Abstract
We demonstrated previously that the protein GEC1 (glandular epithelial cell 1) bound to the human κ opioid receptor (hKOPR) and promoted cell surface expression of the receptor by facilitating its trafficking along the secretory pathway. Here we showed that three hKOPR residues (Phe345, Pro346, and Met350) and seven GEC1 residues (Tyr49, Val51, Leu55, Thr56, Val57, Phe60, and Ile64) are indispensable for the interaction. Modeling studies revealed that the interaction was mediated via direct contacts between the kinked hydrophobic fragment in hKOPR C-tail and the curved hydrophobic surface in GEC1 around the S2 β-strand. Intramolecular Leu44-Tyr109 interaction in GEC1 was important, likely by maintaining its structural integrity. Microtubule binding mediated by the GEC1 N-terminal domain was essential for the GEC1 effect. Expression of GEC1 also increased cell surface levels of the GluR1 subunit and the prostaglandin EP3.f receptor, which have FPXXM and FPXM sequences, respectively. With its widespread distribution in the nervous system and its predominantly hydrophobic interactions, GEC1 may have chaperone-like effects for many cell surface proteins along the biosynthesis pathway.
κ opioid receptor (KOPR)2 is one of the three major types of opioid receptors mediating effects of opioid drugs and endogenous opioid peptides. Stimulation of KOPR generates many effects in vivo, for example antinociception (especially for visceral chemical pain, antipruritis, and water diuresis (1). The KOPR agonist nalfurafine (TRK-820) is used clinically in Sweden for the treatment of uremic pruritus in kidney dialysis patients (2). Because KOPR agonists produce profound sedative effects, it has been proposed that KOPR agonists may be useful in treating mania, antagonists as anti-depressants, and partial agonists for the management of mania depression (3). KOPR antagonists may also be useful for curbing cocaine craving and as anti-anxiety drugs (4, 5).
KOPR, a member of the rhodopsin subfamily of the seven-transmembrane receptor superfamily, is coupled preferentially to pertussis toxin-sensitive G proteins, namely Gi/o proteins (6). KOPR has been found to interact with several non-G protein-binding partners, such as Na+,H+-exchanger regulatory factor-1/ezrin-radixin-moesin-binding phosphoprotein-50 and the δ opioid receptor. These interactions have influence on signal transduction and trafficking of the receptor (7–9). By yeast two-hybrid (Y2H) assay using the hKOPR C-tail to screen a human brain cDNA library, we identified GEC1, also named GABAA receptor-associated protein like 1 (GABARAPL1), to be a binding partner of hKOPR (10).
GEC1 cDNA was first cloned as an early estrogen-regulated mRNA from guinea pig endometrial glandular epithelial cells by Pellerin et al. (11). Subsequently, it was cloned from other species, including human and house mouse (12). Interestingly, the amino acid sequences of GEC1 are completely conserved among all these species except orangutan, in which Arg99 substitutes for His99. Northern blot and immunoblotting analyses revealed that it has widespread tissue distribution (12–14). In particular, GEC1 was found to be abundant in the central nervous system and expressed throughout the rat brain (14, 15). This wide tissue distribution and the high sequence identity across species strongly suggest that GEC1 has important biological functions in mammalian cells.
Based on sequence similarity, GEC1 is classified as a member of microtubule-associated proteins (MAPs), which also include GABAA receptor-associated protein (GABARAP), Golgi-associated ATPase enhancer of 16 kDa (GATE16), GABARAP-like 3 (GABARAPL3), light chain 3 (LC3) of MAP 1A/1B, and the yeast autophagy protein 8 (Atg8) (12, 13). Among these homologues, GEC1 share the highest identity with GABARAPL3 (93%), followed by GABARAP (86%), GATE16 (61%), Atg8 (55%), and LC3 (∼30%).
A growing body of evidence shows that this protein family is closely related to two distinct biological functions. Studies mainly on GABARAP, GATE16, and GEC1 indicate that they promote intracellular protein trafficking by enhancing vesicle fusion (10, 16–21). In addition, they facilitate degradation of proteins and intracellular organelles via autophagy-related pathways, which is bolstered largely by research on Atg8 and LC3 (22, 23).
We previously reported that GEC1 interacted with the hKOPR C-tail and enhanced cell surface levels of hKOPR stably expressed in CHO cells. GEC1 expression enhances hKOPR expression through facilitating its anterograde trafficking along the protein biosynthesis pathway without affecting degradation of the receptor (10). This represented the first biological function reported for GEC1. Mansuy et al. (24) demonstrated that GEC1 interacted with tubulin and promoted microtubule bundling in vitro, and that green fluorescence protein-tagged GEC1 was localized in the perinuclear vesicles with a scattered pattern. Our electron microscopic studies in the rat brain showed that GEC1 was associated with ER, Golgi apparatus, endosome-like vesicles, and plasma membranes and scattered in cytoplasm in neurons (14). In addition, N-ethylmaleimide-sensitive factor, a protein critical for intracellular membrane-trafficking events, binds directly to GEC1 (10).
In this study, we employed Y2H techniques to determine the amino acid residues in both GEC1 and hKOPR C-tail involved in the interaction. Further studies were then carried out in mammalian cells to examine if elimination of the interaction affected the effect of GEC1 on hKOPR expression. In addition, we generated a molecular model of GEC1 based on the x-ray crystal structure of GABARAP and found that the residues involved in hKOPR binding formed hydrophobic patches on the exterior surface of GEC1. Moreover, we found that the cytosolic tail of AMPA receptor subunit GluR1 has the same FPXXM motif as that found in the hKOPR C-tail to be involved in GEC1 binding and that GEC1 expression up-regulated GluR1.
EXPERIMENTAL PROCEDURES
Materials
[15,16-3H]Diprenorphine (∼56 Ci/mmol) was purchased from PerkinElmer Life Sciences. Naloxone and rabbit anti-FLAG polyclonal antibody were purchased from Sigma. Cell media (Dulbecco's modified Eagle's medium/F-12, 1:1), Opti-MEM I reduced serum, fetal bovine serum (FBS), and Lipofectamine transfection reagent were acquired from Invitrogen. QIAquick gel extraction kit, QIAquick PCR purification kit, and QIAprep 8 miniprep kit were acquired from Qiagen (Valencia, CA). Materials for yeast two-hybrid assays such as bait and prey vectors (pGBKT7 and pGADT7, respectively), minimal SD agar base, dropout supplement, and Yeastmaker yeast transformation system 2 kit were purchased from Clontech. The following reagents were purchased from the indicated companies: geneticin (G418) from Cellgro Mediatech (Herndon, VA); horseradish peroxidase-conjugated goat anti-rabbit, IgG horseradish peroxidase-conjugated goat anti-mouse IgG, and Quick Ligation kit from New England Biolabs (Beverly, MA); PfuUltra High Fidelity DNA polymerase from Stratagene (La Jolla, CA); all restriction endonucleases and dNTP from Promega (Madison WI); and monoclonal antibody against HA (HA.11) from Covance (Princeton, NJ).
The cDNA construct of the wild-type rat AMPA receptor subunit GluR1 in pcDNA3.1 and rabbit anti-GluR1 antibody were generous gifts from Dr. Richard Huganir (Department of Neuroscience, The Johns Hopkins University, Baltimore). The cDNA constructs of HA-tagged human prostaglandin receptor EP3.f and EP3.I in pcDNA3.1 were provided by Dr. Barrie Ashby (Department of Pharmacology, Temple University, Philadelphia). Rabbit anti-GEC1 polyclonal antibody (PA629p) was generated previously (10). The following software products were used: Prism 3.0 program from GraphPad Software Inc. (San Diego, CA), ImageJ 1.34S from National Institutes of Health (Bethesda), and OptiQuant (PerkinElmer Life Sciences).
Cell Lines
A clonal CHO cell line stably expressing the FLAG-hKOPR was generated previously (25), and the Bmax value of FLAG-hKOPR was ∼1.9 pmol/mg protein (26). CHO cells with stable expression of FLAG-hKOPR-F346A (residue Phe346 replaced by Ala), FLAG-hKOPR-P347A, FLAG-hKOPR-M350A, rGluR1, HA-hEP3.f, and HA-hEP3.I were established similarly. For cells stably expressing hKOPR mutants, further screening using [3H]diprenorphine binding assay was performed to obtain cells with similar Bmax values as that of FLAG-hKOPR. All cells were cultured in 10-cm culture dishes in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% FBS, 0.2 mg/ml geneticin in a humidified atmosphere consisting of 5% CO2 and 95% air at 37 °C.
Transient Expression of GEC1
Lipofectamine-mediated DNA transfection experiments were performed by following the manufacturer's protocol with some modifications. GEC1-pcDNA3.1/Hygro(+) was previously constructed (10). Twenty four hours before transfection, 1.8–2.0 million cells stably expressing FLAG-hKOPR were seeded on each 10-cm Petri dish. On the experiment day, transfection was carried out with 40 μl of Lipofectamine (1 mg/ml), 10 μg of the DNA construct or the plasmid vector (control), and 6 ml of Opti-MEM medium per 10-cm dish. At 16 h after transfection, medium was replaced by 10 ml of Opti-MEM containing 10% FBS. Forty hours following transfection, the cells were harvested for further experiments.
Co-immunoprecipitation of hKOPR and GEC1-(38–117)
The cDNA construct of wild-type or mutant FLAG-hKOPR or the vector pcDNA3.1 was co-transfected with HA-GEC1-(38–117) at a ratio of 5:5 (μg) into one 100-mm dish of CHO cells. Forty hours after transfection, two dishes of cells (2 × 107) were collected and solubilized in 1 ml of TTSEC (0.5% Triton X-100, 50 mm Tris, pH 7.5, 0.15 m NaCl, 1 mm EDTA, and protease inhibitor mixture from Roche Applied Science) for 1 h at 4 °C. Cell lysates were cleared by centrifugation at 105 × g followed by filtration through a 0.2-μm filter. One ml of supernatant was incubated with 20 μl of anti-FLAG-agarose beads (M2, Sigma) overnight at 4 °C. The beads were then washed three times with TTSEC containing 1% Triton X-100 and extracted in 40 μl of loading buffer (4% SDS, 50 mm Tris, pH 6.25, and 100 mm dithiothreitol). Samples were separated on 12% SDS-PAGE and transferred to Immobilon (Millipore), and immunoblotting of HA-GEC1-(38–117) and FLAG-hKOPR was performed with rabbit anti-HA antibody and rabbit anti-FLAG antibody, respectively, and followed by enhanced chemiluminescence. Transfection and co-immunoprecipitation of HA-GEC1-(38–117) or its mutants with FLAG-hKOPR were performed similarly. The program OptiQuant (PerkinElmer Life Sciences) was used to analyze the immunoblotting results.
[3H]Diprenorphine Binding to hKOPR in Intact Cells
Samplesturation binding was performed with six concentrations of [3H]diprenorphine (0.1, 0.2, 0.4, 0.6, 1.0, and 2.0 nm) and 150,000 cells/tube in duplicate in 1 ml of PBS buffer containing 1 mg/ml bovine serum albumin at room temperature for 60 min. Ten μm naloxone was used to define nonspecific binding. The Kell program (known as EBDA previously) was used to analyze data and to obtain the Bmax and Kd values. Binding with ∼1 nm [3H]diprenorphine was performed with 200,000 cells/tube in a similar manner. Naloxone (10 μm) was employed to define the nonspecific binding for total receptors as described previously (10).
SDS-PAGE and Immunoblotting
Cells were harvested using Versene buffer, solubilized in 2× Laemmli sample buffer, and subjected to Tricine-SDS-PAGE on 8% separating gel as described previously (10). The separated protein bands were transferred to Immobilon-P polyvinylidene difluoride transfer membranes on which immunoblotting was carried out with primary antibodies, horseradish peroxidase-linked secondary antibody, and SuperSignal West Pico Chemiluminescent reagents (10). Primary antibodies used were rabbit polyclonal anti-FLAG (F7425) antibody (0.8 mg/ml, 1:5000), rabbit polyclonal anti-GluR1 antibody (1:5000) or mouse monoclonal anti-HA antibody (1 mg/ml, 1:5000). The protein bands were visualized and then digitalized with Fuji LAS-1000 Plus gel documentation system (Fuji Film, Tokyo, Japan). Expression level of transfected GEC1 protein was evaluated by immunoblotting as described above except using 15% Tricine/SDS-PAGE, boiled (100 °C, 5 min) sample solution, and rabbit polyclonal anti-GEC1 (PA629p) antibody (0.49 μg/ml, 1:7500).
Y2H Assays
The general strategies were to use different hKOPR-C tail (334–380) or GEC1-(38–117) mutants to narrow down (truncation mutants) and then to define (double and single alanine substitution mutants) the amino acid residues that account for GEC1-hKOPR interaction. GEC1-(38–117) is a truncated GEC1, which was the form originally identified to bind the hKOPR C-tail in our previous Y2H studies (10).
For determining the amino acid residues in GEC1 involved in hKOPR binding, GAL4 yeast two-hybrid system was employed to evaluate interaction between GEC1 mutants and hKOPR-(334–380). GEC1 mutant cDNA was generated using PCR and inserted into the prey vector pGADT7 containing a LEU2 selection marker for yeast, which was then transformed into yeast strain Y187 that is auxotrophic for adenine (Ade), tryptophan (Trp), histidine (His), and leucine (Leu). Human KOPR-(334–380) cDNA was inserted into the bait vector pGBKT7 containing a TRP1 nutritional marker for yeast selection that was then transformed into yeast strain AH109 that is also auxotrophic for Ade, Trp, His, and Leu. After selecting transformants on SD/-Leu and SD/-Trp media, respectively, the two positive haploids were mated, and growth status of the diploids on media with different stringency was monitored and evaluated. The amino acids in hKOPR C-tail involved in GEC1 binding were determined with the same method except that the plasmid constructs of hKOPR-(334–380) mutants/pGBKT7 and GEC1-(38–117)/pGADT7 were used.
Plasmid Construction for Y2H—N and C termini of GEC1-(38–117) were truncated by an increment of 10 and 2 residues via a series of PCRs. The PCR products digested with restriction enzymes EcoRI and BamHI were ligated into the prey vector pGADT7. After delineating the hKOPR binding region using aforementioned GEC1-(38–117) truncation mutants, single alanine substitution mutants were generated within the 40–67 and 109–113 fragments of GEC1-(38–117) by overlap PCR to define the residues required for the GEC1-hKOPR interaction. With the same strategy, five-amino acid truncation, double and single alanine substitution mutants of hKOPR-(334–380) in pGBKT7 (NdeI/SalI) were established. All the resulting DNA plasmids were purified using QIAprep Spin Miniprep kit. DNA sequences of the inserts were verified by the University of Pennsylvania DNA sequencing facility.
Transformation of Yeast Cells—Transformation of yeast with constructs was carried out using standard PEG/LiAc protocol as described in our previous publication (10).
Mating of Yeast Cells—The two mating partner strains, Y187 and AH109, were transformed with prey (GEC1/pGADT7) and bait (hKOPR-C/pGBKT7) plasmids, respectively. Transformants were selected using the proper SD dropout medium, and the mating was conducted in 96 six-well plates. For both transformants, a single colony (2–3 mm, less than 1 month old) was suspended in 1 ml of 2× YPDA/kanamycin medium. Twenty μl each of the Y187 and the AH109 cell suspension was added to one well containing 200 μl of 2× YPDA medium with kanamycin (10 μg/ml) and mixed well. The plate was then incubated at 30 °C for 16–18 h on an orbital shaker set at 200 rpm. Mating reactions were monitored for diploid formation using phase-contrast microscope (27). The mating culture was diluted with physiological saline and then plated in a volume of 100 μl onto SD medium with three levels of stringency: double dropout (2DO) SD/-Leu/-Trp, triple dropout (TDO) SD/-Leu/-Trp/-His, and quadruple dropout (QDO) SD/-Leu/-Trp/-His/-Ade. The plates were incubated upside-down at 30 °C for 5–7 days to obtain well separated colonies with a diameter of 1–3 mm. Dilution factor for the mating culture was adjusted so that the resulting colony number of the diploid on the 2DO plate was 200–400.
Assessment of the Protein-Protein Interaction—Throughout the yeast two-hybrid assays, the mating mixture of GEC1-(38– 117)/pGADT7 and hKOPR-C(334–380)/pGBKT7 was used as the positive control and GEC1-(38–117)/pGADT7 combined with pGBKT7(-) as the negative control. Plasmid pGBKT7(-) was derived from the pGBKT7 vector of which the sequence between the NdeI site and SalI site (in the multiple cloning sites) was deleted. Five to 7 days after yeast mating, the colony number on the SD agar plates was counted using software ImageJ 1.34S. For each mating reaction, the diploid number on TDO and QDO plates was normalized against that on 2DO plate. Then the normalized numbers were compared with its counterparts in the positive control group. The protein-protein interaction between GEC1-(38–117) and hKOPR C-tail was considered to be strong (+++), moderate (++), weak (+), or absent (-) if the resulting percentages were larger than 80%, between 30 and 80%, between 5 and 30%, or less than 5%, respectively.
Structural Modeling of GEC1
Initial Model—Although no crystal structure of GEC1 has been published, GEC1 shares 86% identity with GABARAP. So the 1.75-Å resolution GABARAP x-ray crystal structure (Protein Data Bank code 1GNU) (28) served as template here for an initial (united atom) GEC1 model. The GABARAP/GEC1 sequence alignment presented in Chen et al. (10) (also see Fig. 9A) was used as input to Modeler (version 9.1) (29, 30). The GEC1 model with the lowest objective function out of 100 models generated was chosen for further refinement. Hydrogens were added to the model using Maestro version 8.0.110 (Schrödinger, LLC, New York), with a short minimization performed to optimize the hydrogen positions. Energy minimization was performed using Macromodel version 9.5 (31) and the OPLS2005 all atom force field. A distance-dependent dielectric, 8.0-Å extended nonbonded cutoff, 20.0-Å electrostatic cutoff, and 4.0-Å hydrogen bond cutoff were used. The minimization consisted of 500 steps (Polak-Ribier conjugate gradient method) with backbone and side chain heavy atoms fixed.
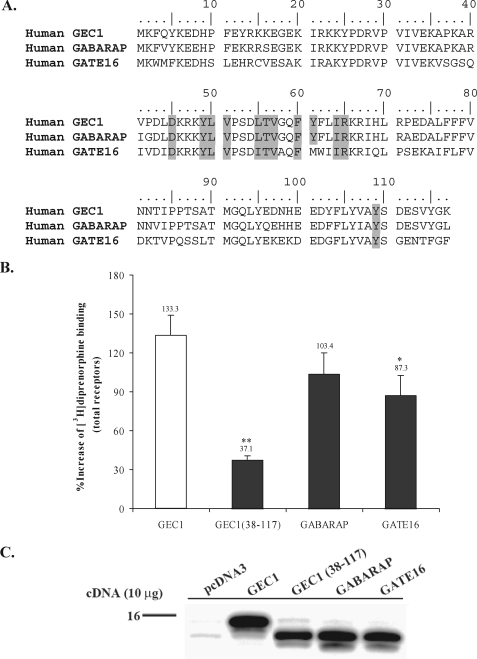
FIGURE 9.
Effects of GEC1 and its analogues on FLAG-hKOPR expression. A, amino acid sequence alignment of GEC1, GABARAP, and GATE16 showing the highly conserved nature of the 12 GEC1 residues involved in interaction with hKOPR. B, GEC1 up-regulated FLAG-hKOPR to a higher extent than GABARAP, GATE16, and GEC1-(38–117). Ten μg of vector, GEC1, GEC1-(38–117), GABARAP, or GATE16 cDNA was transfected, and receptor binding assays were conducted 40 h later. Results are expressed as mean ± S.E. of three experiments. *, p < 0.05, and **, p < 0.01 compared with GEC1 group using one-way ANOVA followed by Tukey's post hoc test. C, immunoblotting of GABARAP family proteins and truncated GEC1. Immunoblotting of proteins at 40 h after transfection was conducted. Two hundred thousand cells in Laemmli sample buffer (per lane) were loaded and resolved by 15% Tricine/SDS-PAGE. Proteins were detected by immunoblotting with polyclonal anti-GEC1 antibody, which cross-reacts with GABARAP and GATE16 in immunoblotting (14).
Molecular Dynamics of Apo-GEC1 in an Aqueous Environment—NAMD2 (32) molecular dynamics simulations were used to relax the initial GEC1 model in an aqueous environment. 10 Å of waters in each direction of the protein were added using the VMD Solvate package, and the VMD Meadionize plugin was employed to achieve electroneutrality. NAMD2 calculations used the CHARMM27 parameter set (33–35) and the TIP3P model for water. Periodic boundary conditions were employed with the long range coulombic electrostatic potential treated with the Particle-Mesh Ewald summation method (36). The Lenard-Jones potential was smoothly cut off between 8.5 and 10 Å. The NAMD2 simulation used the r-RESPA multiple time step algorithm with a time step of 4 fs for long range electrostatic forces, 2 fs for short range nonbonded forces, and 1 fs otherwise. The simulation cell was minimized for 20 ps (5 ps with heavy atoms fixed and 15 ps with all atoms unrestrained). Using Langevin coupling to a heat bath, the cell was warmed to 310 K (NVT ensemble) in 10° increments for 20 ps with the backbone constrained using a 0.5 kcal/mol Å2 harmonic force. 1.25 ns of equilibration (NPT; 310 K/1 atm) was performed in 0.25-ns runs with a decreasing backbone harmonic constraint for each run (0.5, 0.25, 0.10, 0.05, and 0.0 kcal/mol Å2). Once the protein was equilibrated, a 10-ns production run was initiated (NPT; 310 K/1 atm).
Conformational Memories—The Monte Carlo/simulated annealing method, Conformational Memories (CM) (37–39), was used to explore (in the presence of GEC1) the low free energy conformations of an octapeptide from the C terminus of the hKOPR that contains the GEC1-binding motif, FPXXM (CFPLKMRM). The GEC1 model, taken from the MD trajectory, was that frame in which key KOPR C-terminal interaction residues were most exposed. The CHARMM force field was employed for this calculation. In the CM exploratory phase, the peptide was positioned 59 Å above GEC1 at T = 3000 K. Then the peptide was drawn toward the GEC1 surface in 18 steps (3.28 Å/step) as the temperature was decreased to T = 310 K (18 steps). 102 runs were performed per temperature with a run consisting of 50,000 steps applied to all peptide torsion and bond angles, allowing them to vary ±180 and ±8°, respectively. The CM biased-annealing phase, began at T = 749.4 K, cooling to 310 K in nine steps. The peptide was drawn to the GEC1 surface using a planar restraint on the peptide center of mass (k = 0.025 kcal/mol Å2), and the center of mass was confined spatially using a cylindrical restraint (k = 0.02 kcal/mol Å2; 15.7 Å diameter circle at the protein surface, T = 310 K). To account for electrostatic screening in water, a distance-dependent, nonlinear (sigmoidal) screened Coulomb potential as described by Hassan et al. (40, 41) (average screening parameter 1.0367) was employed, with the dielectric constant decreased from 80 to 1, based on the distance between the peptide center of mass and the protein surface.
The 102 octapeptide-GEC1 complexes (holo-GEC1) output at 310 K were screened using the criteria that the GEC1-binding motif (FPXXM) of the hKOPR C-terminal peptide (residues Phe2, Pro3, and Met6) is critical for GEC1 interaction and that the first octapeptide residue (Cys1), being a putative palmitoylation site, needs to be unobstructed. The complex that best met these criteria was then equilibrated in water via NAMD2 MD simulations.
Molecular Dynamics of Holo-GEC1 in Aqueous Environment—The NAMD2 settings used for apo-GEC1 detailed above were used to equilibrate holo-GEC1 in water, with the exception that the water box extended 12 Å from the complex surface in all directions. After minimization (heavy atoms fixed), the simulation cell was warmed to 310 K in 10° increments for 20 ps (heavy atoms fixed), and then run for 1 ns with harmonic constraints to maintain the complex while the system was solvated. Because the equilibrated apo-GEC1 was used for the complex, a heavy atom harmonic constraint of 1.0 kcal/mol Å2 was employed for GEC-1, whereas a 0.5 kcal/mol Å2 harmonic constraint was placed on the following: (i) the peptide Phe2, Pro3, and Met6 C-α atoms; (ii) the most extreme Phe2 and Pro3 carbons; and (iii) the Met6 sulfur and its adjacent methylene carbon to maintain the CM identified complex. The system was minimized (NVT), and then a 1.5-ns equilibration was followed by a 5-ns production run (NPT; 310 K/1 atm) using the same constraints as the warm up and equilibration.
Data Analysis
All quantitative data were present as mean ± S.E. if they were derived from at least 3× experiments. For comparison of multiple groups, the data were analyzed by one-way analysis of variance (ANOVA) followed by a Tukey post hoc test. If the p value less than 0.05, the difference was defined as significant. All statistical analyses were performed using GraphPad Prism 3.0.
RESULTS
Effect of GEC1 on KOPR Expression—Results of saturation binding of [3H]diprenorphine, an antagonist, revealed that GEC1 expression resulted in 85% increase in Bmax value compared with the control, but there was no change in the Kd value (Table 1). When binding was conducted with ∼1 nm [3H]diprenorphine, a concentration close to saturation, the percentage increase in KOPR binding was ∼84%. We therefore performed binding with 1 nm [3H]diprenorphine, rather than saturation binding, in the subsequent experiments.
TABLE 1.
Effect of GEC1 expression on [3H]diprenorphine binding to the KOPR stably transfected into CHO cells Cells were transiently transfected with GEC1 or the vector pcDNA3.1. Forty hours later, saturation binding of [3H]diprenorphine (six concentrations ranging from 0.1 to 2 nm, including 1 nm) to the receptor was performed, and Kd and Bmax values were calculated. The percent change in KORP expression level measured by 1 nm [3H]diprenorphine binding was determined, which is consistent with that in Bmax value. [3H]Diprenorphine (1 nm) binding was therefore chosen to measure the changes of receptor expression in all subsequent experiments. Data are expressed as means ± S.E. (n = 3). ** indicates p < 0.01 compared with the control group by Student's t test.
|
Cells transfected with
|
Kd
|
Bmax fmol/106 cells
|
% increase of control
|
|
|---|---|---|---|---|
| Bmax | 1 nm [3H]diprenorphine | |||
| nm | ||||
| Control vector | 0.13 ± 0.01 | 70.00 ± 0.67 | ||
| GEC1 | 0.17 ± 0.02 | 129.90 ± 0.83** | 85.6 ± 2.3 | 84.4 ± 3.1 |
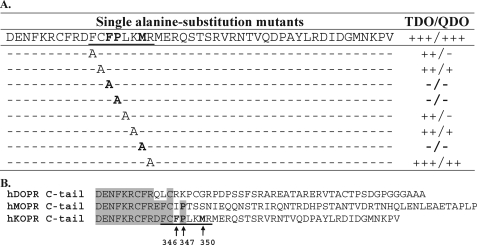
Determination of GEC1-binding Sequence in the hKOPR C-tail—Two series of truncation mutants were generated by deleting five amino acids incrementally from the N terminus and the C terminus, respectively, and their interactions with GEC1-(38–117) were examined. The fragment 344FCFPLKMRMERQSTSRV360 appeared to be necessary for the interaction (supplemental Fig. I). We then generated double alanine substitution mutants of the hKOPR C-tail within this fragment, and their interactions with GEC1-(38–117) were examined. We found that the residues 346FP347 and 350MR351 were essential (supplemental Fig. I). Single alanine substitution mutations were subsequently carried out, and Phe346, Pro347, and Met350 were found to be critical (Fig. 1A). In addition, Phe344, Cys345, Leu348, and Lys349 appeared to be involved in the GEC1-hKOPR interaction, but they were not as important (Fig. 1A). Thus, the GEC1-binding sequence was defined as FPXXM. Sequence alignment (Fig. 1B) of the C-tails of human opioid receptors (μ, δ, and κ) revealed that this sequence was unique to the KOPR among the opioid receptors.
FIGURE 1.
Mapping the GEC1-binding motif in the C-tail of hKOPR using Y2H assays. A, summary of Y2H results. Truncation and double and single alanine substitution mutants of hKOPR C-tail were examined for their abilities to interact with GEC1-(38–117). The hKOPR C-tail mutants were generated by PCR, constructed into GAL4-binding domain vector pGBKT7, and transformed into yeast strain AH109. The GEC1-(38–117) was constructed into GAL4 activation domain vector pGADT7 and transformed into yeast strain Y187. The two transformed yeast strains mated, and the mating mixtures were cultured on SD/-Trp/-Leu (2DO), SD/-Trp/-Leu/-His (TDO), and SD/-Trp/-Leu/-His/-Ade (QDO) agar plates. The resulting diploid colonies were counted, and the ratio of the number of colony-forming units on TDO/QDO plates over colony-forming units on the 2DO plate was used to evaluate the interaction strength of each pair of GEC1-(38–117) and hKOPR C-tail mutant. Assessment details were described under “Experimental Procedures.” The experiments were performed three times with reproducible results. Results on truncation and double alanine substitution mutants are shown in supplemental Fig. 1. B, amino acid sequence alignment of C-tails of the human opioid receptors demonstrating that the defined GEC1 binding region is hKOPR-specific.
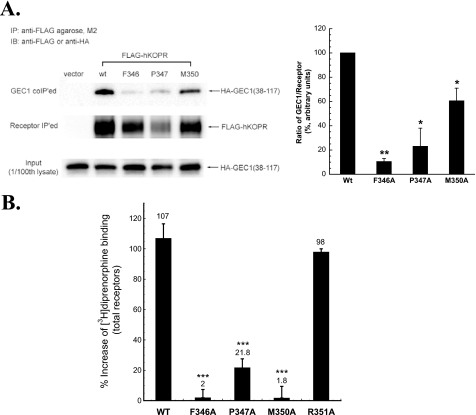
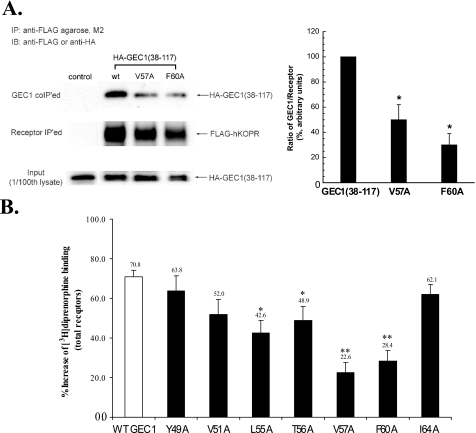
We then investigated if mutations in the FPXXM sequence in the hKOPR C-tail affected interaction of the hKOPR with GEC1 in CHO cells. Three single alanine substitution FLAG-hKOPR mutants, F346A, P347A, and M350A, were constructed, and CHO cells stably expressing each mutant were established. We have previously demonstrated that although GEC1 immunoprecipitated with hKOPR, the signals appeared to be weak (10), most likely because of the strong interaction of GEC1 with microtubules via its N-terminal region. The tubulin-binding domain of GEC1 was mapped to the fragment (amino acids 1–22) (24). We found that the amount of GEC1-(38–117) immunoprecipitated with the hKOPR was higher than that of GEC1 (data not shown); therefore, GEC1-(38–117) was used in the co-immunoprecipitation experiments. GEC1-(38–117) was transiently transfected, and co-immunoprecipitation was examined. As shown in Fig. 2A, substitutions of Phe346, Pro347, or Met350 with Ala significantly reduced the amount of GEC1-(38–117) co-immunoprecipitated with FLAG-hKOPR.
FIGURE 2.
A, F346A, P347A, and M350A mutations in the hKOPR C-tail reduced co-immunoprecipitation of GEC1-(38–117). GEC1-(38–117) was co-transfected into CHO cells with wild-type (wt) or a mutant FLAG-hKOPR or the control vector. The receptors were immunoprecipitated (IP) with anti-FLAG (M2)-agarose beads 40 h after transfection. The SDS-PAGE was performed, and the GEC1-(38–117) (GEC1 co-IPed, 1st row) and hKOPR (receptor IPed, 2nd row) were detected with immunoblotting (IB) with anti-HA antibody and anti-FLAG antibodies, respectively. The ratios of density of GEC1-(38–117) over receptor were normalized against that of wild-type receptor and are shown in the graph at right. Each value represents the mean ± S.E. of three independent experiments. *, p < 0.05, and **, p < 0.01 compared with wild-type group using one-way ANOVA followed by Tukey's post hoc test. B, effects of GEC1 on expression of the wild-type and alanine-substituted mutants (F346A, P347A, M350A, and R351A) of FLAG-hKOPR. Wild-type hKOPR and its alanine-substituted mutants were cloned in pcDNA3.1 and then stably expressed in CHO cells. Wild-type and mutated receptors were expressed at similar levels (∼1.9 pmol/mg protein). GEC1 or the vector pcDNA3.1 (control) was transiently transfected into these cells. [3H]Diprenorphine (1 nm) binding to hKOPR was performed on intact cells 40 h after transfection. Data were expressed as the percentage of increase in [3H]diprenorphine binding in GEC1-transfected cells compared with the control cells. Each value represents the mean ± S.E. of three independent experiments. ***, p < 0.005 compared with wild-type group using one-way ANOVA followed by Tukey's post hoc test.
We next determined whether mutations in the FPXXM sequence in the hKOPR C-tail influenced up-regulation of KOPR by GEC1 in CHO cells. As shown in Fig. 2B, the enhancing effect of GEC1 on hKOPR expression was greatly reduced by F346A, P347A, and M350A mutations (p < 0.005) but not by R351A mutation. In Y2H assay, R351A substitution in the hKOPR C-tail did not affect its interaction with GEC1 (see above). Thus, the data from Y2H assays, co-IP, and functional studies indicate that Phe346, Pro347, and Met350 are critical for the interaction between GEC1 and hKOPR, which results in up-regulation of the hKOPR.
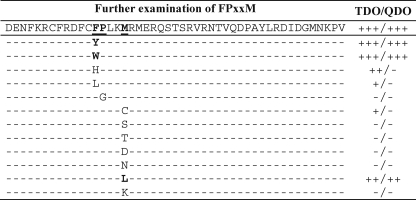
Permutations of the FPXXM Sequence—We further characterized structural requirements of the GEC1-binding motif. We generated the following single mutants in the hKOPR C-tail: mutations of Phe346 to Tyr, Trp, His, and Leu; Pro347 to Gly; and Met350 to Cys, Ser, Thr, Asp, Asn, Leu, and Lys. We found that, at the position 346, having Tyr and Trp substitutions resulted in robust interaction with GEC1, but His or Leu did not (Fig. 3), indicating that the aromatic ring is indispensable. For the 347 position, substitution of Pro with the simplest and most flexible residue glycine totally abolished the binding, demonstrating that that the kink structure is critical for the two proteins to bind. Change of Met350 to acidic, basic, or polar residue eliminated GEC1-hKOPR interaction, but substitution with the hydrophobic residue Leu had little effect on the interaction. Therefore, lipophilic residue at the 350 position is important for GEC1 binding. Therefore, the GEC1-interacting FRXXM sequence can be expanded to (Phe, Tyr, Trp(Pro-XX)Met, Leu), which is a kink-producing hydrophobic fragment. As Leu and Ile are very similar, it is possible that Met350 can be replaced with Ile.
FIGURE 3.
Permutations of the GEC1-binding motif in the hKOPR C-tail. Phe346, Pro347, and Met350 in the hKOPR C-tail were replaced, one at a time, with the indicated amino acids. The interactions of the hKOPR C-tail mutants with GEC1-(38–117) were evaluated in Y2H assays as described in Fig. 1. The experiments were performed three times with similar results.
Determination of hKOPR-binding Domain in GEC1-(38–117)—GEC1-(38–117) was employed as the starting construct because it was found to interact with the hKOPR-C tail in the original Y2H screening (10). With the 10-amino acid truncation mutants of GEC1-(38–117), we found that at least the (38KARVPDLDKRKYLVPSDLTV57) fragment in the N-terminal region and the (108AYSDESVYGK117) fragment in the C-terminal region were required for the interaction (supplemental Fig. II). Serial double-truncation mutants of GEC1-(38–117) within these two regions were generated and examined. We found that, at least, the amino acid residues 44LD45, 50LVPS53, and 108AYSD111 were required for GEC1 to interact with the hKOPR (supplemental Fig. II).
Alanine substitution scanning studies in the fragments 40RVPDLDKRKYLVPSDLTVGQFYFLIRKR67 and 109YSDES113 were performed. The Y2H results showed that residues Tyr49, Val51, Leu55, Thr56, Val57, Phe60, and Ile64 were indispensable for GEC1-(38–117) to bind to the hKOPR C-tail (Fig. 4). In addition, five other residues, Leu44, Leu50, Tyr61, Arg65, and Tyr109, appeared to be involved (Fig. 4).
FIGURE 4.
Mapping of the residues in GEC1 involved in binding to hKOPR using Y2H assays. Serial truncation and substitution mutants of GEC1-(38–117) were examined for their ability to interact with hKOPR C-tail. GEC1-(38–117) mutants were generated by PCR, constructed into GAL4 activation domain vector pGADT7, and transformed into yeast strain Y187. The hKOPR C-tail was constructed into GAL4-binding domain vector pGBKT7 and transformed into yeast strain AH109. The two transformed yeast strains mated, and the mating mixtures were selected, and interaction strength was evaluated as described in Fig. 1. The experiments were performed three times with reproducible results. Results on truncation and double alanine substitution mutants are shown in supplemental Fig. II.
We then investigated if single alanine-substituted full-length GEC1 mutants interacted with the hKOPR in CHO cells, using V57A and F60A as the representatives. As shown in Fig. 5A, alanine substitution of Val57 or Phe60 in GEC1 greatly reduced its interaction with hKOPR. Subsequently, we examined if alanine substitution at Tyr49, Val51, Leu55, Thr56, Val57, Phe60, or Ile64 affected the effect of GEC1 on hKOPR expression. Transient expression of each of the seven mutants resulted in less up-regulation of hKOPR than the wild-type GEC1, and the decreases caused by alanine substitution mutation at Leu55, Thr56, Val57, and Phe60 were statistically significant (Fig. 5B).
FIGURE 5.
A, V57A or F60A mutation in GEC1-(38–117) GEC1 reduced its co-immunoprecipitation with FLAG-hKOPR. Wild-type GEC1-(38–117) or its mutant was co-transfected into CHO cells with wild-type FLAG-hKOPR or the control vector. The receptors were immunoprecipitated (IP) using anti-FLAG (M2)-agarose beads 40 h after transfection. SDS-PAGE was performed, and GEC1-(38–117) and the mutants (GEC1 co-IPed, 1st row) and FLAG-hKOPR (receptor IPed, 2nd row) were detected with immunoblotting (IB). The ratios of density of GEC1-(38–117) or the mutants over FLAG-hKOPR were normalized against that of wild-type GEC1-(38–117) and are shown in the graph at right. Each value represents the mean ± S.E. of three independent experiments. *, p < 0.05 compared with wild-type group using one-way ANOVA followed by Tukey's post hoc test. B, effects of single alanine substitution of the critical residues in the hKOPR-binding motif on the GEC1-induced enhancement in FLAG-hKOPR expression. Wild-type GEC1 and its alanine-substituted mutants were cloned in pcDNA3.1 and then transiently transfected into CHO cells stably expressing FLAG-hKOPR. Forty hours after transfection, [3H]diprenorphine (1 nm) binding to hKOPR was performed on intact cells. Data were expressed as the percentage of increase in [3H]diprenorphine binding in GEC1-transfected cells compared with the control cells that were transfected with the vector pcDNA3.1. Each value represents the mean ± S.E. of five independent experiments. *, p < 0.05, and **, p < 0.01 compared with wild-type group using one-way ANOVA followed by Tukey's post hoc test.
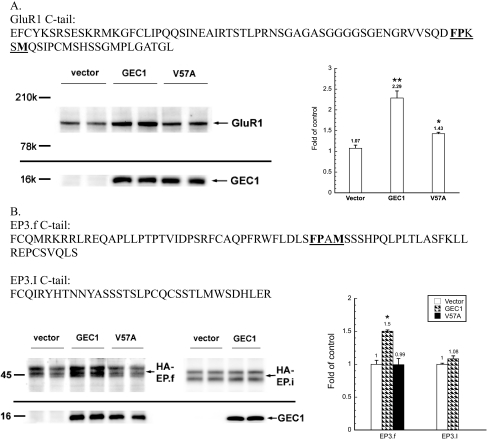
Other Receptors Containing FPXXM or a Similar Sequence— A search of the Swiss Protein Database revealed that many proteins contain FPXXM sequence. The C-tail of AMPA receptor subunit GluR1 contains such a sequence (Fig. 6A). Using CHO cells stably transfected with GluR1, we found that transient expression of the wild-type GEC1, but not V57A GEC1, enhanced GluR1 expression (Fig. 6A).
FIGURE 6.
Effect of GEC1 on expression of GluR1 and EP3.f, which contain the GEC1-binding motif or a similar sequence. Amino acid sequences of the cytosolic C-tails of AMPA receptor subunit GluR1 (A), prostaglandin receptor EP3.f, and EP3.I (B) are shown. The matched patterns (FPXXMorFPXM) are underlined and in boldface. Effects of GEC1 or its V57A mutant on expression of GluR1 (A) and HA-EP3.f and HA-EP3.I prostaglandin (B) receptors stably expressed in CHO cells were examined. Cells were transiently transfected with GEC1 or its V57A mutant. Forty hours after the transfection, immunoblottings were performed in duplicate with 200,000 cells in Laemmli sample buffer (per lane). GluR1, GEC1 and prostaglandin receptors were detected with rabbit anti-GluR1, rabbit anti-GEC1, and mouse monoclonal anti-HA antibodies, respectively. Each value represents the mean ± S.E. of three independent experiments. *, p < 0.05, and **, p < 0.01 compared with wild-type group using one-way ANOVA followed by Tukey's post hoc test.
Prostaglandin receptor EP3.f has a FPXM sequence in its C-terminal domain (Fig. 6B), which is similar, but not identical, to FPXXM. In CHO cells stably transfected with EP3.f, expression of wild-type GEC1, but not the V57A mutant, enhanced the level of the EP3.f receptor (Fig. 6B). In contrast, GEC1 had no impact on expression of the EP3.I, a splice variant of the EP3.f that does not have the FPXM sequence (Fig. 6B). Thus, it is very likely that GEC1 directly interacted with GluR1 and EP3.f through similar sequences in the receptor C-tails.
Molecular Model of GEC1—The x-ray crystal structure of GABARAP (28) (Protein Data Bank code 1GNU) was employed as a template for the GEC1 model given the high sequence similarity (94%) and identity (86%) of GEC1 with GABARAP. Among the 100 models produced by Modeler, the one with the lowest objective function was chosen for further refinement (see Fig. 7A).
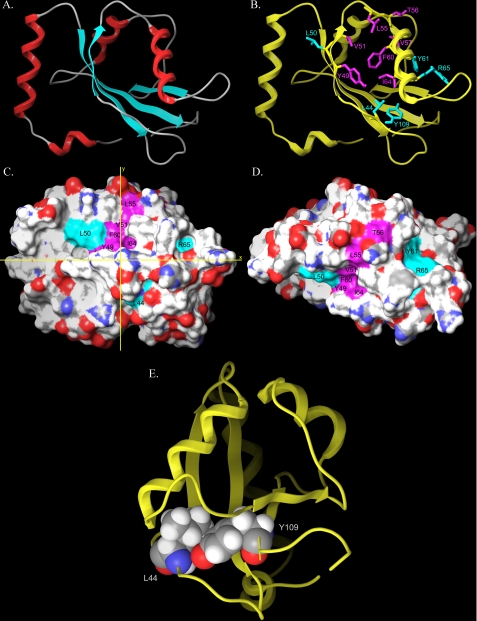
FIGURE 7.
Molecular model of GEC1. A, molecular model of GEC1 created by using the structure of GABARAP as the template. B, view of residues that are critical for (magenta color) and involved in (cyan) interaction with the hKOPR C-tail as determined by the Y2H assays. Spatial orientations of side chains (in stick format in magenta and cyan) of these residues are displayed. C and D, GEC1 model derived from the NAMD2 trajectory. The model represents that frame from the trajectory in which key residues were most exposed. D, model in C has been rotated by 90° about the x axis. C and D, color schemes of residues are identical to B; residues of primary importance are shown in magenta (Tyr49, Val51, Leu55, Thr56, Val57 (not visible in either view), Phe60, Ile64), and residues of secondary importance are shown in cyan (Leu44, Leu50, Tyr61, Arg65, and Tyr109 (not visible in either view)). E, intramolecular interaction between Leu44 and Tyr109. This image results from a 90° clockwise rotation about the vertical y axis from the view in B. Side chains of these two residues may interact through van der Waals forces, which may contribute to structural stability of GEC1.
The GEC1 model is composed of an N-terminal helical region (residues 1–25) that is highly basic and a core structure (residues 27–117) with a typical β-grasp ubiquitin-like folding. The basic N-terminal domain of GEC1 consisting of two α-helices (1 and 2) has been suggested to be responsible for mediating direct interaction with microtubules. Following residue Pro26, the core domain of GEC1 structure includes four β-strands (strands 1–4) and two α-helices (helices 3 and 4), the two middle strands (strands 1 and 4) parallel to each other, and the two outer strands (strands 2 and 4) anti-parallel to their neighboring inner strands, respectively. α-Helix 3 is located between β-strands 2 and 3, whereas α-helix 4 is between β-strands 3 and 4. Fig. 7B shows spatial orientations of residues of primary importance for GEC1 interaction with the hKOPR C terminus in magenta and those of secondary importance in cyan.
Molecular Dynamics of Apo-GEC1 in an Aqueous Environment; KOPR C-tail Binds to Hydrophobic Patches on the Surface of GEC1— The 10-ns production run trajectory of GEC1 in an aqueous environment was analyzed to identify a GEC1 conformation that most exposed key residues identified to be critical for binding. We found that when the Leu63 χ1 torsion angle was in g+ (-60°), the binding site was more exposed than when this torsion was trans (-180°). In addition, GEC1 clearly underwent a “breathing” motion during the simulation, permitting the identification of a conformation that most exposed key residues. This conformer is illustrated in Fig. 7, C and D, which show two views of the GEC1 surface. Residues found to be of primary importance for GEC1 interaction with the KOPR C-tail are colored magenta in Fig. 7, C and D, (Tyr49, Val51, Leu55, Thr56, Val57 (not visible in the views), Phe60 and Ile64), whereas residues found to be of secondary importance (Leu44, Leu50, Tyr61, Arg65, and Tyr109 (not visible in the views)) are colored cyan.
Each residue that is critical for interaction has a hydrophobic side chain or an aromatic ring, except Thr56. As shown in Fig. 7B, all side chains of these amino acids, with the exception of the isopropyl group of Val57, appeared to orient in similar directions, forming a curved hydrophobic surface (around the S2 β-strand) (Fig. 7, C and D). In addition, the lipophilic isobutyl group of Leu50 in β-strand 2 appeared to enlarge this hydrophobic surface of GEC1, although its side chain did not point to the same direction. This curved face in GEC1 likely fits well with the kinked hydrophobic FPXXM motif in hKOPR C-tail. Moreover, the side chains of Tyr61 and Arg65 in α-helix 3 were very close spatially and appeared to be on the same GEC1 surface (Fig. 7, B–D).
The Y2H data revealed that Leu44 and Tyr109 of GEC1 were important for its interaction with the hKOPR C-tail. The model of GEC1 showed that part of the Leu44 side chain was exposed to the surface, whereas the entire residue of Tyr109 was in the interior of the protein (Fig. 7, B–D). Fig. 7E clearly demonstrated that Leu44 and Tyr109 likely have a direct intramolecular interaction between their hydrophobic side chains, which may act to maintain the three-dimensional GEC1 conformation that is essential for interaction.
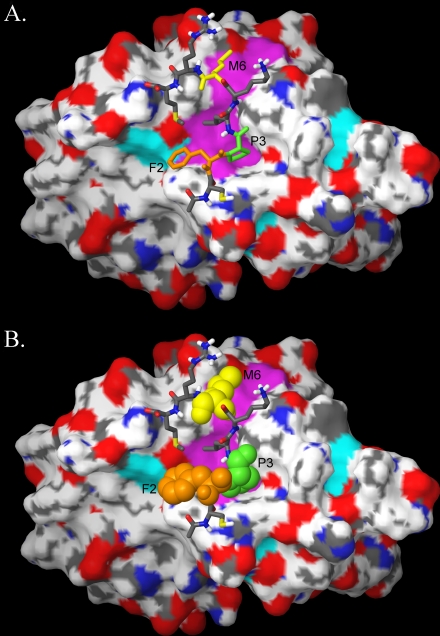
The molecular dynamics output of the hKOPR C-terminal octapeptide (CFPLKMRM)-GEC1 complex identified by Conformational Memories is illustrated in Fig. 8. In this complex, the peptide FPXXM-binding motif residues are illustrated at the top in tube display (Fig. 8A), and below (Fig. 8B) these residues are contoured at their van der Waals radii, with Phe2 colored orange, Pro3 colored green, and Met6 colored yellow. It is clear here that the peptide FPXXM-binding motif residues contact the Tyr49/Val51/Leu55/Thr56/Phe60/Ile64 region of GEC1 (Fig. 8B, colored magenta). The energy of interaction of these important amino acids with the GEC1 model was found to be dominated by van der Waals forces. The interaction energies were -110.25 kcal/mol for Phe2, -68.42 kcal/mol for Pro3, and -109.48 kcal/mol for Met6.
FIGURE 8.
A and B, molecular dynamics output of the hKOPR C-terminal octapeptide-GEC1 complex identified by Conformational Memories. The octapeptide containing the FPXXM GEC1-binding motif is shown on the surface of GEC1 in tube display with F2 in orange, P3 in green, and M6 in yellow. B, those same amino acids are displayed contoured at their van der Waals radii.
Effects of GABARAP and GATE16 on Expression of FLAG-hKOPR—Sequence alignment (Fig. 9A) demonstrated that all seven critical residues were completely conserved between GEC1 and GABARAP, and that, except similar residue at position 55, all the other six amino acids were identical between GEC1 and GATE16. We examined whether these two proteins enhanced FLAG-hKOPR expression like GEC1. As shown in Fig. 9B, both GABARAP and GATE16 greatly enhanced the expression of FLAG-hKOPR. However, GATE16 up-regulated hKOPR to a significantly lower extent than GEC1.
Tubulin-binding Domain Is Important for GEC1 to Promote hKOPR Expression—Microtubule is one type of cytoskeletons that determine organelle positions and control intracellular transport. GEC1-(38–117) does not have the first 37 amino acids and thus lacks the tubulin-binding domain (24). Compared with the full-length GEC1 (Fig. 9), GEC1-(38–117) caused a significantly lower extent of hKOPR up-regulation (p < 0.01), indicating that binding to tubulin cytoskeleton is important for the effect of GEC1 on hKOPR expression. However, the possibility cannot be excluded that truncation causes changes in three-dimensional GEC1 conformation that may affect GEC1-hKOPR binding.
DISCUSSION
Hydrophobic Interactions Mediate GEC1-hKOPR Binding— We found that seven residues, Tyr49, Val51, Leu55, Thr56, Val57, Phe60, and Ile64, spanning from β-strand 2 to α-helix 3, were indispensable for GEC1 to bind to the hKOPR, whereas five other residues, Leu44, Leu50, Tyr61, Arg65, and Tyr109, were involved in the interaction, but to lesser extent. GABARAP-based model of GEC1 demonstrates a hydrophobic patch on the surface of GEC1 formed by Tyr49, Val51, Leu55, Thr56, Phe60, and Ile64 (around the S2 β-strand), which is extended by Leu50 (Fig. 7). Tyr61 and Arg65 formed another small patch. Because of the lipophilic nature of the GEC1-binding sequence in hKOPR C-tail, FPXXM, it is very likely that the interaction resulted from hydrophobic contacts between the two proteins.
Studies on the GABARAP crystal structure also suggest a hydrophobic surface consisting of the residues Ile21, Pro30, Tyr49, Leu50, Val51, Leu55, Phe60, Leu63, and Phe104, which is required for formation of head-to-tail GABARAP oligomer (42). X-ray crystal structure of GATE-16 showed similar exposed hydrophobic patches formed by Ile21, Pro30, Tyr49, Leu50, Val51, Pro52, Ile55, Trp62, Ile63, and Phe104. This is the largest conserved patch of surface between GATE-16 and yeast Atg8 and was predicted to be functionally important in protein-protein interactions (43). Comparison between these three hydrophobic surfaces shows striking conservation of residue components. It is likely that the hydrophobic patches on GABARAP and GATE-16 interact with the lipophilic FPXXM motif in the hKOPR C-tail in a similar manner.
The Y2H data revealed that Leu44 and Tyr109 of GEC1 were important for its interaction with the hKOPR C-tail (Fig. 4), which is likely because of their interaction (Fig. 7E) in maintaining structural integrity. Thus native conformations are essential for GEC1 to bind hKOPR C-tail.
GABARAP and GATE16 Also Enhanced hKOPR Expression— GABARAP and GATE16 up-regulated hKOPR but to lower extents than GEC1. These results indicate that these three proteins are redundant in their functions, at least to some extent. This inference is strongly supported by the finding that deletion of GABARAP in mice did not influence expression and subcellular distribution of the GABAA receptor (44). GEC1 interacts with the γ2 subunit of the GABAA receptor (24) and may have similar effects on this receptor as GABARAP.
GATE16 increased hKOPR expression to a significantly lower extent than GEC1 and GABARAP (Fig. 9). Because the residues in GEC1 critical for binding the KOPR C-tail are highly identical, except Leu55 in GEC1 and Ile55 in GATE16, the difference may stem from dissimilarity in their three-dimensional structures. There are differences between GATE-16 and GABARAP in the putatively flexible C-terminal residues and smaller differences in helix 2 and loop regions (28, 42, 43, 45, 46).
Microtubules Are Involved in GEC1-induced Enhancement in hKOPR Expression—We have demonstrated by pulldown techniques that GEC1 interacts directly with tubulin (10). Mansuy et al. (24) defined the tubulin-binding motif in GEC1 to be the fragment (amino acids 1–22) in the N-terminal domain, similar to the (amino acids 1–27) fragment in GABARAP (42). The x-ray crystal structures of GABARAP, GATE-16, and MAP-LC3 showed that the tubulin-binding domains contain two helices (H1 and H2) (28, 42, 43, 45–47). The regions in all these proteins have high contents of basic residues, which are thought to be important for binding to the highly acidic C-terminal region of tubulin via ionic interaction (48). The observations that GEC1-tubulin binding is salt-sensitive and is abolished in the presence of high salt (400 mm NaCl) are consistent with the ionic nature of GEC1-tubulin interaction (24). GEC1 has been demonstrated to enhance tubulin assembly and microtubules bundling (24).
As shown in Fig. 9, deletion of the first 37 amino acids at the N-terminal region of GEC1 greatly reduced its influence on hKOPR expression. Thus, microtubule-based cytoskeletal structures are important for the effect of GEC1 on hKOPR expression. Similarly, the tubulin binding region of GABARAP is important for its effect on clustering of GABAA receptors (49). Therefore, GEC1 not only interacts with tubulin to promote microtubule bundling but also functions as a linker between hKOPR and cytoskeleton to facilitate anterograde trafficking of the receptor.
Possible Oligomerization of GEC1—The crystal structure of GABARAP indicates there is a head-to-tail GABARAP oligomer formed via interactions between the N-terminal six residues of one molecule and the hydrophobic patch (around the S2 β-strand) of an adjacent molecule (42). Nymann-Andersen et al. (50) found that GABARAP formed homodimers through the amino acids 41–51. These 11 residues are also involved in GABARAP-GABAA receptor interaction (51), and they partially contribute to formation of the highly conserved hydrophobic surface near the S2 β-strand (42, 43). Considering the high degree of homology between GABARAP and GEC1, it is reasonable to postulate that GEC1 may form homodimers or homo-oligomers and heterodimers or hetero-oligomers with GABARAP, which may provide platforms for receptor interactions.
GEC1 Binds to Other Cell Surface Receptors—We defined 347FPXXM351 in the hKOPR C-tail to be critical for interacting with GEC1. Among opioid receptors, only KOPR has this sequence and is the only one of which expression is enhanced by GEC1 (10). Expression of FPXXM-containing AMPA receptor subunit GluR1 and FPXM-containing prostaglandin receptor EP3.f was enhanced by GEC1, but not that of EP3.I, which does not have a similar sequence. These results, coupled with the widespread tissue distribution of GEC1 (14), support the notion that GEC1 may function to promote cell surface expression of many integral membrane proteins that contain FPXXM or a similar sequence in their cytosolic domains.
GEC1 has been shown to bind to the GABAA receptor γ2 subunit (24). The fragment in the GABAA receptor γ2 subunit involved in GABARAP binding was mapped to 394–411 (RTGAWRHGRIHIRIAKMD) in the second intracellular loop of the long form (16). Notably, there is no FPXXM sequence in the GABAA receptor γ2 subunit, in or outside of the GABARAP-binding fragment, suggesting that GEC1 may bind to the γ2 subunit via a different sequence within this fragment or even a different part of the subunit. The binding interface of GEC1 for the GABAA receptor γ2 subunit is likely to involve the hydrophobic patches on the surface. Thus, it can then be inferred that if the interacting protein has a hydrophobic patch that can complement the GEC1 hydrophobic surface, the protein may bind GEC1, regardless whether it has FPXXM sequence or not.
By phage display screening of a randomized peptide library, Mohrluder et al. (52) identified peptides that bound GABARAP. Among the peptides identified, there was a consensus sequence of W(V/I)(F/Y)(V/L)(P/Q). Interestingly, 394–411 of the GABAA receptor γ2 subunit, identified to be the fragment interacting with GABARAP (16), does not contain this sequence. Neither does the KOPR C-tail. These findings further support the notion that hydrophobic interactions, not specific amino acid sequences, are the main driving force for GEC1 and GABARAP to bind their partners.
Conclusion—We have demonstrated that the residues critical for hKOPR binding form a curved hydrophobic patch on the surface of GEC1, which interacts with FPXXM, and that hydrophobic interaction is the major force driving binding between GEC1 and hKOPR C-tail. GEC1 is likely to bind to other molecules by hydrophobic interactions, even molecules without the FPXXM sequence. Our study strongly suggests an expanding set of GEC1-binding cell surface receptors and a broader functional importance than previously expected. Thus, GEC1 may have chaperone-like effects for many molecules. GEC1 mRNA has been shown to be up-regulated by estrogen (11, 53); therefore, GEC1 may be important in sex differences of some biological functions.
Supplementary Material
Acknowledgments
We thank Dr. Richard Huganir and Dr. Barrie Ashby for reagents (see “Experimental Procedures”).
This work was supported, in whole or in part, by National Institutes of Health Grants DA17302 and P30 DA13429. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. I and II.
Footnotes
The abbreviations used are: KOPR, κ opioid receptor; TRK-820, KOPR agonist nalfurafine; hKOPR, human KOPR; GEC1, also named GABAA receptor-associated protein like 1, GABARAPL1; MAP, microtubule-associated protein; GABAA, γ-aminobutyric acid, type A; GABARAP, GABAA receptor-associated protein; GATE16, Golgi-associated ATPase enhancer of 16 kDa; LC3, light chain 3 of MAP 1A/1B; CM, Monte Carlo/simulated annealing method, Conformational Memories; CHO, Chinese hamster ovary; co-IP, co-immunoprecipitation; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazole propionate; Y2H, yeast-two-hybrid; ANOVA, analysis of variance; HA, hemagglutinin; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; 2DO, double dropout; TDO, triple dropout; QDO, quadruple dropout.
References
- 1.Liu-Chen, L.-Y. (2004) Life Sci. 75 511-536 [DOI] [PubMed] [Google Scholar]
- 2.Wikstrom, B., Gellert, R., Ladefoged, S. D., Danda, Y., Akai, M., Ide, K., Ogasawara, M., Kawashima, Y., Ueno, K., Mori, A., and Ueno, Y. (2005) J. Am. Soc. Nephrol. 16 3742-3747 [DOI] [PubMed] [Google Scholar]
- 3.Carlezon, W. A., Jr., Beguin, C., Dinieri, J. A., Baumann, M. H., Richards, M. R., Todtenkopf, M. S., Rothman, R. B., Ma, Z., Lee, D. Y., and Cohen, B. M. (2006) J. Pharmacol. Exp. Ther. 316 440-447 [DOI] [PubMed] [Google Scholar]
- 4.Knoll, A. T., Meloni, E. G., Thomas, J. B., Carroll, F. I., and Carlezon, W. A., Jr. (2007) J. Pharmacol. Exp. Ther. 323 838-845 [DOI] [PubMed] [Google Scholar]
- 5.Beardsley, P. M., Howard, J. L., Shelton, K. L., and Carroll, F. I. (2005) Psychopharmacology 183 118-126 [DOI] [PubMed] [Google Scholar]
- 6.Law, P.-Y., Wong, Y. H., and Loh, H. H. (2000) Annu. Rev. Pharmacol. Toxicol. 40 389-430 [DOI] [PubMed] [Google Scholar]
- 7.Li, J.-G., Chen, C., and Liu-Chen, L.-Y. (2002) J. Biol. Chem. 277 27545-27552 [DOI] [PubMed] [Google Scholar]
- 8.Jordan, B. A., and Devi, L. A. (1999) Nature 399 697-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, P., Steplock, D., Weinman, E. J., Hall, R. A., Ding, Z., Li, J., Wang, Y., and Liu-Chen, L.-Y. (2004) J. Biol. Chem. 279 25002-25009 [DOI] [PubMed] [Google Scholar]
- 10.Chen, C., Li, J. G., Chen, Y., Huang, P., Wang, Y., and Liu-Chen, L. Y. (2006) J. Biol. Chem. 281 7983-7993 [DOI] [PubMed] [Google Scholar]
- 11.Pellerin, I., Vuillermoz, C., Jouvenot, M., Ordener, C., Royez, M., and Adessi, G. L. (1993) Mol. Cell. Endocrinol. 90 R17-R21 [DOI] [PubMed] [Google Scholar]
- 12.Xin, Y., Yu, L., Chen, Z., Zheng, L., Fu, Q., Jiang, J., Zhang, P., Gong, R., and Zhao, S. (2001) Genomics 74 408-413 [DOI] [PubMed] [Google Scholar]
- 13.Nemos, C., Mansuy, V., Vernier-Magnin, S., Fraichard, A., Jouvenot, M., and Delage-Mourroux, R. (2003) Brain Res. Mol. Brain Res. 119 216-219 [DOI] [PubMed] [Google Scholar]
- 14.Wang, Y., Dun, S. L., Huang, P., Chen, C., Chen, Y., Unterwald, E. M., Dun, N. J., Van Bockstaele, E. J., and Liu-Chen, L. Y. (2006) Neuroscience 140 1265-1276 [DOI] [PubMed] [Google Scholar]
- 15.Mansuy-Schlick, V., Tolle, F., Delage-Mourroux, R., Fraichard, A., Risold, P. Y., and Jouvenot, M. (2006) Brain Res. 1073 83-87 [DOI] [PubMed] [Google Scholar]
- 16.Wang, H., Bedford, F. K., Brandon, N. J., Moss, S. J., and Olsen, R. W. (1999) Nature 397 69-72 [DOI] [PubMed] [Google Scholar]
- 17.Kittler, J. T., Rostaing, P., Schiavo, G., Fritschy, J. M., Olsen, R., Triller, A., and Moss, S. J. (2001) Mol. Cell. Neurosci. 18 13-25 [DOI] [PubMed] [Google Scholar]
- 18.Sagiv, Y., Legesse-Miller, A., Porat, A., and Elazar, Z. (2000) EMBO J. 19 1494-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leil, T. A., Chen, Z. W., Chang, C. S., and Olsen, R. W. (2004) J. Neurosci. 24 11429-11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Z. W., and Olsen, R. W. (2007) J. Neurochem. 100 279-294 [DOI] [PubMed] [Google Scholar]
- 21.Kanematsu, T., Mizokami, A., Watanabe, K., and Hirata, M. (2007) J. Pharmacol. Sci. 104 285-292 [DOI] [PubMed] [Google Scholar]
- 22.Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., Noda, T., and Ohsumi, Y. (2000) Nature 408 488-492 [DOI] [PubMed] [Google Scholar]
- 23.Kabeya, Y., Mizushima, N., Yamamoto, A., Oshitani-Okamoto, S., Ohsumi, Y., and Yoshimori, T. (2004) J. Cell Sci. 117 2805-2812 [DOI] [PubMed] [Google Scholar]
- 24.Mansuy, V., Boireau, W., Fraichard, A., Schlick, J. L., Jouvenot, M., and Delage-Mourroux, R. (2004) Biochem. Biophys. Res. Commun. 325 639-648 [DOI] [PubMed] [Google Scholar]
- 25.Li, J., Li, J.-G., Chen, C., Zhang, F., and Liu-Chen, L.-Y. (2002) Mol. Pharmacol. 61 73-84 [DOI] [PubMed] [Google Scholar]
- 26.Zhang, F., Li, J., Li, J.-G., and Liu-Chen, L.-Y. (2002) J. Pharmacol. Exp. Ther. 302 1184-1192 [DOI] [PubMed] [Google Scholar]
- 27.Soellick, T. R., and Uhrig, J. F. (2001) Genome Biol. 2 RESEARCH0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight, D., Harris, R., McAlister, M. S., Phelan, J. P., Geddes, S., Moss, S. J., Driscoll, P. C., and Keep, N. H. (2002) J. Biol. Chem. 277 5556-5561 [DOI] [PubMed] [Google Scholar]
- 29.Marti-Renom, M. A., Stuart, A. C., Fiser, A., Sanchez, R., Melo, F., and Sali, A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29 291-325 [DOI] [PubMed] [Google Scholar]
- 30.Notredame, C., Higgins, D. G., and Heringa, J. (2000) J. Mol. Biol. 302 205-217 [DOI] [PubMed] [Google Scholar]
- 31.Mohamadi, F., Richards, N. G. J., Guida, W. C., Liskamp, R., Lipton, M., Caufield, C., Chang, G., Hendrickson, T., and Still, W. C. (1990) J. Comput. Chem. 11 440-467 [Google Scholar]
- 32.Kale, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K., and Schulten, K. (1999) J. Comput. Phys. 151 283-312 [Google Scholar]
- 33.Feller, S. E., and MacKerell, A. D., Jr. (2000) J. Phys. Chem. B 104 7510-7515 [Google Scholar]
- 34.MacKerell, A. D., Jr., Bashford, D., Bellot, M., Dunbrack, R. L., Jr., Evanseck, J., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., Joseph, D., Kuchnir, L., Kuczera, K., Lau, F. T. K., Mattos, C., Michnick, S., Ngo, T., Nguyen, D. T., Prodhom, B., Reiher, I. W. E., Roux, B., Schlenkrich, M., Smith, J., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz-Kuczera, J., Yin, D., and Karplus, M. (1998) J. Phys. Chem. B 102 3586-3616 [DOI] [PubMed] [Google Scholar]
- 35.Schlenkrich, M., Brickmann, J., MacKerell, A. D. Jr., and Karplus, M. (1996) in Biological Membranes: A Molecular Perspective from Computation and Experiment (Merz, K. M., and Roux, B., eds) Birkhauser Boston, Inc., Cambridge, MA
- 36.Darden, T. A., York, D., and Pedersen, L. (1993) J. Chem. Phys. 98 10089-10092 [Google Scholar]
- 37.Barnett-Norris, J., Hurst, D. P., Buehner, K., Ballesteros, J. A., Guarnieri, F., and Reggio, P. H. (2002) Int. J. Quantum Chem. 88 76-86 [Google Scholar]
- 38.Guarnieri, F., and Weinstein, H. (1996) J. Am. Chem. Soc. 118 5580-5589 [Google Scholar]
- 39.Whitnell, R. M., Hurst, D. P., Reggio, P. H., and Guarnieri, F. (2008) J. Comput. Chem. 29 741-752 [DOI] [PubMed] [Google Scholar]
- 40.Hassan, S. A., Guarnieri, F., and Mehler, E. L. (2000) J. Phys. Chem. B 104 6478-6489 [Google Scholar]
- 41.Hassan, S. A., and Mehler, E. L. (2001) Int. J. Quantum Chem. 83 193-202 [Google Scholar]
- 42.Coyle, J. E., Qamar, S., Rajashankar, K. R., and Nikolov, D. B. (2002) Neuron 33 63-74 [DOI] [PubMed] [Google Scholar]
- 43.Paz, Y., Elazar, Z., and Fass, D. (2000) J. Biol. Chem. 275 25445-25450 [DOI] [PubMed] [Google Scholar]
- 44.O'Sullivan, G. A., Kneussel, M., Elazar, Z., and Betz, H. (2005) Eur. J. Neurosci. 22 2644-2648 [DOI] [PubMed] [Google Scholar]
- 45.Bavro, V. N., Sola, M., Bracher, A., Kneussel, M., Betz, H., and Weissenhorn, W. (2002) EMBO Rep. 3 183-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stangler, T., Mayr, L. M., and Willbold, D. (2002) J. Biol. Chem. 277 13363-13366 [DOI] [PubMed] [Google Scholar]
- 47.Sugawara, K., Suzuki, N. N., Fujioka, Y., Mizushima, N., Ohsumi, Y., and Inagaki, F. (2004) Genes Cells 9 611-618 [DOI] [PubMed] [Google Scholar]
- 48.Nogales, E. (2000) Annu. Rev. Biochem. 69 277-302 [DOI] [PubMed] [Google Scholar]
- 49.Wang, H., and Olsen, R. W. (2000) J. Neurochem. 75 644-655 [DOI] [PubMed] [Google Scholar]
- 50.Nymann-Andersen, J., Wang, H., and Olsen, R. (2002) Neuropharmacology 43 476. [DOI] [PubMed] [Google Scholar]
- 51.Nymann-Andersen, J., Wang, H., Chen, L., Kittler, J. T., Moss, S. J., and Olsen, R. W. (2002) J. Neurochem. 80 815-823 [DOI] [PubMed] [Google Scholar]
- 52.Mohrluder, J., Stangler, T., Hoffmann, Y., Wiesehan, K., Mataruga, A., and Willbold, D. (2007) FEBS J. 274 5543-5555 [DOI] [PubMed] [Google Scholar]
- 53.Malyala, A., Pattee, P., Nagalla, S. R., Kelly, M. J., and Ronnekleiv, O. K. (2004) Neurochem. Res. 29 1189-1200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.