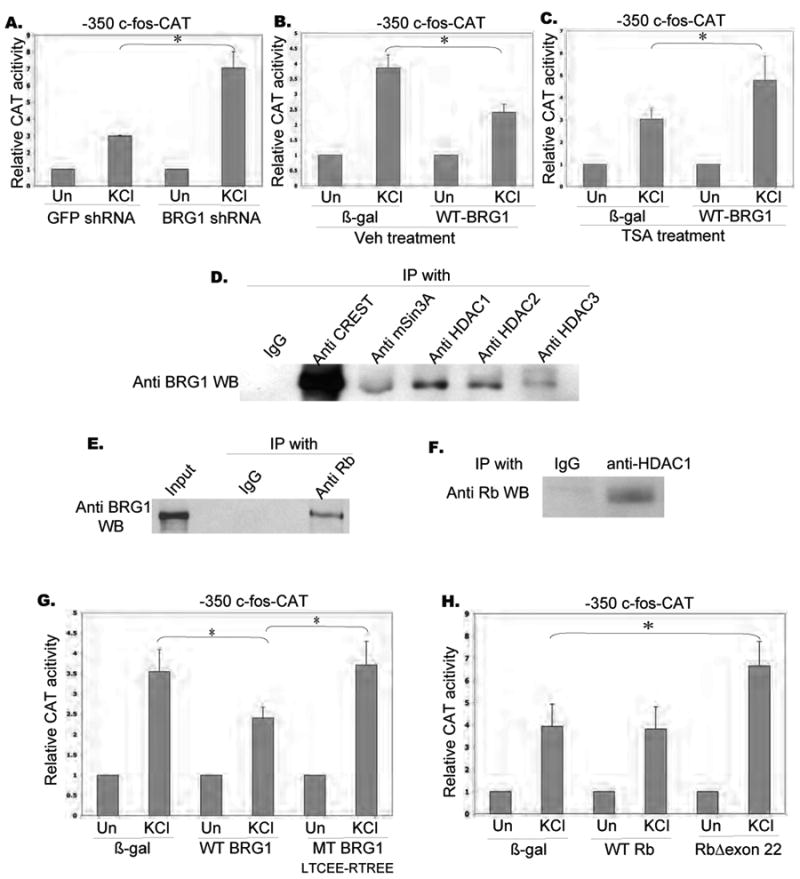
A. Relative CAT activity in E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with shRNA against GFP and rat BRG1 at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
B. Relative CAT activity in rat E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with b-galactosidase and wild type BRG1 constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM) with the presence of DMSO.
C. Relative CAT activity in rat E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with b-galactosidase and wild type BRG1 construct at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM) with TSA pre-treatment (TSA 250ng/ml).
D. Immunoprecipitation of BRG1 with class I HDAC complex. Cortices from postnatal 4 day ratx were dissected, homogenized and immunoprecipitated with antibodies as indicated. The immunoprecipitates were resolved by SDS-PAGE and the Western blot was probed with an anti-BRG1 antibody.
E. Immunoprecipitation of BRG1 with Rb. Nuclear fractions of rat E18 cortical neurons were collected at 5DIV, and immunoprecipitated with IgG and anti-Rb antibody. The immunoprecipitates were resolved by SDS-PAGE and Western blot was probed with an anti-BRG1 antibody.
F. Immunoprecipitation of Rb with HDAC1. Nuclear fractions of rat E18 cortical neurons were collected at 5DIV, and immunoprecipitated with IgG and anti-HDAC1 antibody. The immunoprecipitates were resolved by SDS-PAGE and Western blot was probed with an anti-Rb antibody.
G. Relative CAT activity in rat E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with b-galactosidase, wild type BRG1 and mutated BRG1 (LTCEERTREE) constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
H. Relative CAT activity in rat E18 cortical neurons transfected with mouse -350 c-fos-CAT reporter along with b-galactosidase, wild type Retinoblastoma protein and mutated Rb protein (Δexon 22) constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
Asterisks indicate significance at p<0.05. Error bars represent +SD.
We next investigated the mechanism by which BRG1 recruits the repressor complex to the promoter. Specifically, we decided to consider the possibility that the Retinoblastoma protein (Rb) might mediate recruitment of a repressor complex to BRG1. Rb and BRG1 have been independently reported to suppress c-fos expression, but whether their actions are mechanistically linked is not known (Robbins et al., 1990, Dunaief et al., 1994; Murphy et al. 1999). We found that Rb could be co-immunoprecipitated with both BRG1 and HDAC1 from cortical lysates (Figs. 3 E, F), suggesting that the three proteins may be part of one repressor complex. To determine if association of BRG1 and Rb was involved in BRG1-mediated repression of c-fos activation, we examined the effects of a BRG1 mutant (LXCXE-RXRXE) that prevents association with Rb (Zhang et al., 2000). As shown in Fig. 3G, this BRG1 mutant was unable to inhibit KCl activation of c-fos-CAT, indicating that the repressive effect of BRG1 requires interaction with Rb. We also found that a mutant of Rb that does not bind HDAC (RbΔexon22; Zhang et al., 2000) potentiated KCl activation of c-fos-CAT, further strengthening the evidence that a BRG1-Rb-HDAC complex negatively regulates c-fos expression (Fig. 3H).
To exert a repressive effect, the BRG1-Rb-HDAC complex needs to be recruited to the promoter. The interaction of BRG1 with CREST is unlikely to help with promoter recruitment since CREST does not contain a DNA-binding domain. One possibility is that the BRG1 complex is recruited to the promoter via an Sp1 binding site since BRG1 associates with Rb, and the suppression of c-fos expression by Rb depends on Sp1 binding sites (Robbins et al., 1990, Udvadia et al., 1993, Sohm et al., 1999). To determine if BRG1 associates with the c-fos-CAT promoter in a region that includes the Sp1 site, we carried out chromatin immunoprecipitation (ChIP) experiments in neurons transfected with c-fos-CAT constructs including different promoter elements using a forward primer in the c-fos promoter and a reverse primer in the CAT sequence (Supplemental Fig. 4A). As shown in Supplemental Fig. 4B, BRG1 could be efficiently precipitated with -290 c-fos-CAT but not with -67 c-fos-CAT suggesting that it binds to the c-fos promoter between −67 and -290. This region includes the Sp1 site, but not the CRE or SRE sites. Consistent with the possibility that the BRG1 complex is recruited to the Sp1 site, we were able to co-precipitate BRG1 with Sp1, but not with CREB (Fig. 4A; Kadam and Emerson, 2003). Furthermore we found that BRG1 failed to bind with c-fos-CAT reporter that carried a mutation in the SP1 binding site (-73 CCGCCC to AAATTT) (Fig. 4B). These observations indicate that the BRG1 complex is recruited to the c-fos promoter via its interaction with Sp1.
Figure 4. Role of Sp1 in recruitment of BRG1 on the c-fos promoter, and release of repressor complex from c-fos promoter upon stimulation.
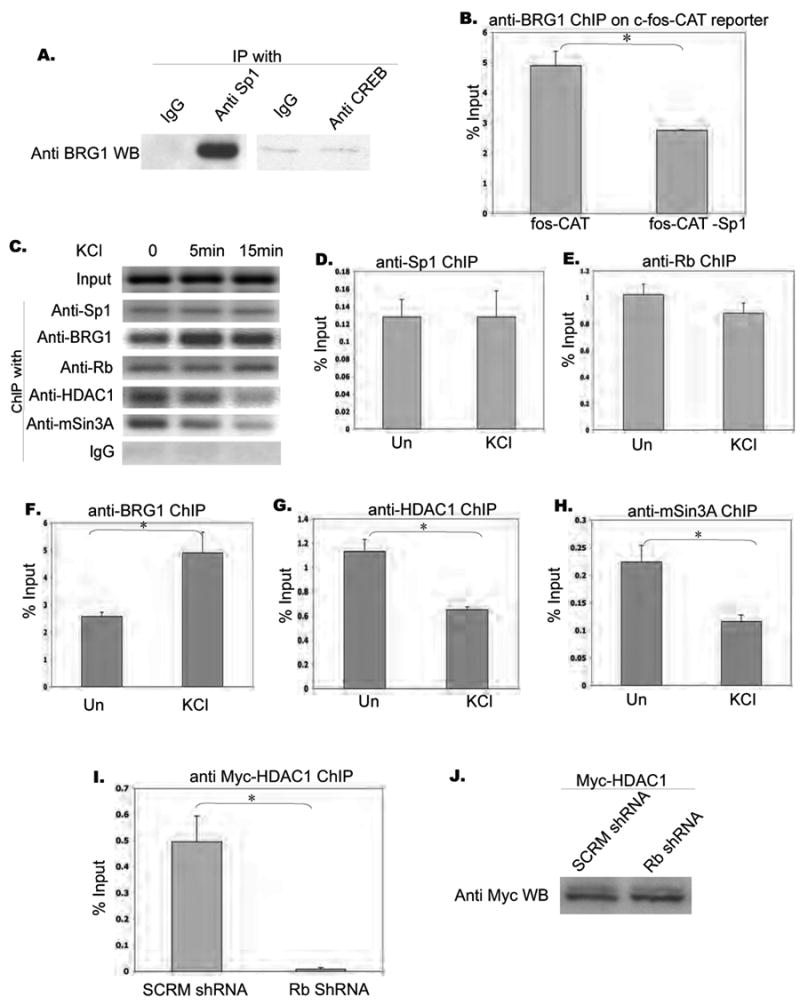
A. Immunoprecipitation of BRG1 with Sp1. Cortices from postnatal 4 day rat were dissected, homogenized, and immunoprecipitated with antibodies as indicated. The immunoprecipitates were resolved by SDS-PAGE and the Western blot was probed with an anti-BRG1 antibody.
B. Chromatin immunoprecipitation of BRG1 with c-fos-CAT reporter. Rat E18 cortical neurons were transfected with mouse -350 c-fos-CAT or -350 c-fos-CAT (-Sp1), and immunoprecipitated with antibodies as indicated. PCR reactions with ChIP F and ChIP R primers were used to amplify reporter-specific DNA segments. Real time PCR was performed and signals were normalized as percentage of input. ChIP with IgG was less than 0.05% of Input. Asterisks indicate significance at p<0.05. Error bars represent +SD.
C. Chromatin immunoprecipitation on c-fos promoter. Rat E18 cortical neurons were stimulated with KCl (50mM) for 5 minutes and 15 minutes at 5DIV, and immunoprecipitated with antibodies as indicated. PCR reactions with endogenous c-fos primers (upper panel) were used to amplify endogenous c-fos promoter-specific DNA segments from -200 to -1.
D. E. F. G. H. Real time PCR results of ChIP experiments. Data are collected from two sets of independent experiments, each with triplicate samples. Asterisks indicate significance at p<0.05. Error bars represent SD.
I. Chromatin immunoprecipitation of myc-HDAC1 with endogenous c-fos promoter. Myc-HDAC1 along with pSilencer 1.0 vector harboring scrambled shRNA or shRNA against rat Rb1 gene were transfected into rat E18 cortical culture at 3 DIV. Cells were collected and ChIPed with anti-Myc antibody at 6 DIV. Endogenous c-fos promoter primers were used to amplify enriched promoter segments that associated with mycHDAC1. Real time PCR was performed and signals were normalized as percentage of input. Asterisks indicate significance at p<0.05. Error bars represent +SD.
J. Input control shows that the same amount myc-HDAC1 was expressed in transfected neurons in the two conditions.
Mechanism of release of the HDAC complex after calcium stimulation
We next investigated whether calcium stimulation leads to a release of the BRG1 repressor complex from the c-fos promoter by examining the association and dissociation of various factors with the promoter before and after depolarization. Chromatin immunoprecipitation experiments showed that Sp1, BRG1, and Rb were associated with the promoter both before and after KCl stimulation (Fig. 4 C, D, E, F, G, H). There was even an increased recruitment of BRG1 after stimulation. In contrast, HDAC1 and mSin3A were associated with the promoter before stimulation, but were released from the promoter upon KCl stimulation (Fig. 4 G, H). Thus calcium stimulation leads to a rapid release of HDAC1 and mSin3A from the c-fos promoter.
To determine whether Rb is required for HDAC1 recruitment onto the c-fos promoter, we expressed myc-HDAC1 together with a scrambled shRNA and shRNA against rat Rb in rat cortical neurons (Liu et al., 2005). Chromatin immunoprecipitation of myc-HDAC1 indicated that binding of HDAC1 to the c-fos promoter was significantly reduced in Rb shRNA-expressing cells (Fig. 4 I, J). This, together with the co-immunoprecipitation data for Rb and HDAC1 (Fig. 3F), indicates that Rb is required for recruitment of HDAC1 onto the c-fos promoter, and implies that calcium influx must lead to a release of the HDAC1 from the c-fos promoter.
In the next set of experiments we investigated the mechanism by which the HDAC1 complex was released from the c-fos promoter upon calcium stimulation. To determine if this might be mediated by a post-translational modification of Rb, we examined whether Rb phosphorylation was regulated by calcium signaling. We decided to focus on Serine 795, as that site has been reported to be phosphorylated by MAP kinase, and MAP kinase has been implicated in calcium-dependent activation of c-fos (Garnovskaya et al., 2004). As shown in Figure 5A, Rb was phosphorylated at Serine 795 in unstimulated neurons. Surprisingly, KCl stimulation did not lead to an increase in Rb phosphorylation at this site and instead led to a rapid dephosphorylation of S795 (Fig. 5A). The dephosphorylation of Rb at S795 suggested that this was mediated by a calcium-dependent phosphatase, and we decided to examine the possibility that Rb dephosphorylation might be mediated by Calcineurin, a major calcium-regulated phosphatase in neurons that has been implicated in transcriptional regulation (Hogan et al., 2003, Kingsbury et al. 2007). To determine if Calcineurin activity was required for calcium-induced dephosphorylation of Rb, we examined the effects of cyclosoprin A, a Calcineurin inhibitor. As shown in Fig. 5A, KCl-induced dephosphorylation of Rb was completely blocked by cyclosporin A, indicating that Calcineurin activity was required for Rb dephosphorylation. In complementary experiments we found that purified Calcineurin was effective in dephosphorylating Rb immunoprecipitated from cortical lysates (Fig. 5B). Calcium-induced dephosphorylation of Rb was not affected by the CaM kinase inhibitor KN93, indicating that calcium regulation of Rb dephosphorylation is independent of CaM kinase activity (Supplemental Fig. 5).
Figure 5. Calcineurin dependent dephosphorylation of Rb and release of HDAC1 from c-fos promoter.
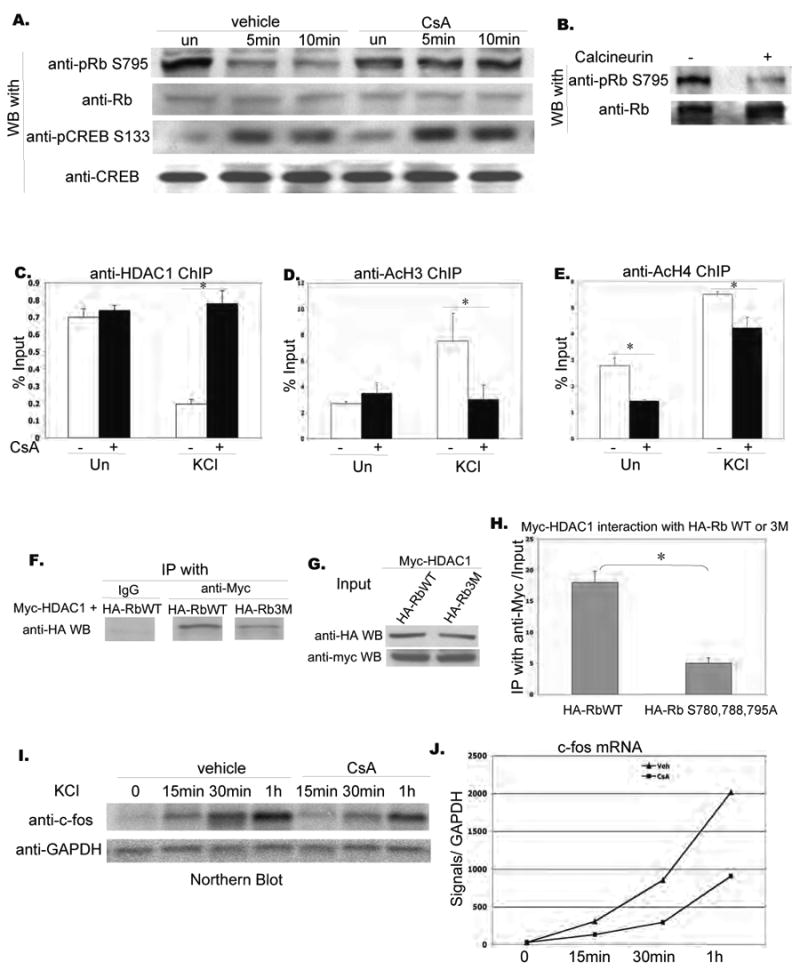
A. Assessment of Rb phosphorylation at S795 in unstimulated and stimulated neurons using a phospho-specific antibody and effect of cyclosporin A on depolarization-induced Rb dephosphorylation. Whole cell lysates from E18 cortical cultures stimulated as indicated with 50mM KCl were resolved by SDS-PAGE and probed with an anti-phospho-Rb S795, anti-pCREB S133 antibody and anti-Rb, anti-CREB antibody sequentially.
B. Effect of calcineurin on Rb phosphorylation at S795. Rb was immmunoprecipitated from rat E18 cortical cultures, and treated with or without activated CaM and human recombinant active calcineurin at 30 degree. Reactions were resolved by SDS-PAGE and probed with antibodies indicated.
C. Effect of cyclosporin A on association of HDAC1 with the c-fos promoter. Rat E18 cortical cultures were pre-treated with vehicle or cyclosporin A for 1 hr and then left unstimulated or stimulated with 50mM KCl for 10 minutes. Cells were lysed for chromatin immunoprecitation, immunoprecipitated with antibodies as indicated, and PCR reactions with endogenous c-fos promoter primers were used to amplify c-fos promoter-specific segments associated with the indicated proteins. Real time PCR was performed and signals were normalized as percentage of input.
D. E. Effect of cyclosporin A on histone acetylation of the c-fos promoter. Rat E18 cortical cultures were pre-treated with vehicle or cyclosporin A for 1 hr and then left unstimulated or stimulated with 50mM KCl for 10 minutes. Cells were lysed for chromatin immunoprecitation, immunoprecipitated with antibodies as indicated, and PCR reactions with endogenous c-fos promoter primers were used to amplify c-fos promoter-specific segments associated with the indicated proteins. Real time PCR was performed and signals were normalized as percentage of input. Asterisks indicate significance at p<0.05. Error bars represent +SD.
F. G. Immunoprecipitation of HDAC1 with Rb WT and phosphorylation mutant. Rat E18 cortical cultures were transfected with myc-HDAC1 with HA-Rb WT and HA-Rb 3M respectively. Cells are harvested 3 days after transfection, immunoprecipitated with anti-myc antibody and resolved on SDS-PAGE, and blotted with anti-HA antibody.
H. Quantification of co-immunoprecipitation experiments. Data are collected from two sets of independent experiments. Asterisks indicate significance at p<0.05. Error bars represent +SD.
I. Effects of calcineurin on endogenous c-fos expression upon KCl stimulation. Rat E18 cortical neurons were cultured and stimulated (KCl 50mM) for time indicated with pre-treated by vehicle and cyclosporin A respectively. Total RNA was collected with Trizol Reagent (Invitrogen), resolved by RNA agarose gel and probed by an antic-fos cDNA probe labeled with P32 followed by radioautography with X-ray films.
J. Quantification of Northern blot. Intensity of blots was measured using Image J.
To determine if Calcineurin activity was required for the release of HDAC1 from the c-fos promoter, we examined the effects of cyclosporin A on association of HDAC1 with the c-fos promoter as measured by chromatin immunoprecipitation. As shown in Fig. 5C, treatment with cyclosporin A completely blocked the release of HDAC1 from the c-fos promoter. Consistent with a role for HDAC1 release in c-fos activation, we found that treatment with cyclosporin A led to a significant decrease in the association of acetylated Histone H3 and H4 with the c-fos promoter (Fig. 5D, E).
We next examined whether the association of HDAC1 with Rb was dependent on the specific phosphorylation sites on Rb. Serine 780, Serine 788 and Serine 795 comprise a set of key phosphorylation sites on the Rb protein (Knudsen and Wang, 1997). Whereas we could effectively co-immunoprecipitate HA-WT-Rb and Myc-HDAC1 from transfected cells, Rb constructs that carried triple mutations in S780, S788, and S795 did not co-precipitate with Myc-HDAC1 with the same efficiency, suggesting that these phosphorylation sites are important for the association of Rb and HDAC1 (Fig. 5F, G, H). Consistent with a role for Calcineurin in calcium activation of the c-fos promoter, we found that treatment with cyclosporin A significantly attenuated KCl-induced expression of endogenous c-fos (Fig. 5I, J), as well as KCl activation of the c-fos-CAT reporter (data not shown).
These observations indicate that calcium influx leads to a Calcineurin-dependent dephosphorylation of Rb, which leads to a release of HDAC1 and activation of gene transcription. In the context of these experiments it is noteworthy that Calcineurin also contributes to calcium-dependent activation of MEF2-mediated transcription by regulating dephosphorylation (Flavell et al., 2006; Shalizi et al., 2006).
Role of CREST in activity-dependent regulation of NR2B expression
Finally, we were interested in determining whether the CREST complex was involved in regulating activity-dependent expression of other genes that have a direct effect on neuronal physiology. To identify activity-dependent genes that may be regulated by CREST, we carried out a microarray analysis of genes induced by treating cortical cultures with bicuculline for 6 hours. Bicuculline blocks GABA-A receptors and leads to increased network activity by suppressing inhibition. Analysis of this dataset showed that expression of the NMDA receptor subunit NR2B was significantly increased by bicuculline treatment (Fig. 6).
Figure 6. CREST mediates activity-dependent regulation of NR2B expression.
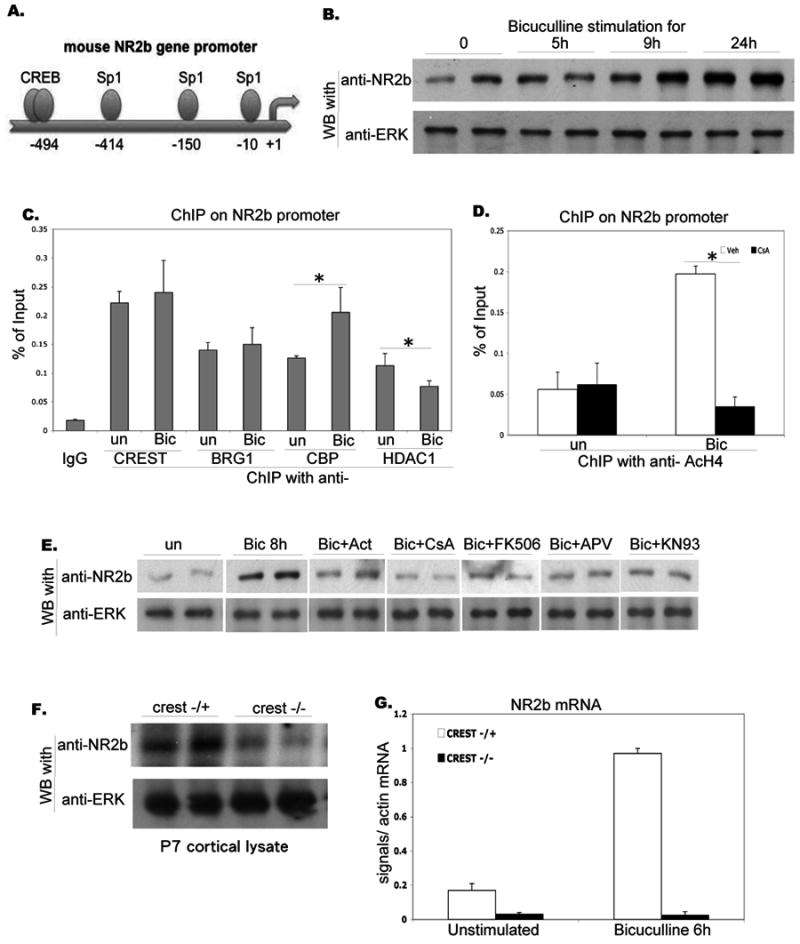
A. Schematic representation of the NR2B promoter. The promoter includes several Sp1 sites and a distal CREB site.
B. Activity-dependent regulation of NR2B expression. Rat E18 cortical neurons were cultured for 16DIV and stimulated with bicuculline for the period indicated. Proteins were separated by SDS-PAGE and blotted with NR2B and ERK antibodies as indicated.
C. Association of the CREST complex with the NR2B promoter. Lysates from unstimulated and bicuculline-stimulated (1 hr) E18 cortical cultures were processed for chromatin immunoprecipitation using antibodies indicated. PCR reactions with NR2B primers were used to amplify endogenous NR2B promoter-specific DNA segments from -200 to -1.
D. Activity-dependent changes in Histone acetylation at the NR2B promoter. Lysates from unstimulated and bicuculline-stimulated (1 hr) E18 cortical cultures were treated with vehicle or Cyclosporine A (CsA) and processed for chromatin immunoprecipitation using anti-acetylated Histone H4 antibodies and NR2B primers.
E. Signaling pathways involved in activity-dependent regulation of NR2B expression. Rat E18 cortical neurons were cultured for 16DIV, pretreated by indicated blockers for 1 hour, and stimulated with bicuculline for 8 hours. Proteins were separated by SDS-PAGE and blotted with NR2B and ERK antibodies as indicated.
F. Levels of NR2B proteins are reduced in CREST null brains. Cortical lysates from P7 crest-/- and crest+/- mice were collected, separated by SDS-PAGE, and blotted with NR2B and ERK antibodies as indicated.
G. CREST is required for activity-dependent regulation of NR2B expression. Embryonic 15 days cortical neurons from crest-/- and crest+/- were cultured for 12DIV and stimulated with bicuculline for 6 hours. Total RNA was collected, reverse-transcribed, and analyzed with NR2B-specific primers using real-time PCR. NR2B-specific signals, normalized with actin mRNA levels, are shown.
The NR2B promoter contains multiple Sp1 sites, suggesting that, as in the case of c-fos, NR2B expression might also be regulated by an BRG1-CREST complex (Sasner et al., 1996; Klein et al., 1998; Fig. 6A). To determine if proteins of the CREST-BRG1 complex associate with the NR2B promoter, we carried out chromatin immunoprecipitation experiments and found that that CREST, BRG1, CBP, and HDAC1 bind to the NR2B promoter (Fig. 6C). As with the c-fos promoter, stimulation led to an increase in CBP recruitment and a decrease in HDAC1 recruitment at the NR2B promoter (Fig. 6C). Consistent with transcriptional activation, chromatin immuniprecipitation experiments revealed a marked increase in the levels of acetylated Histone A4 antibody associated with the NR2B promoter following bicuculline stimulation. This increase was blocked by the Calcineurin inhibitor, Cyclosporin A. Western blot analysis showed that an increase in NR2B proteins levels was detectable by 5 hours after the onset of bicuculline-stimulation, and continued to increase over a period of 24 hours (Fig. 6B). The bicuculline-induced increase in NR2B expression was blocked by actinomycin D (suggesting a requirement for transcription), and by Cyclosporin A and FK506 (suggesting a requirement for Calcineurin) (Fig. 6E). The increase in NR2B expression also required NMDA receptor activation and CaM Kinase II activity (Fig. 6E).
To determine if the CREST complex was involved in regulating NR2B expression in vivo, we examined NR2B expression in crest heterozygous (control) and crest null neurons. Analysis of NR2B expression in cortical lysates showed that NR2B levels were significantly reduced in crest null mice at P7 (Fig. 6F). To determine if activity-dependent up-regulation of NR2B expression required CREST function, we examined NR2B mRNA levels in unstimulated and bicuculline-stimulated cultures in control and crest null cultures, and found that the bicuculline-induced increase in NR2B expression was completely absent in crest null neurons (Fig. 6G). These findings strongly suggest that the activity-dependent increase in NR2B expression involves a switch from a repressor to activator complex and requires CREST function.
Discussion
The observations described here reveal a novel regulatory mechanism involved in calcium dependent transcription. Previous studies on this problem have tended to focus on recruitment of transcriptional activators, with a particular focus on the CREB-CBP complex. Our investigation of CREST-mediated transcription reveals a much more intricate mechanism of transcriptional regulation. In resting neurons the CREST mediated transcription complex is negatively regulated the BRG1-Rb-HDAC1 complex. Upon calcium stimulation HDAC1 is released from the complex and CBP is recruited, which facilitates transcriptional activation. The contribution of CREST to calcium-dependent transcription is independent of the CRE, and represents a major new mechanism for calcium-dependent transcriptional activation.
An important contribution of this work is the identification of a novel role of a BRG1 complex in regulating activity-dependent transcription. The BRG1 complex has been extensively studied in the past few years as a core component of a chromatin remodeling complex. Here we find that the BRG1 complex plays a major role in negatively regulating transcriptional activation of specific promoters, that is subject to regulation by calcium signaling. The BRG1 complex associates with the c-fos promoter via its interaction with Sp1. In unstimulated neurons, BRG1 recruits Rb, which in turn recruits HDAC1 and mSin3A to the promoter to suppress gene expression. Depolarization-dependent calcium influx leads to a Calcineurin-dependent dephosphorylation of Rb at S795, and a release of HDAC1 from the promoter. This de-repression mechanism works in concert with calcium-dependent activation of a CREST-CBP complex to stimulate transcription. Based on these observations we suggest that a calcium dependent switch from a BRG1-Rb-HDAC1 repressor complex to a BRG1-CREST-CBP activator complex plays a critical role in transcriptional activation (Fig. 7).
Figure 7. Proposed mechanism for calcium-dependent activation of the proximal c-fos promoter.
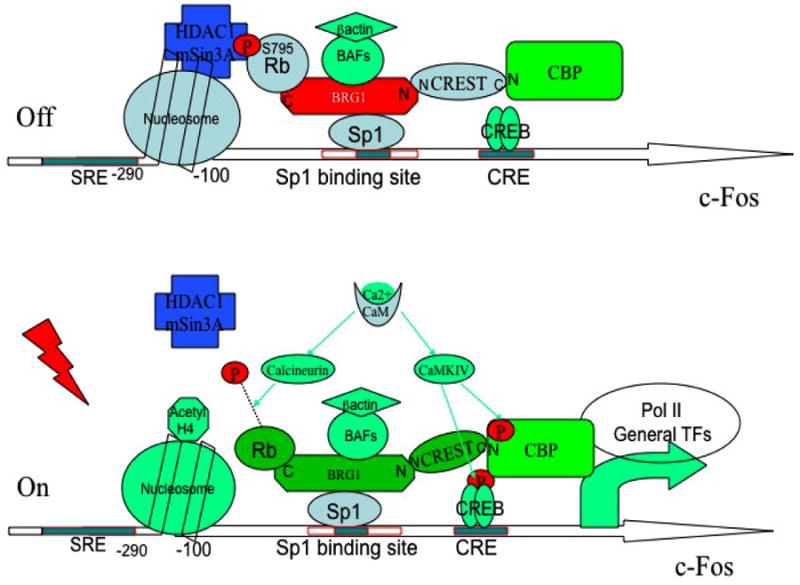
In unstimulated neurons transcription is repressed due to recruitment of an HDAC complex to the Sp1 site via BRG1 and Rb. Calcium stimulation leads to dephosphorylation of Rb via a calcineurin-dependent mechanism and a release of the HDAC complex. At the same time, the transcriptional co-activator CBP (which is recruited to the promoter by CREST and CREB) is phosphorylated by CaM kinase IV and mediates transcriptional activation.
Interaction of CREST with BRG1 and characterization of the BRG1-associated repressor complex
CREST is 62% identical in protein sequence with a proto-oncoprotein SYT, and the two proteins share several molecular interactions. We had previously shown that CREST interacts with CBP (Aizawa et al., 2004), and here we show that CREST directly binds BRG1. Recent results from the Crabtree group also indicate that CREST is tightly associated with the BRG1 complex (Wu et al. 2007). Similar to CREST, SYT can directly interact with N terminus of BRG1 (Perani et al. 2003). Moreover SYT is found to interact with p300, a transcription activator similar with CBP (Eid et al., 2000). The similarities between the CREST and SYT complexes suggest that SYT-mediated transcription might also be bidirectionally regulated by BRG1 and CBP/p300. Whether SYT can also mediate calcium-dependent transcription remains to be determined.
BRG1 is the core component of an ATPase-dependent chromatin remodeling complex. ATPase-dependent chromatin remodeling complexes use the energy from ATP hydrolysis to move nucleosomes, unfold DNA and facilitate transcription. They are conserved from yeast to mammals. BRG1 has ATPase activity, and is the human homolog of SNF2b, which is a component of SWI/SNF complex in yeast. Besides SWI/SNF family, there are two other ATPase dependent chromatin remodeling complexes, Chromodomain and helicase-like domain (CHD) family and ISWI family (de la Serna et al., 2006). These three families use different ATPases as core enzymes. Although the BRG1 chromatin remodeling complex has been studied mainly with regard to its role in transcriptional activation, previous observations hint at a negative role in regulating gene expression. For example, after inactivating SWI/SNF chromatin remodeling complex in yeast, it was found that most of the genes whose expression is altered are upregulated, suggesting that they are negatively regulated by the SWI/SNF complex (Holstege et al., 1998). Similarly, a previous study reported that over-expression of BRG1 could inhibit basal level c-fos expression in various in vitro cell lines via the existence of Rb protein although the mechanism was not identified (Murphy et al., 1999). The observations presented here clearly show that the BRG1 complex can directly repress transcriptional activation by recruitment of a repressor complex to the promoter.
Our experiments strongly suggest a repressive role of BRG1 at the c-fos promoter. Inhibition of BRG1 expression by a BRG1 shRNA increases calcium-dependent activation of the c-fos promoter, and over-expressing BRG1 inhibits calcium-dependent activation of the promoter. Furthermore, BRG1 associates with the c-fos promoter and can associate with Retinoblastoma protein and an HDAC complex to regulate promoter activation. The association of BRG1with an HDAC complex was first reported by Sif and colleagues (Sif et al., 2001; Battaglioli et al., 2002), but the mechanism of recruitment was not known. Our observations indicate that recruitment of HDAC1 by BRG1 is mediated by the Rb protein. Interestingly the inhibitory effect of BRG1 requires ATPase activity, which is consistent with a previous study (Qiu and Ghosh, unpublished data; Murphy et al., 1999). It will be of interest to determine if recruitment of Rb and HDAC1 by BRG1 requires chromatin remodeling.
Since BRG1, Rb, and HDAC1 do not have sequence-specific DNA-binding domains, they must be recruited to specific promoter sites via an indirect mechanism. Our results indicate that the transcription factor Sp1 associates with BRG1, providing a potential mechanism for recruitment of the BRG1 complex to the c-fos promoter. This interpretation is consistent with previous observations that Rb can negatively regulate c-fos expression via the so-called Rb control element (RCE), which corresponds to the Sp1 binding site CCG/ACCC (Udvadia et al., 1993). The Sp1 binding site is directly upstream of the cAMP responsible element (CRE), TGACGTAG. Although the Sp1 site was identified a long time ago, its role in calcium-dependent transcription had not been explored. We have found that both the CRE and the Sp1 site contribute to calcium dependent activation of the c-fos promoter (Supplemental Fig. 6). ChIP experiments indicate that both CREB and Sp1 are bound to the c-fos promoter before and after KCl stimulation. Following calcium influx there is a rapid release of HDAC1 from the promoter and an increase in CBP recruitment. This switch is likely to be critical for the transcription activation program.
Release of the repressor complex by calcium/Calcineurin signaling
What is the mechanism by which calcium influx leads to recruitment of CBP and the release of HDAC1? CBP is recruited to the promoter by association with phosphorylated CREB, as well as by interaction with CREST. The significance of the CREST-CBP interaction for transcription activation is highlighted by the fact that calcium activation of Gal4-CREST is completely blocked by expression of E1A cxdl domain, and association of CBP with the c-fos promoter is significantly reduced in CREST null neurons. As previously described, the association of CREST and CBP is mediated by the C-terminal of CBP (Aizawa et al., 2004).
The recruitment and release of HDAC1 to the promoter is mediated by BRG1. BRG1 interacts with Rb, which in turn interacts with HDAC1. Calcium-dependent release of HDAC1 appears to be mediated by calcium-induced dephosphorylation of Rb. We find that Rb is phosphorylated at Serine 795 in resting neurons. This site becomes rapidly dephosphoryated upon calcium stimulation. Inhibiting Calciuneurin activity prevents dephosphorylation of Rb, as well as release of HDAC1 from the promoter. These observations reveal a novel mechanism by which Calcineurin can regulate association of a Histone deacetylase with a promoter and show that the recruitment and release of histone modifiers can be regulated by calcium signaling.
Implication for mechanisms of plasticity
Much of our understanding of molecular mechanism that mediate adaptive responses in the nervous system has come from investigations of activity-dependent gene expression (Ghosh and Greenberg, 1995). For example, CREB was identified as a mediator of cAMP- and calcium-dependent transcription before it was implicated a mediator of learning and memory processes (Silva et al., 1998). It is likely that the transcription regulatory mechanisms identified here will also play key regulatory roles in mediating activity-dependent development and plasticity. Consistent with such a possibility, we find that CREST is associated with the NR2B promoter, and activity-dependent increases in NR2B levels require CREST function. We also find that CREST and BRG1 are associated with promoters of several plasticity-related genes, including Arc and zif268, suggesting that activity-dependent transcription of these genes might also depend on the BRG1-CREST complex (Supplemental Figure 7 A,B). The NR2B, Arc, and zif268 promoters all contain Sp1 sites, and it will be interesting to determine if BRG1 is recruited to these promoters via Sp1, and if activity-dependent transcription of these promoters involves release of HDAC1, as we find to be the case for c-fos expression.
The potential importance of the BRG1-CREST complex in adaptive responses is also supported by the fact that this complex recruits CBP, which has been implicated in regulation of cognitive functions such as learning and memory (Alarcon et al., 2004; Korzus et al., 2004). Our observation that calcium signaling regulates release of HDAC1 from the BRG1-Rb suggests that this is an important regulatory step in activity-dependent gene expression, and may be important for learning and memory. Consistent with such a possibility, it was recently reported that intracerebroventricular injections of HDAC inhibitors sodium butyrate and TSA significantly facilitated associative learning, such as fear conditioning and spatial learning (Fischer et al., 2007). Our findings suggest that the calcium-dependent switch in BRG1-CREST-associated complexes could be generally involved in mediating adaptive changes in neurons that underlie neural development and long-term plasticity.
Experimental Procedures
Plasmids
The following plasmids used in this study have been previously described: UAS-CAT, Gal4-CREST and deletion mutants, HA-CREST, Myc-CREST and E1A cxdl (Aizawa, et al., 2004, Hu, et al., 1999). pcDNA3-WT BRG1-flag construct was made by enzymatic digestion-based subcloning from original human BRG1 constructs from Dr. Anthony Imbalzano at University of Massachusetts. pcDNA3-BRG1N(1-282)-flag, GSTBRG1N(1-283) and GST-BRG1C(1371-1573) was obtained from Dr. Beverly Emerson at the Salk institute. pcDNA3-MT BRG1 (LTCEE-RTREE)-Flag was made from pcDNA3-wtBRG1-flag using the QuikChange II kit from Stratagene. Retinoblastoma protein wild type and Δexon 22 constructs were from Dr. James DeCaprio at Dana-Farber Cancer Institute. Human HDAC1 cDNA was obtained from Dr. M.G. Rosenfeld at UCSD. The c-fos-CAT reporter used included the mouse c-fos gene from -350 to +50 fused with the CAT gene. -107 c-fos-CAT contains sequences from -107 to +50 fused with CAT gene. Sp1 mutation changes -73 CCGCCC to AAATTT. CRE mutation changes -65 TGACGTAG to TGGGAGTG.
Molecular Biology reagents
general molecular biology reagents, competent cells, real-time PCR mix and reverse transcription kits are from Biopioneer. Inc. San Diego, CA. http://www.biopioneerinc.com.
Antibodies
Antibodies were obtained from the following sources: BRG1 G7, CREB, CBP C22, HDAC1, HDAC2, HDAC3, mSin3A, Rb IF8, Sp1 and control rabbit IgG (Santa Cruz Biotechnology); HA 16B12 (Covance); c-fos ab5 clone (EMD PC38), Rb phospho-specific S795 (Assay Bio Tech); myc 9E10 (Roche) and Flag M2 (Sigma). Antibodies against CREST were generated in our lab (Aizawa et al., 2004).
Cell culture
E18 rat cortical cells were cultured and transfected in 12-well plates as previously described (Song and Ghosh, 2004). Cultures were used at 3-5 DIV. We used 50mM KCl depolarization to activate calcium-dependent transcription. For TSA treatment, cells were pretreated either trichostatin A (250ng/ml) or ethanol (for the vehicle group) prior to stimulation for 2 hours. For CsA treatment, cells were pre-treated with cyclosporin A (Calbiochem)(1μM) or DMSO prior to stimulation for 1 hour. For VT-11R treatment, cells were pre-treated with 11R-VIVIT (Calbiochem, 480401) (1μM) or DMSO prior to simulation for 1 hour.
Immunoprecipitation
For endogenous IP, cortices from postnatal 4 day rat were dissected in cold HBSS buffer, homogenized in RIPA buffer with protease inhibitor cocktail (Roche) and immunoprecipitated with 2μg antibodies for each reactions in 4°C for overnight and followed by 30 μl of a 50% slurry of mixed protein-A/G agarose (Santa Cruz) for 1 hour for each reaction. The precipitates were then washed four times with the lysis buffer, then eluted by boiling-SDS lysis and resolved by the 6% SDS-PAGE. The gel was transferred to nitrocellulose membranes, and the membrane was blocked with 5%milk in TBST buffer for 1 hour. It was then incubated overnight at 4°C with the antiBRG1 G7 antibody (Santa Cruz Biotechnology) at 1:2,000 dilution, washed three times in TBST, and the signals were revealed by HRP reaction using the Super signal chemiluminescent substrate (Pierce).
For nuclear fraction IP, nuclear fractions from 5DIV cultured cortical neurons are collected with Nuclear Complex co-IP kit (Active Motif, Cat#54001) as manual described. Gentle procedures were applied to all steps.
For co-IP in 293T cells, HEK 293T cells were plated on 60 mm plates pre-coated with poly-D-lysine in 3 ml of high-glucose DMEM supplemented with fetal bovine serum (10%), and transfection was performed when the cells reached 50% confluence. Lipofectamine 2000 (Invitrogen) was used for transfection. A total of 5 μg of DNA was used per well in 6-wells plates at a molar ratio of 1:1 for myc-tagged and HA-tagged constructs. Cells were harvested 24 hours later. The cells were rinsed with cold PBS, harvested, and lysed for 20 min at 4°C in a modified RIPA buffer. 10 percent of the supernatant was saved for the input control, and the rest was incubated with 2 μg anti-HA antibody 16B12 (covance) or anti-myc 9E10 (Roche) overnight at 4°C. The immune complex was isolated by addition of 30 μl of a 50% slurry of mixed protein-A/G agarose (Santa Cruz) for 1 hour, washed three times with the lysis buffer, then eluted by boiling-SDS lysis and resolved by the 10%SDS-PAGE. The gel was transferred to nitrocellulose membranes, and the membrane was blocked with 5% milk in TBST buffer for 1 hour. It was then incubated overnight at 4°C with the anti-Flag M2 (Sigma) at 1:5,000 dilution, washed three times in TBST, and the signals were revealed by HRP reaction using the Supersignal chemiluminescent substrate (Pierce).
GST pull down assay
GST pull down assay was performed as described before (Aizawa et al., 2004). Overnight express TB medium from Novagen (Cat#71491-4) was used to generate recombinant protein from E.coli.
Transactivation assay
Detailed procedures for CAT assay had been described before (Hu et al., 1999). Cells are harvested 20 hours after stimulation unless other indicated. CAT assay experiments were done with triple duplicates. Paired t tests were performed using GraphPad InStat version 3.0a for Macintosh, GraphPad Software, San Diego California USA, www.graphpad.com.
Chromatin immunoprecipitation
ChIP assays were performed as originally described with minor modifications (Song and Ghosh, 2004). Primers: c-fos F: ATCCTACATGCGGAGGGTCCAGGA, c-fos R: AGTAGTAGGCGCCTCAGCTGGCCG. For reporter-based chromatin IP, a differentprimer pairs was used. ChIP F: AGTGACGTAGGAAGTCCATC, ChIPR: CTCCCGGGGATCCTCTAGAG. Egr1 promoter ChIP primers are -149 to +23 of mouse Egr1 gene. Arc promoter primers amplify -200 to -1 of mouse Arc gene.
For real time PCR experiments, SYBR Green PCR master mix from Applied Biotechnology was used. ABI 7000 was used to perform experiments. Data set are from at least two independent experiments, each of which has triple duplicates. Paired t tests were performed using GraphPad InStat version 3.0a for Macintosh, GraphPad Software, San Diego California USA, www.graphpad.com.
In vitro dephosphorylation assay
Immunoprecipitation purified Rb proteins from 5DIV E18 rat cortical culture combine reaction assay buffer, CaM and with or without recombinant active calcineurin (BIOMOL), incubating at 30 degree for 2 hours. Reaction products are analyzed by Western blot with antibody indicated.
Short hairpin RNA target design
pSuper retro neo GFP vector from Oligoengine was used to harbor CREST specific Short hairpin RNA. Workstation software from Oligoengine was used to design short hairpins. CREST specific shRNA target sequence is GGTCAGCAGTATGGAAGCT. Scrambled sequence (control) is ATAGACGCGCACGCACACT. Rescue construct was designed to replace original DNA bases by introducing same sense mutations, which are resistant to short hairpin RNA and don't change the amino acid sequences. Replacing sequence is GGACAACAATACGGTAGTT. Target sequence for rat BRG1 is CCAAAGCAACCATCGAACT. Target sequence for rat Rb1 is GGAGCACGAGTGTAATGTA. pSilencer 1.0 vector from Ambion is also used for harboring rat Rb1 shRNA.
Measurements of dendritic growth
GFP expressing plasmid was transfected into rat E18 cortical cultures at 3DIV, pretreatment and KCl stimulation were done at 5DIV. Immunostaining and imaging experiments were carried out at 24 hours after stimulation. Image J software from NIH was used for measuring dendritic growth. Sholl analysis plug-in could be downloaded from http://wwwbiology.ucsd.edu/labs/ghosh/software/index.html. Paired t tests were performed using GraphPad InStat version 3.0a for Macintosh, GraphPad Software, San Diego California USA, www.graphpad.com.
Supplementary Material
Supplemental Figure 1. Determination of efficacy of BRG1 shRNA
Rat E18 cortical culture transfected with pSuper vectors harboring scrambled shRNA or shRNA against rat BRG1. Neurons were immunostained with anti-BRG1 antibody 3 days after transfection, as shown in lower panel. Quantification is shown at upper panel. N=6, Asterisks indicate significance at p<0.05. Error bars represent +SD.
Supplemental Figure 2. Specificity of the transcriptional repressive effect of BRG1
A. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREST and UAS-CAT along with pSuper vector harboring short hairpin RNA against GFP and rat BRG1 constructs at 3DIV. The same amounts of b-galactosidase and wild type human BRG1 constructs were co-transfected to perform rescue experiments. Transfected neurons were stimulated as indicated at 5 DIV (KCl 50 mM).
B. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREB and UAS-CAT along with pSuper vector harboring short hairpin RNA against GFP and rat BRG1 constructs at 3DIV. The same amounts of b-galactosidase and wild type human BRG1 constructs were co-transfected to perform rescue experiments. Transfected neurons were stimulated as indicated at 5 DIV (KCl 50 mM).
C. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREST full length and UAS-CAT along with b-galactosidase and wild type human BRG1 constructs at 3DIV, and stimulated with KCl (50mM) as indicated at 5 DIV.
D. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREST ΔN1 and UAS-CAT along with b-galactosidase and wild type human BRG1 constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
E. Relative CAT activity in E18 cortical neurons transfected with Gal4-NeuroD1 and UAS-CAT reporter along with b-galactosidase and wild type BRG1 constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM). CAT assay experiments were done with triple duplicates. Asterisks indicate significance at p<0.05. Error bars represent +SD.
Supplemental Figure 3. Specificity of TSA treatment
A, B. Relative CAT activity in E18 cortical neurons transfected with -350 c-fos-CAT reporter along with β galactosidase and ACREB at 3DIV. Cells were pre-treated with vehicle or TSA for 1 hour and stimulated with KCl (50mM) at 5DIV. TSA reverses effect of BRG1 transfection (Fig. 3C), but not ACREB transfection.
Supplemental Figure 4. Sp1 binding site is responsible for BRG1 recruitment on cfos promoter
A. Strategy of reporter-based chromatin immunoprecipitation. PCR primers used here are indicated as ChIP F and ChIP R.
B. Chromatin immunoprecipitation of BRG1 with c-fos-CAT reporter. Rat E18 cortical neurons were transfected with mouse -290 c-fos-CAT or -67 c-fos-CAT, and immunoprecipitated with antibodies as indicated. PCR reactions with ChIP F and ChIP R primers were used to amplify reporter-specific DNA segments.
Supplemental Figure 5. CaM kinase inhibitors do not affect calcium dependent Rb dephosphorylation
Rat E18 cortical neurons were cultured and stimulated for 10 minutes (50mM KCl) at 5DIV, with pre-treated by KN92 and KN93 respectively. Neurons were lysed by boiling SDS lysis buffer, resolved by SDS-PAGE, and probed with antibodies indicated.
Supplemental Figure 6. Sp1 binding site contributes to calcium-dependent c-fos-CAT activation.
Relative CAT activity in E18 cortical neurons transfected with mouse -107 c-fos-CAT, -107 c-fos-CAT (-Sp1) and -107 c-fos-CAT (-CRE) reporter constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
Supplemental Figure 7. Presence of CREST, BRG1, CBP and HDAC at multiple activity-dependent promoters.
A, B. Recruitment of CREST, BRG1, CBP and HDAC1 at the Arc, Zif268/Egr1 promoter. Rat E18 cortical cultures at 5DIV were lysed for chromatin immunoprecitation, immunoprecipitated with antibodies as indicated, and PCR reactions with endogenous promoter primer sets were used to amplify Arc, Zif268/Egr1 promoter-specific segments associated with the indicated proteins. Real time PCR was performed and signals were normalized as percentage.
Acknowledgments
We would like to thank Drs. A. Imbalzano, Jean.Y.Wang, B. Emerson, J. DeCaprio and Xiang-Ting (Tina) Wang, Ping Zhu and M.G. Rosenfeld for providing plasmids, Drs. Stefanie Otto, Yang Xu and Hoseok Song for helping with real time PCR experiments, Dr. Megan Williams for helping imaging experiments, Drs. M. Montminy, G. R. Crabtree, Ping Zhu and members of the Ghosh Lab for valuable discussion and comments on the manuscript, and Mrs. Fang Gao for instructions about Word and Excel software. We would like to give special thanks to Drs. G.R. Crabtree and J Wu for sharing data prior to publication. This work was supported by NIH grant MH068578 (AG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Battaglioli E, Andres ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G. REST repression of neuronal genes requires components of the hSWI.SNF complex. J Biol Chem. 2002;277:41038–41045. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Eid JE, Kung AL, Scully R, Livingston DM. p300 interacts with the nuclear proto-oncoprotein SYT as part of the active control of cell adhesion. Cell. 2000;102:839–848. doi: 10.1016/s0092-8674(00)00072-6. [DOI] [PubMed] [Google Scholar]
- Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Garnovskaya MN, Mukhin YV, Vlasova TM, Grewal JS, Ullian ME, Tholanikunnel BG, Raymond JR. Mitogen-induced rapid phosphorylation of serine 795 of the retinoblastoma gene product in vascular smooth muscle cells involves ERK activation. J Biol Chem. 2004;279:24899–24905. doi: 10.1074/jbc.M311622200. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/s0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hu SC, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN. Early exploration of the visual cortex. Neuron. 1998;20:401–412. doi: 10.1016/s0896-6273(00)80984-8. [DOI] [PubMed] [Google Scholar]
- Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kingsbury TJ, Bambrick LL, Roby CD, Krueger B. Calcineurin activity is required for depolarization-induced, CREB-dependent gene transcription in cortical neurons. J Neurochem. 2007;103:761–70. doi: 10.1111/j.1471-4159.2007.04801.x. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Ladanyi M. Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene. 2001;20:5755–5762. doi: 10.1038/sj.onc.1204601. [DOI] [PubMed] [Google Scholar]
- Liu DX, Nath N, Chellappan SP, Greene LA. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev. 2005;19:719–732. doi: 10.1101/gad.1296405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Hardy S, Engel DA. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perani M, Ingram CJ, Cooper CS, Garrett MD, Goodwin GH. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene. 2003;22:8156–8167. doi: 10.1038/sj.onc.1207031. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Riccio A, Alvania RS, Lonze BE, Ramanan N, Kim T, Huang Y, Dawson TM, Snyder SH, Ginty DD. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Robbins PD, Horowitz JM, Mulligan RC. Negative regulation of human c-fos expression by the retinoblastoma gene product. Nature. 1990;346:668–671. doi: 10.1038/346668a0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Sohm F, Gaiddon C, Antoine M, Boutillier AL, Loeffler JP. The retinoblastoma susceptibility gene product/Sp1 signalling pathway is modulated by Ca2+/calmodulin kinases II and IV activity. Oncogene. 1999;18:2762–2769. doi: 10.1038/sj.onc.1202634. [DOI] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Watanabe T, Seki T, Kimura T, Sawa H, Minami A, Akagi T, Isobe K, Nagashima K, Tanaka S. Induction of p21(WAF1/CIP1) by human synovial sarcoma-associated chimeric oncoprotein SYT-SSX1. Oncogene. 2005;24:7984–7990. doi: 10.1038/sj.onc.1208942. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Rogers KT, Higgins PD, Murata Y, Martin KH, Humphrey PA, Horowitz JM. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci U S A. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J Neurosci. 2001;21:5484–5493. doi: 10.1523/JNEUROSCI.21-15-05484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Dean DC. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene. 2001;20:3134–3138. doi: 10.1038/sj.onc.1204338. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Determination of efficacy of BRG1 shRNA
Rat E18 cortical culture transfected with pSuper vectors harboring scrambled shRNA or shRNA against rat BRG1. Neurons were immunostained with anti-BRG1 antibody 3 days after transfection, as shown in lower panel. Quantification is shown at upper panel. N=6, Asterisks indicate significance at p<0.05. Error bars represent +SD.
Supplemental Figure 2. Specificity of the transcriptional repressive effect of BRG1
A. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREST and UAS-CAT along with pSuper vector harboring short hairpin RNA against GFP and rat BRG1 constructs at 3DIV. The same amounts of b-galactosidase and wild type human BRG1 constructs were co-transfected to perform rescue experiments. Transfected neurons were stimulated as indicated at 5 DIV (KCl 50 mM).
B. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREB and UAS-CAT along with pSuper vector harboring short hairpin RNA against GFP and rat BRG1 constructs at 3DIV. The same amounts of b-galactosidase and wild type human BRG1 constructs were co-transfected to perform rescue experiments. Transfected neurons were stimulated as indicated at 5 DIV (KCl 50 mM).
C. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREST full length and UAS-CAT along with b-galactosidase and wild type human BRG1 constructs at 3DIV, and stimulated with KCl (50mM) as indicated at 5 DIV.
D. Relative CAT activity in rat E18 cortical neurons transfected with Gal4 DBD-CREST ΔN1 and UAS-CAT along with b-galactosidase and wild type human BRG1 constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
E. Relative CAT activity in E18 cortical neurons transfected with Gal4-NeuroD1 and UAS-CAT reporter along with b-galactosidase and wild type BRG1 constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM). CAT assay experiments were done with triple duplicates. Asterisks indicate significance at p<0.05. Error bars represent +SD.
Supplemental Figure 3. Specificity of TSA treatment
A, B. Relative CAT activity in E18 cortical neurons transfected with -350 c-fos-CAT reporter along with β galactosidase and ACREB at 3DIV. Cells were pre-treated with vehicle or TSA for 1 hour and stimulated with KCl (50mM) at 5DIV. TSA reverses effect of BRG1 transfection (Fig. 3C), but not ACREB transfection.
Supplemental Figure 4. Sp1 binding site is responsible for BRG1 recruitment on cfos promoter
A. Strategy of reporter-based chromatin immunoprecipitation. PCR primers used here are indicated as ChIP F and ChIP R.
B. Chromatin immunoprecipitation of BRG1 with c-fos-CAT reporter. Rat E18 cortical neurons were transfected with mouse -290 c-fos-CAT or -67 c-fos-CAT, and immunoprecipitated with antibodies as indicated. PCR reactions with ChIP F and ChIP R primers were used to amplify reporter-specific DNA segments.
Supplemental Figure 5. CaM kinase inhibitors do not affect calcium dependent Rb dephosphorylation
Rat E18 cortical neurons were cultured and stimulated for 10 minutes (50mM KCl) at 5DIV, with pre-treated by KN92 and KN93 respectively. Neurons were lysed by boiling SDS lysis buffer, resolved by SDS-PAGE, and probed with antibodies indicated.
Supplemental Figure 6. Sp1 binding site contributes to calcium-dependent c-fos-CAT activation.
Relative CAT activity in E18 cortical neurons transfected with mouse -107 c-fos-CAT, -107 c-fos-CAT (-Sp1) and -107 c-fos-CAT (-CRE) reporter constructs at 3DIV, and stimulated as indicated at 5 DIV (KCl 50 mM).
Supplemental Figure 7. Presence of CREST, BRG1, CBP and HDAC at multiple activity-dependent promoters.
A, B. Recruitment of CREST, BRG1, CBP and HDAC1 at the Arc, Zif268/Egr1 promoter. Rat E18 cortical cultures at 5DIV were lysed for chromatin immunoprecitation, immunoprecipitated with antibodies as indicated, and PCR reactions with endogenous promoter primer sets were used to amplify Arc, Zif268/Egr1 promoter-specific segments associated with the indicated proteins. Real time PCR was performed and signals were normalized as percentage.


