Abstract
Efflux of cytosolic Ca2+ mediated by plasma membrane Ca2+-ATPases (PMCA) plays a key role in fine tuning the magnitude and duration of Ca2+ signaling following activation of G-protein-coupled receptors. However, the molecular mechanisms that underpin the trafficking of PMCA to the membrane during Ca2+ signaling remain largely unexplored in native cell models. One potential mechanism for the recruitment of proteins to the plasma membrane involves PDZ interactions. In this context, we investigated the role of PMCA interactions with the Na+/H+ exchanger regulatory factor 2 (NHERF-2) during muscarinic-induced Ca2+ mobilization in the HT-29 epithelial cell line. GST pull-downs in HT-29 cell lysates showed that the PDZ2 module of NHERF-2 bound to the PDZ binding motif on the C terminus of PMCA. Co-immunoprecipitations confirmed that PMCA1b and NHERF-2 associated under normal conditions in HT-29 cells. Cell surface biotinylations revealed significant increases in membrane-associated NHERF-2 and PMCA within 60 s following muscarinic activation, accompanied by increased association of the two proteins as seen by confocal microscopy. The recruitment of NHERF-2 to the membrane preceded that of PMCA, suggesting that NHERF-2 was involved in nucleating an efflux complex at the membrane. The muscarinic-mediated translocation of PMCA was abolished when NHERF-2 was silenced, and the rate of relative Ca2+ efflux was also reduced. These experiments also uncovered a NHERF-2-independent PMCA retrieval mechanism. Our findings describe rapid agonist-induced translocation of PMCA in a native cell model and suggest that NHERF-2 plays a key role in scaffolding and maintaining PMCA at the cell membrane.
A change in the concentration of cytosolic Ca2+ ([Ca2+]i) is a key initiator of a multitude of cellular responses. Low resting [Ca2+]i is maintained by the sequestration of Ca2+ into intracellular stores or the extrusion of Ca2+ across the plasma membrane by transporters such as the ubiquitous plasma membrane Ca2+-ATPase (PMCA)3 (1). Many G-protein-coupled receptors (GPCRs) (such as purinergic and muscarinic receptors) signal via transient increases in [Ca2+]i to effect physiological responses (2). The increase in [Ca2+]i is biphasic, with an initial mobilization of Ca2+ from intracellular stores which in turn triggers Ca2+ influx via ion channels in the plasma membrane. Following termination of receptor signaling, [Ca2+]i returns rapidly to resting levels due to refilling of the sarcoplasmic/endoplasmic reticulum as well as Ca2+ efflux mediated by PMCA and/or Na+-Ca2+ exchange (3). Thus the magnitude and duration of the increase in [Ca2+]i during the activation of a GPCR signaling cascade reflects a dynamic equilibrium between influx, reuptake, and efflux.
Since the discovery of PMCA in the erythrocyte membrane in the mid-1960s (4), this ubiquitous enzyme has been well-defined as a primary mechanism for Ca2+ efflux from eukaryotic cells to maintain low resting Ca2+ levels (5). PMCA is encoded by four separate genes that give rise to four distinct variants; PMCA1, -2, -3, and -4 (6). PMCA1 and -4 are thought to be ubiquitous, while the expression of PMCA2 and -3 is much more restricted predominantly in the brain and striated muscle (7, 8). Alternate splicing of the original transcripts indicates that there are numerous PMCA variants in the mammalian proteome (9). The diversity of splice variants of PMCA is thought to allow for the complex and dynamic, spatio-temporal regulation of cell-specific Ca2+ handling (6).
The level of complexity of Ca2+ signaling may be increased by spatially constraining various regulatory components associated with the movement of Ca2+. Such supramolecular signaling platforms are already described for store-operated Ca2+ channels as well as their upstream, downstream, and regulatory components (10), and PMCAs are also reported to be part of such store-operated signaling complexes (11). PMCAs are known to interact with a number of cytosolic accessory and scaffold proteins that can direct the formation of Ca2+ signaling microdomains. These scaffolds include PSD-95/Dlg/Zo-1 (PDZ) proteins that are now recognized as playing key roles in aggregating receptors, ion channels, and transporters into functionally specific complexes in membrane microdomains (12-14). Due to alternate splicing of the mRNA transcripts at the C-terminal tail splice site C, there are 2 main variants of the PMCA isoforms denoted PMCAa and PMCAb. Importantly, all PMCAb splice variants have a PDZ consensus binding motif at their C terminus that has been shown to bind a wide variety of PDZ proteins (15-19). Some of these interactions have been shown to be physiologically relevant; for example, PDZ-mediated interactions between PMCA4b and nitric-oxide synthase I (NOS1) are essential for the regulation NO synthesis in the heart (20, 21).
One family of PDZ proteins are the Na+/H+ exchanger regulatory factors, denoted NHERF-1 to -4 (22, 23). NHERF-1 was the first PDZ scaffold identified for an epithelial transporter. It was shown to direct the formation of a multi-protein complex that mediated cAMP-regulated protein kinase (PKA) phosphorylation and inhibition of the Na+/H+ exchanger 3 (NHE3) (24-27). NHERF-1 and -2 contain two PDZ modules (PDZ1 and PDZ2) that bind the PDZ binding motifs in the C terminus of target ion channels, receptors, and transporters (23). NHERF-1 and -2 also contain a C-terminal ezrin-radixin-moesin (ERM) binding domain that interacts with ERM proteins to link to the actin cytoskeleton. NHERF-1 and -2 can also nucleate the formation of macromolecular complexes by physically linking plasma membrane proteins to the cytoskeleton and recruiting signaling molecules such as PKA (26), protein kinase C (PKC) (28), and phospholipase Cβ (PLCβ) (29) to the complex. Examples include the NHE3-NHERF-1-ezrin and NaPi-IIa-NHERF-1-ezrin complexes anchored to the actin cytoskeleton in the brush border membrane of renal proximal tubule cells (30) and the CFTR-NHERF-1/2-ezrin complex found in airway epithelial cells (31, 32). NHERF-2 has been reported to interact with PMCA2b when it was transiently transfected in (Madine Darby Canine Kidney) MDCK cells (18). However, the physiological relevance of the interaction between PMCAs and NHERF-2, particularly in the context of endogenous GPCR-mediated increases in [Ca2+]i remain unexplored. Many of the intracellular kinases that have been shown to bind NHERF-2 such as PKC and PKA are also regulators of PMCA (33). Coordinated signaling via NHERF-2 and modulation of PMCA-mediated efflux introduces a level of complexity that may account for the differences observed in the morphology of Ca2+ responses following GPCR activation (34).
The human colonic epithelial cell line (HT-29) is an accepted model for studying Ca2+ signaling in epithelial cells and expresses only the M3 isoform of the muscarinic receptor (35). Muscarinic activation of HT-29 cells increases [Ca2+]i following activation of PLCβ by the βγ-subunits and release of Ca2+ stores and subsequent Ca2+ influx (34, 36). Although the mechanisms of muscarinic receptor-mediated Ca2+ mobilization in these cells have been well established (34, 36, 37), little is known about the role of PMCA-mediated Ca2+ efflux in this pathway. The current study was undertaken to determine the functional relevance of the interactions between NHERF-2 and PMCA in the context of muscarinic receptor-mediated Ca2+ signaling in HT-29 cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and siRNA—The human colorectal adenocarcinoma cell line HT-29 (ATCC) was maintained in standard RPMI 1640 medium (Invitrogen), 10% turbo calf serum (Invitrogen) and 1% (v/v) penicillin/streptomycin (Invitrogen) at 37 °C, 5% CO2. Cells used in this study were between passages 20-40. Human embryonic kidney HEK293 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (Invitrogen), 10% turbo calf serum (Invitrogen), and 1% (v/v) penicillin/streptomycin (Invitrogen) at 37 °C, 5% CO2. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For the experiments to silence NHERF-2, HT-29 cells were transfected with plasmids encoding siRNA directed against two different regions of NHERF-2 (Shag-274 and Shag-361) or control (Shag-FF). These plasmids have been shown previously to effectively suppress the endogenous levels of NHERF-2 (38, 39). All experiments were performed 48-h post-transfection.
Antibodies—Monoclonal antibodies 5F10 and JA9 (Affinity Bioreagents), have been previously characterized (40, 41). 5F10 is a pan-PMCA antibody that recognizes a highly conserved epitope near the PMCA active site common to all PMCA isoforms, while JA9 is specific for PMCA4. Generation of the specific antibodies against NHERF-1 and -2 was previously described (27). The monoclonal GFP antibody was from Molecular Probes (Invitrogen).
RNA Isolation and PCR—Total RNA was extracted from HT-29 cells using the TRIzol reagent following the manufacturer's instructions (Invitrogen). RNA (100 ng) was reverse-transcribed using the One-Step RT-PCR kit (Qiagen, Hilden, Germany) with primers based on mouse NHERF-1 and -2 (39). β-Actin was used as a housekeeping gene using commercially available primers (Promega). Samples without the reverse transcriptase were also run in parallel as a control. The PCR was run for 20 cycles at 94, 55, and 72 °C, 1 min each. RT-PCR products were electrophoresed on a 2% agarose gel and visualized with ethidium bromide.
Cell Surface Biotinylation and Western Blotting—Flasks (75 cm2) of confluent HT-29 cells were washed three times in PBS at room temperature. Cells were according to experimental conditions first preincubated with 50 μm BAPTA-AM (1,2-bis(o-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid-acetoxymethylester, Invitrogen) for 30 min, then exposed to either acetylcholine (ACh, 10 μm) or cyclopiazonic acid (CPA, 5 μm). At the given time points, the flasks were immediately placed on ice and washed three times in ice-cold PBS. Cells were then exposed to Sulfo-NHS-biotin (Pierce Biotechnology Inc.) (1.22 μg/ml) for 45 min at 4 °C. This was followed by a wash of 100 μm glycine in PBS at 4 °C for 5 min to quench excess biotin, followed by three more washes of cold PBS. Cells were solubilized in 150 mm NaCl, 20 mm HEPES, 1 mm EDTA, 1% Nonidet P-40 (Roche Applied Science), complete protease inhibitor (Roche Applied Science) for 30 min at 4 °C. Insoluble cell debris was removed by centrifugation. Protein quantification was done on the supernatants then equal amounts of lysate were mixed with immunopure streptavidin (Pierce Biotechnology Inc.) for 30 min with end-to-end rotation at 4 °C. Protein was eluted off the beads using 6× SDS loading dye and run on 10% SDS-PAGE gel. Gels were transferred to nitrocellulose membrane and stained with Ponceau-S to visually confirm equal loading of membrane fraction. Blots were then blocked for 1 h in 5% skim milk powder in Tris-NaCl-Tween buffer (TBS-T). Primary antibody was incubated overnight at 4 °C (5F10 1:2000, JA9 1:1000, and NHERF-1-2, 1:5000) in TBS-T. Blots were washed four times with TBS-T, incubated for 1 h with horseradish peroxidase-conjugated secondary antibody (Pierce Biotechnology Inc.) and washed again. Immunoreactive proteins were detected by enhanced chemiluminescence using the SuperSignal kit (Pierce Biotechnology Inc).
Expression and Purification of Fusion Proteins—Full-length NHERF-1, NHERF-2, and individual domains of NHERF-2 (PDZ1, PDZ2, and COOH) cloned into pGEX4T-1 have been described previously (42). Glutathione S-transferase (GST) fusion proteins were expressed in the Escherichia coli strain BL21 (Stratagene). Optimal expression of fusion proteins was achieved by adding a final concentration of 2 mm isopropyl-ß-d-thiogalactopyranoside to the bacterial culture, with overnight incubation at 22 °C. The bacteria were harvested by centrifugation and resuspended in PBS. After sonication, GST fusion proteins were purified using glutathione-Sepharose 4B beads (Amersham Biosciences) following the manufacturer's instructions.
GST Pull-downs—HT-29 cells were solubilized in lysis buffer (150 mm NaCl, 50 mm Tris-Cl, 0.5% Nonidet P-40, protease inhibitors) for 1 h at 4 °C, lysates were centrifuged at 4 °C, and the supernatant collected. Lysates for the pull-down experiment investigating the Ca2+ dependence of PMCA-NHERF-2 interactions had an additional 100 μm BAPTA free acid. GST fusion proteins (10 μg) were bound to Sepharose in GST binding buffer (lysis buffer, + 5 mm dithiothreitol) for 3 h at 4 °C; the beads were washed three times in wash buffer (GST binding buffer, + 0.0075% SDS) and once in lysis buffer. The cell lysate (5-10 mg) was then incubated with GST fusion protein beads in lysis buffer at 4 °C overnight with gentle rotation. Bound proteins were washed three times with lysis buffer (no protease inhibitor) and resuspended in SDS sample buffer. The samples were resolved by SDS-polyacrylamide gel electrophoresis, Western-blotted and probed with the appropriate antibodies.
Co-immunoprecipitation—HT-29 cell lysates were prepared following activation with 10 μm ACh 60 s. The supernatant was collected and incubated with 50 μl of protein A-agarose (Roche Applied Science) for 3 h at 4 °C. Cleared cell lysate (3 mg) was incubated with NHERF-1 and NHERF-2 antibodies overnight at 4 °C. Protein complexes were recovered using protein A-agarose beads for 4 h, and subsequent complexes recovered in SDS loading buffer. Samples were run on an 10% SDS-PAGE gel and Western blots performed using appropriate antibodies.
In Vitro Binding Assay—The 130-amino acid C-terminal tail of PMCA1b and the tail minus the PDZ binding motif were cloned into pGFP-C1N (Clontech), using the BamH1/Sal1 site and were PCR-amplified using the following primers: PMCA1b fwd-gcgtcgacgagattgatcacgctgaaacc, PMCA1b: rev-cgcggatccttcagagtgatgtttcc, PMCA1b-PDZΔ: rev-ctggatccctcaaactatgtagtggg. PCR amplicons were ligated into pGFP-C1N to make the constructs GFP-PMCA1b-CT and GFP-PMCA1b-PDZΔ. Constructs were transfected into HEK cells and left for 48 h to allow optimal transgene expression. Cells were solubilized in GST lysis buffer (as described above). Using the same GST pull-down protocol as previously described, GST-NHERF-2 bound to Sepharose beads were incubated with the cell lysates produced. Complexes were recovered, run on a Western blot, and probed with the anti-GFP antibody (Invitrogen).
Measurement of Cytosolic [Ca2+]—[Ca2+]i measurements were performed using the fluorescent ratiometric Ca2+ dye Fura-2 (Fura-2 AM, Invitrogen). The efflux of free [Ca2+]i was determined using a fluorescence microplate reader to monitor the rate of decline in [Ca2+]i after the generation of ACh-evoked Ca2+ transients. Cells were seeded in tissue culture-treated black 96-well plates (PerkinElmer Life and Analytical Sciences, VIC, Australia). Before experimentation, cells were washed twice in HEPES buffered salt solution (HBSS, 137 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm d-glucose, 10 mm HEPES; pH adjusted to 7.4 with NaOH), and loaded with 5 μm Fura-2 in the same buffer for 1 h at 37 °C with gentle shaking. After loading, cells were washed three times in HBSS to remove excess dye. All experiments were performed in a BMG Fluostar Optima (BMG Lab Technologies, Offenburg, Germany) using 340 nm, 380 nm excitation, and 520 nm emission filters (BMG Lab Technologies). An automated injection of agonist was made at 20 s to give final well concentrations of ACh(10 μm) and CPA (5 μm). The ratio of the emission at 520 nm relative following 348/380 nm excitation represent the relative changes in [Ca2+]i.
Relative PMCA-mediated Ca2+ efflux was assessed based on previously described methods (43). For Ca2+ efflux measurements the HBSS buffer was removed, and cells were washed once with loading buffer and then twice with Na+/Ca2+-free HBSS in which Na+ was replaced by N-methyl-d-glucamine and which contained 100 μm BAPTA-tetra-K+ salt (cell impermeant BAPTA, Sigma Aldrich). The solution in which the agonist was injected also contained 5 μm CPA to prevent Ca2+ reuptake by the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA). This Na+/Ca2+-free HBSS buffer also prevents any possible contribution of Na+/Ca2+ exchange. The rate of decline in [Ca2+]i measured under these conditions was predominately a measure of PMCA-mediated efflux of cytosolic Ca2+. In some experiments in which NHERF-2 was silenced, Ca2+ store release was determined without the inclusion of CPA in the agonist solution. The relative rate of recovery from the peak Ca2+ transient was measured by fitting the declining phase of the [Ca2+]i transient with a monoexponential curve over 30 s, commencing 20 s after the peak.
Immunohistochemistry and Confocal Microscopy—HT-29 cells were grown on 12-mm glass coverslips. Cells were incubated in HBSS ± 10 μm ACh for 60 s, then fixed in 4% paraformaldehyde for 15 min. Cells were washed four times in PBS and blocked for 1 h at room temperature in blocking buffer (3% normal horse serum, 0.5% bovine serum albumin, and 0.05% Triton X-100). Cells were then washed two more times with PBS, then primary antibodies (pan-PMCA 1:1000 and NHERF-2 1:500) were applied in blocking buffer for 1 h at room temperature. Cells were washed four times in PBS, and secondary antibody was applied (Alexa-Fluor 488 and 546, Invitrogen, 1:500) for 30 min. Cells were washed again four times in PBS and mounted onto glass slides using Prolong-Gold mounting medium (Invitrogen) according to the manufacturer's instructions. Cells were visualized using a Zeiss LSM 510 meta confocal microscope using a 488-nm argon and 543-nm HeNe laser (Carl Zeiss, Oberkochen, Germany). Images were taken using Plan-Apochromat 63× and 100×, 1.4 oil immersion objectives. All images were captured and analyzed using Zeiss LSM image viewer.
Statistical Analysis—Statistical significance was assessed using ANOVA with a Tukey-Neuman post-test. Data were considered statistically significant when p < 0.05. Unless otherwise specified, results are presented as mean ± S.E. Data from microplate assays are representative of 3-4 independent assays where each group was measured from 6-8 wells.
RESULTS
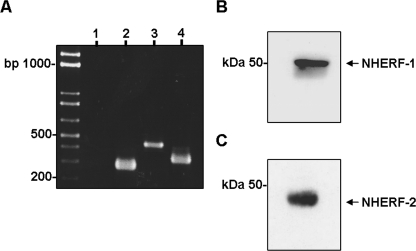
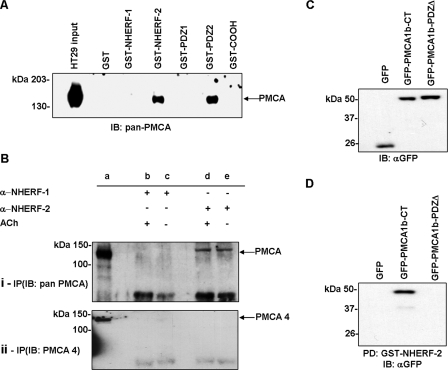
NHERF/PMCA Interactions in HT-29 Cells—We have previously reported that HT-29 cells express only PMCA1 and PMCA4 isoforms (44). RT-PCR confirmed that the HT-29 cells used in this study expressed the mRNA for NHERF-1 and NHERF-2 (Fig. 1A). Western blotting also confirmed that HT-29 cells expressed readily detectable levels of both NHERF-1 and NHERF-2 protein (Fig. 1, B and C). It has been shown recently, using heterologous expression systems that the CNS isoform PMCA2b interacts with NHERF-2 (18). Importantly, PMCA1b has a Class 1 PDZ binding motif identical to that of PMCA2b (6). HT-29 cell lysates were incubated with GST fusion proteins of either full-length NHERF-1 or NHERF-2, the bound proteins were eluted, and then run on Western blots and probed with the 5F10 antibody. We found that NHERF-2 but not NHERF-1 interacted with PMCA (Fig. 2A). In parallel we performed these experiments using three GST fusion proteins that encoded the individual binding domains of NHERF-2; PDZ1, PDZ2, and the C-terminal ezrin binding domain. These data revealed that PMCA in HT-29 cells bound the PDZ2 module of NHERF-2 (Fig. 2A). GST pull-downs rely on an added excess of fusion proteins and can therefore unmask low affinity interactions. To confirm that the interaction between PMCA and NHERF-2 occurred in the presence of endogenous levels of each protein we also performed co-immunoprecipitation experiments. These experiments were also performed in the presence of ACh to determine whether the PMCA/NHERF-2 interaction was altered during muscarinic receptor activation. Cell lysates were prepared from cells treated ± ACh for 60 s, the lysates then incubated with anti-NHERF-1 or NHERF-2 antibodies, and the resultant complexes resolved by Western blotting. Under these conditions, NHERF-2, but not NHERF-1, interacted with PMCA (Fig. 2B, panel i) and interestingly the level of interaction was not altered in the presence of ACh.
FIGURE 1.
HT-29 cells express NHERF-1 and NHERF-2. A, RT-PCR showing mRNA in HT-29 cells for both NHERF-1 (lane 3, 461 bp) and NHERF-2 (lane 4, 364 bp), no reverse-transcriptase control (lane 1) and β-actin control (lane 2). B, Western blot for NHERF-1 in HT-29 cells showing a single band at ∼50 kDa. C, Western blot for NHERF-2 in HT-29 cells showing a band at ∼45 kDa. Western blots are representative of at least three individual experiments.
FIGURE 2.
PMCA1 binding to NHERF-2 in HT-29 cells is mediated by the C-terminal PDZ motif. A, GST pull-downs using pan-PMCA antibody showing that PMCA interacted with NHERF-2 but not NHERF-1 in HT-29 cells, and the interaction was mediated by the second PDZ module (PDZ2) of NHERF-2. B, co-immunoprecipitations investigating the interaction between NHERF1/2 and PMCA. Lane a, HT-29 cell lysate control; lanes b and c, immunoprecipitation with anti-NHERF-1 antibody; lanes d and e, immunoprecipitation with anti-NHERF-2 antibody. B, panel i, blot probed with pan-PMCA antibody. B, panel ii, blot probed with anti-PMCA4 antibody. NHERF-2 interacted with PMCA, and this interaction occurred in the presence and absence of ACh. C, Western blot showing HEK293 cells transfected with GFP-tagged PMCA1b C terminus ± PDZ binding motif expressed proteins of the predicted size (∼50 kDa). D, pull-downs using GFP constructs of the C terminus of PMCA1b ± PDZ binding motif together with GST-NHERF-2. The interaction with NHERF-2 was abolished when the C-terminal ETSL motif was deleted (GFP-PMCA1b-DPDZ). Western blots are representative of at least three individual experiments.
The above experiments used a pan-PMCA antibody as PMCA1-specific antibodies were not found to be suitable for these experiments. However, HT-29 cells express only PMCA1 and PMCA4; therefore, we repeated the co-immunoprecipitations using a specific PMCA4 antibody. In this case we were unable to demonstrate any interaction between NHERF-2 and PMCA4 (Fig. 2B, panel ii). Thus we can conclude that in HT-29 cells, PMCA1b is the only PMCA isoform interacting with NHERF-2.
PDZ interactions are mediated typically by a 4-amino acid C-terminal PDZ binding motif on the target protein. To confirm that the C-terminal PDZ motif was required for PMCA1b binding to NHERF-2, we made plasmids with GFP tagged to the C-terminal 130 amino acids of PMCA1b with or without the last 4 C-terminal amino acids (ETSL); GFP-PMCA1b-CT and GFP-PMCA1b-PDZΔ. These plasmids were transfected into HEK293 cells, and the lysates run on Western blots with the GFP antibody to confirm the proteins expressed correctly (Fig. 2C). We then used GST-NHERF-2 to pull-down the GFP fusion proteins from HEK293 cell lysates. As predicted, GFP-PMCA1b-CT bound to NHERF-2 and this interaction was abolished when the PDZ binding motif was deleted (GFP-PMCA1b-PDZΔ) (Fig. 2D).
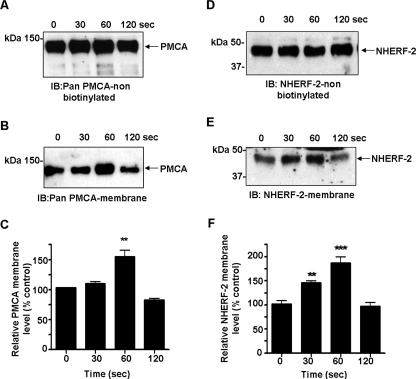
Agonist-induced Trafficking of PMCA and NHERF-2 to the Plasma Membrane—For many membrane proteins only a small fraction of the total cellular pool of the protein is expressed at the plasma membrane, with the rest stored in intracellular recycling pools from which they can be rapidly recruited to or from the membrane (45, 46). For example, it has been shown in the ileum that NHE3 exists in multiple dynamic pools and activation with ACh causes a rapid translocation from the membrane to early endosomes (47). Significant levels of PMCA are also found in the non-biotinylated fraction (see Fig. 3A), so it can be predicted that similar mechanisms exist for the trafficking of PMCA to the membrane from these pools. We therefore hypothesized that (i) muscarinic mobilization of Ca2+ would result in the translocation of PMCA to the plasma membrane and (ii) that the recruitment of PMCA to the membrane requires a macromolecular complex that may involve NHERF-2 as a molecular scaffold. Cell surface biotinylation was used to determine changes in cell surface-associated PMCA and NHERF-2 during a time course of muscarinic receptor activation. When cells were exposed to ACh, the cell surface level of PMCA increased by 55 ± 11% above control levels (n = 4; **, p < 0.01) within 60 s (Fig. 3, A-C). Interestingly, the levels of PMCA returned to control levels within 120 s despite the continued presence of the agonist. Importantly, we found that the cell surface associated levels of NHERF-2 also increased in response to ACh. The translocation of NHERF-2 to the membrane preceded that of PMCA. NHERF-2 levels increased significantly by 45 ± 6% above control levels (n = 4; **, p < 0.01) within 30 s of exposure to ACh and reached a peak at 80 ± 11% above control after 60 s (n = 4; ***, p < 0.001) (Fig. 3, D-F). Similar to PMCA, the surface levels of NHERF-2 also returned to resting levels by 120 s while ACh was still present. The blots were stripped and re-probed for PMCA4. No significant changes were observed in the levels of cell surface PMCA4 (data not shown), indicating that PMCA1 was the isoform that translocated and interacted with NHERF-2.
FIGURE 3.
Changes in plasma membrane-associated PMCA and NHERF-2 following muscarinic activation. HT-29 cells were exposed to ACh (10 μm), and cell surface biotinylation was used to determine surface levels of PMCA and NHERF-2. A, representative Western blot of non-biotinylated PMCA fraction. B, representative Western blot showing the time-dependent increase of PMCA at the plasma membrane after muscarinic activation. C, ACh caused a transient increase by 55 ± 11% (n = 4; **, p < 0.01) in plasma membrane PMCA at 60 s. D, representative Western blot of non-biotinylated NHERF-2 fraction. E, representative Western blot showing the time-dependent increase of NHERF-2 at the plasma membrane after muscarinic activation. F, ACh caused a transient increase by 45 ± 6% (n = 4; **, p < 0.01) of plasma membrane-associated NHERF-2 within 30 s and reached a peak to 85 ± 13% above control levels (n = 4; ***, p < 0.001) after 60 s. Data are expressed as mean ± S.E.
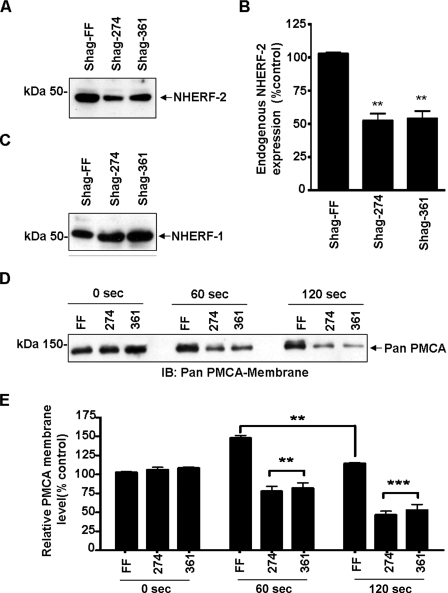
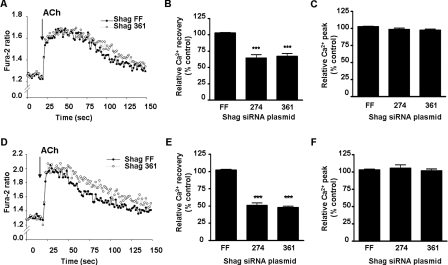
Trafficking of PMCA in Response to Muscarinic Stimulation Is Dependent on NHERF-2—The above data were consistent with NHERF-2 playing a key role in scaffolding/nucleating the assembly of a PMCA Ca2+ efflux complex at the membrane. We hypothesized that silencing NHERF-2 would therefore prevent the translocation of PMCA to the membrane. HT-29 cells were transfected with silencing constructs (Shag-274 and Shag-361) directed against two different regions of NHERF-2 or empty vector control (Shag-FF) (39). Both silencing constructs caused a significant decrease in endogenous NHERF-2 protein to 57 ± 5% and 54 ± 6% for Shag-274 and 361, respectively (n = 4; **, p < 0.01) compared with the Shag-FF control (Fig. 4, A and B). Silencing of NHERF-2 was specific as the levels of NHERF-1 were unaltered (n = 4) (Fig. 4C). We then investigated the dynamics of ACh-mediated PMCA translocation in HT-29 cells that NHERF-2 had been silenced. When endogenous NHERF-2 was knocked down, the translocation of PMCA to the membrane at the 60-s time point was effectively abolished (Fig. 4, D and E). Furthermore at the later time points, the levels of PMCA dropped significantly below control (unstimulated) levels to 46 ± 5% (Shag-274) and 53 ± 7% (Shag-361) at 120 s (n = 3; ***, p < 0.001) (Fig. 4, D and E). These data indicated that NHERF-2 was responsive to muscarinic receptor activation and that the subsequent translocation of PMCA to the membrane required NHERF-2. Importantly, the reduction of PMCA below control levels suggested that a different mechanism, that did not involve NHERF-2, was responsible for the retrieval of membrane-associated PMCA.
FIGURE 4.
Silencing of NHERF-2 abolishes muscarinic-mediated translocation of PMCA. NHERF-2 was silenced in HT-29 cells using siRNA plasmids Shag-FF (vector control) and Shag-274 or Shag-361, exposed to ACh (10 μm), and cell surface biotinylation was used to determine surface levels of PMCA and NHERF-2. A, representative Western blot demonstrating silencing of NHERF-2. B, Shag-274 and Shag-361 reduced endogenous NHERF-2 protein levels to 57 ± 5% and 54 ± 6% (n = 3; **, p < 0.01). C, representative Western blot showing endogenous NHERF-1 was not altered. D, representative Western blot showing changes in membrane-associated PMCA following muscarinic activation in HT-29 cells silenced for NHERF-2. E, densitometry showing changes in membrane-associated PMCA during muscarinic stimulation in NHERF-2 knock-down cells. The ACh-mediated insertion of PMCA at 60 s was abolished. Furthermore, during stimulation the levels of PMCA in silenced cells reduced at 120 s to 46 ± 5% (Shag-274) and 53 ± 7% (Shag-361) compared with control (n = 3; ***, p < 0.001). Data are expressed as mean ± S.E.
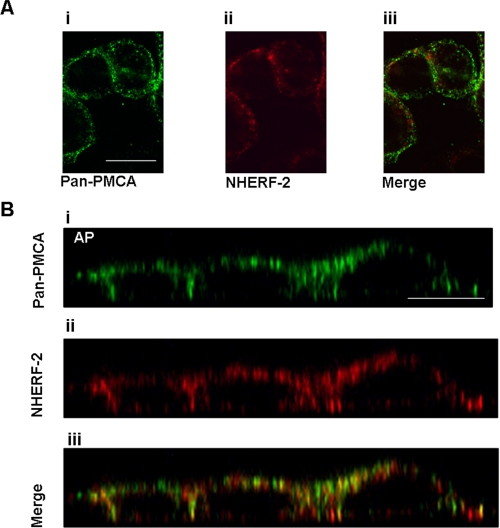
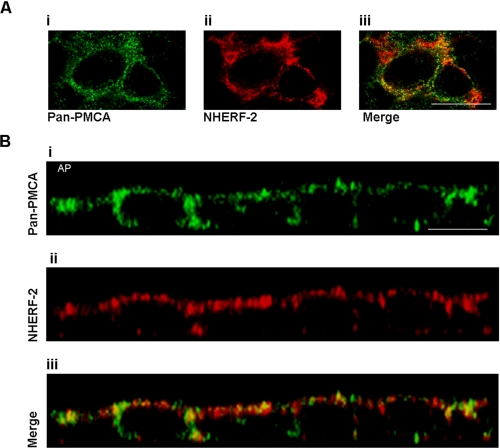
Co-localization of PMCA and NHERF-2—If NHERF-2 and PMCA functionally interact, they must also exhibit spatio-temporal colocalization. We used confocal immunofluorescence microscopy to monitor changes in the subcellular distribution of PMCA and NHERF-2. In control HT-29 cells (unstimulated), both PMCA and NHERF-2 were widely distributed throughout the cytosol, with some colocalization observed (Fig. 5A, panels i-iii). Z-scans revealed that the two proteins were distributed largely at the apical and lateral domains. (Fig. 5B, panels i-iii). Merging of both channels revealed that there was significant co-localization of the proteins even in unstimulated cells, a finding that is consistent with the co-immunoprecipitation data. When the cells were exposed to ACh for 60 s, there was a pronounced increase in co-localization (Fig. 6A, panels i-iii). This was more apparent in the z-scan, where a significant redistribution of both PMCA and NHERF-2 into large distinct large puncta was observed, with a higher level of co-localization of the two proteins (Fig. 6B, panels i-iii).
FIGURE 5.
PMCA and NHERF-2 in HT-29 cells. A, X-Y scans of pan-PMCA (panel i) and NHERF-2 (panel ii) revealing punctuate distribution and some degree of co-localization (panel iii). B, Z-scan of HT-29 cells showed an apical-lateral distribution of both PMCA (panel i) and NHERF-2 (panel ii) with a significant degree of colocalization (panel iii). Scale bar represents 10 μm. These images are representative of four separate experiments.
FIGURE 6.
PMCA and NHERF-2 distribution in HT-29 cells exposed to ACh. A, X-Y scans of pan-PMCA exposed to ACh for 60 s. This resulted in a more pronounced punctuate distribution of PMCA (panel i) and NHERF-2 (panel ii) and a higher degree of colocalization (panel iii). B, Z-scan of the same cells more clearly demonstrates the association of PMCA (panel i) and NHERF-2 (panel ii) into larger punctuate structures (panel iii). Scale bar represents 10 μm. These images are representative of four separate experiments.
Silencing NHERF-2 Attenuates PMCA-mediated Ca2+ Efflux during Muscarinic Signaling—Our data suggest a highly dynamic model for PMCA translocation during acute receptor activation. The next question was whether the NHERF-2-dependent trafficking of PMCA was physiologically relevant. We therefore measured the parameters of Ca2+ signaling using Fura-2 in a microplate-based assay to determine the effect of silencing NHERF-2 on muscarinic-induced Ca2+ handling. HT-29 cells were transfected with Shag-274, Shag-361, or Shag-FF, then loaded with Fura-2, and assays performed in a microplate reader. We first isolated the Ca2+ recovery component predominately due to PMCA (43). Here we observed a significant reduction in the rate of PMCA-mediated Ca2+ recovery of 33 ± 3% (Shag-274) and 36 ± 4% (Shag-361) compared with the control Shag-FF (n = 4;***, p < 0.001) (Fig. 7, A and B). There was no effect of silencing NHERF-2 on the peak increase in [Ca2+]i (Fig. 7C). The data are consistent with reduced translocation of PMCA to the plasma membrane when NHERF-2 is silenced and suggests that the interaction between PMCA and NHERF-2 plays a role in PMCA-mediated Ca2+ efflux. Reduced rate of recovery was also observed when SERCA was not inhibited (50 ± 3% Shag-274 and 53 ± 3% Shag-361; n = 4; ***, p < 0.001) (Fig. 7, D and E). In this case, the difference in recovery rate was also not due to changes in the peak [Ca2+]i level attained (Fig. 7F). Finally, silencing of NHERF-2 had no significant effect on the rate of Ca2+ influx as measured by Mn2+ quenching of Fura-2 (data not shown).
FIGURE 7.
NHERF-2 silencing attenuates relative Ca2+ recovery in HT-29 cells following exposure to ACh. Cells were transfected with NHERF-2 silencing constructs as described and [Ca2+]i assessed using a Fura-2-based fluorometric assay. For all experiments, agonist injection was at 20 s. A, representative Fura-2 tracings showing relative PMCA-mediated [Ca2+]i recovery following an ACh-evoked Ca2+ transient (bath solutions; zero Ca2+, 5 μm CPA, extracellular BAPTA). B, relative PMCA-mediated Ca2+ efflux was significantly reduced by 32 ± 8% and 31 ± 6% (n = 4; **, p < 0.01) for Shag 274 and Shag 361, respectively. C, no significant change in peak [Ca2+]i was observed. D, representative Fura-2 tracings showing relative [Ca2+]i recovery following an ACh-evoked Ca2+ transient in the absence of CPA (bath solution; zero Ca2+, extracellular BAPTA). E, relative Ca2+ efflux was significantly reduced by 50 ± 3% and 53 ± 3% (n = 4; ***, p < 0.001) for Shag 274 and Shag 361, respectively. F, no significant change in peak [Ca2+]i was observed. All data are representative of 3-4 independent assays where each group was measured from 6-8 wells and are expressed as mean ± S.E.
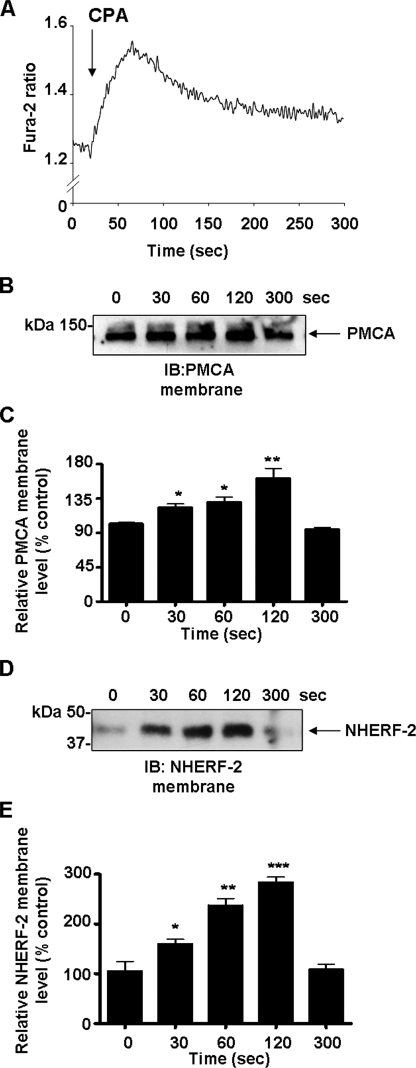
Store Release of Ca2+ Is a Trigger for PMCA and NHERF-2 Translocation—We then investigated whether Ca2+ leak from intracellular stores was sufficient to initiate translocation of PMCA and NHERF-2 in the absence of GPCR activation. Cells bathed in a Ca2+-free extracellular solution were exposed to the SERCA pump inhibitor CPA, and changes in Fura-2 ratio were monitored. This resulted in a characteristic release of Ca2+ from the intracellular stores (Fig. 8A) with the peak Ca2+ occurring at ∼50-60 s after CPA application. We then performed cell surface biotinylation experiments to determine the effects of CPA on the translocation of both PMCA and NHERF-2 to the membrane. Store release of Ca2+ with CPA resulted in a time dependent translocation of PMCA to the plasma membrane that reached a maximum at 120 s to 61 ± 12% above resting levels (n = 3; **, p < 0.01) and then returned to baseline by 300 s (Fig. 8B). Similarly, store release resulted in a time-dependent translocation of NHERF-2 reaching a maximum at 120 s at 180 ± 11% above control levels (n = 3; ***, p < 0.001) that also returned to resting levels within 300 s (Fig. 8C). The movement of both proteins paralleled the increase in [Ca2+]i, and interestingly, the movement of NHERF-2 appeared to precede that of PMCA, similar to the effect observed following muscarinic activation. Thus the translocation of both NHERF-2 and PMCA to the plasma membrane can be triggered by store release of Ca2+ independent of GPCR activation.
FIGURE 8.
Depletion of Ca2+ stores triggers PMCA and NHERF-2 trafficking to the cell membrane. HT-29 cells were exposed to CPA (5 μm) in the absence of extracellular Ca2+. A, representative Fura-2 tracing showing changes in [Ca2+]i following the addition of CPA (5 μm) to the bathing solution at 20 s. B, representative Western blot showing changes in membrane-associated PMCA during exposure to CPA. C, densitometry showing time-dependent changes in membrane-associated PMCA during exposure to CPA. Membrane levels of PMCA peaked to 61 ± 12% (n = 3; **, p < 0.01) above control levels at 120 s and returned to baseline levels by 300 s. D, representative Western blot showing changes in membrane-associated NHERF-2 during exposure to CPA. E, densitometry showing changes in membrane-associated NHERF-2 during exposure to CPA. Membrane levels of NHERF-2 peaked by 180 ± 11% (n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001) and returned to resting levels by 300 s. All data are expressed as mean ± S.E.
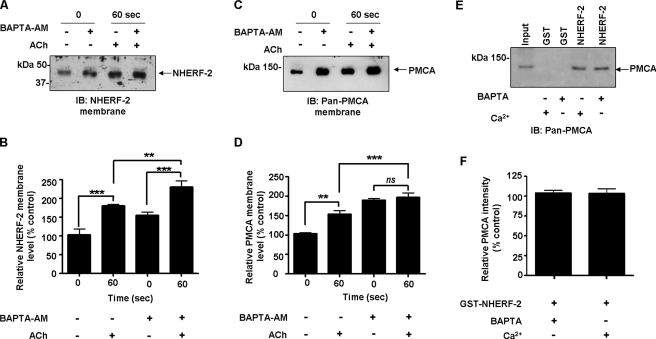
ACh-evoked Trafficking of PMCA and NHERF-2 Is Altered in the Presence of BAPTA-AM—There are other potential signals, in addition to changes in [Ca2+]i that could trigger the movement of NHERF-2 and PMCA to the membrane. To determine if an increase in [Ca2+]i was the sole stimulus for translocation during muscarinic activation, we used cell permeable BAPTA-AM to chelate intracellular Ca2+. The concentration of BAPTA-AM was optimized to ensure inhibition of the ACh-induced [Ca2+]i transient (data not shown). Cells were exposed to ACh for 60 s and cell surface biotinylations performed. In unstimulated cells, chelation of intracellular free Ca2+ resulted in a significant increase in the levels of membrane-associated NHERF-2 to 54 ± 5% above control cells (n = 3; **, p < 0.01) (Fig. 9, A and B). In cells treated with BAPTA-AM, exposure to ACh further increased membrane-associated NHERF-2 by 130 ± 16% over the unstimulated BAPTA-AM-treated cells (n = 3; ***, p < 0.001) (Fig. 9, A and B). This suggests that the muscarinic-mediated increase in membrane-associated NHERF-2 may involve a Ca2+-independent pathway. In the case of PMCA, in unstimulated cells, treatment with BAPTA-AM resulted in a pronounced increase in cell surface PMCA to 89 ± 5% over control levels (n = 3; ***, p < 0.001) (Fig. 9, C and D). However, when BAPTA-AM-treated cells were exposed to ACh, there was no increase in cell surface-associated PMCA (Fig. 9, C and D). This then raised the question as to the Ca2+ dependence of the binding between PMCA to NHERF-2. Pull-downs were performed in HT-29 cell lysates in the presence of 100 μm extracellular BAPTA. The interaction between GST-NHERF2 and PMCA was unaltered in the absence of Ca2+ (Fig. 9, E and F). Taken together, these data suggest that although the association of NHERF-2 and PMCA is Ca2+-independent, Ca2+ is required for the nucleation of the efflux complex at the membrane in response to muscarinic stimulation.
FIGURE 9.
Chelation of intracellular Ca2+ alters ACh-evoked trafficking of PMCA and NHERF-2, but does not alter the association of the proteins in vitro. HT-29 cells were preincubated with BAPTA-AM (50 μm), exposed to ACh (10 μm), and membrane-associated NHERF-2 and PMCA detected. Whole cell lysates were incubated with BAPTA and GST pull-down experiments performed with NHERF-2 to evaluate if the association between NHERF-2 and PMCA was Ca2+-dependent. A, representative Western blot of membrane-associated NHERF-2 ± BAPTA-AM and ACh. B, densitometry of membrane-associated NHERF-2. Under control conditions, NHERF-2 increased by 80 ± 6% over control after 60 s of exposure to ACh (n = 3; ***, p < 0.001). When Ca2+ was chelated with BAPTA-AM, there was a significant increase in NHERF-2 to 54 ± 5% over control unstimulated cells (n = 3; **, p < 0.01). Addition of ACh resulted in a further significant increase in membrane-associated NHERF-2 after 60 s to 130 ± 16% (n = 3; **, p < 0.001). C, representative Western blot of cell surface-associated PMCA ± BAPTA-AM and ACh. D, densitometry of membrane-associated PMCA. Under control conditions, PMCA increased by 53 ± 5% over control after 60 s of exposure to ACh (n = 3; ***, p < 0.001). When Ca2+ was chelated with BAPTA-AM, there was a significant increase in membrane-associated PMCA to 89 ± 5% above control (n = 3; ***, p < 0.001). Addition of ACh failed to increase the levels of membrane-associated PMCA in BAPTA-AM-treated cells. E, representative Western blot of GST-NHERF-2 pull-downs in HT-29 cells in the absence of Ca2+. F, densitometry of relative PMCA intensity for GST pull-down in vitro binding assay with NHERF-2. The association between NHERF-2 and PMCA was not altered when Ca2+ was chelated. All data are expressed as mean ± S.E.
DISCUSSION
PMCA-mediated Ca2+ efflux plays a key role in regulating basal [Ca2+]i levels and the rates of recovery of [Ca2+]i, thereby shaping the Ca2+ responses following activation of GPCRs. One mechanism to regulate PMCA is through accessory/scaffold proteins, and there has been recent interest in splice variants that involve C-terminal PDZ interactions. Here we describe the molecular and physiological basis of the interaction between endogenous PMCA1b and NHERF-2 in HT-29 cells.
Our finding that PMCA1b binds to NHERF-2 builds on the previous study by DeMarco et al. (18) who showed in overexpression studies that PMCA2b but not PMCA4b bound to NHERF-2. We also showed that, like PMCA2b, NHERF-1 did not bind PMCA1b and also demonstrate that the in vitro interaction between PMCA and NHERF-2 does not require of Ca2+ and that these two proteins have a constitutive association. The previous study on NHERF-2 relied on exogenously expressed PMCA2b and NHERF-2. The significance of the current study is that we used HT-29 cells that endogenously express NHERF-2 and PMCA1b to define the physiological relevance of PMCA-NHERF-2 interactions.
NHERF-2 interacts with proteins involved in Ca2+ signaling via its PDZ2 module, such as the P2Y1 receptor (48). NHERF-2 also associates with metabotropic glutamine receptor 5 (mGluR5) to prolong intracellular Ca2+ mobilization by mGluR5 and thereby promoting mGluR5-mediated Ca2+ toxicity (49). As a scaffold, NHERF-2 has been shown to stabilize or tether membrane proteins in functionally distinct complexes particularly in polarized epithelial cells. This is very apparent in the apical surfaces of gastric epithelia, where NHERF-2 forms the functional basis for the coupling between lysophosphatidic acid signaling and CFTR-mediated chloride secretion (50). This tethering also reduces the lateral mobility in the cell membrane of these proteins by anchoring the complex to the underlying actin cytoskeleton via accessory proteins such as ezrin, which is highly expressed in gastric epithelia (51). It has already been shown that NHERF-2 is important for the trafficking of membrane transporters such as NHE3 (47) and ion channels such as ClC-5 and TRPV5 (39, 52). We have recently also shown that NHERF-1 and NHERF-2 differentially regulate albumin uptake in renal proximal tubule cells by mechanisms that involve altering the cell-surface levels of ClC-5 in different domains of the apical membrane (39).
These data shed new light as to how PMCA responds to rapid increases in [Ca2+]i. The interaction between PMCA and NHERF-2 does not require Ca2+. The muscarinic-mediated translocation of NHERF-2 to the membrane does not require Ca2+ but does depend on store-depletion. As the Ca2+ levels increase, PMCA moves to the membrane to form an efflux complex with NHERF-2. This translocation of PMCA has an absolute requirement for Ca2+ and the presence of NHERF-2. In contrast, treatment of cells with BAPTA-AM caused a significant increase in both NHERF-2 and PMCA at the membrane. The mechanisms underlying this effect are unclear. Elevated cell surface levels of a protein can be achieved by increased rates of insertion or reduced rates of removal of the protein. In the case of NHERF-2, chelation of Ca2+ simply increased the levels of NHERF-2 at the membrane but did not prevent the ACh-induced increase. Thus although there was a shift in the baseline levels, there was no effect on the muscarinic-evoked translocation. However, in the absence of Ca2+, PMCA levels increased to levels above that observed in control cells treated with ACh. Furthermore, there was no change in the cell surface levels of PMCA in response to ACh when the Ca2+ transient was abolished. This may relate to the internalization pathway that was revealed in cells silenced for NHERF-2. Under these conditions the ACh-induced increase in membrane PMCA was abolished; however, the levels of PMCA were also reduced below baseline levels. One possibility is that the membrane-associated PMCA is subject to constitutive internalization by a process dependent on [Ca2+]i. Chelation of Ca2+ may abolish the internalization step resulting in an accumulation of PMCA at the membrane and preventing the normal trafficking. PMCA retrieval may be particularly sensitive to [Ca2+]i chelation because the C-terminal tail of PMCA is less exposed in unstimulated pumps (autoinhibited state) (53). An increase in [Ca2+]i may be required to expose the C-terminal tail, and in its absence, recycling of PMCA might be turned off and trafficking disrupted. Thus PMCA appears to exist in pools that can be rapidly mobilized in response to increases in Ca2+ and that multiple and independent pathways exist for its insertion and removal.
PMCA1b binds to PDZ2 yet the role of PDZ1 remains to be determined in this context. One possibility is that NHERF-2 itself could dimerize (via the unoccupied PDZ1) (54) thereby increasing the density of PMCA at the membrane. Another possibility is that at rest, PMCA is concentrated in subapical pools by homodimers of NHERF-2 but upon stimulation, PMCA associates with NHERF-2 in a complex where PDZ1 is occupied by another protein required for Ca2+ efflux. Indeed, shuffling of transporters between NHERF-1 and NHERF-2 has been implicated for NHE3 in the renal proximal tubule (55). NHERF-2 is also known to interact with proteins involved in Gαq-mediated signaling, including Gαq itself, PKCα, and PLCβ. Thus, NHERF-2 may control PMCA activity during muscarinic signaling by acting as a scaffold to link these key signaling proteins in the vicinity of the muscarinic receptor. NHERF-2 is known to up-regulate the PLC-β3 activity coupled to muscarinic receptor activation (56). In addition, the PLC substrate phosphatidylinositol 4,5-bisphosphate (PIP2) is a known activator of PMCA (6). Thus NHERF-2 may act to ensure the close proximity of PMCA and PLC-β3 and the sharing of a common local pool of PIP2. In this context, it has been recently demonstrated that the regulation of TRPC4 by PIP2 requires the presence of the PDZ motif on TRPC4. The authors proposed a novel mechanism in which NHERF-cytoskeletal interactions are necessary to stabilize the PIP2 binding site close to the inner surface of the plasma membrane (57). Importantly, this mechanism may also underlie the PIP2 regulation of the TRPC4-like muscarinic activated current in ileal myocytes (58). The fact that NHERF-2 has been shown to associate with TRPC4 in the rat vasa-recta provides further support for such a role for NHERF-2 (59). Our data provide further support for the agonist-induced assembly of supramolecular complexes that contain the components required for spatio-temporal fine-tuning of the Ca2+ signal.
The overall shape of the Ca2+ signal is critical, and up-regulation of Ca2+ efflux during the rising phase of the Ca2+ transient would reduce the efficiency of Ca2+ signaling (60, 61). Thus the transient nature of PMCA trafficking in the case of muscarinic activation, suggests that PMCA1b is mobilized to fine tune [Ca2+]i only at the time when the peak phase of the Ca2+ signal is reached. The recruitment of PMCA to the membrane is temporally constrained by NHERF-2 to correspond to the Ca2+ peak. This ensures that Ca2+ efflux is only augmented after the peak has been achieved and that Ca2+ levels do not reach excessive levels during the peak. This in turn influences the duration and shape of the overall Ca2+ responses. The absence of sustained insertion of PMCA at the plasma membrane also ensures that excess levels of PMCA at the membrane during the plateau phase of the response do not prematurely terminate the sustained Ca2+ signal and subsequent downstream responses. Such downstream responses could include the level of NFATc1-mediated transcription which is regulated by the amount of net Ca2+ entry across the plasma membrane (62). NHERF-2 may also recruit modulator proteins such as PKA, PKC PLCβ3, and tyrosine phosphatases (63) to the complex with PMCA to further fine tune Ca2+ levels within the membrane microdomains. Indeed PMCA1 has been reported to be the isoform most sensitive to activation by PKA (64, 65). Such mechanisms could help to encode the diversity of cellular responses mediated by increases in [Ca2+]i.
In summary, this study presents a new insight into the complex roles of PMCA in the fine-tuning of intracellular Ca2+ transients during agonist-induced Ca2+ signaling. We also reveal the highly dynamic turnover of PMCA at the plasma membrane and its association with specific protein complexes responsible for Ca2+ efflux in response to specific agonists. Future studies will identify the other proteins that may be involved in such Ca2+ efflux complexes and the role of scaffold proteins in conferring specificity of G-protein-mediated Ca2+ signaling. Our demonstration of rapid agonist-induced translocation of PMCA in a native cell type reveals a new mechanism for regulating Ca2+ efflux. The recent identification of NHERFs in the brain and other excitable cells (66), and the fact that the brain specific PMCA 2b also binds NHERF-2 opens up the possibility that such mechanisms of regulation may be more widely operative.
This work was supported, in whole or in part, by Grant DK061418 from the National Institutes of Health (to C. C. Y.). This work was also supported by grants from the Australian Research Council and National Health and Medical Research Council of Australia (to P. P.) and a University of Queensland Development Grant (to P. P. and G. R. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PMCA, plasma membrane Ca2+-ATPase; GST, glutathione S-transferase; GPCR, G-protein-coupled receptor; PKC, protein kinase C; PKA, cAMP-dependent protein kinase; PBS, phosphate-buffered saline; NHERF-2, Na+/H+ exchanger regulatory factor 2; BAPTA-AM, 1,2-bis(o-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid-acetoxymethylester; ACh, acetylcholine; CPA, cyclopiazonic acid; GFP, green fluorescent protein; PDZ, PSD-95/Dlg/Zo-1.
References
- 1.Strehler, E. E., Caride, A. J., Filoteo, A. G., Xiong, Y., Penniston, J. T., and Enyedi, A. (2007) Ann. N. Y. Acad. Sci. 1099 226-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carafoli, E. (2003) Nat. Rev. Mol. Cell Biol. 4 326-332 [DOI] [PubMed] [Google Scholar]
- 3.Duman, J. G., Chen, L., and Hille, B. (2008) J. Gen. Physiol. 131 307-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatzmann, H. J. (1966) Experientia 22 364-365 [DOI] [PubMed] [Google Scholar]
- 5.Wanaverbecq, N., Marsh, S. J., Al-Qatari, M., and Brown, D. A. (2003) J. Physiol. 550 83-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strehler, E. E., and Zacharias, D. A. (2001) Physiol. Rev. 81 21-50 [DOI] [PubMed] [Google Scholar]
- 7.Zylinska, L., Kawecka, I., Lachowicz, L., and Szemraj, J. (2002) Cell Mol. Biol. Lett. 7 1037-1045 [PubMed] [Google Scholar]
- 8.Stauffer, T. P., Guerini, D., Celio, M. R., and Carafoli, E. (1997) Brain Res. 748 21-29 [DOI] [PubMed] [Google Scholar]
- 9.Brini, M., Coletto, L., Pierobon, N., Kraev, N., Guerini, D., and Carafoli, E. (2003) J. Biol. Chem. 278 24500-24508 [DOI] [PubMed] [Google Scholar]
- 10.Ambudkar, I. S. (2006) Trends Pharmacol. Sci. 27 25-32 [DOI] [PubMed] [Google Scholar]
- 11.Kim, J. Y., Zeng, W., Kiselyov, K., Yuan, J. P., Dehoff, M. H., Mikoshiba, K., Worley, P. F., and Muallem, S. (2006) J. Biol. Chem. 281 32540-32549 [DOI] [PubMed] [Google Scholar]
- 12.Weinman, E. J., Hall, R. A., Friedman, P. A., Liu-Chen, L. Y., and Shenolikar, S. (2006) Annu. Rev. Physiol. 68 491-505 [DOI] [PubMed] [Google Scholar]
- 13.Pawson, T., and Scott, J. D. (1997) Science 278 2075-2080 [DOI] [PubMed] [Google Scholar]
- 14.Hung, A. Y., and Sheng, M. (2002) J. Biol. Chem. 277 5699-5702 [DOI] [PubMed] [Google Scholar]
- 15.Schuh, K., Uldrijan, S., Gambaryan, S., Roethlein, N., and Neyses, L. (2003) J. Biol. Chem. 278 9778-9783 [DOI] [PubMed] [Google Scholar]
- 16.Zabe, M., and Dean, W. L. (2001) J. Biol. Chem. 276 14704-14709 [DOI] [PubMed] [Google Scholar]
- 17.DeMarco, S. J., and Strehler, E. E. (2001) J. Biol. Chem. 276 21594-21600 [DOI] [PubMed] [Google Scholar]
- 18.DeMarco, S. J., Chicka, M. C., and Strehler, E. E. (2002) J. Biol. Chem. 277 10506-10511 [DOI] [PubMed] [Google Scholar]
- 19.Goellner, G. M., DeMarco, S. J., and Strehler, E. E. (2003) Ann. N. Y. Acad. Sci. 986 461-471 [DOI] [PubMed] [Google Scholar]
- 20.Schuh, K., Uldrijan, S., Telkamp, M., Rothlein, N., and Neyses, L. (2001) J. Cell Biol. 155 201-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oceandy, D., Stanley, P. J., Cartwright, E. J., and Neyses, L. (2007) Biochem. Soc. Trans. 35 927-930 [DOI] [PubMed] [Google Scholar]
- 22.Shenolikar, S., Voltz, J. W., Cunningham, R., and Weinman, E. J. (2004) Physiology (Bethesda) 19 362-369 [DOI] [PubMed] [Google Scholar]
- 23.Brone, B., and Eggermont, J. (2005) Am. J. Physiol. Cell Physiol. 288 C20-C29 [DOI] [PubMed] [Google Scholar]
- 24.Weinman, E. J., Steplock, D., and Shenolikar, S. (1993) J. Clin. Investig. 92 1781-1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinman, E. J., Steplock, D., Wang, Y., and Shenolikar, S. (1995) J. Clin. Investig. 95 2143-2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun, C. H., Oh, S., Zizak, M., Steplock, D., Tsao, S., Tse, C. M., Weinman, E. J., and Donowitz, M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3010-3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamprecht, G., Weinman, E. J., and Yun, C. H. (1998) J. Biol. Chem. 273 29972-29978 [DOI] [PubMed] [Google Scholar]
- 28.Liedtke, C. M., Yun, C. H., Kyle, N., and Wang, D. (2002) J. Biol. Chem. 277 22925-22933 [DOI] [PubMed] [Google Scholar]
- 29.Tang, Y., Tang, J., Chen, Z., Trost, C., Flockerzi, V., Li, M., Ramesh, V., and Zhu, M. X. (2000) J. Biol. Chem. 275 37559-37564 [DOI] [PubMed] [Google Scholar]
- 30.Wade, J. B., Liu, J., Coleman, R. A., Cunningham, R., Steplock, D. A., Lee-Kwon, W., Pallone, T. L., Shenolikar, S., and Weinman, E. J. (2003) Am. J. Physiol. Cell Physiol. 285 C1494-C1503 [DOI] [PubMed] [Google Scholar]
- 31.Sun, F., Hug, M. J., Lewarchik, C. M., Yun, C. H., Bradbury, N. A., and Frizzell, R. A. (2000) J. Biol. Chem. 275 29539-29546 [DOI] [PubMed] [Google Scholar]
- 32.Naren, A. P., Cobb, B., Li, C., Roy, K., Nelson, D., Heda, G. D., Liao, J., Kirk, K. L., Sorscher, E. J., Hanrahan, J., and Clancy, J. P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 342-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zylinska, L., Guerini, D., Gromadzinska, E., and Lachowicz, L. (1998) Biochim. Biophys. Acta 1448 99-108 [DOI] [PubMed] [Google Scholar]
- 34.Poronnik, P., Cummins, M. M., O'Mullane, L. M., and Cook, D. I. (2002) Cell Biochem. Biophys. 36 221-233 [DOI] [PubMed] [Google Scholar]
- 35.Kopp, R., Lambrecht, G., Mutschler, E., Moser, U., Tacke, R., and Pfeiffer, A. (1989) Eur. J. Pharmacol. 172 397-405 [DOI] [PubMed] [Google Scholar]
- 36.Cummins, M. M., O'Mullane, L. M., Barden, J. A., Cook, D. I., and Poronnik, P. (2002) Pflugers Arch. 444 644-653 [DOI] [PubMed] [Google Scholar]
- 37.Cummins, M. M., Poronnik, P., O'Mullane, L. M., and Cook, D. I. (2000) Immunol. Cell Biol. 78 375-386 [DOI] [PubMed] [Google Scholar]
- 38.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J., and Conklin, D. S. (2002) Genes Dev. 16 948-958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hryciw, D. H., Ekberg, J., Ferguson, C., Lee, A., Wang, D., Parton, R. G., Pollock, C. A., Yun, C. C., and Poronnik, P. (2006) J. Biol. Chem. 281 16068-16077 [DOI] [PubMed] [Google Scholar]
- 40.Kessler, F., Bennardini, F., Bachs, O., Serratosa, J., James, P., Caride, A. J., Gazzotti, P., Penniston, J. T., and Carafoli, E. (1990) J. Biol. Chem. 265 16012-16019 [PubMed] [Google Scholar]
- 41.Caride, A. J., Filoteo, A. G., Enyedi, A., Verma, A. K., and Penniston, J. T. (1996) Biochem. J. 316 353-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fouassier, L., Yun, C. C., Fitz, J. G., and Doctor, R. B. (2000) J. Biol. Chem. 275 25039-25045 [DOI] [PubMed] [Google Scholar]
- 43.Lee, W. J., Robinson, J. A., Holman, N. A., McCall, M. N., Roberts-Thomson, S. J., and Monteith, G. R. (2005) J. Biol. Chem. 280 27076-27084 [DOI] [PubMed] [Google Scholar]
- 44.Aung, C. S., Kruger, W. A., Poronnik, P., Roberts-Thomson, S. J., and Monteith, G. R. (2007) Biochem. Biophys. Res. Commun. 355 932-936 [DOI] [PubMed] [Google Scholar]
- 45.Li, X., Galli, T., Leu, S., Wade, J. B., Weinman, E. J., Leung, G., Cheong, A., Louvard, D., and Donowitz, M. (2001) J. Physiol. 537 537-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murtazina, R., Kovbasnjuk, O., Donowitz, M., and Li, X. (2006) J. Biol. Chem. 281 17845-17855 [DOI] [PubMed] [Google Scholar]
- 47.Li, X., Zhang, H., Cheong, A., Leu, S., Chen, Y., Elowsky, C. G., and Donowitz, M. (2004) J. Physiol. 556 791-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fam, S. R., Paquet, M., Castleberry, A. M., Oller, H., Lee, C. J., Traynelis, S. F., Smith, Y., Yun, C. C., and Hall, R. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8042-8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paquet, M., Asay, M. J., Fam, S. R., Inuzuka, H., Castleberry, A. M., Oller, H., Smith, Y., Yun, C. C., Traynelis, S. F., and Hall, R. A. (2006) J. Biol. Chem. 281 29949-29961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, C., Dandridge, K. S., Di, A., Marrs, K. L., Harris, E. L., Roy, K., Jackson, J. S., Makarova, N. V., Fujiwara, Y., Farrar, P. L., Nelson, D. J., Tigyi, G. J., and Naren, A. P. (2005) J. Exp. Med. 202 975-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingraffea, J., Reczek, D., and Bretscher, A. (2002) Eur. J. Cell Biol. 81 61-68 [DOI] [PubMed] [Google Scholar]
- 52.Padanyi, R., Paszty, K., Penheiter, A. R., Filoteo, A. G., Penniston, J. T., and Enyedi, A. (2003) J. Biol. Chem. 278 35798-35804 [DOI] [PubMed] [Google Scholar]
- 53.Corradi, G. R., and Adamo, H. P. (2007) J. Biol. Chem. 282 35440-35448 [DOI] [PubMed] [Google Scholar]
- 54.Shenolikar, S., Minkoff, C. M., Steplock, D. A., Evangelista, C., Liu, M., and Weinman, E. J. (2001) FEBS Lett. 489 233-236 [DOI] [PubMed] [Google Scholar]
- 55.Cha, B., Kenworthy, A., Murtazina, R., and Donowitz, M. (2004) J. Cell Sci. 117 3353-3365 [DOI] [PubMed] [Google Scholar]
- 56.Suh, P. G., Hwang, J. I., Ryu, S. H., Donowitz, M., and Kim, J. H. (2001) Biochem. Biophys. Res. Commun. 288 1-7 [DOI] [PubMed] [Google Scholar]
- 57.Otsuguro, K., Tang, J., Tang, Y., Xiao, R., Freichel, M., Tsvilovskyy, V., Ito, S., Flockerzi, V., Zhu, M. X., and Zholos, A. V. (2008) J. Biol. Chem. 283 10026-10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ambudkar, I. S., and Ong, H. L. (2007) Pflugers Arch. 455 187-200 [DOI] [PubMed] [Google Scholar]
- 59.Lee-Kwon, W., Wade, J. B., Zhang, Z., Pallone, T. L., and Weinman, E. J. (2005) Am. J. Physiol. Cell Physiol. 288 C942-C949 [DOI] [PubMed] [Google Scholar]
- 60.Monteith, G. R., and Roufogalis, B. D. (1995) Cell Calcium 18 459-470 [DOI] [PubMed] [Google Scholar]
- 61.Bautista, D. M., Hoth, M., and Lewis, R. S. (2002) J. Physiol. 541 877-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Timmerman, L. A., Clipstone, N. A., Ho, S. N., Northrop, J. P., and Crabtree, G. R. (1996) Nature 383 837-840 [DOI] [PubMed] [Google Scholar]
- 63.Dean, W. L., Chen, D., Brandt, P. C., and Vanaman, T. C. (1997) J. Biol. Chem. 272 15113-15119 [DOI] [PubMed] [Google Scholar]
- 64.Guerini, D., Pan, B., and Carafoli, E. (2003) J. Biol. Chem. 278 38141-38148 [DOI] [PubMed] [Google Scholar]
- 65.Bruce, J. I., Yule, D. I., and Shuttleworth, T. J. (2002) J. Biol. Chem. 277 48172-48181 [DOI] [PubMed] [Google Scholar]
- 66.Paquet, M., Kuwajima, M., Yun, C. C., Smith, Y., and Hall, R. A. (2006) J. Comp. Neurol. 494 752-762 [DOI] [PMC free article] [PubMed] [Google Scholar]