Abstract
The conversion of TGF-β from a tumor suppressor to a tumor promoter occurs frequently during mammary tumorigenesis, yet the molecular mechanisms underlying this phenomenon remain undefined. We show herein that TGF-β repressed NF-κB activity in normal NMuMG cells, but activated this transcription factor in their malignant counterparts, 4T1 cells, by inducing assembly of TAB1:IKKβ complexes, which led to the stimulation of a TAK1:IKKβ:p65 pathway. TAB1:IKKβ complexes could only be detected in NMuMG cells following their induction of epithelial-mesenchymal transition (EMT), where upon TGF-β treatment activated NF-κB. Expression of a truncated TAB1 mutant (i.e., TAB1(411)) reduced basal and TGF-β-mediated NF-κB activation in NMuMG cells driven to undergo EMT by TGF-β, and in 4T1 cells stimulated by TGF-β. TAB1(411) expression also inhibited TGF-β-stimulated TNF-α and COX-2 expression in 4T1 cells. Additionally, the ability of human MCF10A-CA1a breast cancer cells to undergo invasion in response to TGF-β absolutely required the activities of TAK1 and NF-κB. Moreover, siRNA-mediated TAK1-deficiency restored the cytostatic activity of TGF-β in MCF10A-CA1a cells. Finally, expression of truncated TAB1(411) dramatically reduced the growth of 4T1 breast cancers in syngeneic Balb/C, as well as in nude mice, suggesting a potentially important role of NF-κB in regulating innate immunity by TGF-β. Collectively, our findings have defined a novel TAB1:TAK1:IKKβ:NF-κB signaling axis that forms aberrantly in breast cancer cells, and consequently, enables oncogenic signaling by TGF-β.
Keywords: Breast Cancer, NF-κB, TAB1, TAK1, TGF-β
Introduction
TGF-β is a pleiotropic cytokine that mediates a diverse array of physiological activities in responsive cells and tissue through all stages of the metazoan lifespan (1, 2). The biological actions of TGF-β are communicated across the plasma membrane through the combined actions of the TGF-β type I (TβR-I) and type II (TβR-II) Ser/Thr protein kinase receptor complexes, which phosphorylate Smad2 and 3 and stimulate their association with the Co-Smad, Smad4 (1, 3). Nuclear translocation of these transcription factor complexes promotes their physical interaction with a variety of transcriptional activators and repressors that ultimately regulate the expression of TGF-β-responsive genes in a cell- and promoter-specific manner (3). The complexity of TGF-β signaling is increased through its activation of mitogen-activated protein kinases (MAPKs; e.g., ERK1/2, JNK, and p38 MAPK), which modulate the activity of Smad2/3 or other downstream transcription factors (4-7). Phosphorylation and activation of MAPKs are regulated by MAPK kinases (MAPKK/MKK), which in turn are phosphorylated and activated by upstream MAPK kinase kinases (MAPKKK/MKKK) (8).
TGF-β-activated kinase 1 (TAK1) is a MAPKKK that is activated by receptors for TGF-β, TNF-α, and IL-1 (9-11). TAK1 activation is regulated via its association with the C-terminal binding domain of TAK1-binding protein 1 (TAB1), and via its association with TAB2 and TAB3, which target TAK1 binding sites distinct from those bound by TAB1 (12-14). Activated TAK1 phosphorylates and activates MKK3/MKK6 and MKK4, which then mediate stimulation of p38 MAPK or JNK, respectively (6, 15). TAK1 also interacts with and activates the IκB kinase (IKK) complex (i.e., IKKα, IKKβ, and IKKγ/NEMO), which mediates NF-κB transcription factor activation (16-18). Upon TAK1-mediated activation, the IKK complex phosphorylates the IκBα inhibitory protein, leading to its degradation and subsequent activation of the p65 NF-κB transcription factor. Gene ablation studies in mice revealed that TAK1, but not is binding partner TAB1, is essential for TNF-α- and IL-1-mediated activation of NF-κB (19). These studies also established a critical role for TAB1 and TAK1 during embryonic development, and in mediating inflammatory gene expression, while recent evidence points to their involvement in mediating dysregulated signaling in cancer cells (20-27). Currently, relatively little is known regarding the roles of TAK1 and TAB1 in regulating the response of normal and malignant mammary epithelial cells (MECs) to TGF-β. This knowledge deficit is medically relevant because inappropriate and constitutive NF-κB activity has been linked to the development and progression of human cancers (28), and to the conversion of TGF-β from a suppressor to a promoter of mammary tumorigenesis (29, 30). The aim of this study was to further our understanding of the molecular events that contribute to dysregulated TGF-β and NF-κB signaling in developing and progressing breast cancers.
Materials and Methods
Materials
Recombinant human TGF-β1 and mouse TNF-α were obtained from R&D Systems (Minneapolis, MN). All constructs used in this work were kindly provided by: (i) kinase-dead TAK1 [pcDNA3-HA-TAK1(K63M)] by Dr. Gary L. Johnson (University of North Carolina, Chapel Hill, NC); (ii) human TAB1α cDNA provided by Dr. Jiahuai Han (The Scripps Research Institute, La Jolla, CA); (iii) mammalian IκBα-NULL (pMSCV-IκBα-NULL-GFP) and NF-κB promoter-driven luciferase reporter by Dr. John M. Routes (Medical College of Wisconsin, Milwaukee, WI); (iv) GST-IκBα by Dr. Robert Scheinman (University of Colorado Health Sciences Center, Denver, CO); (v) human TNF-α promoter-driven luciferase reporter (containing bases -639 to -162 from start site) by Dr. Harvey F. Lodish (Whitehead Institute, Cambridge, MA); and (vi) COX-2 promoter-driven luciferase reporter by Dr. Bharat B. Aggarwal (University of Texas M. D. Anderson Cancer Center, Houston, TX). All additional supplies or reagents were routinely available.
Cell Culture and Retroviral Infections
Normal murine NMuMG mammary gland and malignant 4T1 breast cancer cells were obtained from ATCC (Manassas, VA), while human metastatic MCF10A-CA1a breast cancer cells were provided by Dr. Fred Miller (Barbara Ann Karmanos Cancer Institute, Detroit, MI). Cell lines were maintained and cultured in a constant atmosphere of 5% CO2 at 37°C as described previously (31, 32).
The following murine ecotropic retroviral constructs were used herein: (i) pMSCV-IRES-GFP or pMSCV-IRES-YFP (i.e., control vectors); (ii) pMSCV-DNTβR-II-GFP (i.e., truncated TβR-II); (iii) pMSCV-TAK1(K63M)-YFP (i.e., kinase-dead TAK1); (iv) pMSCV-IκBα-NULL-GFP; and (v) pMSCV-TAB1(1-411)-YFP (i.e., truncated TAB1). Retroviral supernatants were produced by EcoPack2 retroviral packaging cells (Clontech, Mountain View, CA) and used to infect NMuMG and 4T1 cells, and MCF10A-CA1a cells engineered to express the ecotropic receptor as previously described (33). Forty-eight h post-infection, the infected cells were analyzed and isolated on a MoFlo cell sorter (Cytomation, Fort Collins, CO), and subsequently were expanded to yield stable polyclonal populations of control and transgene-expressing cells that were ≥90% for expression of GFP or YFP.
Luciferase Reporter Gene Assays
Analysis of TGF-β-stimulated luciferase activity driven by the synthetic NF-κB, SBE, 3TP, TNF-α, or COX-2 promoters was performed as described previously (34). NMuMG and 4T1 MECs (25-30,000 cells/well) were cultured overnight onto 24-well plates, and subsequently were transfected the following morning by overnight exposure to LT1-liposomes (Mirus, Madison, WI) containing 300 ng/well of individual luciferase reporter cDNA and 50 ng/well of CMV-β-gal cDNA, which was used to control for differences in transfection efficiency. The cells were washed twice with PBS and stimulated overnight in serum-free media with TGF-β1 (0-5 ng/ml) or TNF-α (0-20 ng/ml) as indicated. Afterward, luciferase and β-gal activities contained in detergent-solubilized cell extracts were determined. Data are the mean (± SE) luciferase activities of at least three independent experiments normalized to untreated cells.
NF-κB Biotinylated Oligonucleotide Capture Assay
DNA-binding activity of NF-κB was monitored in quiescent 4T1 cells before and after their activation by TGF-β as indicated. Afterward, 4T1 cells were collected and fractionated into cytoplasmic and nuclear extracts using a Nuclear Extraction Kit according to manufacturer's instructions (Chemicon, Temecula, CA). NF-κB binding activity was determined by incubating 600 μg of nuclear extract with 1 μg of biotinylated double-stranded DNA oligonucleotides that contained a NF-κB consensus sequence site under continuous rotation at 4°C (forward probe: 5'-GATCTAGGGACTTTCCGCTGGGGACTTTCCAGTCGA; reverse probe: 5'-TCGACTGGAAAGTCCCCAGCGGAAAGTCCCTAGATC). The resulting NF-κB:oligonucleotide complexes were captured by addition streptavidin-agarose beads (Pierce, Rockford, IL) and collected by microcentrifugation. Washed complexes were fractionated through 10% SDS-PAGE prior to their immobilization to nitrocellulose membranes, which subsequently were probed with anti-phopho-p65 antibodies (1:1000; Cell Signaling, Danvers, MA) and anti-p65 antibodies (1:500; Santa Cruz Biotechnology, Santa Cruz, CA). Differences in extract loading were monitored by immunoblotting 25 μg of resolved nuclear extract aliquots with antibodies against histone H1 (1:200; Santa Cruz Biotechnology).
Western Blotting
Control and TGF-β-stimulated NMuMG and 4T1 cells were lysed and solubilized in Buffer H/Triton X-100 (35) for 30 min on ice. Clarified cell extracts were resolved on 10% SDS-PAGE gels, transferred electrophoretically to nitrocellulose membranes, and blocked in 5% milk prior to incubation with the following primary antibodies (dilutions): (i) anti-Flag M2 (1:1000; Sigma, St. Louis, MO); (ii) anti-β-actin (1:5000; Sigma); (iii) anti-COX-2 (1:2000; Cayman Chemical Company, Ann Arbor, MI); (iv) anti-Histone H1 (1:200); (v) anti-IKKβ (1:500; Cell Signaling); (vi) anti-phospho-p65 (1:1000); (vii) anti-TAB1 (1:500; Cell Signaling and Novus Biologicals, Littleton, CO); (viii) anti-TAK1 (1:500; Cell Signaling); and (ix) and anti-xIAP (1:500; Cell Signaling). The resulting immunocomplexes were visualized by enhanced chemiluminescence. Differences in protein loading were monitored by reprobing stripped membranes with anti-β-actin antibodies (1:1000; Rockland Immunology, Gilbertsville, PA).
IKKβ Kinase Assay
The ability of TGF-β to activate IKKβ was determined using an in vitro protein kinase assay that measured the extent of immunoprecipitated IKKβ to phosphorylate recombinant GST-IκBα. Briefly, NMuMG and 4T1 cells (350,000 cells/well) were stimulated with TGF-β1 (5 ng/ml) for 0-3 h at 37°C as indicated, and subsequently were harvested and solubilized on ice in Buffer H/1% Triton X-100. Clarified whole cell extracts were prepared and incubated under continuous rotation with anti-IKKβ antibodies (2 μl/tube) for 4 h at 4°C. Immunocomplexes were recovered by brief microcentrifugation, and subsequently were washed thrice in lysis buffer and twice in Buffer H. Phosphotransferase reactions were performed in a final volume of 40 μl consisting of IKKβ immunocomplexes and 2.5 μg of GST-IκBα, and were initiated by addition of 10 μl of 4X assay buffer and allowed to proceed for 30 min at 30°C (35). The phosphorylation reactions were stopped by addition of 4X sample buffer and boiled for 5 min prior to their fractionation through 10% SDS-PAGE gels. Phosphorylation of recombinant IκBα by IKKβ was detected by exposure of the dried gels to a phosphoscreen. Differences in protein loading were monitored by immunoblotting whole cell extract aliquots (i.e., 10% of that subjected to immunoprecipitation) with antibodies against IKKβ and β-actin.
TAB1:IKKβ Co-immunoprecipitation Assay
Detergent-solubilized NMuMG and 4T1 whole cell extracts (350,000 cells/tube) were prepared and incubated under continuous rotation with anti-TAB1 antibodies (2 ml/tube; Santa Cruz Biotechnology) for 6 h at 4°C. The resulting immunocomplexes were collected by microcentrifugation, washed, and fractionated through 10% SDS-PAGE gels prior to their immobilization to nitrocellulose membranes, which subsequently were probed with anti-IKKβ antibodies (1:500). Differences in protein loading were monitored by immunoblotting whole cell extract aliquots with antibodies against β-actin as above.
TAK1 siRNA Knockdown
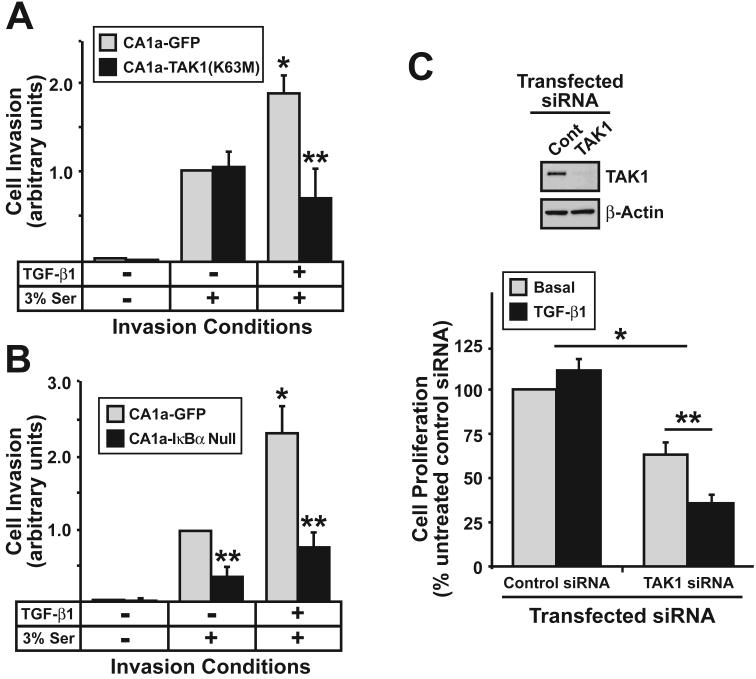
The creation of MCF10A-CA1a cells lacking TAK1 was accomplished using SMARTpool siRNA (Dharmacon) according to the manufacturer's recommendations, and as described previously (31, 32). Briefly, MCF10A-CA1a cells (10,000 cells/well) were plated onto 96-well plates and cultured overnight in antibiotic-free media. The following morning, the cells were transiently transfected overnight with DharmaFECT One reagent (Dharmacon, Lafayette, CO) supplemented with TAK1 siRNAs (50 nM), and subsequently were stimulated with TGF-β1 (5 ng/ml) for varying times at 37°C. Upon completion of agonist stimulation, the cells were harvested and prepared for [3H]thymidine incorporation assays as described below. The extent of siRNA-mediated TAK1-deficiency was monitored by immunoblotting whole cell extracts with antibodies against TAK1.
Cell Biological Assays
The effect of manipulating TAB1, TAK1, or IκBα function on various TGF-β-stimulated activities in normal and malignant MECs was determined as follows: (i) cell proliferation using 10,000 cells/well in a [3H]thymidine incorporation assay as described previously (33); (ii) cell invasion induced by 3% serum using 100,000 cells/well in a modified Boyden-chamber coated with Matrigel matrices (diluted 1:25 in serum-free DMEM) as described previously (34); and (iii) EMT induced by TGF-β1 (5 ng/ml) treatment as described previously (31, 32).
Tumor Growth Study
Control (YFP) and truncated TAB1(411)-expressing 4T1 cells were resuspended in sterile PBS and injected (12,500 cells/mouse) orthotopically into the mammary fat pad of 6 week old female Balb/C and Nude mice (4 mice/condition; Jackson Labs, Bar Harbor, ME). Mice were monitored daily and primary tumors were measured with digital calipers (Fisher Scientific, Pittsburg, PA) between days 10 and 24. Tumor volumes were calculated using the following equation: Tumor volume = (x2)(y)(0.5), where “x” is the tumor width and “y” is the tumor length. Twenty-four days post-inoculation, the mice were sacrificed and their primary tumors were excised and weighed.
Animal studies were performed two (i.e., Balb/C) or three (i.e., nude) times in accordance with the animal protocol procedures approved by the Institutional Animal Care and Use Committee of University of Colorado.
Results
Mammary tumorigenesis alters TGF-β coupling to NF-κB activity
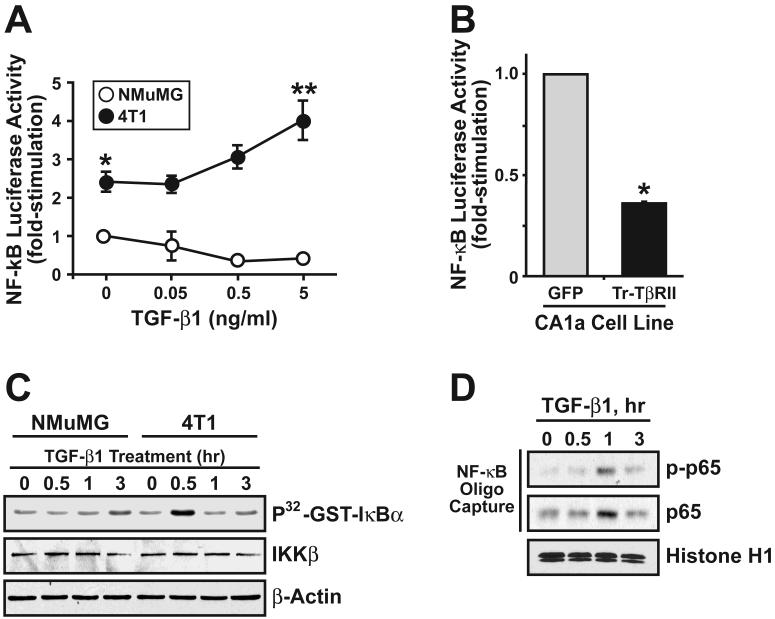
Activation of the TGF-β signaling system typically represses NF-κB activity in normal epithelial cells, including those of the breast (36). In using a NF-κB-driven luciferase reporter gene, we too find that TGF-β stimulation of normal NMuMG cells significantly repressed (by 58%) their activation of NF-κB (Fig. 1A). When identical analyses were performed in malignant, metastatic 4T1 breast cancer cells, basal NF-κB-driven transcriptional activity was significantly higher when compared to NMuMG cells (Fig. 1A). Moreover, administration of TGF-β to 4T1 cells was observed to induce significant NF-κB-driven transcriptional activity. Importantly, the relative difference in NF-κB activation regulated by TGF-β between normal and malignant MECs was ~10-fold. The human MCF10A breast cancer system has been proposed to represent a model of mammary tumorigenesis regulated by TGF-β (37). Our own analyses of these MCF10A derivatives also showed that increasing MEC malignancy did indeed convert TGF-β from an inhibitor to a stimulator of NF-κB-driven luciferase activity (data not shown), and of NF-κB mediated DNA binding activity (data not shown). TGF-β treatment readily induced the nuclear accumulation of phospho-p65/RelA in MCF10A-CA1a cells (Supplementary Fig. S1A). Comparison of NF-κB-driven luciferase activity in control (i.e. GFP) and truncated TβR-II-expressing (i.e., dominant-negative TβR-II) MCF10A-CA1a cells revealed that these cells are subjected to a significant amount of autocrine TGF-β signaling contributes to NF-κB activation (Fig. 1B). Additionally, TGF-β stimulated the phosphotransferase activity of IKKβ against recombinant IκBα in malignant 4T1 cells, but not in normal NMuMG cells (Fig. 1C). Likewise, TGF-β treatment of 4T1 cells induced phosphorylation and binding of nuclear p65/RelA to a biotinylated oligonucleotide containing a NF-κB consensus sequence site (Fig. 1D). We also monitored Smad2/3 activation induced by TGF-β in normal and malignant MECs. These analyses showed that TGF-β stimulation of either NMuMG, 4T1, or MCF10-CA1a cells rapidly induced the phosphorylation of Smad2/3, as well as the expression of SBE-luciferase (Supplementary Fig. S1B). Thus, these findings indicate that mammary tumorigenesis does indeed convert TGF-β from an inhibitor to a stimulator of NF-κB activity.
Figure 1.
Mammary tumorigenesis alters TGF-β coupling to NF-κB activity. (A) TGF-β1 treatment of normal NMuMG cells significantly repressed the expression of luciferase activity driven by the NF-κB promoter. In stark contrast, this same stimulation protocol resulted in the significant activation of NF-κB-driven luciferase activity in malignant metastatic 4T1 cells. Data are the mean (± SE; n=3) luciferase activities relative to unstimulated NMuMG cells. (*, P < 0.05; Student's T-Test). (B) Control (i.e., GFP) and dominant-negative TβR-II-expressing MCF10-CA1a cells were transiently transfected with NF-κB-luciferase and β-gal, and subsequently were processed for determination of luciferase and β-gal activities contained in detergent-solubilized whole cell extracts. Data are the mean (± SE; n=3) luciferase activities relative to GFP-expressing cells. (*, P < 0.05; Student's T-Test). (C) Quiescent NMuMG and 4T1 cells were stimulated with TGF-β1 (5 ng/ml) for 0-3 h as indicated, and IKKβ subsequently was isolated by immunoprecipitation with anti-IKKβ antibodies. IKKβ phosphotransferase activity in the resulting immunocomplexes was monitored using in vitro GST-IκBα protein kinase assay. Data are from a representative experiment that was repeated three times with similar results. (D) Quiescent 4T1 cells stimulated with TGF-β1 (5 ng/ml) as above, and nuclear extracts were prepared and incubated with biotinylated NF-κB oligonucleotide probes. Afterward, p65/RelA:oligonucleotide complexes were captured with streptavidin-agarose beads, and subsequently were visualized by immunoblotting with antibodies against phospho-p65/RelA or total p65/Rel A as indicated. Data are from a representative experiment that was repeated twice with similar results.
TGF-β Stimulation of NF-κB Activity Requires TAK1 and the IKK complex
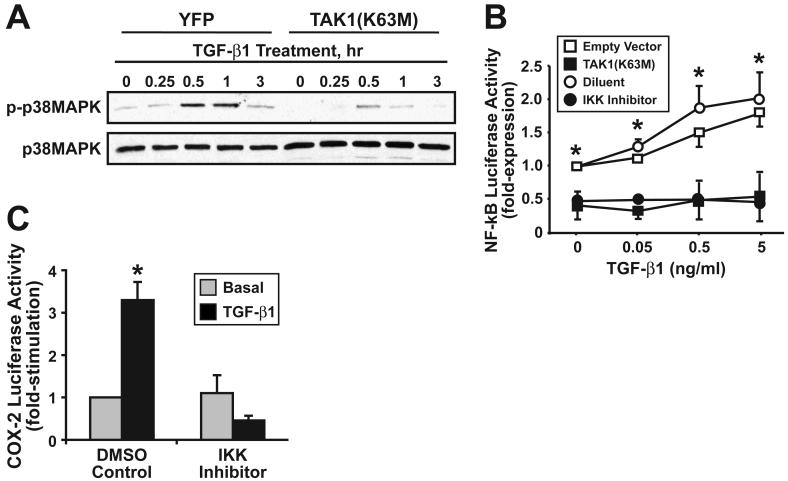
Activation of TAK1 by TGF-β has been linked to the stimulation of NF-κB activity in certain cancer models, including those of the prostate and liver (26, 27). Whether TAK1 couples TGF-β to NF-κB activation in malignant MECs remains unknown. We addressed this question by expressing in 4T1 cells a kinase-dead TAK1(K63M), which functions in a dominant-negative manner to inhibit TAK1-mediated signaling (10), to monitor its effects on TGF-β stimulation of p38 MAPK and NF-κB activity. As expected, TAK1(K63M) expression inhibited TGF-β-mediated activation of p38 MAPK in 4T1 cells (Fig. 2A). Moreover, expression of TAK1(K63M) in 4T1 cells not only reduced their basal level of NF-κB activity, but also uncoupled TGF-β from activation of NF-κB in these malignant MECs (Fig. 2B). Interestingly, pharmacological inhibition of IKKβ activity (i.e., IKK-2 VI inhibitor administration) wholly mimicked the uncoupling of TGF-β to NF-κB in 4T1 cells (Fig. 2B), suggesting that IKKβ lies downstream of TAK1 during TGF-β stimulation of NF-κB and its pro-tumorigenic target genes. TGF-β treatment of 4T1 cells stimulated COX-2 promoter driven luciferase activity, an effect that was blocked by treatment with the IKK-2 VI inhibitor (Fig. 2C). Thus, the ability of TGF-β to stimulate NF-κB and target gene expression in malignant MECs requires the activity of TAK1 and IKKβ, which promote proinflammatory gene expression in malignant MECs.
Figure 2.
TGF-β stimulation of NF-κB activity requires TAK1 and the IKKβ activity. (A) Quiescent control (i.e., YFP) or TAK1(K63M)-expressing 4T1 cells were stimulated with TGF-β1 (5 ng/ml) for 0-3 h as indicated. Afterward, the phosphorylation status of p38 MAPK was monitored by immunoblotting with phospho-specific p38 MAPK antibodies. Differences in protein loading were monitored using anti-p38 antibodies. Images are from a single experiment that was repeated twice with similar results. (B) Transient TAK1(K63M) expression or pharmacological inhibition of IKKβ (i.e., IKK inhibitor VI, 1 μM) significantly inhibited basal and TGF-β1-stimulated NF-κB activity in 4T1 cells. Data are the mean (± SE; n=3) luciferase activities relative to corresponding unstimulated controls. (*, P < 0.05; Student's T-Test). (C) Murine 4T1 cells were transiently transfected with COX-2 luciferase, together with β-gal as above, and subsequently were stimulated overnight with TGF-β1 (5 ng/ml) in the absence or presence of the IKKβ antagonist (1 μM). Data are the mean (± SE; n=2) luciferase activities relative to unstimulated controls. (*, P < 0.05; Student's T-Test).
TAB1:IKK Complexes Form Solely in Breast Cancer Cells and Mediate Their Activation of NF-κB by TGF-β
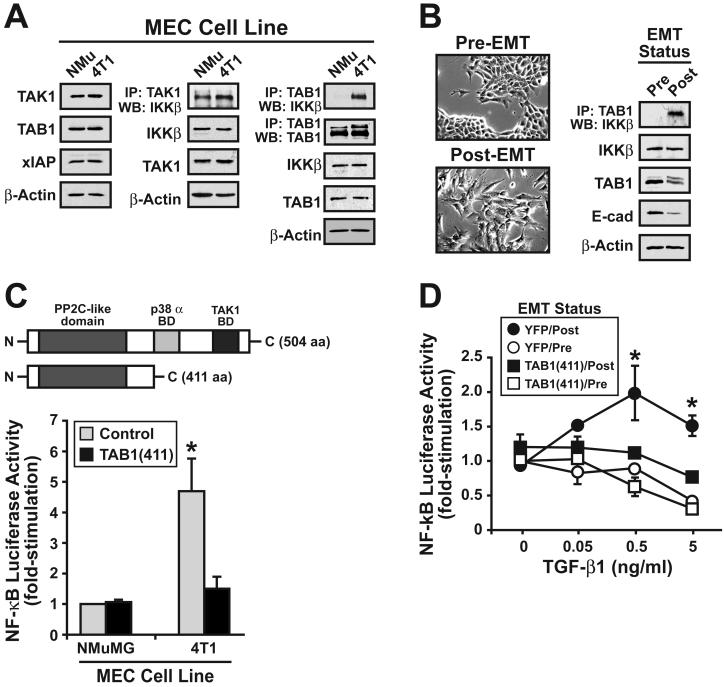
Our findings thus far show that mammary tumorigenesis converts TGF-β from a repressor to a promoter of NF-κB activation via a TAK1:IKKβ-dependent mechanism (Figs. 1 and 2). Although currently unknown, altered expression of various components of the TAK1 signaling complex could underlie the initiation of this aberrant signaling phenomenon in malignant MECs. We tested this hypothesis by comparing the cellular levels of TAK1, TAB1, and xIAP in normal and malignant MECs. As shown in Fig. 3A (left panel), normal NMuMG and malignant 4T1 cells express similar quantities of TAK1, TAB1, or xIAP. Identical findings were observed when comparing the cellular levels of these proteins in derivatives of the human MCF10A cell system (data not shown). TAK1 binds and phosphorylates IKKβ, leading to its activation (38). Thus, altered formation of TAK1:IKKβ complexes could underlie the differential coupling to TGF-β to NF-κB in malignant MECs. We tested this hypothesis by monitoring the formation of TAK1:IKKβ complexes in NMuMG and 4T1 cells and found that both cell lines housed similar quantities of TAK1:IKKβ complexes (Fig. 3A, middle panel). Although mammary tumorigenesis had no effect on the expression of TAK1 signaling components, this pathological process led to the formation of TAB1:IKKβ complexes solely in malignant, metastatic 4T1 cells (Fig. 3A, right panel). This findings suggests that incorporation of TAB1 into TAK1:IKKβ complexes couples TGF-β to stimulation of NF-κB. Along these lines, the interaction between TAB1:IKKβ was undetectable in normal MECs, but the formation of TAB1:IKKβ complexes was readily apparent in these same cells following their stimulation with TGF-β to induce EMT (Fig. 3B), which manifested in their acquisition of a fibroblastoid morphology and reduced E-cadherin expression (Fig. 3B). Collectively, these findings suggest that the interaction of TAB1 with IKKβ mediates TGF-β stimulation of NF-κB solely in malignant MECs, or in normal MECs that have undergone an EMT in response to TGF-β.
Figure 3.
TAB1:IKKβ complexes form solely in breast cancer cells, or in normal MECs following their induction of EMT by TGF-β. (A) Detergent-solubilized whole cell extracts prepared from NMuMG and 4T1 cells were immunoblotted with antibodies against TAK1, TAB1, xIAP, and β-actin as indicated (left panel), or were immunoprecipitated with antibodies against either TAK1 (middle panel) or TAB1 (right panel), followed by immunoblotting with antibodies against IKKβ. Differences in protein loading were monitored by reprobing stripped membranes with β-actin antibodies. Images are from representative experiments that were repeated three times with identical results. (B) NMuMG cells were incubated for 36 hr in the absence or presence of TGF-β1 (5 ng/ml) to induce EMT (left panel), and subsequently were subjected to TAB1 co-immunoprecipitation analysis. Induction of EMT by TGF-β1 was monitored by immunoblotting for diminished E-cadherin expression, while differences in protein loading were monitored by β-actin immunoblotting. Images are from representative experiments that were repeated three times with identical results. (C) Schematic depicting the modular structure of full-length TAB1, as well as that of the truncated TAB1 mutant [i.e., TAB1(411)] that lacks the binding domains (BD) for p38 MAPK and TAK1 (top panel). NMuMG and 4T1 cells were transiently transfected with NF-κB-luciferase and β-gal, together with either empty vector or TAB1(411) cDNA as indicated. Afterward, luciferase and β-gal activities contained in detergent-solubilized whole cell extracts were measured (lower panel). Data are the mean (± SE; n=3) luciferase activities relative to empty vector-expressing, unstimulated NMuMG cells. (*, P < 0.05; Student's T-Test).
TAB1α is a modular adapter protein comprised of three distinct functional domains: (i) an N-terminal PP2C-like domain; (ii) a central p38α MAPK binding domain; and (iii) a C-terminal TAK1 binding domain [(39); Fig. 3C, upper panel]. TAB1-deficiency in mice alters some transcriptional responses regulated by TGF-β (19), and as such, we generated a truncated, dominant-negative TAB1 mutant [i.e., TAB1(411)] that lacked the binding domains for p38α MAPK and TAK1 to assess its effects on NF-κB activity in normal and malignant MECs. As expected, transient TAB1(411) expression failed to effect basal NF-κB activity in NMuMG cells, but significantly reduced that observed in 4T1 cells to a level of similar to that in normal NMuMG cells (Fig. 3C, lower panel). This finding suggests that TAB1 does indeed play an important role in regulating NF-κB activity in malignant MECs. Along these lines, our finding that TGF-β stimulation of EMT in normal MECs promoted the formation of TAB1:IKKβ (Fig. 3B) implies that this event may be sufficient in coupling TGF-β to NF-κB activation in post-EMT MECs. We tested this hypothesis by stably expressing TAB1(411) in NMuMG cells and subsequently monitoring their ability to undergo EMT and activate NF-κB in response to TGF-β. Figure 3D shows that following their induction of EMT by TGF-β administration, NMuMG cells readily acquired the ability to activate NF-κB when stimulated by TGF-β. Interestingly, although TAB1(411) expression had no effect on the ability of NMuMG cells to undergo EMT stimulated by TGF-β (data not shown), its expression did uncouple TGF-β from NF-κB activation in post-EMT NMuMG cells (Fig. 3D). Collectively, these findings indicate that incorporation of TAB1 into TAK1:IKKβ complexes mediates activation of NF-κB by TGF-β in MECs. Our results also dissociate NF-κB activity from the ability of TGF-β to stimulate EMT in normal MECs.
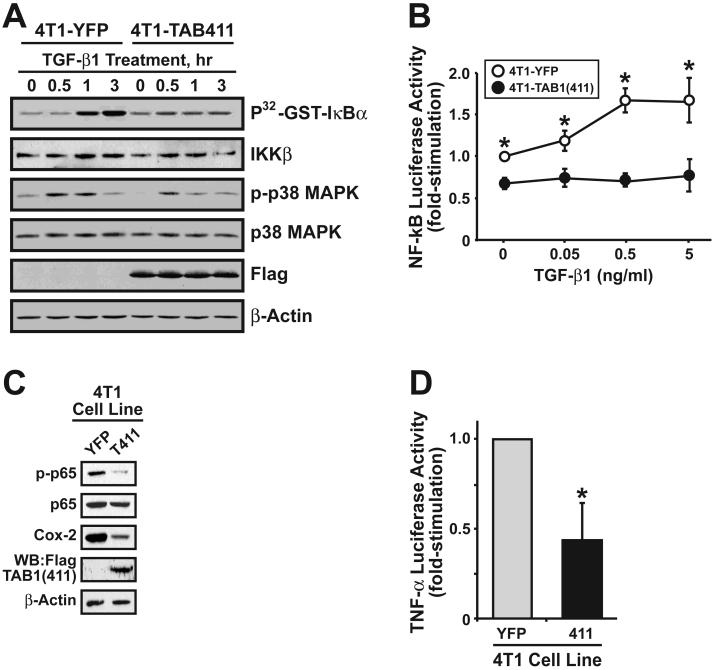
We also examined the effects of stable TAB1(411) expression on the ability of TGF-β to activate NF-κB in 4T1 cells. Stable TAB1(411) expression in 4T1 cells resulted in a distorted, more rounded cell morphology characteristic of altered cellular adhesion (Supplementary Fig. S2A). Similar to its full-length counterpart, truncated TAB1(411) interacted physically with TβR-I, indicating that the molecular determinants that mediate TAB1 binding to TβR-I are located N-terminal to its p38α MAPK binding domain (Supplementary Fig. S2B). Expression of TAB1(411) in 4T1 cells prevented the ability of TGF-β to induce their (i) activation of IKKβ (Fig. 4A); (ii) expression of luciferase driven by NF-κB (Fig. 4B); (iii) phosphorylation of p65/RelA (Fig. 4C); and (iv) expression of the proinflammatory genes, Cox-2 (Fig. 4C) and TNF-α (Fig. 4D). The inhibitory effects of TAB1(411) expression on TGF-β signaling were specific for the NF-κB pathway and failed to alter the ability of TGF-β to activate Smad2/3-mediated gene expression (Supplementary Fig. S2C) and p38 MAPK phosphorylation (Fig. 4A). Finally, consistent with its lack of involvement in TNF-α signaling (19, 40), TAB1(411) expression failed to impact TNF-α stimulation of NF-κB activity in 4T1 cells (Supplementary Fig. S2D). Thus, these findings indicate that TAB1:IKKβ complexes form and function specifically in mediating activation of NF-κB and its downstream effectors in response to TGF-β treatment of post-EMT normal and malignant MECs.
Figure 4.
Truncated TAB1(411) expression selectively blocks TGF-β-mediated NF-κB activation in 4T1 cells. (A) Quiescent control (i.e., YFP) and TAB1(411)-expressing 4T1 cells were stimulated with TGF-β1 (5 ng/ml) as indicated. Afterward, IKKβ was isolated by immunoprecipitation and analyzed for GST-IκBα phosphotransferase activity. The phosphorylation status of p38 MAPK also was monitored by immunoblotting with phospho-specific p38 MAPK antibodies. Differences in protein loading were monitored using antibodies against p38 MAPK. Data are from a representative experiment that was repeated three times with similar results. (B) Control (i.e., YFP) and TAB1(411)-expressing 4T1 cells were transiently transfected with NF-κB-luciferase and β-gal as above, and subsequently were stimulated with increasing concentrations of TGF-β1 for 24 h. Data are the mean (± SE; n=4) luciferase activities relative to unstimulated control cells. (C) Detergent-solubilized whole cell extracts prepared from control (i.e., YFP) and TAB1(411)-expressing 4T1 cells were immunoblotted for phospho-p65/RelA, p65/RelA, or COX-2 as indicated. TAB1(411) expression was detected by anti-FLAG immunoprecipitation, followed by immunoblotting with anti-TAB1 antibodies as indicated. Differences in protein loading were monitored by β-actin immunoblotting. Images are from a representative experiment that was repeated three times with identical results. (D) Control (i.e., YFP) and TAB1(411)-expressing 4T1 cells were transiently transfected with TNF-α-luciferase and β-gal, and subsequently were processed for determination of luciferase and β-gal activities contained in detergent-solubilized whole cell extracts. Data are the mean (± SE; n=3) luciferase activities relative to YFP-expressing cells.
TAK1 is Essential for TGF-β Stimulation of NF-κB and Invasion in Metastatic Breast Cancer Cells
Our findings thus far have identified a novel TAB1:TAK1:IKKβ signaling axis that selectively couples TGF-β to NF-κB in malignant MECs. We next addressed the contribution of this signaling axis to breast cancer cell proliferation and invasion stimulated by TGF-β. Similar to 4T1 cells, the formation of TAB1:IKKβ complexes also were readily detected in metastatic human MCF10A-CA1a cells (data not shown), indicating that the formation of TAB1:IKKβ complexes was not a phenomenon unique to murine 4T1 cells. Introduction of kinase-dead TAK1(K63M) (Fig. 5A) or dominant-negative IκBα (Fig. 5B) into MCF10A-CA1a cells abrogated their invasion through synthetic basement membranes in response to TGF-β. Finally, MCF10A-CA1a cells are refractory to the cytostatic activities of TGF-β [Fig. 5C; (37)]. Surprisingly, TAK1-deficiency not only reduced the proliferative capacity of MCF10A-CA1a cells, but also partially restored their cytostatic response to TGF-β (Fig. 5C). Collectively, these findings indicate that activation of the TAB1:TAK1:IKKβ signaling axis is essential in mediating oncogenic signaling by TGF-β, particularly its ability to inhibit TGF-β-mediated growth inhibition and mediated TGF-β stimulated breast cancer cell invasion.
Figure 5.
TAK1 is essential for TGF-β stimulation of NF-κB and invasion in metastatic breast cancer cells. MCF10A-CA1a cells were engineered to stably express either kinase-dead TAK1(K63M) (A) or dominant-negative IκBα null (B), and subsequently were induced to invade through synthetic basement membranes by serum (3%) in the presence or absence of TGF-β1 (5 ng/ml) as indicated. Data are the mean (± SE; n=4) invasion relative to that induced by serum in GFP-expressing cells. (*, **, P < 0.05; Student's T-Test). (C) MCF10ACA1a cells were rendered TAK1-deficient by siRNA transfection, and subsequently were stimulated with TGF-β1 (5 ng/ml) for 48 h as indicated. Cellular DNA was radiolabeled with [3H]thymidine and quantified by scintillation counting. Data are the mean (± SE; n=3) [3H]thymidine incorporation normalized to unstimulated control transfected cells. (*, **, P < 0.05; Student's T-test). TAK1-deficiency was monitored by immunoblotting whole cell extracts with the TAK1-specific antibodies. Differences in protein loading were monitored using anti-β-actin antibodies. Images are from a single experiment that was repeated thrice with similar results.
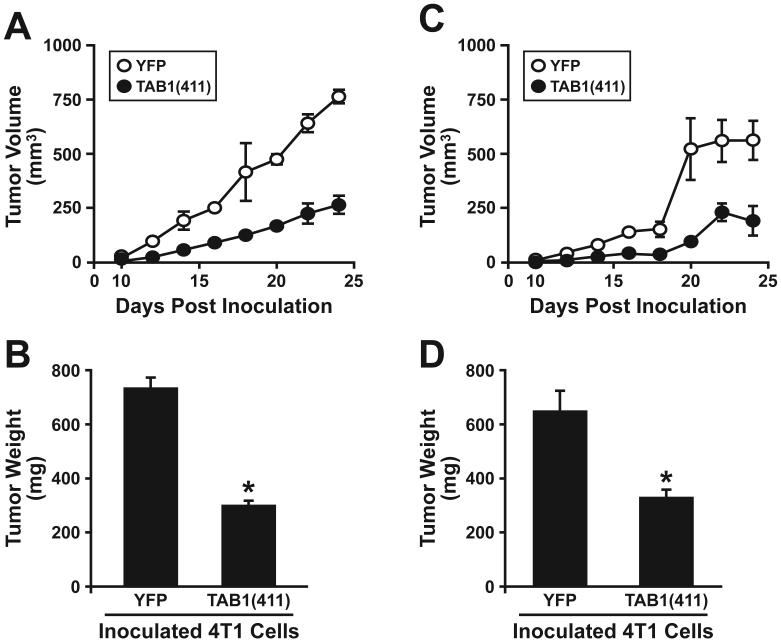
TAB1(411) Expression Inhibits Mammary Tumor Growth in Mice
We tested that above hypothesis by orthotopically injecting the mammary fat pads of Balb/C mice with syngeneic 4T1 breast cancer cells that expressed control (YFP) or truncated TAB1(411) (Figs. 3 and 4). Interestingly, stable TAB1(411) expression failed to alter the proliferation of 4T1 cells in vitro (data not shown); however, the growth of 4T1 tumors in Balb/C mice was decreased significantly by their expression of TAB1(411) (Fig. 6). Indeed, over the course of these 24 day experiments, TAB1(411) expression significantly reduced 4T1 tumor volume by ~65% (Fig. 6A) and their resulting weights at the time of necropsy by ~59% (Fig. 6B). We also examined the effects of TAB1(411) expression on 4T1 tumor growth in nude mice. Contrary to our expectations, we again observed TAB1(411) expression to significantly reduce growth of 4T1 tumors in nude mice (Figs. 6C & D). Thus, the specific incorporation of TAB1 into TAK1:IKKβ complexes in malignant MECs plays a significant role in promoting TGF-β stimulation of breast cancer development and progression, presumably in part via NF-κB-mediated activation of the innate immune system and its ability to promote tumor progression (41).
Figure 6.
TAB1(411) expression inhibits mammary tumor growth in mice. Parental (i.e., YFP) or TAB1(411)-expressing 4T1 cells were injected orthotopically into the mammary fat pads of either syngeneic Balb/C (A and C) or nude mice (B and D). Tumor volumes were measured every other day beginning at day 10 and continued through day 24. Data are the mean (± SE) tumor volumes (A and C) or weights (B and D) at the time of necropsy observed in two (i.e., Balb/C mice) or three (i.e., nude mice) independent experiments (3 mice/condition/experiment). (*, P < 0.05; Student's T-Test).
Discussion
The conversion of TGF-β from a tumor suppressor to a tumor promoter plays a significant role in determining how developing and progressing tumors interact with and respond to changes in their microenvironments. Intense research efforts over the last decade have identified a number of aberrant genetic and epigenetic events that potentially contribute to oncogenic signaling by TGF-β in malignant MECs (1). Included in this list oncogenic mediators of TGF-β signaling is NF-κB, whose stimulation normally is repressed by TGF-β, but instead becomes activated by this cytokine during the course of carcinogenesis (26, 27, 42, 43). Unfortunately, precisely how carcinogenesis converts the cellular response to TGF-β and its coupling to NF-κB remains to be determined definitively. This question is medically relevant because NF-κB activation has been associated with tumor inflammation, as well as with elevated tumor angiogenesis, invasion, and resistance to apoptotic stimuli (28, 41), and with TGF-β-mediated EMT (44). Thus, chemotherapeutic targeting of these molecular events may afford novel avenues to alleviate oncogenic signaling by TGF-β in patients with metastatic breast cancer.
By comparing the NF-κB activity profiles regulated by TGF-β in normal and malignant MECs, we defined a novel TAB1:TAK1:IKKβ signaling axis that forms specifically in breast cancer cells and mediates NF-κB activation by TGF-β. This cellular response contrasted sharply with that observed in normal MECs, where TGF-β typically inhibits NF-κB activation by inducing the expression of the NF-κB inhibitor, IκBα (36). Although p38 MAPK and AKT have been linked to NF-κB activation in cancer cells (45, 46), our results indicate that the formation of TAB1:IKKβ complexes represents a key molecular event that enables TGF-β to activate NF-κB in malignant MECs (Figs. 3 & 4), and in normal MECs induced to undergo EMT in response to TGF-β. It is interesting to note that TAB1(411) expression, which inhibited the coupling of TGF-β to NF-κB activation (Figs. 3 & 4), had no effect on the ability of TGF-β to induce EMT in NMuMG cells, suggesting that NF-κB activation in NMuMG cells is dissociated from their ability to undergo EMT in response to TGF-β. This finding contradicts those of Huber et al (44) who found NF-κB to be essential for the ability of TGF-β to induce and stabilize EMT in EpRas-transformed MECs, and for the ability of these cells to colonize the lung during the performance of tail vein injection assays. Discordance between our respective studies may reflect differences in the MECs studied, or in the relative contribution of oncogenic Ras, whose activity clearly cooperates with the TGF-β (1) and NF-κB (47) signaling systems. However, both studies show the overall importance of NF-κB in promoting the growth and development of mammary tumors in mice.
The ability of TAB1:IKKβ complexes to link TGF-β to NF-κB signaling in malignant MECs likely occurs through the interaction of TAB1 with TβR-I, a binding reaction dependent upon structural determinants located in the N-terminus of TAB1 (Fig. S2B). Once bound to TβR-I, C-terminal TAB1 sequences coordinate the binding and activation of TAK1:IKKβ, and consequently, the induction of NF-κB specifically by TGF-β, but not by TNF-α (Fig. S2C). The specificity of TAB1 for TGF-β signaling is consistent with gene targeting experiments that established the requirement for TAB2 and TAB3 in mediating intracellular signaling by IL-1 and TNF-α (19, 40). Future studies need to identify the molecular mechanisms whereby TAB1:IKKβ complexes only form in malignant MECs.
A potentially important observation of our study concerns the connection between the coupling of TGF-β to NF-κB activation and its consequential induction of proinflammatory cytokines, whose production can promote tumor progression via activation of the innate immune system (41). Indeed, while the link between NF-κB and inflammation in mediating cancer progression has clearly been established (28), the molecular mechanisms that underlie this pathological sequence of events has yet to be fully elucidated. We observed TAB1(411) expression to prevent TGF-β stimulation of NF-κB and its production of the proinflammatory genes COX-2 and TNF-α in 4T1 cells, and of COX-2, TNF-α, IL-6, and GMCSF in post-EMT NMuMG cells (data not shown). These findings are reminiscent of the gene expression profiles detected in response to either TAK1 inactivation (25) or TAB1-deficiency (20), and as such, further support the role of TAB1:TAK1:IKKβ complexes in mediating malignant MEC expression of proinflammatory genes when stimulated by TGF-β. Once released into the tumor milieu, these proinflammatory cytokines function in recruiting innate immune effectors, such as immature and tumor-associated macrophages, neutrophils, and mast cells, which collectively induce the remodeling of tumor microenvironments to favor the growth, metastasis, and angiogenesis of developing and progressing mammary tumors (28, 41). Our finding that TAB1(411) expression retains its ability to suppress the growth of mammary tumors in nude mice (Fig. 6) suggests that abrogating TGF-β-mediated stimulation of NF-κB preferentially prevents the activation of the innate immune system, not that of the adaptive immune system. Future studies need to further investigate the linkage between TGF-β and NF-κB in mediating activation of the innate immune system during the development and progression of mammary tumors.
Finally, an important and somewhat surprising finding presented herein was the essential requirement of TAK1 and NF-κB in mediating breast cancer cell invasion. In addition, TAK1-deficiency not only inhibited breast cancer proliferation, but also partially restored the cytostatic function of TGF-β in malignant MECs resistant to its growth inhibitory activities. These findings, together with those linking the TAB1:TAK1:IKKβ signaling axis to inflammatory gene expression, suggest that measures capable of antagonizing this oncogenic pathway may prevent the development and progression of mammary tumors. Accordingly, overexpression of TAB1(411) in 4T1 cells significantly impaired their growth relative to parental 4T1 cells when implanted into the mammary fat pads of mice (Fig. 6). Thus, the selective antagonism of TAB1:TAK1:IKKβ:NF-κB signaling activated by TGF-β holds the potential to one day improve the clinical course of patients with metastatic breast cancer.
Supplementary Material
Acknowledgements
Members of the Schiemann Laboratory are thanked for critical reading of the manuscript. The technical expertise of members of the Flow Cytometry at the University of Colorado Cancer Center is gratefully acknowledged. Research support was provided in part by the National Institutes of Health (CA095519 and CA114039) to WPS.
Research Support: Research support was provided in part by the National Institutes of Health (CA095519 and CA114039) to W.P.S.
References
- 1.Galliher AJ, Neil JR, Schiemann WP. Role of TGF-β in cancer progression. Future Oncology. 2006;2:743–63. doi: 10.2217/14796694.2.6.743. [DOI] [PubMed] [Google Scholar]
- 2.Blobe GC, Schiemann WP, Lodish HF. Role of TGF-β in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 4.Kamaraju AK, Roberts AB. Role of Rho/ROCK and p38 MAP kinase pathways in TGF-β-mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J Biol Chem. 2005;280:1024–36. doi: 10.1074/jbc.M403960200. [DOI] [PubMed] [Google Scholar]
- 5.Hartsough MT, Mulder KM. TGF-β activation of p44MAPK in proliferating cultures of epithelial cells. J Biol Chem. 1995;270:7117–24. doi: 10.1074/jbc.270.13.7117. [DOI] [PubMed] [Google Scholar]
- 6.Hanafusa H, Ninomiya-Tsuji J, Masuyama N, et al. Involvement of the p38 mitogen-activated protein kinase pathway in TGF-β-induced gene expression. J Biol Chem. 1999;274:27161–7. doi: 10.1074/jbc.274.38.27161. [DOI] [PubMed] [Google Scholar]
- 7.Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent SMAD and JNK signaling in TGF-β-mediated transcription. J Biol Chem. 1999;274:37413–20. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 8.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–12. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi K, Shirakabe K, Shibuya H, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–11. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 10.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–6. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 11.Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J Mol Biol. 2003;326:105–15. doi: 10.1016/s0022-2836(02)01404-3. [DOI] [PubMed] [Google Scholar]
- 12.Ono K, Ohtomo T, Sato S, et al. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J Biol Chem. 2001;276:24396–400. doi: 10.1074/jbc.M102631200. [DOI] [PubMed] [Google Scholar]
- 13.Brown K, Vial SC, Dedi N, Long JM, Dunster NJ, Cheetham GM. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. J Mol Biol. 2005;354:1013–20. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- 14.Besse A, Lamothe B, Campos AD, et al. TAK1-dependent signaling requires functional interaction with TAB2/TAB3. 2006;282:3918–28. doi: 10.1074/jbc.M608867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriguchi T, Kuroyanagi N, Yamaguchi K, et al. A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J Biol Chem. 1996;271:13675–9. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 16.Sakurai H, Miyoshi H, Toriumi W, Sugita T. Functional interactions of TGF-β-activated kinase 1 with IκB kinases to stimulate NF-κB activation. J Biol Chem. 1999;274:10641–8. doi: 10.1074/jbc.274.15.10641. [DOI] [PubMed] [Google Scholar]
- 17.Sakurai H, Nishi A, Sato N, Mizukami J, Miyoshi H, Sugita T. TAK1-TAB1 fusion protein: a novel constitutively active mitogen-activated protein kinase kinase kinase that stimulates AP-1 and NF-κB signaling pathways. Biochem Biophys Res Commun. 2002;297:1277–81. doi: 10.1016/s0006-291x(02)02379-3. [DOI] [PubMed] [Google Scholar]
- 18.Greten FR, Karin M. The IKK/NF-κB activation pathway - A target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–9. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Shim JH, Xiao C, Paschal AE, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;31:31. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertelsen M, Sanfridson A. TAB1 modulates IL-1α mediated cytokine secretion but is dispensable for TAK1 activation. Cell Signal. 2007;19:646–57. doi: 10.1016/j.cellsig.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Shibuya H, Iwata H, Masuyama N, et al. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 1998;17:1019–28. doi: 10.1093/emboj/17.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu Y, Shibuya H, Takeda N, et al. Targeted disruption of the TAB1 gene causes embryonic lethality and defects in cardiovascular and lung morphogenesis. Mech Dev. 2002;119:239–49. doi: 10.1016/s0925-4773(02)00391-x. [DOI] [PubMed] [Google Scholar]
- 23.Ninomiya-Tsuji J, Kajino T, Ono K, et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem. 2003;278:18485–90. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 24.Ono K, Ohtomo T, Ninomiya-Tsuji J, Tsuchiya M. A dominant negative TAK1 inhibits cellular fibrotic responses induced by TGF-beta. Biochem Biophys Res Commun. 2003;307:332–7. doi: 10.1016/s0006-291x(03)01207-5. [DOI] [PubMed] [Google Scholar]
- 25.Thiefes A, Wolter S, Mushinski JF, et al. Simultaneous blockade of NF-κB, JNK, and p38 MAPK by a kinase-inactive mutant of the protein kinase TAK1 sensitizes cells to apoptosis and affects a distinct spectrum of tumor necrosis factor target genes. J Biol Chem. 2005;280:27728–41. doi: 10.1074/jbc.M411657200. [DOI] [PubMed] [Google Scholar]
- 26.Park J-I, Lee M-G, Cho K, et al. TGF-β1 activates interleukin-6 expression in prostate cancer cells through the synergistic collaboration of the Smad2, p38-NF-κB, JNK, and Ras signaling pathways. Oncogene. 2003;22:4314–32. doi: 10.1038/sj.onc.1206478. [DOI] [PubMed] [Google Scholar]
- 27.Arsura M, Panta GR, Bilyeu JD, et al. Transient activation of NF-κB through TAK1/IKK kinase pathway by TGF-β1 inhibits AP-1/SMAD signaling and apoptosis: Implication in liver tumor formation. Oncogene. 2003;22:412–25. doi: 10.1038/sj.onc.1206132. [DOI] [PubMed] [Google Scholar]
- 28.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–83. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagarajan RP, Chen F, Li W, et al. Repression of TGF-β-mediated transcription by NF-κB. Biochem J. 2000;348:591–6. [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Rovira T, Chalaux E, Rosa JL, Bartrons R, Ventura F. Interaction and functional cooperation of NF-κB with Smads. Transcriptional regulation of the JunB promoter. J Biol Chem. 2000;275:28937–46. doi: 10.1074/jbc.M909923199. [DOI] [PubMed] [Google Scholar]
- 31.Galliher AJ, Schiemann WP. β3 integrin and Src facilitate TGF-β-mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 2006;8:R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galliher AJ, Schiemann WP. Src phosphorylates Tyr284 in TβR-II and regulates TGF-β stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res. 2007;67:3752. doi: 10.1158/0008-5472.CAN-06-3851. [DOI] [PubMed] [Google Scholar]
- 33.Schiemann BJ, Neil JR, Schiemann WP. SPARC inhibits epithelial cell proliferation in part through stimulation of the TGF-β-signaling system. Mol Biol Cell. 2003;14:3977–88. doi: 10.1091/mbc.E03-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiemann WP, Blobe GC, Kalume DE, Pandey A, Lodish HF. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by TGF-β and affects protein kinase cascades. J Biol Chem. 2002;277:27367–77. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- 35.Schiemann WP, Graves LM, Baumann H, et al. Phosphorylation of the human leukemia inhibitory factor (LIF) receptor by mitogen-activated protein kinase and the regulation of LIF receptor function by heterologous receptor activation. Proc Natl Acad Sci USA. 1995;92:5361–5. doi: 10.1073/pnas.92.12.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sovak MA, Arsura M, Zanieski G, Kavanagh KT, Sonenshein GE. The inhibitory effects of TGF-β1 on breast cancer cell proliferation are mediated through regulation of aberrant NF-κB/Rel expression. Cell Growth Differ. 1999;10:537–44. [PubMed] [Google Scholar]
- 37.Tang B, Vu M, Booker T, et al. TGF-β switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest. 2003;112:1116–24. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 39.Ge B, Xiong X, Jing Q, et al. TAB1β (TGF-β-activated protein kinase 1-binding protein 1β), a novel splicing variant of TAB1 that interacts with p38α but not TAK1. J Biol Chem. 2003;278:2286–93. doi: 10.1074/jbc.M210918200. [DOI] [PubMed] [Google Scholar]
- 40.Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor R, Matsumoto K. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 2003;22:6277–88. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karin M, Greten FR. NF-κB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 42.Kim DW, Sovak MA, Zanieski G, et al. Activation of NF-κB/Rel occurs early during neoplastic transformation of mammary cells. Carcinogenesis. 2000;21:871–9. doi: 10.1093/carcin/21.5.871. [DOI] [PubMed] [Google Scholar]
- 43.Rayet B, Gelinas C. Aberrant Rel/NF-κB genes and activity in human cancer. Oncogene. 1999;18:6938–47. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 44.Huber MA, Azoitei N, Baumann B, et al. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–81. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter AB, Knudtson KL, Monick MM, Hunninghake GW. The p38 mitogen-activated protein kinase is required for NF-κB-dependent gene expression. The role of TATA-binding protein (TBP) J Biol Chem. 1999;274:30858–63. doi: 10.1074/jbc.274.43.30858. [DOI] [PubMed] [Google Scholar]
- 46.Madrid LV, Wang CY, Guttridge DC, Schottelius AJ, Baldwin AS, Mayo MW. AKT suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol Cell Biol. 2000;20:1626–38. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orlowski RZ, Baldwin AS. NF-κB as a therapeutic target in cancer. Trends Mol Med. 2002;8:385–9. doi: 10.1016/s1471-4914(02)02375-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.