Abstract
In a manner unique among activity-regulated immediate early genes (IEGs), mRNA encoded by Arc (also known as Arg3.1) undergoes rapid transport to dendrites and local synaptic translation. Despite this intrinsic appeal, relatively little is known about the neuronal and behavioral functions of Arc or its molecular mechanisms of action. Here, we attempt to distill recent advances on Arc spanning its transcriptional and translational regulation, the functions of the Arc protein in multiple forms of neuronal plasticity [long-term potentiation (LTP), long-term depression (LTD), and homeostatic plasticity], and its broader role in neural networks of behaving animals. Worley and colleagues have shown that Arc interacts with endophilin and dynamin, creating a postsynaptic trafficking endosome that selectively modifies the expression of AMPA-type glutamate receptors at the excitatory synapses. Both LTD and homeostatic plasticity in the hippocampus are critically dependent on Arc-mediated endocytosis of AMPA receptors. LTD evoked by activation of metabotropic glutamate receptors depends on rapid Arc translation controlled by elongation factor 2. Bramham and colleagues have shown that sustained translation of newly induced Arc mRNA is necessary for cofilin phosphorylation and stable expansion of the F-actin cytoskeleton underlying LTP consolidation in the dentate gyrus of live rats. In addition to regulating F-actin, Arc synthesis maintains the activity of key translation factors during LTP consolidation. This process of Arc-dependent consolidation is activated by the secretory neurotrophin, BDNF. Moore and colleagues have shown that Arc mRNA is a natural target for nonsense-mediated mRNA decay (NMD) by virtue of its two conserved 3′-UTR introns. NMD and other related translation-dependent mRNA decay mechanisms may serve as critical brakes on protein expression that contribute to the fine spatial-temporal control of Arc synthesis. In studies in behaving rats, Guzowski and colleagues have shown that location-specific firing of CA3 and CA1 hippocampal neurons in the presence of theta rhythm provides the necessary stimuli for activation of Arc transcription. The impact of Arc transcription in memory processes may depend on the specific context of coexpressed IEGs, in addition to posttranscriptional regulation of Arc by neuromodulatory inputs from the amygdala and other brain regions. In sum, Arc is emerging as a versatile, finely tuned system capable of coupling changes in neuronal activity patterns to diverse forms of synaptic plasticity, thereby optimizing information storage in active networks.
Keywords: synaptic plasticity, long-term potentiation, long-term depression, glutamate receptor, nonsense-mediated RNA decay, actin cytoskeleton, translation control, learning, memory
Introduction
The immense capacity and specificity of memory storage in the mammalian CNS is thought to depend on the plasticity of neuronal communication at synapses. Dysfunction of synaptic plasticity is implicated in a range of disorders from Alzheimer's disease to mental retardation and development of chronic pain states. Understanding how neural activity patterns are translated into lasting changes in synaptic connectivity that shape neural network functions and behavior is therefore a major goal. Glutamatergic synapses are capable of expressing diverse forms of activity-dependent potentiation and depression of synaptic efficacy. Persistent forms of synaptic change, as seen in long-term potentiation (LTP) and long-term depression (LTD), typically require rapid new gene expression. Although these complex genomic responses are increasingly understood, it has proven difficult to show how changes in specific activity-regulated genes contribute to functional and structural modifications of synapses.
The mRNA encoded by the immediate early gene (IEG) Arc (activity-regulated cytoskeleton-associated protein), also known as activity-regulated gene 3.1 (Arg3.1), traffics to dendrites and specifically accumulates at sites of synaptic activity (Link et al., 1995; Lyford et al., 1995; Steward et al., 1998). Arc protein also accumulates in dendrites and becomes enriched at the site of local synaptic activity suggesting that Arc protein is locally synthesized (Steward et al., 1998; Yin et al., 2002; Moga et al., 2004; Rodríguez et al., 2005). These unique features implied a function for Arc in coupling synaptic activity to protein synthesis-dependent synaptic plasticity. The goal of the symposium was to collate recent advances in this area, spanning the transcriptional and translational regulation of Arc, its function in multiple forms of neuronal plasticity (LTP, LTD, and homeostatic plasticity), and its role in neural networks and behavior. In writing this minireview, we have attempted to integrate the findings presented at the symposium with other recent advances in the field.
Mechanisms and cellular functions of Arc in LTD and homeostatic plasticity
Arc is a single copy gene that encodes a single protein that is highly conserved in vertebrates and is induced in divergent behavioral paradigms in many species. Initial studies of predicted it to be a cytosolic protein, and noted a weak homology with spectrin (Lyford et al., 1995). Biochemical studies demonstrated that Arc protein coprecipitates with polymerized actin. Moreover, the intracellular distribution of Arc was noted to be coextensive with polymerized actin in dendritic spines, suggesting that it may contribute to actin-dependent cell biology. However, Arc did not coprecipitate with pure actin, and the relevant protein for this interaction remains unknown. Subsequent studies examined Arc protein interactions using the yeast two-hybrid system and identified two proteins, dynamin and endophilin, that are known to play a role in endocytosis (Chowdhury et al., 2006). The interaction between Arc and endophilin is notable in that it is selective for endophilin 2 and endophilin 3, which are enriched with Arc in the postsynaptic compartment. The Arc-endophilin interaction requires the C terminus of the BAR domain of endophilin, which is thought to contact the curved lipid bilayer of the endosome. The Arc–dynamin interaction requires the PH domain of dynamin, which also is thought to be important for dynamin association with charged lipids. When coexpressed in heterologous cells, Arc, endophilin2/3, and dynamin coassociate on the surface of endosomes that traffic transferrin and EGFR. When coexpressed in neurons, Arc and endophilin associate with vesicles that selectively traffic AMPA-type glutamate receptors (AMPARs). Expression of Arc transgene increases the rate of surface AMPAR endocytosis and reduces the level of surface AMPAR. Arc similarly reduces synaptic AMPAR responses as revealed by analysis of spontaneous miniature excitatory synaptic currents (mEPSCs). In this process, Arc appears to be the rate-limiting molecule, because overexpression of endophilin alone does not alter AMPAR trafficking. The molecular interactions that allow for selective trafficking of AMPAR remain unknown, but Arc, endophilin, and dynamin do not directly bind AMPAR. These observations suggest that natural fluctuations of Arc protein that occur in neurons as a consequence of changes in neuronal activity would be linked to changes in surface/synaptic AMPAR. Consistent with this model, AMPAR levels at synapses of Arc knock-out (KO) neurons exhibit markedly reduced endocytosis, increased steady-state surface AMPAR levels, and increased amplitude of spontaneously evoked mEPSCs.
The tight association between Arc expression and the strength of excitatory synapses suggested it plays a role in homeostatic plasticity (Rial Verde et al., 2006; Shepherd et al., 2006). Homeostatic plasticity is hypothesized to compensate for Hebbian forms of synaptic plasticity, such as LTP and LTD, by scaling neuronal output without changing the relative strength of individual synapses (Turrigiano, 2007). Homeostatic scaling can be modeled in neuronal cultures by changing the steady-state level of network activity. Activity is reduced by addition of tetrodotoxin, and activity can be increased by addition of bicuculline to block GABA type A receptors. Under these conditions, the steady-state level of Arc protein is decreased by tetrodotoxin and increased by bicuculline, consistent with a role in the reciprocal changes in surface AMPAR. Expression of Arc transgene blocks the homeostatic increases in AMPAR function induced by chronic neuronal inactivity. Conversely, loss of Arc results in increased AMPAR function and abolishes homeostatic scaling of AMPARs. These observations reveal the importance of Arc's dynamic expression as it exerts continuous and precise control over synaptic strength and cellular excitability.
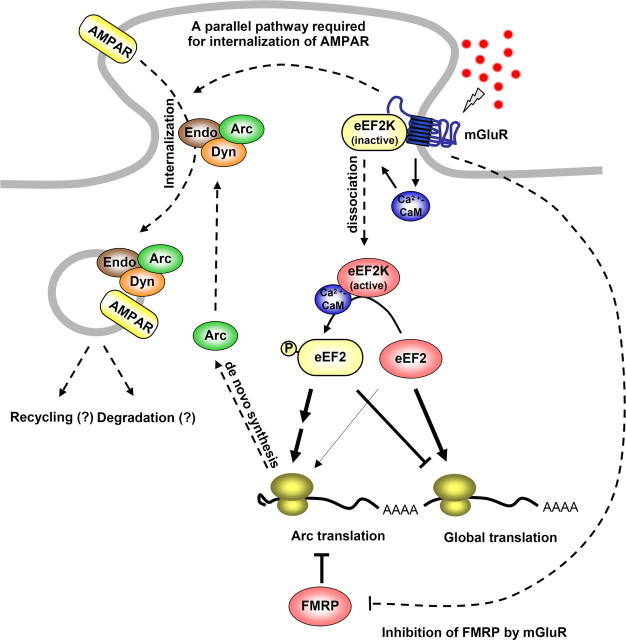
It was anticipated that Arc would contribute exclusively to transcription-dependent late-phase plasticity. However, studies that examined the mechanism of rapid, local translation of Arc mRNA reveal an essential role of Arc in a form of NMDA receptor-independent Hebbian plasticity that requires rapid, de novo protein synthesis in dendrites (Park et al., 2008; Waung et al., 2008). Arc is translationally induced within 5 min of activation of group 1 metabotropic glutamate receptors (mGluR), and this response is essential for mGluR-dependent LTD. The increase in Arc translation requires eukaryotic elongation factor 2 kinase (eEF2K), a Ca2+/calmodulin-dependent kinase that binds mGluR and dissociates after mGluR activation, whereupon it phosphorylates eukaryotic elongation factor 2 (eEF2) (Park et al., 2008). Phospho-eEF2 acts to slow the elongation step of translation and inhibits general protein synthesis, but simultaneously increases Arc translation (Chotiner et al., 2003; Kanhema et al., 2006). In the report of Park and colleagues, genetic deletion of eEF2K resulted in a selective deficit in rapid mGluR-dependent Arc translation and mGluR-LTD. This rapid translational mechanism is disrupted in the Fragile X disease mouse (Fmr1 KO), in which mGluR-LTD does not require de novo protein synthesis, but does require Arc. These observations suggest that eEF2K-eEF2 and FMRP coordinately control the dynamic translation of Arc mRNA in dendrites that is critical for synapse-specific LTD (Fig. 1).
Figure 1.
Model of Arc function in mGluR-LTD. Group I mGluRs activate eEF2K via calcium–calmodulin (CaM). eEF2K phosphorylates eEF2, which inhibits elongation generally but rapidly increases de novo Arc translation. Arc forms a complex with endophilin2/3 (Endo) and dynamin (Dyn) and induces the internalization of AMPAR. FMRP inhibits the translation of Arc at the basal state. Arc induction alone is not sufficient for mGluR-LTD, indicating that mGluR activates another pathway that is required to internalize AMPAR. In Fmr1 KO mice, the synthesis of Arc protein is constitutively de-repressed, and de novo synthesis of Arc is not required for mGluR-LTD. This figure was adapted with permission from Park et al. (2008), their Figure 8. FMRP, Fragile-X mental retardation protein.
Mechanisms and cellular function of Arc in LTP consolidation
Physiological studies in Arc KO mice have given important insights into the overall function of Arc in long-term synaptic plasticity. Early-phase LTP is enhanced, whereas late-phase LTP is blocked, in both the dentate gyrus in vivo and in the CA1 region of acute hippocampal slices (Plath et al., 2006). NMDA receptor-dependent LTD of the Schaffer collateral–CA1 synapse is also reduced in acute hippocampal slice recordings (Plath et al., 2006). Consistent with its role in plasticity, activity-dependent refinement of visual projections to the primary visual cortex is disrupted in Arc KO mice (Wang et al., 2006).
Messaoudi et al. (2007) sought to resolve the dynamic functions of Arc in LTP in vivo. Arc antisense (AS) oligodeoxynucleotides were transiently infused at various times after high-frequency stimulation (HFS) of the medial perforant path input to the dentate gyrus of anesthetized rats. The study revealed distinct temporal requirements for Arc synthesis. Surprisingly, early Arc synthesis is necessary for expression of LTP, whereas late synthesis is required for LTP consolidation. Infusion of Arc AS during early-phase LTP (15 min post-HFS) resulted in transient inhibition of LTP, paralleled by inhibition and recovery of enhanced Arc synthesis. In contrast, application of AS at 2 h (but not 4 h) after HFS resulted in a rapid and permanent reversal of LTP and inhibition of dendritic Arc mRNA and protein expression. Together, the study suggests a critical role for sustained translation of newly induced, dendritically transported Arc mRNA in LTP consolidation.
In addition to gene expression, formation of late-phase LTP involves enduring structural changes, including expansion of the postsynaptic density and enlargement of postsynaptic dendritic spines (Matsuzaki et al., 2004; Bourne and Harris, 2008). These structural changes depend on accumulation of F-actin as well as new protein synthesis (Fukazawa et al., 2003; Bramham, 2008; Tanaka et al., 2008). One of the major regulators of F-actin dynamics in spines is cofilin, which in its phosphorylated state promotes actin polymerization. In the study of Messaoudi et al. (2007), inhibition of LTP consolidation was associated with rapid dephosphorylation of hyperphosphorylated cofilin and corresponding loss of nascent F-actin at medial perforant path synapses. Importantly, the ability of Arc AS to reverse LTP was blocked by the F-actin stabilizing drug, jasplakinolide. Thus, Arc effectively couples new gene expression to F-actin expansion underlying stable LTP. Interestingly, local F-actin formation is necessary for selective localization of Arc mRNA to activated synapses (Huang et al., 2007), raising the possibility of a two-way interaction between Arc synthesis and actin polymerization. Although it is not known how Arc synthesis regulates cofilin phosphorylation, a recent study using yeast two-hybrid and protein pull-down strategies has identified the actin-binding protein WAVE3 as an Arc binding partner (Peebles et al., 2008).
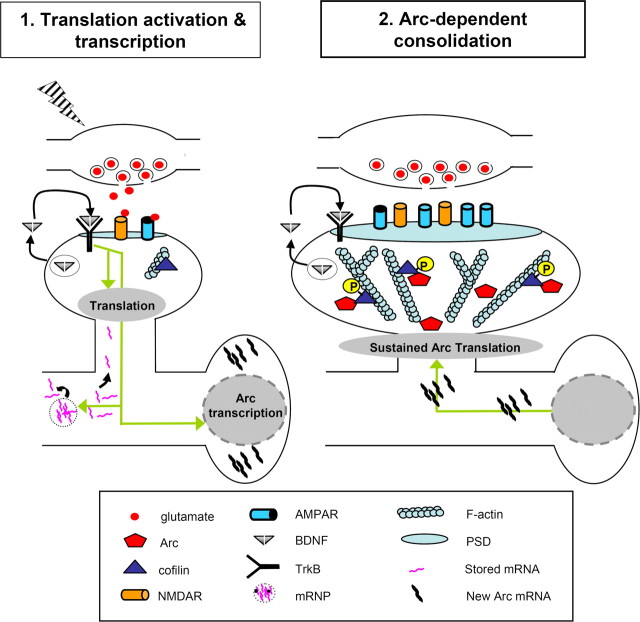
LTP consolidation is strongly modulated by signaling events initiated after LTP induction. One of the major regulators of this process is the secretory peptide brain-derived neurotrophic factor (BDNF) (Bramham and Messaoudi, 2005; Lynch et al., 2007). HFS of excitatory input triggers release of BDNF, leading to activation of postsynaptic TrkB receptors, which can mobilize further BDNF secretion. Stimulus protocols generating late-phase LTP are associated with a period of sustained BDNF release, and disruption of the BDNF-TrkB interaction blocks late-phase LTP (Kang et al., 1997; Aicardi et al., 2004; Santi et al., 2006). Exogenous application of BDNF induces a lasting potentiation of excitatory synaptic transmission (BDNF-LTP) in several brain structures. In the dentate gyrus, BDNF-LTP is transcription dependent, occluded by prior expression of late-phase LTP, and associated with dendritic transport of Arc mRNA (Messaoudi et al., 2002; Ying et al., 2002; Wibrand et al., 2006). BDNF-LTP induction is completely abolished by prior treatment with Arc AS (Messaoudi et al., 2007). The maintenance of BDNF-LTP and the associated phosphorylation of cofilin is also rapidly reversed by Arc AS application during a critical time window, as seen for HFS-LTP. Together, the evidence suggests that BDNF is capable of directly activating Arc-dependent LTP consolidation. Furthermore, BDNF signaling phosphorylates cofilin in dendritic spines and is necessary for expansion of dendritic spines during LTP (Rex et al., 2007; Tanaka et al., 2008). A model of Arc-dependent LTP consolidation is shown in Figure 2.
Figure 2.
Model of Arc function in LTP consolidation. In this two-stage model, translation activation is followed by Arc-dependent consolidation. In Translation activation: HFS (lightning bolt) causes activation of postsynaptic NMDAR receptors and TrkB receptors, leading to local translation activation as well as Arc transcription. Translation is modulated through regulation of translation factor activity, mobilization of mRNA (e.g., αCaMKII) from mRNPs and fine-localization of the translational machinery. Arc-dependent consolidation: Arc mRNA is transported to dendrites and translated in activated spines. Sustained translation of dendritically transported Arc is necessary for cofilin phosphorylation and local F-actin expansion. This figure was adapted with permission from Bramham and Wells (2007), their Figure 3. mRNP, Messenger ribonucleoprotein particle; TrkB, Tropomyosin-related kinase B; αCaMKII, α-subunit calcium/calmodulin-dependent protein kinase II.
Protein synthesis-dependent modification of synaptic strength is tightly controlled at the posttranscriptional level, and local regulation in dendrites provides a compelling mechanism for coupling synaptic input patterns to local changes in protein composition (Sutton and Schuman, 2006; Bramham and Wells, 2007). Modulation of translation factor activity through regulated phosphorylation is a major means of adjusting translation rates. LTP in the dentate gyrus is associated with rapid, ERK-dependent phosphorylation of eukaryotic initiation factor 4E (eIF4E) and eEF2 along with enhanced Arc protein synthesis (Kanhema et al., 2006). However, the mechanisms that sustain eIF4E and eEF2 phosphorylation during LTP maintenance are unknown. F-actin formation has long been implicated in the fine-localization of the translational machinery and the activity of certain F-actin-associated translation factors (Smart et al., 2003; Gross and Kinzy, 2007). This prompted investigation of whether Arc synthesis contributes to the regulation of translation factor activity (Tiron and Bramham, 2008). Infusion of Arc AS reversed ongoing LTP and abolished the persistent hyperphosphorylation of the eIF4E and eEF2, without affecting LTP-evoked hyperphosphorylation of ribosomal protein S6. Thus, in addition to regulating F-actin dynamics, Arc appears to play a prominent role in regulating the activity of the translational machinery during LTP consolidation.
Regulation of Arc expression by translation-dependent mRNA decay
Whereas the mechanisms involved in localization and translational repression/activation of Arc and other dendritic mRNAs have received considerable attention in recent years (Czaplinski and Singer, 2006; Bramham and Wells, 2007), how their expression is kept in check once it is activated is much less understood. One means for limiting synaptic protein abundance is through activity-dependent recruitment and sequestration of proteasomes at activated synapses (Bingol and Schuman, 2005; Bingol and Schuman, 2006). On-site ubiquitin-dependent degradation by such proteasomes has been proposed to play a key role in sculpting local protein composition in response to synaptic activity. Another potential means for achieving tight control of gene expression at synapses is through the regulation of transcript abundance via translation-dependent mRNA decay. Because such decay pathways are thought to work exclusively on translationally active mRNAs, they would not be expected to interfere with the transport and local accumulation of translationally repressed mRNPs at distal reaches of the cytoplasm such as dendritic spines. Once translation is activated, however, the mRNA can be quickly degraded, resulting in a tightly controlled burst of protein synthesis
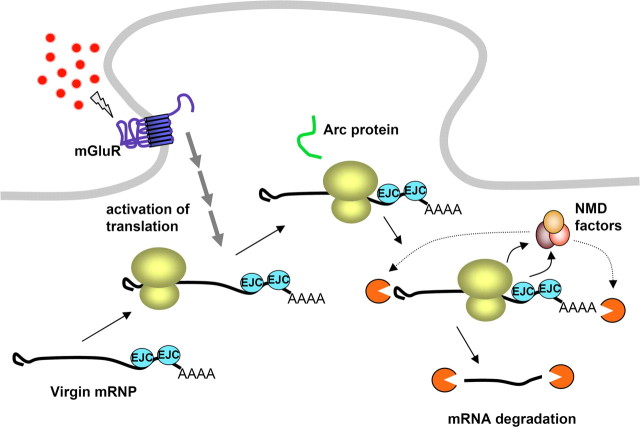
One well known translation-dependent mRNA degradation pathway is nonsense-mediated mRNA decay (NMD) (Fig. 3). Traditionally, NMD has been viewed as a quality control mechanism for the rapid elimination of mRNAs with premature stop codons. Prevailing wisdom holds that such aberrant mRNAs, the products of either genetic mutation or faulty pre-mRNA processing, must be destroyed because of their potential for encoding deleterious truncated proteins (Isken and Maquat, 2007). Recently, however, it has become apparent that numerous wild type mRNAs, including Arc, are also subject to this pathway (Wittmann et al., 2006; Giorgi and Moore, 2007; Ni et al., 2007).
Figure 3.
Translation-dependent degradation of Arc mRNA via the NMD pathway. Virgin (not yet translated) Arc mRNA is stable and can accumulate in dendrites. However, juxtaposition of the ribosome with two EJCs in the 3′-UTR at the termination of translation leads to activation of NMD and rapid mRNA degradation. This mechanism could potentially limit each Arc mRNA to producing just a single copy of Arc protein.
NMD of mammalian transcripts is thought to occur during the first or “pioneer” round of translation (Maquat, 2004). Thus natural substrates for NMD potentially give rise to just a single molecule of protein before their demise. One trigger for NMD in mammalian cells is the presence of an exon junction complex (EJC) in the 3′-UTR. EJCs are stably deposited on mRNAs upstream of exon-exon junctions as a consequence of pre-mRNA splicing. Structurally, the EJC consists of a stably bound tetrameric core that serves as a binding platform for other more transiently associated factors. Within this core, the DEAD-box protein eIF4AIII serves as the main RNA-binding constituent.
Recent work of Giorgi et al. (2007) has shown that Arc is a natural target for NMD by virtue of two conserved introns in its 3′-UTR. In cortical neuronal cultures, eIF4AIII colocalizes with Arc as well as the RNA-binding proteins FMRP and STAU1 in dendritic RNA granules. This colocalization strongly suggests that a major fraction of Arc mRNA in dendrites has not yet undergone a pioneer round of translation. Inhibition of NMD by knockdown of eIF4AIII resulted in elevated expression of Arc mRNA and protein combined with an increase in excitatory synaptic transmission (selective increase in mEPSC amplitude). Bioinformatic analysis also revealed a number of other potential synaptic NMD targets, including two cadherins, a neurexin, a neuregulin, a synaptotagmin, and several ion channels.
Functionally, translation-dependent decay of Arc and other dendritic mRNAs could act as a crucial brake to create a strictly limited burst of protein synthesis (Fig. 3). In the case of Arc, NMD could allow a time-limited, quantity-limited, and mRNA-specific translational response. This has several important implications for synaptic function and plasticity. For example, sustained translation of Arc during LTP consolidation may be strictly limited by the delivery of newly induced Arc mRNA to synapses.
Arc, network functions, and behavior
The regulation of Arc gene expression by behavioral experience has received considerable attention, due, in large part, to the dendritic localization of Arc mRNA/protein and its requirement in synaptic plasticity and long-term memory (Guzowski et al., 2000; Steward and Worley, 2001; Plath et al., 2006; Messaoudi et al., 2007) These behavioral studies show Arc to be dynamically regulated in many regions of the brain critical for learning and memory (for review, see Miyashita et al., 2008). The most compelling evidence that Arc transcription is tightly and specifically linked to neural activity associated with information processing (Guzowski et al., 2004, 2005; Vazdarjanova and Guzowski, 2004; Kubik et al., 2007), and not a nonspecific response to stress or behavioral arousal, was provided by the “catFISH” approach [cellular compartment analysis of temporal activity by fluorescence in situ hybridization (Guzowski et al., 1999)]. By exploiting the precise timing of IEG transcription and mRNA processing, catFISH allows cellular activity maps, for two or three discrete behavioral experiences, to be compared within the same brain (Guzowski et al., 1999, 2005; Marrone et al., 2008). Using fluorescence in situ hybridization (FISH) and confocal microscopy, the subcellular distribution of Arc alone, or Arc and another IEG, Homer 1a, provides activity history “time stamps” for individual neurons throughout the forebrain (Guzowski et al., 2005) (Fig. 4a). catFISH studies show that Arc transcription is activated in neural ensembles of hippocampal subfields in a highly context-specific manner, in a manner that is both quantitatively and qualitatively similar to hippocampal cell firing activity recorded in electrophysiological studies (Guzowski et al., 2004, 2006; Kubik et al., 2007).
Figure 4.
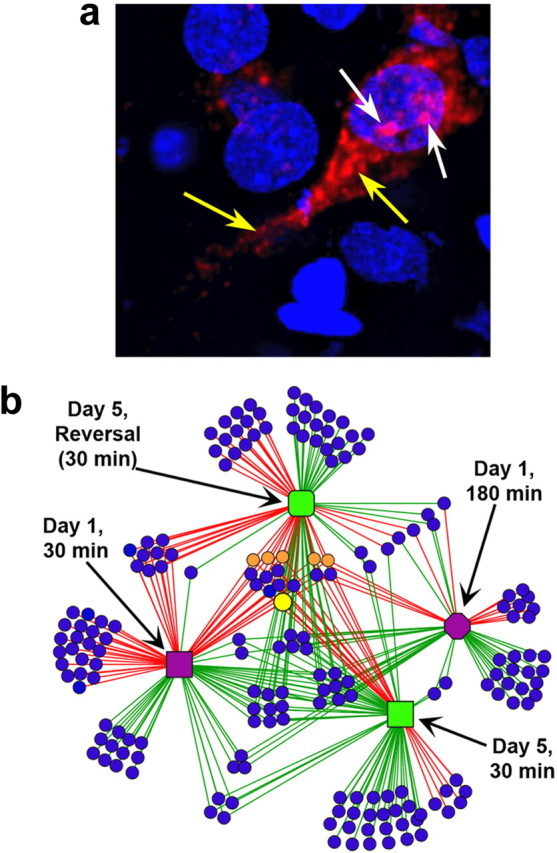
Network regulation and function of Arc. a, Confocal projection image from CA3 of rat hippocampus showing the subcellular distribution of Arc RNA (red) using high-sensitivity FISH [Guzowski et al. (2005), their Fig. 1, reproduced with permission]. Note the presence of the two intense sites of Arc synthesis (transcription foci; white arrows) in the nucleus (DAPI; blue color) and the somatic/dendritic Arc mRNA (yellow arrows). The subcellular distribution of Arc RNA provides a “time stamp” of neural activity history for two behavioral epochs and provides the basis for the catFISH imaging approach. b, Complex network analysis (CNA) of gene expression changes associated with distinct stages of learning and memory [Miyashita et al. (2008), their Fig. 4, reproduced with permission]. Rats were trained in the spatial water maze task, and RNA from dissected dorsal hippocampi was used for microarray analysis. Details of the analysis are provided elsewhere (Miyashita et al., 2008b), but the data shown here represent a subset of differentially regulated genes (at p < 0.05 with fold changes of <1.5 or >1.5 relative to caged control values). The groups included the following: caged control (rats killed from the home cage); day 1, 30 min (rats killed 30 min after the first training session; early learning); day 1, 180 min (rats killed 180 min after an initial training session; early learning); day 5, 30 min (rats killed 30 min on the fifth day of training; stable reference memory retrieval); and day 5, reversal, 30 min (rats killed 30 min after spatial reversal learning on the fifth day of training; extinction of previous reference memory and new reversal learning). The CNA graph shows gene expression differentially regulated for all groups: day 1, 30 min (purple square); day 1, 180 min (purple octagon); day 5, 30 min (green square); and day 5, reversal, 30 min (green rounded-square). The lines (“edges”) connect behavioral groups with the genes (blue circle nodes) that are differentially regulated in that group. Red and green edges indicate upregulation and downregulation of gene expression, respectively, relative to caged control baseline levels. Note that the several genes in the center of the network are regulated across multiple stages of learning, as demonstrated by the high connectivity of these nodes. Of these “core” genes, several known IEGs are indicated as orange circle nodes (c-fos, Nr4a1, Homer 1a, junB, and zif268), and Arc is shown as a larger yellow circle node. In contrast, low-connectivity genes, represented by blue circles connected to only one group, are regulated only by a single behavior (i.e., in a distinct stage of learning and memory: “state specific”). The degree of similarity or difference of the gene expression networks between any two of the behavior groups (stages of learning and memory) can be culled from the number of shared and distinct regulated genes. Note that whereas Arc is upregulated 30 min after training on day 1, day 5, and day 5, reversal, the cohort of differentially regulated genes is distinct for each behavioral group. Thus, the “molecular context” of Arc RNA expression changes across stages of spatial water maze learning.
A recent study has more precisely defined the relationship between neural activity and Arc transcriptional activation. Using a closed track paradigm, as used in electrophysiological studies of hippocampal “place cells” (T. Myashita, S. Kubik, N. Haghighi, O. Steward, and J. F. Guzowski, unpublished observations), showed that behavioral experience as minimal as walking a single initial lap around the track was sufficient to activate Arc transcription in the full CA3 ensemble, consistent with a role for CA3 in rapid encoding of episodic information. In contrast, activation of Arc transcription in a complete CA1 ensemble required additional experience on the track. To determine whether location-specific place cell activity is sufficient for activating Arc transcription, rats were tested on the track after infusion of tetracaine or vehicle into the medial septum. Previous studies have demonstrated that intraseptal tetracaine infusions block hippocampal theta rhythm, impair LTP, and disrupt hippocampal-dependent learning, without affecting place specific firing of CA1 neurons. Behaviorally driven Arc induction was abolished in both CA3 and CA1 neurons of the tetracaine-treated rats (Miyashita, Kubik, Haghighi, Steward, and Guzowski, unpublished observations). Together with past catFISH studies, these data support the hypothesis that location-specific firing of CA3 and CA1 neurons in the presence of theta rhythm provide the necessary stimuli for activation of Arc transcription.
The tight coupling of Arc transcription to hippocampal place cell activity, however, is not static, but instead plastic. Guzowski et al. (2006) showed that recent, but not remote, firing history can strongly inhibit further Arc transcriptional activation. This “electro-transcriptional coupling” represents a form of metaplasticity and may function to maintain proper levels of Arc for regulating synaptic homeostasis or plasticity. In addition, the coupling of Arc mRNA expression to functional protein expression can also be subject to modulation (McIntyre et al., 2005). In rats trained in inhibitory avoidance and given posttraining intra-amygdala infusions of drugs that either enhanced or impaired long-term memory, hippocampal Arc protein levels were increased or decreased (respectively) relative to controls. These changes in Arc protein expression were not accompanied by similar drug-dependent changes in mRNA expression, suggesting posttranscriptional regulation of Arc protein expression by amygdala-dependent neuromodulatory processes. Thus, functional Arc gene expression enables the integration of multiple neural signals to optimize information storage in active networks.
Although Arc is among the most dynamically regulated genes in the forebrain, other IEGs share similar robust transcriptional regulation by behavioral stimuli. In a recent study, hippocampal expression of Arc and other known IEGs (e.g., zif268, c-fos, and Homer 1a) remained strongly coupled to ongoing neural activity in rats trained in a spatial water maze task, regardless of the stage of learning (Fig. 4b) (Miyashita, Kubik, Haghighi, Steward, and Guzowski, unpublished observations). However, distinct and overlapping patterns of experience-dependent gene expression were seen during initial learning, in overtraining, and in reversal learning. Thus, the contribution of Arc to specific neuroplastic mechanisms may be dependent on other activity-regulated genes present in a neuron, at a given time. In this view, the function of Arc or any other IEG to behavioral plasticity can only be fully understood in the “molecular context” of other coexpressed genes.
Conclusion and open questions
Arc is emerging as a versatile, finely tuned system capable of translating neural activity patterns into various forms of protein synthesis-dependent synaptic plasticity. Current evidence suggests a critical role for Arc in AMPAR endocytosis underlying LTD and homeostatic plasticity as well as in F-actin formation underlying LTP consolidation. These new insights have spawned a host of questions that are likely to fuel further discovery. For one, what are the mechanisms that selectively engage newly synthesized Arc in LTP or LTD? Do the various forms of Arc-dependent plasticity coexist in the same neuron and experimental preparation? For example, local Arc translation underlying F-actin expansion and LTP consolidation could be followed by a delayed, dendrite-wide homeostatic plasticity that globally resets synaptic excitability while preserving the relative differences in synaptic strength. Recent evidence also suggests that Arc RNA synthesis, localization, translation, and metabolism are all highly regulated. Differential regulation of these processes, for example through activation of TrkB and mGluR receptors, might provide the requisite cellular contexts for Arc-dependent LTP or LTD. Surprisingly, posttranslational regulation of Arc has so far received little attention. Posttranslational modification of Arc or one of its binding partners might dictate the functional mode of Arc at synapses. Another looming question, not addressed at the symposium, is the possible role of Arc in the nucleus, where it accumulates and binds to promyelocytic leukemia nuclear bodies (Bloomer et al., 2007).
The versatile role of Arc in synaptic plasticity coupled with behavioral studies of Arc transcription in physiologically defined neural networks suggests a dedicated role for this immediate early gene in optimizing information storage. A major challenge of future research is to ascribe specific neural network functions to Arc-dependent regulation of LTP, LTD, and homeostatic synaptic plasticity.
Footnotes
C.R.B. was supported by the Norwegian Research Council and European Union Research Training Network Grant GENE-MEMORY (504321). P.F.W. was supported by National Institutes of Health (NIH) Grants MH053608 and DA266. M.J.M. was supported by the Howard Hughes Medical Institute. J.G. was supported by NIH Grant R01 MH060123.
References
- Aicardi G, Argilli E, Cappello S, Santi S, Riccio M, Thoenen H, Canossa M. Induction of long-term potentiation and depression is reflected by corresponding changes in secretion of endogenous brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 2004;101:15788–15792. doi: 10.1073/pnas.0406960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Synaptic protein degradation by the ubiquitin proteasome system. Curr Opin Neurobiol. 2005;15:536–541. doi: 10.1016/j.conb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Bloomer WA, VanDongen HM, VanDongen AM. Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia (PML) bodies. Brain Res. 2007;1153:20–33. doi: 10.1016/j.brainres.2007.03.079. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advance online publication. Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin Neurobiol. 2008 doi: 10.1016/j.conb.2008.09.013. Retrieved October 22, 2008. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76:99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Chotiner JK, Khorasani H, Nairn AC, O'Dell TJ, Watson JB. Adenylyl cyclase-dependent form of chemical long-term potentiation triggers translational regulation at the elongation step. Neuroscience. 2003;116:743–752. doi: 10.1016/s0306-4522(02)00797-2. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Gross SR, Kinzy TG. Improper organization of the actin cytoskeleton affects protein synthesis at initiation. Mol Cell Biol. 2007;27:1974–1989. doi: 10.1128/MCB.00832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Knierim JJ, Moser EI. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron. 2004;44:581–584. doi: 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr Opin Neurobiol. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci U S A. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Chotiner JK, Steward O. Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J Neurosci. 2007;27:9054–9067. doi: 10.1523/JNEUROSCI.2410-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Kang H, Welcher AA, Shelton D, Schuman EM. Neurotrophins and time: different roles for TrkB signaling in hippocampal long-term potentiation. Neuron. 1997;19:653–664. doi: 10.1016/s0896-6273(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Håvik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- Kubik S, Miyashita T, Guzowski JF. Using immediate-early genes to map hippocampal subregional functions. Learn Mem. 2007;14:758–770. doi: 10.1101/lm.698107. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Lynch G, Rex CS, Gall CM. LTP consolidation: substrates, explanatory power, and functional significance. Neuropharmacology. 2007;52:12–23. doi: 10.1016/j.neuropharm.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Marrone DF, Schaner MJ, McNaughton BL, Worley PF, Barnes CA. Immediate-early gene expression at rest recapitulates recent experience. J Neurosci. 2008;28:1030–1033. doi: 10.1523/JNEUROSCI.4235-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre CK, Miyashita T, Setlow B, Marjon KD, Steward O, Guzowski JF, McGaugh JL. Memory-influencing intra-basolateral amygdala drug infusions modulate expression of Arc protein in the hippocampus. Proc Natl Acad Sci U S A. 2005;102:10718–10723. doi: 10.1073/pnas.0504436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi E, Ying SW, Kanhema T, Croll SD, Bramham CR. Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J Neurosci. 2002;22:7453–7461. doi: 10.1523/JNEUROSCI.22-17-07453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi E, Kanhema T, Soulé J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Kubik S, Lewandowski G, Guzowski JF. Networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiol Learn Mem. 2008;89:269–284. doi: 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11. doi: 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles CL, Rao V, Breck J, Finkbeiner S. Arc mediates structural plasticity in neuronal dendrites and spines. Soc Neurosci Abstr. 2008;34:736–20. [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Rex CS, Lin CY, Kramár EA, Chen LY, Gall CM, Lynch G. Brain-derived neurotrophic factor promotes long-term potentiation-related cytoskeletal changes in adult hippocampus. J Neurosci. 2007;27:3017–3029. doi: 10.1523/JNEUROSCI.4037-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez JJ, Davies HA, Silva AT, De Souza IE, Peddie CJ, Colyer FM, Lancashire CL, Fine A, Errington ML, Bliss TV, Stewart MG. Long-term potentiation in the rat dentate gyrus is associated with enhanced Arc/Arg3.1 protein expression in spines, dendrites and glia. Eur J Neurosci. 2005;21:2384–2396. doi: 10.1111/j.1460-9568.2005.04068.x. [DOI] [PubMed] [Google Scholar]
- Santi S, Cappello S, Riccio M, Bergami M, Aicardi G, Schenk U, Matteoli M, Canossa M. Hippocampal neurons recycle BDNF for activity-dependent secretion and LTP maintenance. EMBO J. 2006;25:4372–4380. doi: 10.1038/sj.emboj.7601303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart FM, Edelman GM, Vanderklish PW. BDNF induces translocation of initiation factor 4E to mRNA granules: evidence for a role of synaptic microfilaments and integrins. Proc Natl Acad Sci U S A. 2003;100:14403–14408. doi: 10.1073/pnas.2436349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Worley PF. A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proc Natl Acad Sci U S A. 2001;98:7062–7068. doi: 10.1073/pnas.131146398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiron A, Bramham CR. Arc/Arg3.1 dynamically regulates translation factor activity during LTP maintenance in the adult rat brain in vivo. Soc Neurosci Abstr. 2008;34:433–15. [Google Scholar]
- Turrigiano G. Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol. 2007;17:318–324. doi: 10.1016/j.conb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF. Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci. 2004;24:6489–6496. doi: 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K, Messaoudi E, Håvik B, Steenslid V, Løvlie R, Steen VM, Bramham CR. Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in the adult rat dentate gyrus in vivo. Eur J Neurosci. 2006;23:1501–1511. doi: 10.1111/j.1460-9568.2006.04687.x. [DOI] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jäck HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW. The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proc Natl Acad Sci U S A. 2002;99:2368–2373. doi: 10.1073/pnas.042693699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neurosci. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]