Abstract
The integrin α6β4 is associated with carcinoma progression by contributing to apoptosis resistance, invasion, and metastasis, due in part to the activation of select transcription factors. To identify genes regulated by the α6β4 integrin, we compared gene expression profiles of MDA-MB-435 cells that stably express integrin α6β4 (MDA/β4) and vector-only-transfected cells (MDA/mock) using Affymetrix GeneChip® analysis. Our results show that integrin α6β4 altered the expression of 538 genes (p < 0.01). Of these genes, 36 are associated with pathways implicated in cell motility and metastasis, including S100A4/metastasin. S100A4 expression correlated well with integrin α6β4 expression in established cell lines. Suppression of S100A4 by small interference RNA resulted in a reduced capacity of α6β4-expressing cells to invade a reconstituted basement membrane in response to lysophosphatidic acid. Using small interference RNA, promoter analysis, and chromatin immunoprecipitation, we demonstrate that S100A4 is regulated by NFAT5, thus identifying the first target of NFAT5 in cancer. In addition, several genes that are known to be regulated by DNA methylation were up-regulated dramatically by integrin α6β4 expression, including S100A4, FST, PDLIM4, CAPG, and Nkx2.2. Notably, inhibition of DNA methyltransferases stimulated expression of these genes in cells lacking the α6β4 integrin, whereas demethylase inhibitors suppressed expression in α6β4 integrin-expressing cells. Alterations in DNA methylation were confirmed by bisulfate sequencing, thus suggesting that integrin α6β4 signaling can lead to the demethylation of select promoters. In summary, our data suggest that integrin α6β4 confers a motile and invasive phenotype to breast carcinoma cells by regulating proinvasive and prometastatic gene expression.
Integrins are receptors for the extracellular matrix, which have two major functions. The first is an adhesive function that secures cells to the surrounding extracellular matrix or, in the case of cell motility, provides traction for locomotion. Their second function is to transduce signals that are essential for cells to sense and integrate cues from the extracellular matrix, which include signals for directed cell motility, anchorage-dependent survival, and growth (1). As a result, integrin signaling and function are critical for most biological events in higher eukaryotes, both under normal and pathological conditions. In recent years, one integrin species, the α6β4 integrin, has garnered much attention for its ability to promote an invasive and metastatic phenotype in carcinomas.
In cells of epithelial origin, the integrin α6β4 nucleates the formation of hemidesmosomes that link the cytokeratin cytoskeleton to the laminins found in the basement membrane, which are essential for epithelial integrity (2). During wound healing or the epithelial to mesenchymal transition (EMT),2 the α6β4 integrin is phosphorylated, is released from hemidesmosomes, and then binds the actin cytoskeleton (3). Under these conditions, the α6β4 integrin promotes cell motility (4). Increased expression of the α6β4 integrin is a poor prognostic factor for breast cancer (5, 6) as well as various solid tumors (7, 8) and is associated with an invasive (9, 10) and metastatic phenotype (11). Exogenous expression of the α6β4 integrin in MDA-MB-435 cells substantially increased the ability of these cells to form lamellae, polarize, migrate (12), and invade a reconstituted basement membrane (Matrigel) (10). Importantly, these observations have been extended to the MDA-MB-231 (13) and Sum159 (14) cell lines and have been validated in vivo in the ErbB2 breast cancer mouse model, where targeted deletion of the β4 subunit reduces tumor invasion and progression (15).
Tumor invasion can be controlled by a number of factors. A growing list of these factors converge on the α6β4 integrin to mediate an invasive phenotype, including androgen independence (16), p63 expression (17), and c-Met receptor signaling (18–20). Dissecting the signaling pathways enhanced by α6β4 has revealed that the α6β4 integrin promotes the signaling from several proinvasive molecules (21). Of particular interest here is the observation that the α6β4 integrin can affect gene transcription through the activation of proinvasive transcription factors, such as nuclear factor of activated T-cells (NFAT) (22), NFκB (23, 24), and AP-1 (15). To determine the effect of integrin α6β4 on gene expression, we performed Affymetrix GeneChip® analysis on MDA-MB-435 clones that stably express the α6β4 integrin and compared these cells to vector-only-transfected clones. We find that several hundred genes are regulated by integrin α6β4 by more than 2-fold (99% confidence level). Of these genes, autotaxin/ENPP2 (25) and S100A4/metastasin, genes associated with breast cancer metastasis, are highly up-regulated. Here, we examine how the α6β4 integrin controls the expression of S100A4 and how this regulation extends to other genes.
EXPERIMENTAL PROCEDURES
Cell Lines, Immunoblotting, and Reagents—MDA-MB-435 breast carcinoma cells that were stably transfected with vector only (MDA/mock, clones 6D2 and 6D7) or the integrin β4 subunit cDNA (MDA/β4, clones 5B3 and 3A7) were obtained from Arthur M. Mercurio (University of Massachusetts Medical School, Worchester, MA) (10); MDA-MB-468 and BT-20 cells from Janet Price (University of Texas M. D. Anderson Cancer Center, Houston, TX); and all other breast cancer cell lines from ATCC. Cells were cultured as described previously (12, 26). For all studies, cells were given fresh growth medium the day prior to harvest and harvested at 70% confluence. For stable knockdown of β4 integrin expression, cells were stably transfected with pLKO.1-puro lentiviral constructs (Sigma) containing one of two short hairpin RNAs targeting β4 (number 4, CCGGGAGGGTGTCATCACCATTGAACTCGAGTTCAATGGTGATGACACCCTCTTTTTG; number 5, CCGGCGAGGTCACATGGTGGGCTTTCTCG AGAAAGCCCACCATGTGACCTCGTTTTTG) or a control sequence (number 2, CCGGCCCATGAAGAAAGTGCTGGTTCTCGAGAACCAGCACTTTCTTCATGGGTTTTTG).
For DNA methylation studies, cells were treated with 0.1 or 1 μm 5-aza-2′-deoxycytidine (DAC) or 80 μm S-adenosylmethionine (SAM) in fresh medium daily, as noted. Tricostatin A (1 μm) treatment was given 24 h before harvest. For protein analysis, cells were harvested using radioimmune precipitation buffer (150 mm NaCl, 0.5 mm EGTA, 0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 50 mm Tris-HCl, pH 7.4) containing 15 μg/ml protease inhibitor mixture (Sigma) and 1 mm phenylmethanesulfonyl fluoride. Total protein was then electrophoresed on a gel with the appropriate percentage of acrylamide (SDS-PAGE, reducing conditions), transferred to nitrocellulose, blocked with 5% nonfat dry milk, and probed with the indicated antibody. Stripping solution (Pierce) was utilized to clear antibodies for reprobing membranes. Antibodies used in this study are myristoylated alanine-rich C kinase substrate (MARCKS; catalog number sc-6454; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), CAPG (capping protein G; Genway; catalog number A22527), Elmo (catalog number sc20965; Santa Cruz Biotechnology), integrin β4 clone 439-9B (Chemicon-Millipore), integrin α6 clone GoH3 (Chemicon-Millipore), NFAT1 (catalog number sc-7296; Santa Cruz Biotechnology), NFAT5 (catalog number PA1-023; Affinity Bio-Reagents), and S100A4 (27). Actin (catalog number A5441; Sigma) was used as a loading control.
RNA Isolation and Affymetrix GeneChip Analysis—For routine quantitative real-time PCR (Q-PCR) analysis, RNA was extracted from cells using TRIzol reagent (Invitrogen), purity was confirmed by OD 260:280 ratio, and RNA was analyzed using 0.7% agarose with formaldehyde gel electrophoresis. For GeneChip analysis, the RNAqueous kit from Ambion was used for RNA purification. Gene profiling was performed using Affymetrix Human Genome U133A and -B oligonucleotide arrays. Final statistical analyses are reported only on the U133A chip.
Bioinformatic Processing of Affymetrix GeneChip Data—The results from the Affymetrix GeneChip arrays were processed using the statistical package S-PLUS® Array Analyzer (Insightful Corp., Seattle, WA) (S-PLUS). Expression of each gene was determined by first converting the raw probe level intensities to expression summaries by correcting overall background and probe-specific background prior to normalization. Normalization was performed using G-C content Robust Multichip Analysis, as published previously (28). Differential expression testing was determined using the local pooled error test, a statistical test designed for low replicates (three to five replicates), to determine genes differentially expressed to a 99% confidence level. Multitest comparison tests were then performed using both the Benjamin and Hochberg and the more stringent Bonferroni methods of filtering out false positives.
Q-PCR—cDNA was prepared using the SuperScript first strand synthesis system for reverse transcription-PCR (Invitrogen) prior to Q-PCR analysis. Expression of various genes was then assessed using ABI70000 sequence detector, reagents, and commercially available probes, as described by the manufacturer (Applied Biosystems). The expression level of each gene was normalized by 18 S RNA and reported as relative level.
Fluorescence-activated Cell Sorting (FACS)—Suspended cells were treated with 1 μg of primary antibody for 30 min at room temperature, rinsed three times with phosphate-buffered saline, incubated with Cy2-conjugated goat anti-mouse IgG (Jackson Immunoresearch), rinsed with phosphate-buffered saline, and then analyzed on a FACS Canto analytical cell sorter (BD Biosciences). As a control, primary antibody was omitted. Data are reported as the -fold difference in mean fluorescence compared with secondary antibody alone control for that cell line.
Invasion Assays—Matrigel (5 μg; BD Biosciences) was dried onto the upper side of Transwell chambers (6.5-mm diameter, 8-μm pore size; Corning Glass). One hour before the assay, Matrigel was rehydrated with Dulbecco's modified Eagle's medium, and the bottom surface was coated with 10 μg/ml laminin-1. 100 nm LPA or 50 ng/ml HGF in Dulbecco's modified Eagle's medium/bovine serum albumin or Dulbecco's modified Eagle's medium/bovine serum albumin was added to the lower chamber. Cells (5 × 104) were placed in the top chamber and allowed to invade for 4 h. Noninvading cells in the top chamber were removed using a cotton swab, and cells in the bottom chambers were fixed with methanol, stained with 1% crystal violet, and quantified visually. Values for triplicate membranes are reported as a mean ± S.D., as described previously (12).
Small Interference RNA Treatment—Cells from 70% confluent cultures were suspended by trypsinization and rinsed three times with Dulbecco's modified Eagle's medium. Cells (3 × 106) were electroporated with 200 nm siRNAs specific for an individual target or a control (nontargeting) sequence (Dharmacon, Inc.), as reported previously (25). Individual sequences for NFAT5 are CAACAUGCCUGGAAUUCAAUU (sequence 3) and CAGAGUCAGUCCACAGUUUUU (sequence 5). Dharmacon SMARTPool siRNAs were used for all other targets. Cells were then kept in normal growth medium for 24–96 h and then assessed for target gene expression using Q-PCR and immunoblot analysis as indicated.
Chromatin Immunoprecipitation Assays—Cells under normal culturing conditions were cross-linked with 1% formaldehyde, which was then terminated with 0.125 m glycine. Nuclei were isolated, sonicated to fragment DNA (average length of 500–700 bp), and centrifuged to pellet debris. Extracts were incubated with 1 μg of control rabbit IgG or anti-NFAT5 rabbit polyclonal Ab and protein A/G-Sepharose beads (Amersham Biosciences) at 4 °C overnight. Washed immunoprecipitates were digested with proteinase K, followed by a 65 °C incubation to reverse the cross-linking. DNA was then purified and assessed for the S100A4 promoter using PCR and the following primers: GAGATCCAGATGTGAGATTC (+208/+227) and GGGTTGGAAGAGAAGCTGCA (+565/+584).
Bisulfate Sequencing—Identification of methylated CpG residues was determined by bisulfate conversion and pyrosequencing of the first intron region of the S100A4 promoter (+203 to +662; accession number Z33457). This procedure was performed by EpigenDx.
RESULTS
NFAT and AP-1 are transcription factors that can promote tumor invasion that are known to signal downstream from the α6β4 integrin (15, 22). However, the extent of the changes in gene transcription and what genes are altered as a result of integrin α6β4 signaling have not been established. We hypothesized that the α6β4 integrin can regulate the expression of genes that can promote a motile and invasive phenotype. To test this hypothesis, we performed Affymetrix GeneChip® analysis on MDA-MB-435 clones that were mock-transfected (MDA/mock, clones 6D2 and 6D7) or stably transfected with the β4 integrin (MDA/β4, clones 3A7 and 5B3). Two sets of RNA were prepared for each clone on separate occasions and processed for Affymetrix GeneChip® analysis (n = 4 for each condition, +α6β4 or –α6β4). The results were then processed using the statistical package S-Plus Array Analyzer (S-PLUS), as published previously (28). Differential expression testing was determined using the local pooled error test, a statistical test designed for low replicates (three to five replicates), to determine genes differentially expressed to a 99% confidence level. Multitest comparison tests were performed using both the Benjamin and Hochberg and Bonferroni methods of filtering out false positives. Concentrating on the data from the Affymetrix HG-U133A chip, we found that 538 genes are regulated by integrin α6β4 expression using the local pooled error t test with Benjamin and Hochberg corrections and 239 genes using the Bonferroni corrections. A partial list of these genes is found in Table 1, with the full list of genes located in Tables S1 and S2.
TABLE 1.
Genes regulated by the expression of the α6β4 integrin in MDA-MB-435 cells
Gene symbols in boldface type denote genes that are found in more than one category.
| Function | Number | Up-regulated | Down-regulated |
|---|---|---|---|
| Motility | 36 | SFRP1, MARCKS, ENPP2, CAPG, FST, PTPRZ1, PDLIM4, S100A4, ARHGEF3, CHL1, KIF5C, MYH10, CTGF, FSCN1, SEMA3A, PTPN22, ELMO1, NRCAM, EDG2, HMGCR | MYLK, FOS, MAP1B, ADM, IL8, S100A2, EGR1, FGF13, RAC2, LIF, GADD45A, ITGB1BP1, FOSL1, SDC1, Cortactin, CYR61 |
| Apoptosis | 56 | SFRP1, CRYAB, FST, PEG10, S100A4, MAP2K6, DMD, HDAC4, GPR37, DNAJB6, RAD51C, CHL1, SATB1, QKI, IVNS1ABP, CTGF, TXNIP, TPD52L1, SEMA3A, SGCD, NRCAM, PRKD1, HELLS, EDG2, CFDP1, PTGER4 | BCL2A1, CSNK1E, GADD45B, CCND1, RIPK2, SPP1, BCL6, FOS, MYLK, TGFBI, TNFAIP3, MAP1B, ADM, IL8, FN1, EGR1, MMP1, THBS1, RAC2, LIF, GADD45A, FOSL1, CARD10, NGFRAP1, IGFBP7, DKK1, UCP2, CEBPD, CYR61, IL7R |
| Transcription regulation | 10 | SOX4, NKX2-2, HDAC4, TCFL5, CSRP2, TRIM9, MEF2C | FOS, FOSL1, CEBPD |
| Extracellular matrix, cell adhesion, cytoskeleton | 32 | CHRDL1, SPARCL1, DSG2, COL9A3, DMD, HAPLN1, HS3ST3A1, TUBB6, COL6A2, CHL1, ANK2, COL1A2, CALD1, FSCN1, TUBB4, ITGA10, NRCAM, COL5A2, CDH19 | MUC1, TNC, EVL, MCAM, FN1, THBS1, RAC2, TUBA1, SDC1, CDC42EP1, MYL9, COL13A1, CYR61 |
| Proteases, inhibitors | 9 | ADAMTS1, PRSS11, MMP14, CAPN3, CTSB | MMP1, PRSS7, SERPINB1, SERPINA3 |
| Metabolism | 29 | TYRP1, MAOA, BCHE, DCT, ENO2, AKR1C1, AKR1C2, AKR1C3, ASPA, GALNT11, ALDH1A1, TDG, EPHX1, LOXL2, AGPAT5, TNKS, CA8, NMT2, MRPS6, SLC5A3, ATP5I, MTUS1 | ASNS, KYNU, PDE2A, GLUL, GSTT2, WARS, NNMT |
| Other signaling molecules | 17 | PHACTR1, EPHA3, PRKCBP1, PTPLA, BMP1, FZD7, SKP2 | AKAP2, SHB, RAB27A, PPM2C, IL1RAP, KCNS3, KDELR3, DUSP5, P2RX5, KCTD14 |
| Antigen presentation | 8 | HLA-B, HLA-C, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB4 | |
| Protein stability | 8 | CRYAB, DNAJB6, DNAJC12, USP9X, HSPA1A | UBE2H, SMURF2, TMAP1 |
We identified several classes of genes that are altered by expression of the α6β4 integrin, including extracellular matrix and cell adhesion, transcription, metabolism, antigen presentation, and protein stability, including several ubiquitin ligases. The integrin α6β4 has been well documented to promote tumor cell invasion and survival. Accordingly, genes governing these processes represent the major classes of genes that are altered by integrin α6β4 expression. These include 36 genes reported to affect cell motility and 56 genes that control apoptosis and cell survival, such as the down-regulation of the death matrix CYR61 (Table 1). Here, we chose the genes associated with cell motility, invasion, and metastasis for further investigation.
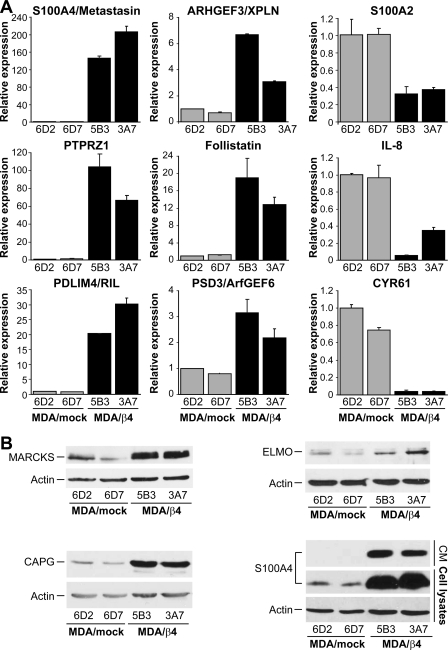
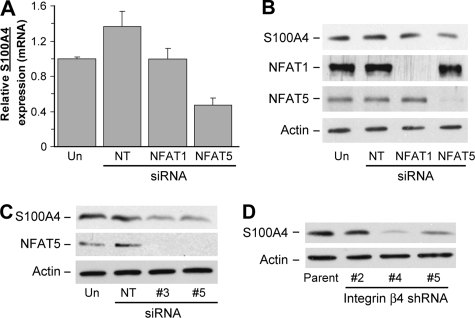
As shown in Fig. 1, we validated changes in expression for several of the genes regulated by integrin α6β4 by Q-PCR and/or immunoblot analysis. A comparison of the -fold differences from the Q-PCR results with those computed from the Affymetrix/Ingenuity pathway analysis (Table S3) shows that the data from the GeneChips are in close agreement with our Q-PCR and immunoblot results, although very high inductions are generally underrepresented in the GeneChip analyses. Of those genes, the most highly up-regulated are S100A4, PTPRZ1, PDLIM4, CAPG, and FST (follistatin).
FIGURE 1.
Analysis of MDA-MB-435 clones for select genes altered by α6β4 integrin expression. Total RNA (A) or protein (B) was isolated from the MDA-MB-435 clones 6D2 and 6D7 (MDA/mock; null for the β4 subunit) and 5B3 and 3A7 (MDA/β4; expressing the α6β4 integrin) and submitted for Q-PCR assessment of the indicated genes or immunoblot analysis for the indicated protein, respectively. For extracellular S100A4 (B), conditioned media (CM) represents 50 μl of growth medium removed from cultures just prior to harvesting the cells for protein. For Q-PCR, expression was normalized to 18 S rRNA levels and reported as a value relative to the clone 6D2. Values represent the mean ± S.D.
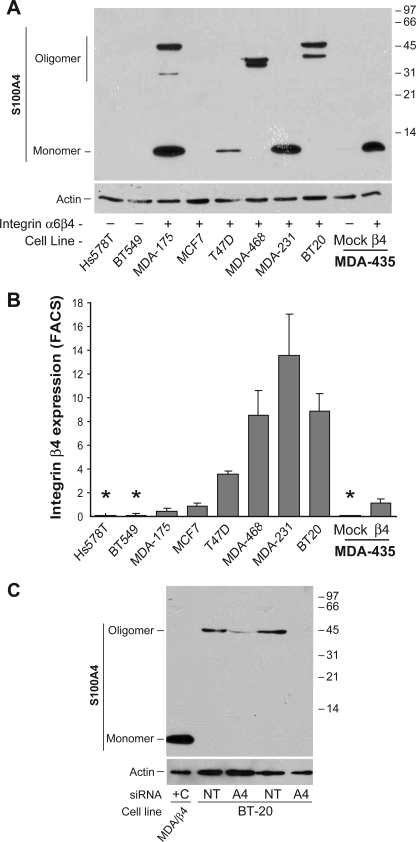
S100A4, also known as fibroblast-specific protein or metastasin, is a metastasis-associated protein documented to promote the metastatic process in several types of cancer, including breast, gastric, pancreatic, and thyroid cancers (29). As shown in Fig. 1, S100A4 expression in the β4-expressing cells is over 140-fold higher than nonexpressers, as determined by Q-PCR. This overexpression extends to both increased intracellular and extracellular protein levels. To determine if S100A4 expression correlated with integrin α6β4, we assessed various breast carcinoma cell lines for S100A4 by immunoblot analysis (Fig. 2A) and the cell surface expression of the β4 integrin by FACS (Fig. 2B). Notably, expression of S100A4 is found in all cell lines that express the α6β4 integrin with the exception of MCF7 cells. Of these cells, MCF7 is the only cell line that does not display a mesenchymal phenotype. Interestingly, some cell lines showed higher molecular weight bands that run at a molecular mass equivalent to a trimer (∼35 kDa) or tetramer (∼47 kDa) of S100A4. To confirm conclusively that these bands are specific for S100A4, we electroporated BT-20 cells with siRNA specific for S100A4 or a nontargeting control prior to immunoblot analysis for S100A4. As shown in Fig. 2B, the S100A4 siRNA effectively reduced the expression of the higher molecular weight bands, thus confirming that these bands represent S100A4. Therefore, these data demonstrate that expression of S100A4 correlates well with expression of integrin α6β4.
FIGURE 2.
S100A4 expression correlates with integrin α6β4 expression. A, the indicated breast carcinoma cell lines and MDA-MB-435 clones were harvested at 70% confluence under normal culturing conditions. Cleared whole cell lysates were submitted to SDS-PAGE and immunoblotted for S100A4 (top) or actin (bottom). B, cells were assessed for β4 integrin content by FACS analysis. Data are reported as the average fold difference in mean fluorescence as compared with secondary antibody-only control ± S.D. from three separate experiments. *, cell lines also determined to be negative for β4 integrin expression by immunoblot analysis (data not shown). C, BT-20 cells were electroporated with 200 nm siRNA specific for S100A4 (A4) or nontargeting siRNA (NT), as noted, and then cultured for 48 h. Cells were then harvested, and cell lysates were immunoblotted for S100A4. Cell extract from a MDA/β4 transfectant serves as a positive control for the monomeric form (+C).
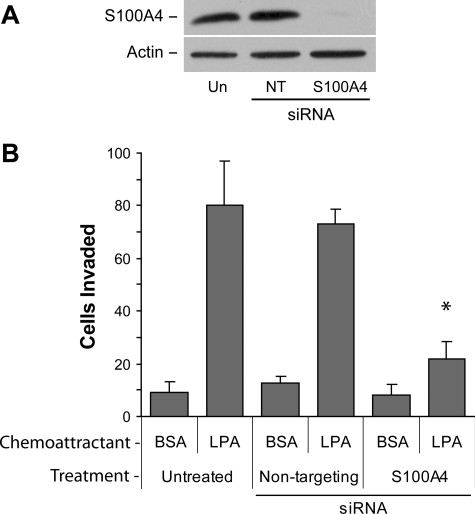
Next we sought to determine whether S100A4 contributes to the invasive phenotype mediated by the α6β4 integrin. For these experiments, we utilized MDA-MB-231, which are highly invasive breast carcinoma cells that have been previously shown to utilize the α6β4 integrin for chemoinvasion. S100A4 expression was suppressed using specific siRNAs, and the chemoinvasion of treated cells to nontargeting or untreated cells was compared. LPA and HGF, two chemoattractants that cooperate with the α6β4 integrin (12, 18, 19), were used as chemoattractants. The loss of S100A4 expression (Fig. 3A) reduced the invasion of MDA-MB-231 cells toward LPA by ∼70% compared with untreated or nontargeting siRNA-transfected cells (Fig. 3B). Similar results were obtained with HGF (data not shown). These data indicate that S100A4 is important for tumor cell invasion, an activity that can predispose cells for metastasis.
FIGURE 3.
S100A4 is important for chemoinvasion of breast carcinoma cells. MDA-MB-231 cells were electroporated with nothing (Un), nontargeting siRNA (NT), or siRNA targeting S100A4. After 48 h, cells were assessed for S100A4 expression by immunoblot analysis (A) or chemoinvasion toward 100 nm LPA (B) as described under “Experimental Procedures.” *, p < 0.002 for treated compared with untreated control and p < 0.0001 for treated compared with nontarget control. BSA, bovine serum albumin.
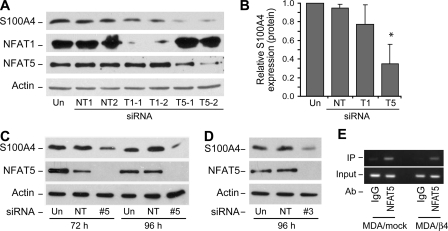
Although the biochemical mechanisms governing how S100A4 contributes to an invasive and metastatic phenotype are becoming clear (27, 29), how S100A4 expression is regulated on the transcriptional level is poorly understood. Using bioinformatic analysis of the promoter region, we find that the S100A4 promoter contains multiple NFAT consensus binding sites. Since NFAT is known to function downstream of the α6β4 integrin, we tested the potential role of NFAT1 and NFAT5 in the regulation of S100A4 expression using specific siRNAs to target their down-regulation in the MDA/β4 transfectants. As shown in Fig. 4, A and B, effective silencing of NFAT5, but not NFAT1, by specific siRNAs reduced S100A4 expression in the MDA/β4 cells. The reduction in S100A4 expression due to loss of NFAT5 was confirmed using two individual siRNAs to target NFAT5 (Fig. 4, C and D). Of note, reduction of NFAT1 expression with the siRNA used here was shown previously to reduce autotaxin expression (25). Using a similar approach, we find that PTPRZ1, but not Nkx2.2, is also an NFAT5 target gene (data not shown).
FIGURE 4.
NFAT5, but not NFAT1, controls the transcriptional regulation of S100A4 in MDA/β4 cells. A and B, MDA/β4 clone 5B3 cells were left untreated (Un) or transfected with either 200 nm (1) or 400 nm (2) Dharmacon siRNA SMARTPools that are nontargeting (NT) or directed against either NFAT1 (T1) or NFAT5 (T5). After 48 h, cell lysates were harvested and immunoblotted for S100A4, NFAT1, NFAT5, and actin, as indicated (A). Blots from two separate experiments were quantified by densitometry and averaged (B). Bars in B, mean expression ± S.D. *, p value < 0.05 compared with either untreated or nontargeting controls. C and D, MDA/β4 cells were treated with individual siRNAs targeting NFAT5 for 72 or 96 h, and then cell lysates were immunoblotted for S100A4, NFAT5, and actin. E, MDA/mock and MDA/β4 cells under normal culturing conditions were cross-linked with formaldehyde. Nuclei were then isolated, DNA was fragmented, and NFAT5-containing chromatin was immunoprecipitated (IP). The S100A4 promoter associated with NFAT5 was then amplified as described under “Experimental Procedures” and compared with an IgG control.
To verify the results with the MDA/β4 cells, we treated MDA-MB-231 cells, which endogenously express the α6β4 integrin, with siRNA targeting NFAT1, NFAT5, or a nontargeting siRNA. Previous studies demonstrated that MDA-MB-231 cells exhibit α6β4 integrin-dependent migration and invasion (13), a process facilitated by NFAT molecules (22). We find that S100A4 expression is reduced by siRNA targeting of NFAT5 in the MDA-MB-231 cells but not by targeting NFAT1 using both Q-PCR (Fig. 5A) and immunoblot analysis (Fig. 5B). These data were confirmed using single duplexes targeting NFAT5 as performed with the MDA/β4 cells (Fig. 5C). To confirm the role of integrin α6β4 in mediating S100A4 expression, we stably transfected MDA-MB-231 cells with commercially available lentiviral short hairpin RNA constructs targeting the β4 integrin subunit. Constructs 4 and 5, which reduce the cell surface expression of β4 integrin by 3- and 2-fold, respectively (data not shown), significantly decreased S100A4 expression (Fig. 5D). This is in contrast with construct 2, which was unable to reduce β4 integrin expression. Collectively, these data indicate that integrin α6β4 expression leads to the NFAT5-dependent transcriptional up-regulation of S100A4.
FIGURE 5.
S100A4 expression is controlled by NFAT5 and integrinα6β4 in MDA-MB-231 cells. A and B, MDA-MB-231 cells were left untreated (Un) or transfected with 200 nm siRNA SMARTPools that are nontargeting (NT) or directed against either NFAT1 or NFAT5. Duplicate cells cultures were then harvested 48 h later and analyzed by Q-PCR for S100A4 mRNA expression (A) or protein expression by immunoblot analysis (B). Blot was stripped and reprobed for NFAT1, NFAT5, and actin. For Q-PCR, p values for NFAT5 samples compared with untreated or nontargeting controls were <0.001. C, MDA-MB-231 cells were treated with individual siRNAs targeting NFAT5 for 72 h, and then cell lysates were immunoblotted for S100A4, NFAT5, and actin. D, MDA-MB-231 cells were stably transfected with lentiviral short hairpin RNA constructs that target the integrin β4 subunit (#4 and #5) or that were ineffective in reducing β4 expression (#2). S100A4 expression of these cell populations was compared with the parental cell line by immunoblot analysis. Reduction in integrin β4 expression by short hairpin RNA #4 and #5 was confirmed by FACS analysis (data not shown).
To determine definitively whether NFAT5 binds the S100A4 promoter, we performed chromatin immunoprecipitation analysis on the MDA/mock and MDA/β4 cells. The second intron region 3′ to the transcriptional start site (equivalent to the first intron in mice) contains a transcriptional enhancer that is critical for the regulation of S100A4. This region contains key CpG residues that suppress S100A4 expression when methylated (30). Notably, this region also contains two NFAT consensus binding sites. Here, we immunoprecipitated NFAT5 and assessed whether it was associated with this region of the S100A4 promoter. As shown in Fig. 4E, immunoprecipitation of NFAT5, but not an IgG control, brought down this regulatory region of the S100A4 promoter in both the MDA/mock and the MDA/β4 cells. These data show that NFAT5 is definitively associated with the S100A4 promoter and suggest that negative regulators suppress NFAT5 action in the MDA/mock cells.
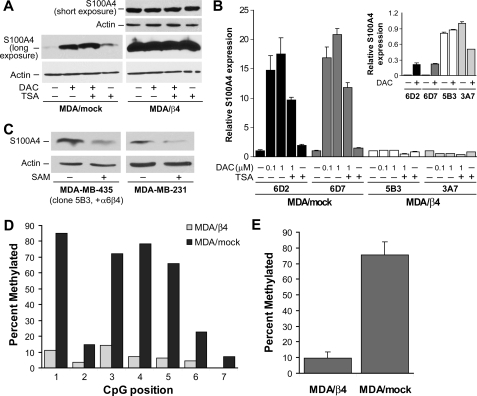
S100A4 message levels are up-regulated by the α6β4 integrin over 140-fold in the MDA-MB-435 cells (Table S3). However, the observations that siRNA-mediated knockdown of NFAT5 reduces S100A4 message levels by only 40% and that NFAT5 is present on the S100A4 promoter in the absence of α6β4 expression suggest that additional mechanisms regulate S100A4 expression. Previous studies suggest that the methylation status of the S100A4 promoter regulates S100A4 expression (30, 31). Given the high degree of S100A4 up-regulation by the α6β4 integrin, we tested the hypothesis that the α6β4 integrin also modulates S100A4 expression by affecting DNA demethylation. For these experiments, we treated MDA-MB-435 clones with inhibitor of DNA methyltransferases (DAC). Inhibition of DNA methyltransferases, but not inhibition of histone deacetylation with tricostatin A, elevated S100A4 protein and mRNA in the MDA/mock cells but did not affect MDA/β4 cells (Fig. 6, A and B). These data are consistent with the concept that the S100A4 promoter in the MDA/β4 transfectants is demethylated and, therefore, unaffected by DAC treatment. In MDA/mock transfectants, the S100A4 promoter would normally be methylated, and DAC treatment results in the removal of repressive methyl groups from the S100A4 promoter and a dramatic up-regulation of S100A4 expression. To confirm that active demethylation functions in S100A4 regulation downstream of the α6β4 integrin, MDA/β4 transfectants or MDA-MB-231 cells were treated with an inhibitor of DNA demethylases (SAM). As shown in Fig. 6C, inhibition of DNA demethylation by SAM treatment led to a decrease in S100A4 expression in both the MDA/β4 transfectants and in MDA-MB-231 cells.
FIGURE 6.
Effect of DNA methyltransferase inhibitor, DAC, and demethylation inhibitor, SAM, on S100A4 expression. A and B, MDA/mock and MDA/β4 transfectants were cultured in the presence or absence of 0.1 or 1 μm DAC for 3 days, as noted. Where indicated, 1 μm tricostatin A (TSA) was added for the final 24 h of culture. Duplicate cultures of each clone were then harvested to assess the level of S100A4 by immunoblot (A) or Q-PCR (B) analysis. Immunoblots in A are from the same gel with the same exposure time. A shorter exposure of the S100A4 blot from a smaller amount of the same samples (1-s exposure) showed that the loading between the MDA/β4 samples was similar. Q-PCR values are reported as -fold change relative to control for each clone. The inset in B represents relative S100A4 level between clones using 3A7 (MDA/β4) control cells as a value of 1. C, MDA-MB-435 clone 5B3 and MDA-MB-231 cells were treated with SAM (80 μm), a methyl donor known to inhibit demethylases, for 3 days under normal culturing conditions prior to harvest and immunoblotting cell lysates for S100A4 and actin. D, genomic DNA from MDA/mock and MDA/β4 transfectants containing the first intron region of the S100A4 promoter (+203 to +662) was assessed for CpG residue methylation by bisulfate conversion and PCR pyrosequencing. The levels of methylation of each of the seven CpG residues in this region are reported. E, the percentages of methylation of CpGs at positions 1, 3, 4, and 5 were averaged and reported as mean ± S.D. TSA, tricostatin A.
To demonstrate that the α6β4 integrin alters DNA methylation, we assessed the first intron region of the S100A4 promoter, which is known to contain a transcriptional enhancer that is regulated by DNA methylation. Using bisulfate pyrosequencing, we analyzed the +208 to +662 region of the S100A4 promoter for methylated CpG residues. As shown in Fig. 6D, MDA-MB-435 clones that express the α6β4 integrin reduced CpG methylation content in the seven CpG residues present in the enhancer. Four of the CpG residues in this region, specifically at positions 1, 3, 4, and 5, show a high level of methylation in the MDA/mock cells that is collectively 8-fold higher than the MDA/β4 transfectants (Fig. 6E). Together, these observations demonstrate that demethylation of the S100A4 promoter is an active process and a key regulator of S100A4 expression that is stimulated by the α6β4 integrin.
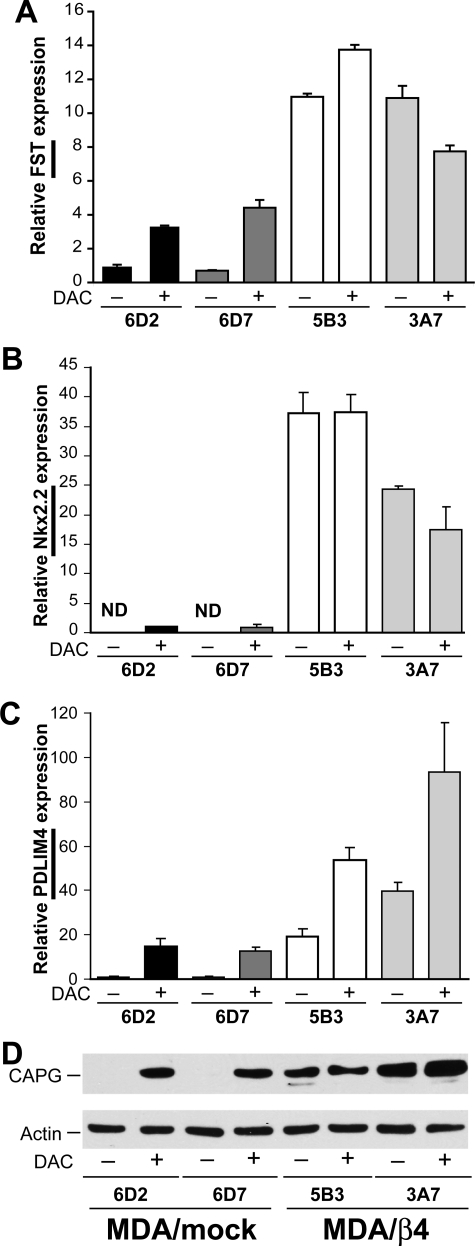
Several genes are highly up-regulated by integrin α6β4 expression in addition to S100A4, including FST, Nkx2.2, PDLIM4, CAPG (Fig. 1), and autotaxin (25). Of these genes, FST (32) and PDLIM4 (33) are known to be regulated by DNA methylation. Here, we examined the expression of these genes in DAC-treated cells to evaluate the consequences of inhibiting DNA methyltransferases. Here, we find that FST, Nkx2.2, PDLIM4, and CAPG (Fig. 7), but not autotaxin (data not shown), are substantially up-regulated by DAC treatment in the MDA/mock transfectants. Notably, both Nkx2.2 (Fig. 7B) and CAPG (Fig. 7D) expression are undetectable in the MDA/mock cells in the absence of DAC, suggesting that their promoters are fully repressed by DNA methylation. Much like S100A4, DAC treatment did not affect the expression levels of FST, Nkx2.2, and CAPG in the MDA/β4 transfectants. A different pattern is noted with PDLIM4 expression. Although DAC treatment dramatically enhances PDLIM4 expression in the MDA/mock cells, it also stimulates expression in the MDA/β4 transfectants, albeit to a lesser degree (Fig. 7C). One possibility is that the α6β4 integrin promotes demethylation of select CpG residues, which leaves others methylated and able to repress associated transcriptional elements. In contrast, DAC treatment is not selective. Collectively, these data indicate that DNA demethylation of select promoters is an important component of α6β4 integrin-mediated gene regulation. Notably, these observations are not based on clonal variation, since the observation extends to multiple promoters. Importantly, this is the first evidence that an integrin can affect the methylation status of a promoter.
FIGURE 7.
DAC treatment induces expression of several genes in MDA-MB-435 mock transfectants but does not alter expression in MDA/β4 cells. MDA/mock and MDA/β4 transfectants were cultured in the presence of 1 μm DAC for 3 days. Duplicate cultures of each condition were then harvested for RNA to assess the levels of FST (A), Nkx2.2 (B), and PDLIM4 (C) by Q-PCR or protein for CAPG (D). ND in B denotes that message was not detected. Values represent the mean ± S.D.
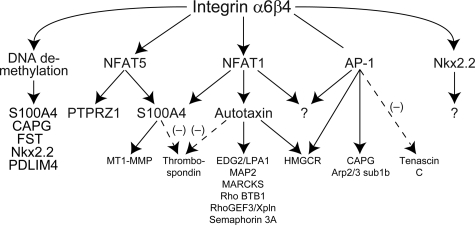
In our final analysis of the gene array data, we surveyed the literature for genes that are transcriptionally regulated by genes that lie downstream of the α6β4 integrin, more specifically by S100A4, autotaxin, and AP-1. As shown in Fig. 8, we find several genes involved in cell motility and invasion that are regulated by the α6β4 integrin in our gene array analysis and are also targets of S100A4 (29), autotaxin (34), and AP-1 (35). In summary, we find that transcriptional control of genes downstream of the α6β4 integrin is multifaceted in which multiple transcription factors contribute to these changes in gene expression, and many of these genes may be controlled by these factors indirectly.
FIGURE 8.
Multilayered regulation of genes downstream of the integrin α6β4. Several transcription factors, including NFAT1, NFAT5, and AP-1 function downstream of the α6β4 integrin. These observations are extended in the current study by identifying genes regulated downstream of NFAT5 (S100A4 and PTPRZ1) and genes regulated by DNA methylation and the α6β4 integrin using the MDA-MB-435 model and using previous analyses of genes regulated by S100A4 (29), autotaxin (34), and AP-1 (35), several of their target genes that are involved in cell motility and invasion that were found regulated by the α6β4 integrin in our gene array analysis. Dashed and solid arrows, negative and positive regulation, respectively. Of note, Cox-2 has been identified as a NFAT1 target; however, this association was not found in our gene array analysis.
DISCUSSION
The ability of the integrin α6β4 to promote an invasive phenotype is well documented. Several pathways have been implicated in this phenomenon, including cooperation with receptors for growth factors, such as epidermal growth factor (36), HGF (18, 19), and LPA (12), and the subsequent activation of phosphatidylinositol 3-kinase, Akt, Rac, Rho, and phosphodiesterases. Despite the mechanistic delineation of immediate downstream signaling events, how transcriptional events downstream of the integrin α6β4 affect these signaling events and subsequent tumor cell invasion has received little attention. Previous studies have shown that the α6β4 integrin can signal to multiple transcription factors. Here, we expand on these observations and define to what extent signaling through the α6β4 integrin can affect gene transcription. We further show that the α6β4 integrin regulates a coordinated program of genes that predispose the cell to a migratory and invasive phenotype, including genes such as metastasis-associated S100A4. Importantly, we demonstrate for the first time that an integrin can affect the DNA methylation pattern of the promoters of select genes, including S100A4.
S100A4 is a member of the S100 family of calcium binding proteins and has been given several names, including metastasin, fibroblast-specific protein, and CAPL. Analysis of S100A4 expression has revealed that it is associated with a metastatic phenotype in multiple types of carcinoma, including breast, prostate, pancreatic, gastric, and thyroid (29). Interestingly, the α6β4 integrin is associated with an invasive phenotype in each of these types of carcinomas (4, 37). In breast cancer, S100A4 can promote hormone-independent growth and metastasis of MCF-7 breast carcinoma cells in nude mice, which are normally nonmetastatic (38). Conversely, crossing mice that overexpress S100A4 in the mammary epithelium with mouse models of metastasis (e.g. MMTV-neu or GRS/A) dramatically increases the incidence of metastasis (39, 40); however, by itself, S100A4 is nontumorigenic (40). Intracellularly, S100A4 can induce cell motility (41), in part through its ability to interact with myosin-IIA (27, 42). In addition, extracellular S100A4 can stimulate MMP-13 activity, possibly contributing to tumor invasion (29). In some breast tumor cell lines, we observe a form of S100A4 that runs at a higher molecular weight than expected for the 11.7-kDa monomer, which may be oligomers of S100A4. Attempts to reduce these bands to a monomer using strong reducing agents, urea, or excessive heat were unsuccessful.3 Although the nature of these oligomers is undefined, interestingly, other members of the S100 family can be cross-linked by transglutaminases (43). Therefore, one possibility is that transglutaminases cross-link S100A4 to form oligomers. At this time, it is unclear how S100A4 oligomers may contribute to tumor biology. Typically, S100A4 forms a noncovalent symmetric homodimer, and it is this dimeric form that is expressed by metastatic and invasive cell lines (e.g. MDA-MB-231) (29).
Despite the strong data supporting a role for S100A4 in tumor metastasis, little is known about the regulation of S100A4 other than it can be up-regulated during EMT (44), by ErbB2 signaling (45), and through promoter demethylation (30, 31). This study demonstrates that the α6β4 integrin can stimulate the dramatic up-regulation of S100A4 expression. Importantly, we determined that S100A4 expression correlates well with the expression of the α6β4 integrin in breast carcinoma cell lines, with the exception of MCF7. Of all the cell lines examined, MCF7 is the only cell line that does not display a mesenchymal phenotype and has been used previously to model EMT downstream of exogenously expressed Snail (46). S100A4 is a well accepted marker for EMT. The ability of integrin α6β4 signaling to activate the S100A4 promoter suggests that the integrin α6β4 may control the expression of a subset of genes during EMT and thus be an integral part of the process. This is an intriguing concept, considering that the α6β4 integrin, and thus its oncogenic potential and ability to regulate proinvasive genes, is released from hemidesmosomes during EMT. However, more work is needed to determine how much the α6β4 integrin contributes to EMT.
Downstream of the α6β4 integrin, S100A4 expression is stimulated through two distinct modes: through NFAT5 and by altering the DNA methylation status of the S100A4 promoter. The removal of methyl groups from CpG residues initially opens the promoter for activation but itself does not activate the promoter. Transcription factors are needed for activation to occur. Here, we implicate NFAT5 in the activation of the S100A4 promoter. NFAT was first identified in T-cells, where, upon T-cell activation and nuclear transport, NFAT promotes specific transcription to promote mobilization of T-cells and elicit an immune response (47). Jauliac et al. (22) were the first to identify the importance of NFAT1 and NFAT5 in the invasion and motility of carcinoma cells. Importantly, this role for NFAT was identified in the MDA-MB-435 cell model, where integrin α6β4 promotes the transcriptional up-regulation of NFAT1 and NFAT5 and the subsequent activation of these factors (22). Few targets of NFAT transcription factors have been identified in carcinoma cells, which includes autotaxin as defined by our group (25) and Cox2 (48, 49), both of which are NFAT1 targets. Here, we extend these studies by identifying S100A4 as a target of NFAT5. Interestingly, our data show that NFAT5 is present on the S100A4 promoter in the absence of signaling from the α6β4 integrin, thus suggesting that other conditions controlled by integrin α6β4 determine whether NFAT5 present on the promoter can drive promoter activity.
Importantly, we find that DNA methylation is a major contributor to gene expression downstream of the α6β4 integrin. Specifically, we show that expression of several genes, namely S100A4, FST, PDLIM4, CAPG, and Nkx2.2, are dramatically enhanced by integrin α6β4 expression. Treatment of cells with methyltransferase inhibitors, such as DAC, in the absence of this integrin can recapitulate the effect of α6β4 integrin expression. Finally, we uncovered evidence that the S100A4 promoter is hypomethylated in MDA-MB-435 cells expressing integrin α6β4 but hypermethylated in the absence of this integrin. Methylation of CpG sites within a promoter is controlled by the balance of DNA methyltransferases and demethylases; however, the exact mechanisms governing the selectiveness toward specific promoters are unknown. Once methylated, promoters are generally silenced either by disruption of transcription factor binding sites or binding of methyl binding proteins, such as MBDs and MeCPs, which recruit histone-modifying agents to the promoter for effective chromatin silencing (50). Certainly, our results demonstrate that the α6β4 integrin can affect the expression of genes normally silenced by promoter methylation and that the S100A4 promoter specifically is hypomethylated in MDA-MB-435 cells expressing the α6β4 integrin. Whether the α6β4 integrin stimulates DNA demethylases directly by altering specific signaling pathways or indirectly through the up-regulation or inhibition of key genes is not clear. However, these results indicate that this cell model is ideal to study how selective promoter demethylation is achieved and will be the focus of future studies.
As expected from the known functions for the α6β4 integrin, the genes most dramatically altered are associated with cell motility and cell survival/apoptosis. However, there are several other classes of genes that deserve mention. We find that eight HLA genes are down-regulated, suggesting that theα6β4 integrin could function in immune modulation by reducing MHC class II and thereby decreasing antigen presentation by the tumor cell. Heat shock proteins, such as αB-crystallin, and E3 ligases, such as SMURF2, are altered, which implies that the stability of select proteins could be affected by theα6β4 integrin. Expression of multiple extracellular matrix proteins are altered, including the up-regulation of several collagen isoforms and lysyl oxidase, an enzyme involved in cross-linking collagens to elastins. Certainly, these data collectively signify a potential involvement of the α6β4 integrin in a diverse array of functions that may be involved in promoting tumor progression.
In a recent publication from the Mercurio laboratory (51), several of the published gene array data bases from breast cancer patients were mined for correlations with β4 integrin subunit mRNA expression. They confirmed the prevalence of α6β4 integrin overexpression in basal intrinsic subtype of breast cancer and defined what they refer to as a “β4 signature.” Notably, few of these genes identified in our analysis are found in the β4 integrin signature. There are several likely reasons for this observation. Their analysis was made using whole tissue homogenates, which includes gene expression profiles from the cells of the tumor microenvironment, such as immune infiltrates and stromal cells. Therefore, this β4 signature incorporates the genes expressed by cancer cells and cells from the microenvironment as well as genes altered due to the interactions between the two cell populations. Our analysis takes into account only those genes expressed by the cancer cells under controlled in vitro conditions. Second, many of the genes identified in our study are genes whose expression levels are increased during EMT, such as S100A4. Stromal cells, including fibroblasts and macrophages, are known to express S100A4 (52). In the presence of a desmoplastic stroma and immune infiltrate, genes in this class are likely to be masked by expression in the stroma and appear not to be significantly altered. Finally, the α6β4 integrin is well documented to cooperate with growth factor signaling to mediate its effects (18, 19, 36, 53). Therefore, it is likely that gene expression augmented by the α6β4 integrin will depend on cellular context, which growth factor receptors are stimulated, and cell origin.
Investigations using the MDA-MB-435 cell line cannot escape the controversy surrounding this line stemming from reports suggesting that it is may be derived from a melanoma (54, 55). Extensive work from the M. D. Anderson Cancer Center shows that these cells express breast-specific (nonmelanocyte) markers and can be induced to secrete milk proteins and lipids (56). Furthermore, these cells preferentially grow when implanted into mammary fat pads compared with subcutaneous injection (57), similar to other breast cancers but unlike melanoma cell lines.4 Many of the markers that MDA-MB-435 cells share with melanomas are typically found in neuroendocrine cells. Therefore, MDA-MB-435 cells may actually be derived from a tumor of neuroendocrine origin, a tumor type not well recognized in the breast cancer literature, rather than of melanocyte origin. However, should the MDA-MB-435 cells conclusively be shown to be of melanoma origin through more reliable methods, such as DNA footprinting, the studies presented here would have important implications for melanoma metastasis. The α6β4 integrin has been shown to be expressed in more aggressive melanomas (58), where, like in breast and other carcinomas, it promotes an invasive and metastatic phenotype.
In summary, our study reveals an important role for the α6β4 integrin in the regulation of genes that promote an invasive and metastatic phenotype. Using siRNA and promoter analysis, we find that S100A4 is the first target of NFAT5 reported in cancer. We also determine that several genes that are known to be regulated by DNA methylation (S100A4, FST, Nkx2.2, PDLIM4, and CAPG) were dramatically up-regulated by integrin α6β4 expression and that the α6β4 integrin promotes the demethylation of the S100A4 promoter. Together, our data suggest integrin α6β4 confers a motile and invasive phenotype in breast carcinoma cells, in part, by regulating transcription factors, including NFAT and chromatin remodeling, such as promoter demethylation to modulate the expression of proinvasive genes. Importantly, this is the first report that an integrin can affect gene transcription through chromatin remodeling.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Janet Price and Art Mercurio for cell lines and reagents; Drs. Adriana Paulucci, Sarita Sastry, and Tao Sheng for helpful discussions and critical evaluation of the manuscript; L. Nicole Towers for technical assistance; Karen Martin for graphics assistance; and Dr. William E. Mitch (Baylor College of Medicine) for use of his laboratory during Hurricane Ike recovery.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA109136. This work was also supported by the University of Texas Medical Branch Gastrointestinal Research Interdisciplinary Research Program, the Center for Interdisciplinary Research in Women's Health, and the Sealy and Smith Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The array data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE11466 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11466).
The on-line version of this article (available at http://www.jbc.org) contains Tables S1–S3.
Footnotes
The abbreviations used are: EMT, epithelial to mesenchymal transition; LPA, lysophosphatidic acid; NFAT, nuclear factor of activated T-cells; DAC, 5-aza-2′-deoxycytidine; SAM, S-adenosylmethionine; Q-PCR, quantitative PCR; FACS, fluorescence-activated cell sorting; HGF, hepatocyte growth factor; siRNA, small interference RNA; Ab, antibody; E3, ubiquitin-protein isopeptide ligase.
M. Chen and K. L. O'Connor, unpublished observations.
J. E. Price, personal communication.
References
- 1.Hynes, R. O. (2002) Cell 110 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Borradori, L., and Sonnenberg, A. (1999) J. Invest. Dermatol. 112 411–418 [DOI] [PubMed] [Google Scholar]
- 3.Rabinovitz, I., Toker, A., and Mercurio, A. M. (1999) J. Cell Biol. 146 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercurio, A. M., and Rabinovitz, I. (2001) Semin. Cancer Biol. 11 129–141 [DOI] [PubMed] [Google Scholar]
- 5.Tagliabue, E., Ghirelli, C., Squicciarini, P., Aiello, P., Colnaghi, M. I., and Menard, S. (1998) Clin. Cancer Res. 4 407–410 [PubMed] [Google Scholar]
- 6.Friedrichs, K., Ruiz, P., Franke, F., Gille, I., Terpe, H.-J., and Imhof, B. A. (1995) Cancer Res. 55 901–906 [PubMed] [Google Scholar]
- 7.Tennenbaum, T., Weiner, A. K., Belanger, A. J., Glick, A. B., Hennings, H., and Yuspa, S. H. (1993) Cancer Res. 53 4803–4810 [PubMed] [Google Scholar]
- 8.Grossman, H. B., Lee, C., Bromberg, J., and Liebert, M. (2000) Oncol. Rep. 7 13–16 [PubMed] [Google Scholar]
- 9.Jones, J. L., Royall, J. E., Critchley, D. R., and Walker, R. A. (1997) Exp. Cell Res. 235 325–333 [DOI] [PubMed] [Google Scholar]
- 10.Shaw, L. M., Rabinovitz, I., Wang, H. H.-F., Toker, A., and Mercurio, A. M. (1997) Cell 91 949–960 [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay, R., Theriault, R. L., and Price, J. E. (1999) Clin. Exp. Metastasis 17 325–332 [DOI] [PubMed] [Google Scholar]
- 12.O'Connor, K. L., Shaw, L. M., and Mercurio, A. M. (1998) J. Cell Biol. 143 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipscomb, E. A., Dugan, A. S., Rabinovitz, I., and Mercurio, A. M. (2003) Clin. Exp. Metastasis 20 569–576 [DOI] [PubMed] [Google Scholar]
- 14.Yoon, S. O., Shin, S., and Mercurio, A. M. (2006) Cancer Res. 66 6288–6295 [DOI] [PubMed] [Google Scholar]
- 15.Guo, W., Pylayeva, Y., Pepe, A., Yoshioka, T., Muller, M. J., Inghirami, G., and Giancotti, F. G. (2006) Cell 126 489–502 [DOI] [PubMed] [Google Scholar]
- 16.Bonaccorsi, L., Carloni, V., Muratori, M., Adriana, S., Giannini, A., Carini, M., Serio, M., Forti, G., and Baldi, E. (2000) Endocrinology 141 3172–3182 [DOI] [PubMed] [Google Scholar]
- 17.Carroll, D. K., Carroll, J. S., Leong, C. O., Cheng, F., Brown, M., Mills, A. A., Brugge, J. S., and Ellisen, L. W. (2006) Nat. Cell Biol. 8 551–561 [DOI] [PubMed] [Google Scholar]
- 18.Trusolino, L., Bertotti, A., and Comoglio, P. M. (2001) Cell 107 643–654 [DOI] [PubMed] [Google Scholar]
- 19.Chung, J., Yoon, S.-O., Lipscomb, E. A., and Mercurio, A. M. (2004) J. Biol. Chem. 279 32287–32293 [DOI] [PubMed] [Google Scholar]
- 20.Bertotti, A., Comoglio, P. M., and Trusolino, L. (2005) Cancer Res. 65 10674–10679 [DOI] [PubMed] [Google Scholar]
- 21.Lipscomb, E. A., and Mercurio, A. M. (2005) Cancer Metastasis Rev. 24 413–423 [DOI] [PubMed] [Google Scholar]
- 22.Jauliac, S., Lopex-Rodriguez, C., Shaw, L. M., Brown, L. F., Rao, A., and Toker, A. (2002) Nat. Cell Biol. 4 540–544 [DOI] [PubMed] [Google Scholar]
- 23.Zahir, N., Lakins, J. N., Russell, A., Ming, W., Chatterjee, C., Rozenberg, G. I., Marinkovich, M. P., and Weaver, V. M. (2003) J. Cell Biol. 163 1397–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver, V. M., Lelievre, S., Lakins, J. N., Chrenek, M. A., Jones, J. C., Giancotti, F., Werb, Z., and Bissell, M. J. (2002) Cancer Cell 2 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, M., and O'Connor, K. L. (2005) Oncogene 24 5125–5130 [DOI] [PubMed] [Google Scholar]
- 26.Chen, M., Towers, L. N., and O'Connor, K. L. (2007) Am. J. Physiol. 292 C1927–C1933 [DOI] [PubMed] [Google Scholar]
- 27.Li, Z. H., and Bresnick, A. R. (2006) Cancer Res. 66 5173–5180 [DOI] [PubMed] [Google Scholar]
- 28.Bao, X., Sinha, M., Liu, T., Hong, C., Luxon, B. A., Garofalo, R. P., and Casola, A. (2008) Virology 374 114–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrett, S. C., Varney, K. M., Weber, D. J., and Bresnick, A. R. (2006) J. Biol. Chem. 281 677–680 [DOI] [PubMed] [Google Scholar]
- 30.Tulchinsky, E., Ford, H. L., Kramerov, D., Reshetnyak, E., Grigorian, M., and Zain, S. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 9146–9150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura, N., and Takenaga, K. (1998) Clin. Exp. Metastasis 16 471–479 [DOI] [PubMed] [Google Scholar]
- 32.Liang, G., Robertson, K. D., Talmadge, C., Sumegi, J., and Jones, P. A. (2000) Cancer Res. 60 4907–4912 [PubMed] [Google Scholar]
- 33.Vanaja, D. K., Ballman, K. V., Morlan, B. W., Cheville, J. C., Neumann, R. M., Lieber, M. M., Tindall, D. J., and Young, C. Y. (2006) Clin. Cancer Res. 12 1128–1136 [DOI] [PubMed] [Google Scholar]
- 34.Noh, J. H., Ryu, S. Y., Eun, J. W., Song, J., Ahn, Y. M., Kim, S. Y., Lee, S. H., Park, W. S., Yoo, N. J., Lee, J. Y., Lee, S. N., and Nam, S. W. (2006) Mol. Cell Biochem. 288 91–106 [DOI] [PubMed] [Google Scholar]
- 35.Bahassi, E. M., Karyala, S., Tomlinson, C. R., Sartor, M. A., Medvedovic, M., and Hennigan, R. F. (2004) Clin. Exp. Metastasis 21 293–304 [DOI] [PubMed] [Google Scholar]
- 36.Mainiero, F., Pepe, A., Yeon, M., Ren, Y., and Giancotti, F. G. (1996) J. Cell Biol. 134 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz-Monserrate, Z., Qiu, S., Evers, B. M., and O'Connor, K. L. (2007) Mod. Pathol. 20 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grigorian, M., Ambartsumian, N., Lykkesfeldt, A. E., Bastholm, L., Elling, F., Georgiev, G., and Lukanidin, E. (1996) Int. J. Cancer 67 831–841 [DOI] [PubMed] [Google Scholar]
- 39.Davies, M. P., Rudland, P. S., Robertson, L., Parry, E. W., Jolicoeur, P., and Barraclough, R. (1996) Oncogene 13 1631–1637 [PubMed] [Google Scholar]
- 40.Ambartsumian, N. S., Grigorian, M. S., Larsen, I. F., Karlstrom, O., Sidenius, N., Rygaard, J., Georgiev, G., and Lukanidin, E. (1996) Oncogene 13 1621–1630 [PubMed] [Google Scholar]
- 41.Jenkinson, S. R., Barraclough, R., West, C. R., and Rudland, P. S. (2004) Br. J. Cancer 90 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, E. J., and Helfman, D. M. (2003) J. Biol. Chem. 278 30063–30073 [DOI] [PubMed] [Google Scholar]
- 43.Ruse, M., Lambert, A., Robinson, N., Ryan, D., Shon, K. J., and Eckert, R. L. (2001) Biochemistry 40 3167–3173 [DOI] [PubMed] [Google Scholar]
- 44.Xue, C., Plieth, D., Venkov, C., Xu, C., and Neilson, E. G. (2003) Cancer Res. 63 3386–3394 [PubMed] [Google Scholar]
- 45.Hernan, R., Fasheh, R., Calabrese, C., Frank, A. J., Maclean, K. H., Allard, D., Barraclough, R., and Gilbertson, R. J. (2003) Cancer Res. 63 140–148 [PubMed] [Google Scholar]
- 46.Lien, H. C., Hsiao, Y. H., Lin, Y. S., Yao, Y. T., Juan, H. F., Kuo, W. H., Hung, M. C., Chang, K. J., and Hsieh, F. J. (2007) Oncogene 26 7859–7871 [DOI] [PubMed] [Google Scholar]
- 47.Macian, F. (2005) Nat. Rev. Immunol. 5 472–484 [DOI] [PubMed] [Google Scholar]
- 48.Yiu, G. K., and Toker, A. (2006) J. Biol. Chem. 281 12210–12217 [DOI] [PubMed] [Google Scholar]
- 49.Duque, J., Fresno, M., and Iniguez, M. A. (2005) J. Biol. Chem. 280 8686–8693 [DOI] [PubMed] [Google Scholar]
- 50.Wade, P. A. (2001) Oncogene 20 3166–3173 [DOI] [PubMed] [Google Scholar]
- 51.Lu, S., Simin, K., Khan, A., and Mercurio, A. M. (2008) Clin. Cancer Res. 14 1050–1058 [DOI] [PubMed] [Google Scholar]
- 52.Cabezón, T., Celis, J. E., Skibshøj, I., Klingelhöfer, J., Grigorian, M., Gromov, P., Rank, F., Myklebust, J. H., Maelandsmo, G. M., Lukanidin, E., and Ambartsumian, N. (2007) Int. J. Cancer 121 1433–1444 [DOI] [PubMed] [Google Scholar]
- 53.O'Connor, K. L., Nguyen, B.-K., and Mercurio, A. M. (2000) J. Cell Biol. 148 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross, D. T., Scherf, U., Eisen, M. B., Perou, C. M., Rees, C., Spellman, P., Iyer, V., Jeffrey, S. S., VandeRijn, M., Waltham, M., Pergamenschikov, A., Lee, L. C., Lashkari, D., Shalon, D., Myers, T. G., Weinstein, J. N., Botstein, D., and Brown, P. O. (2000) Nat. Genet. 24 227–235 [DOI] [PubMed] [Google Scholar]
- 55.Ellison, G., Klinowska, T., Westwood, R. F., Docter, E., French, T., and Fox, J. C. (2002) Mol. Pathol. 55 294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sellappan, S., Grijalva, R., Zhou, X., Yang, W., Eli, M. B., Mills, G. B., and Yu, D. (2004) Cancer Res. 64 3479–3485 [DOI] [PubMed] [Google Scholar]
- 57.Price, J. E., Polyzos, A., Zhang, R. D., and Daniels, L. M. (1990) Cancer Res. 50 717–721 [PubMed] [Google Scholar]
- 58.Raymond, K., Kreft, M., Song, J. Y., Janssen, H., and Sonnenberg, A. (2007) Mol. Biol. Cell 18 4210–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.