Abstract
During apoptosis the Golgi apparatus undergoes irreversible fragmentation. In part, this results from caspase-mediated cleavage of several high molecular weight coiled-coil proteins, termed golgins. These include GM130, golgin 160, and the Golgi vesicle tethering protein p115, whose caspase cleavage generates a C-terminal fragment (CTF) of 205 residues. Here we demonstrate that early during apoptosis, following the rapid cleavage of p115, endogenous CTF translocated to the cell nucleus and its nuclear import was required to enhance the apoptotic response. Expression of a series of deletion constructs identified a putative α-helical region of 26 amino acids, whose expression alone was sufficient to induce apoptosis; deletion of these 26 residues from the CTF diminished its proapoptotic activity. This region contains several potential SUMOylation sites and co-expression of SUMO together with the SUMO ligase, UBC9, resulted in SUMOylation of the p115 CTF. Significantly, when cells were treated with drugs that induce apoptosis, SUMOylation enhanced the efficiency of p115 cleavage and the kinetics of apoptosis. A construct in which a nuclear export signal was fused to the N terminus of p115 CTF accumulated in the cytoplasm and surprisingly, its expression did not induce apoptosis. In contrast, treatment of cells expressing this chimera with the antibiotic leptomycin induced its translocation into the nucleus and resulted in the concomitant induction of apoptosis. These results demonstrate that nuclear import of the p115 CTF is required for it to stimulate the apoptotic response and suggest that its mode of action is confined to the nucleus.
In mammalian cells the Golgi apparatus is a highly polarized organelle comprising a series of stacked cisternae, which form a lace-like network in the perinuclear region of the cell. It receives de novo synthesized secretory and membrane proteins, as well as lipids from the endoplasmic reticulum (ER)2; these cargo molecules are then modified, sorted, and transported to lysosomes, endosomes, secretory granules, and the plasma membrane. Although it is well established that the Golgi apparatus undergoes reversible disassembly during mitosis (1, 2), indeed this appears to be a prerequisite for mitosis (3), studies from several laboratories including our own, have also established a link between the Golgi apparatus and apoptosis (programmed cell death). During apoptosis, the Golgi apparatus undergoes extensive and irreversible fragmentation (4), the ER vesiculates (5) and secretion is inhibited (6).
Golgi disassembly during apoptosis results, in part, from caspase-mediated cleavage of several golgins (7). Proteolysis of golgin 160 by caspase-2, as well as GRASP65, GM130, p115, syntaxin5, and giantin by caspases-3 and -7 contributes significantly to Golgi fragmentation (6, 8–13). Consistent with this idea, overexpression of caspase-resistant forms of golgin 160, GRASP65, or p115 has been shown to delay the kinetics of Golgi fragmentation during apoptosis (8–10). In addition, immunoreactive caspase-2, an upstream caspase, localizes to the Golgi apparatus (9) and caspase-2-mediated cleavage of golgin 160 also appears to be an early event during apoptosis. Depending on the apoptotic stimulus, expression of a golgin 160 triple mutant resistant to caspase cleavage delays the onset of apoptosis (12). Recently, our laboratory demonstrated that Golgi fragmentation is an early apoptotic event that occurs close to or soon after release of cytochrome c from mitochondria, an early indicator of apoptosis (13). Together these observations demonstrate that specific Golgi proteins may function early during apoptosis, although their role in this process and the detailed molecular mechanism by which Golgi fragmentation occurs is not well understood.
A key molecule in mediating Golgi fragmentation during apoptosis is the vesicle tethering protein p115 (10), a 962-residue peripheral membrane protein. p115 is an elongated homodimer consisting of two globular “head” domains, an extended “tail” region reminiscent of the myosin-II structure (14), and 4 sequential coil-coil domains distal to the globular head region, the first of which, CC1, has been implicated in soluble NSF attachment protein receptors (SNARE) binding (15). Earlier in vitro studies on mitotic Golgi reassembly demonstrated that p115 interacts with GM130 and giantin and implicated it in Golgi cisternal stacking (16). Consistent with this idea, microinjection of anti-p115 antibodies caused Golgi fragmentation (17). Based on data demonstrating p115 binding to GM130, giantin, GOS28, and syntaxin-5, Shorter et al. (15) suggested that p115 promotes formation of a GOS28-syntaxin-5 (v-/t-SNARE) complex and hypothesized that it coordinates the sequential tethering and docking of COPI vesicles to Golgi membranes. Interestingly, p115 has also been shown to be a Rab-1 effector that binds Rab-1-GTP directly and cross-linking experiments showed that it interacts with Syntaxin5, sly1, membrin, and rbet1 on microsomal membranes and COPII vesicles suggesting that p115-SNARE interactions may facilitate membrane “docking” (18).
More recent in vivo studies showed that inhibition of GM130 or giantin binding to p115 had little effect on Golgi morphology or reassembly following mitosis, suggesting its role in maintaining Golgi structure might be independent of GM130 binding (19, 20). Thus post-mitotic Golgi reassembly could be rescued by p115 lacking the C-terminal GM130 binding motif (residues 935–962) but not by a mutant lacking the SNARE interacting CC1 domain (20). In addition, other studies have implicated GM130 and GRASP65 in Golgi ribbon formation and suggested that this may occur independently of interactions with p115 (21). Most significantly, knockdown of p115 using siRNA demonstrated that it is essential for maintaining Golgi structure, compartmentalization, and cargo traffic to the plasma membrane (20, 22).
Earlier work from our laboratory demonstrated that p115 is cleaved in vitro by caspase-8, an initiator caspase, as well as by the executioner caspase-3 (10, 13). In response to apoptosis inducing drugs, p115 is cleaved in vivo at Asp757 to generate a 205-residue C-terminal fragment and an N-terminal polypeptide of 757 amino acids. Most significantly, expression of the p115 C-terminal fragment in otherwise healthy cells results in its translocation to the nucleus and the induction of apoptosis suggesting that this polypeptide plays a role in potentiating the apoptotic response. To further dissect p115 function during cell death, we have now determined the minimal domain in its C terminus that mediates apoptosis efficiently and analyzed the requirement of nuclear translocation in triggering the apoptotic response.
MATERIALS AND METHODS
Antibodies—Mouse monoclonal antibodies to p115 (7D1) were a gift from Dr. Gerry Waters (Merck, Rahway, NJ). Dr. Adam Linstedt (Carnegie Melon University, Pittsburgh, PA) provided a polyclonal antibody specific for the C terminus of p115. Rabbit anti-PARP (poly(ADP-ribose) polymerase) serum, specific for the cleaved p85 form of PARP was purchased from Cell Signaling (Danvers, MA). The following mouse monoclonal antibodies were used: anti-GM130 (BD Transduction Laboratories, San Diego, CA); anti-FLAG M2-peroxidase and anti-cytochrome c (BD Pharmingen, San Diego, CA); anti-HA-peroxidase (Roche Applied Science); and anti-Fas clone CH-11 (MBL International Corporation). Anti-active caspase-3 was purchased from Promega (San Luis Obispo, CA). EZview™ Red Anti-FLAG® M2 Affinity Gel was purchased from Sigma. Alexa Fluor goat anti-mouse and anti-rabbit antibodies were purchased from Molecular Probes, Inc. (Eugene, OR).
A human p115 fragment (amino acids 645–962) was fused to the C terminus of glutathione S-transferase in expression vector pGEX2T. The recombinant protein was expressed in and purified from bacteria. The glutathione S-transferase motif was cleaved using thrombin, and the p115 fragment was injected into rabbits to raise the anti-p115 antibody, AE800. This antibody recognized full-length human p115 as well as the N-terminal (1–636) and C-terminal (758–962) polypeptides by both Western blot and immunofluorescence microscopy; it did not recognize rat p115 by immunofluorescence microscopy.
Plasmid Constructs—The pSG5-FLAG-human p115 construct was provided by Dr. Yukio Ikehara (Fukuoka University School of Medicine, Fukuoka, Japan) and was used as a template to generate p115 constructs. For RNA interference, p115 was targeted by siRNA oligos as described (20). pEGFPC1 plasmid was purchased from Clontech (Mountain View, CA). p3XFLAG-CMV™-7.1 expression vector was purchased from Sigma. pGEX2T vector was purchased from Amersham Biosciences. A GFP-NES-CTF (GFP-nuclear export signal tagged CTF) plasmid was generated in the laboratory using the nuclear export signal LQLPPLERLTL from the Rev protein of HIV-1.
Cell Culture and Induction of Apoptosis—HeLa and COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm glutamine, 50 units/ml penicillin G sodium, 50 μg/ml streptomycin sulfate at 37 °C in a humidified incubator containing 5% CO2. Apoptosis was induced by treating cells with Anti-Fas clone CH-11 antibody (13), etoposide (100 μm), or anisomycin (5 μg/ml) for the indicated times.
Preparation of Cell Extracts and Western Blotting—Cells were lysed in medium containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 0.25% aprotinin, and a mixture of protease inhibitors (Roche). Thirty μg of each sample was loaded on a 10% polyacrylamide gel, followed by transfer to Immobilon-P membranes (Millipore Corp). Membranes were blocked in 5% nonfat milk in phosphate-buffered saline containing 0.1% Tween 20 (PBST) for 1 h. Subsequently, they were probed with appropriate antibodies in PBST, washed extensively, and the immunoreactive bands were visualized by Enhanced Chemiluminescence (Amersham Biosciences).
Immunofluorescence Microscopy—Cells were grown on coverslips and treated as indicated, after which they were fixed in 3% paraformaldehyde and processed for immunofluorescence microscopy as described previously (13). Confocal images were acquired by capturing Z-series images with a 0.25-μm step size on a Leica TCS SP2 AOBS confocal microscope (Leica, Dearfield, IL) using a ×63 oil immersion objective (1.4 N.A.). Laser lines at 405, 488, 546, and 633 nm were provided by 20 milliwatt Diode, 100 milliwatt Ar, 1.5 milliwatt HeNe, and 10 milliwatt HeNe lasers; sequential excitation by line and detection range settings were used to eliminate cross-talk between fluorophores. The images (1024 × 1024 pixel, 8 bit) were saved as TIFF files. The entire Z-series was projected using the maximum intensity method. Background was removed and contrast adjusted using Image J and Adobe Photoshop. Threshold background level for each antibody was selected, saved, and loaded in all experiments.
SUMOylation Experiments—HeLa cells were grown on 10-cm dishes and transfected with 2 μg of plasmids encoding (FLAG)3-CTF, (HA)3-SUMO, and UBC9. At 18 h post-transfection, cells were lysed in 100 μl of lysis buffer (above) containing 5 mm N-ethylmaleimide to prevent de-SUMOylation and centrifuged at 1000 × g for 2 min. The postnuclear supernatant was pre-cleared three times for 1 h each at 4 °C with 30 μl of Protein G-Sepharose beads in a total volume of 500 μl of RIPA buffer containing 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 50 mm Tris-HCl, pH 8.0, 0.1% SDS and protease inhibitor mixture. Precleared lysates were incubated with 20 μl of EZview Red Anti-FLAG M2 Affinity Gel overnight at 4 °C. Following incubations, the beads were washed extensively in wash buffer containing 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.05% Triton X-100, and analyzed by SDS-PAGE and Western blotting.
RESULTS
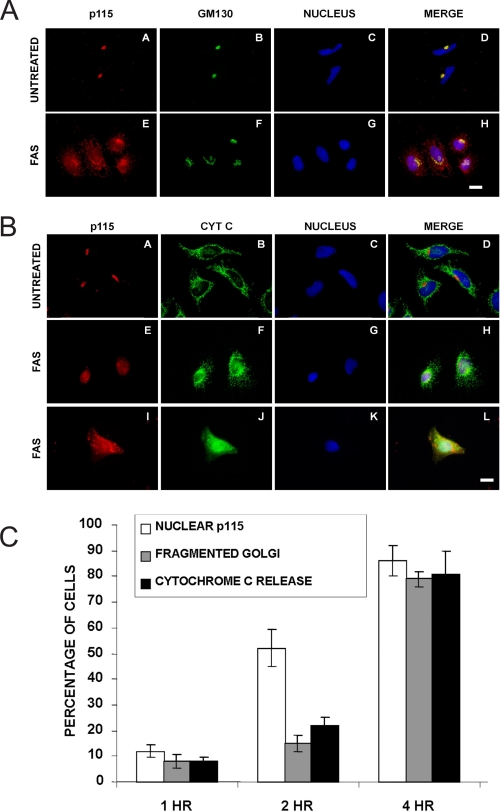
Intracellular Localization of Endogenous p115 CTF—Previous data from our laboratory showed that in cells overexpressing FLAG-tagged p115 CTF, the polypeptide accumulated almost exclusively in nuclei and its expression was sufficient to induce Golgi fragmentation and apoptosis (10). However, it was unknown whether nuclear accumulation of CTF was a consequence of overexpression, if the endogenous polypeptide were present in cell nuclei early during apoptosis, and if its nuclear translocation was a requirement for apoptosis. To address these questions, HeLa cells were treated with Fas antibody that cross-links the Fas receptor, after which they were analyzed by immunofluorescence microscopy using a polyclonal antibody specific for the C terminus of p115 (residues 758–962) and a monoclonal antibody to GM130 (Fig. 1). As expected, p115 and GM130 exhibited significant Golgi co-localization in control cells (Fig. 1, panels A–D). Upon Fas activation by the CH-11 antibody, a high level of immunoreactive p115 CTF was present in nuclei prior to significant fragmentation of the Golgi apparatus (Fig. 1A). To confirm the nuclear localization of p115 during the induction of apoptosis, at different times after Fas antibody treatment HeLa cells were co-stained for mitochondrial cytochrome c release, a characteristic marker of early apoptosis (23), as well as for immunoreactive-p115 (Fig. 1B). Control cells stained with anti-cytochrome c antibodies exhibited the typical reticular staining of mitochondria and no nuclear localization of p115 was observed (Fig. 1, B, panels A–D, and C). In contrast, following 2 or 4 h of Fas activation, cytochrome c staining became quite diffuse consistent with its release from mitochondria, and nuclear p115 staining was evident (Fig. 1B, panels E–L). Most significantly, quantitation of the immunofluorescence microscopy data (Fig. 1C) showed that at 2 h following Fas activation, the cells showed little evidence of mitochondrial cytochrome c release or Golgi fragmentation, whereas ∼50% of treated cells exhibited immunoreactive-p115 in their nuclei, suggesting that the nuclear localization of p115 preceded the onset of apoptosis. As expected, at 4 h following treatment with Fas activating antibody, ∼80 to 90% of cells had characteristics of apoptosis, namely highly diffuse cytochrome c staining, condensed nuclei, and a disrupted Golgi apparatus (Fig. 1B); our previous data showed that PARP is also cleaved at this time (13).
FIGURE 1.
Nuclear localization of endogenous immunoreactive-p115 precedes major changes in Golgi structure early during Fas-mediated apoptosis. A, HeLa cells (panels A–H) were treated with 0.5 μg/ml Fas antibody CH-11 in the presence of 10 μg/ml cycloheximide for 2 h. Cells were then stained with an antibody to the C terminus of p115 (red), GM130 (green), and nuclei were stained with Hoechst 33238 (blue). Bar, 10 μm. Note the accumulation of p115-immunoreactive material in nuclei prior to significant Golgi fragmentation (panels E–H). B, HeLa cells (panels A–L) were treated as above for 2 (panels E–H) or 4 h (panels I–L). Cells were then stained with an antibody to the C terminus of p115 (red), cytochrome c (green), and nuclei were stained with Hoechst 33238 (blue). Bar, 10 μm. C, quantitation of the morphological phenotypes in response to Fas activation. Approximately 100 cells were counted each by two separate individuals; data are the average of two experiments.
To demonstrate that the early nuclear localization of p115 was not confined only to Fas-activated HeLa cells, COS-7 cells were treated with two well characterized activators of apoptosis, etoposide or anisomycin (supplement Fig. S1). In response to etoposide treatment, p115 had a diffuse, punctate appearance, whereas GM130 maintained its normal juxtanuclear localization; notably, p115 staining was evident in nuclei although the cells were not significantly apoptotic at this time. In cells treated with anisomycin the Golgi apparatus had a loose appearance; furthermore, p115 staining was evident in nuclei. Together, these results demonstrated that the endogenous p115 CTF translocated to the cell nucleus and in response to Fas or etoposide treatment, this preceded Golgi fragmentation. Because immunoreactive-GM130 was not observed in nuclei at this time (supplement Fig. S1), we concluded that the appearance of CTF in nuclei was a specific transport event and not a consequence of nuclear envelope breakdown. Together with our previous observations (13), these data are consistent with the hypothesis that p115 cleavage and CTF nuclear translocation are early apoptotic events.
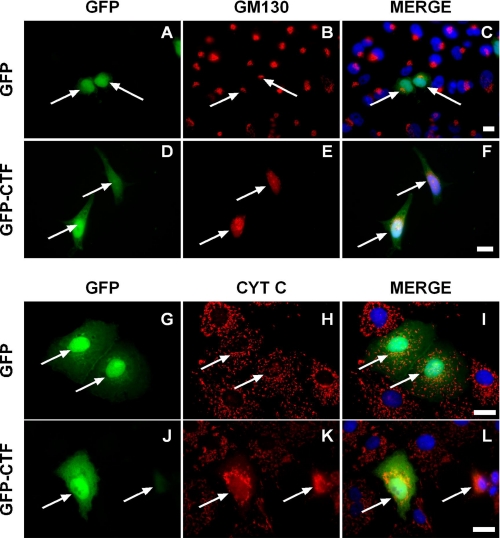
Having demonstrated that the endogenous p115 CTF accumulated in cell nuclei during activation of apoptosis, our goal was to determine whether nuclear translocation was required for CTF-induced apoptosis. Consequently, it was important to determine whether CTF expression induced cytochrome c release or if it functioned independently of the mitochondrial pathway. To address these questions, COS-7 cells were transfected with either GFP alone or GFP-CTF and analyzed by immunofluorescence microscopy using a monoclonal antibody to GM130 or cytochrome c (Fig. 2). Cells transfected with GFP alone showed normal Golgi and cytochrome c staining, whereas ∼60% of those expressing GFP-CTF had a disrupted Golgi apparatus (Fig. 2, panels D–F; see also Fig. 7). Significantly, in these cells cytochrome c staining was quite diffuse consistent with its release into the cytoplasm and the mitochondria aggregated significantly (Fig. 2, panels J–L), indicating that the cells were in an early phase of apoptosis. This result was consistent with observations that mitochondrial aggregation precedes release of cytochrome c and is an upstream apoptotic event (13, 24).
FIGURE 2.
Expression of the p115 CTF induces Golgi fragmentation and disrupts mitochondrial morphology. COS-7 cells were transfected with either GFP alone (panels A–C and G–I) or GFP-CTF (panels D–F and J–L). Cells were fixed 16 h post-transfection and stained with either a monoclonal antibody to GM130 (panels B and E) or cytochrome c (panels H and K) and Hoechst (blue). Bar, 10 μm. Note: in cells expressing full-length CTF, cytochrome c staining was either diffuse, consistent with its release into the cytoplasm, or mitochondria were aggregated and distinct from their punctate, lacy appearance in control cells.
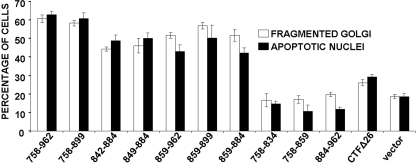
FIGURE 7.
Quantitation of Golgi fragmentation and apoptosis in response to expression of various p115 CTF truncation mutants. COS-7 cells were transfected with the indicated GFP-tagged CTF constructs and stained with GM130 and Hoechst to identify the Golgi apparatus and nuclei, respectively. Golgi fragmentation and apoptosis were quantified by counting ∼300 cells expressing each construct; each set of cells was counted by two individuals. Data are the average of three separate experiments.
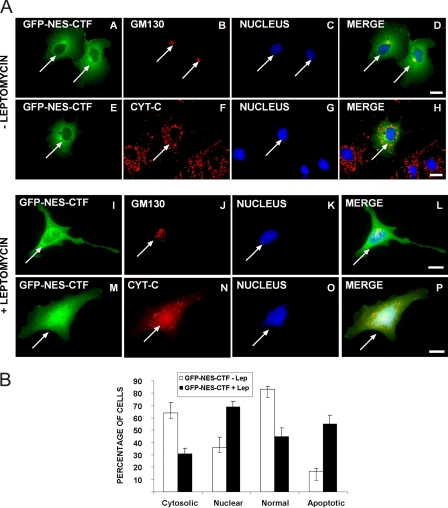
Based on these observations we speculated that the CTF might act on mitochondria directly to release cytochrome c thereby activating the apoptosome. However, because the data of Fig. 1 and our previous studies (10) showed that CTF was present in the nuclei of apoptotic cells, it was likely that the effect on mitochondria occurred as a consequence of nuclear translocation. To distinguish between these possibilities, we expressed a chimera comprising GFP-CTF fused to the HIV-1 Rev protein nuclear export signal (GFP-NES-CTF; Fig. 3). This construct, which was excluded from nuclei, accumulated in the perinuclear region of the cell and had a diffuse reticular appearance characteristic of the ER. Despite its abundant expression this chimera had no effect on the morphology of either the Golgi apparatus or mitochondria and we observed few apoptotic cells (Fig. 3, panels A–D and E--H). If CTF nuclear translocation were required to induce apoptosis, abrogation of the NES by treating these cells with the antibiotic leptomycin would lead to nuclear accumulation of CTF, induction of apoptosis, and Golgi fragmentation. Consistent with this idea, in cells treated with leptomycin (Fig. 3, panels I–L and M--P) the CTF accumulated in the nucleus, and within 4 h the Golgi apparatus was fragmented, cytochrome c staining became quite diffuse and mitochondrial aggregation was evident. Quantitative analysis (Fig. 3B) showed that less than 20% of GFP-NES-CTF expressing cells were apoptotic in the absence of leptomycin, whereas following drug treatment ∼55 to 60% became apoptotic within 4 h. Importantly, these results suggested that CTF did not interact with mitochondria directly to induce cytochrome c release, but were in agreement with our observation that CTF nuclear translocation was required to induce apoptosis.
FIGURE 3.
The presence of a nuclear export signal results in cytoplasmic accumulation of p115 CTF and lack of apoptosis. A, COS-7 cells were transfected with cDNA encoding a fusion protein consisting of GFP, the nuclear export signal of HIV Rev protein, and full-length CTF (GFP-NES-CTF). Approximately 18 h after transfection the cells were stained with monoclonal antibodies to either GM130 (red, panels B and J) or cytochrome c (red, panels F and N) and Hoechst to identify nuclei (blue). Treatment of cells with 10 ng/ml of leptomycin for 4 h (panels I–P) led to nuclear import of the GFP-NES-CTF chimera and induction of apoptosis (panels I–P). Bar, 10 μm. B, quantitation of the effects of leptomycin. The localization of GFP-NES-CTF and morphological changes in response to leptomycin treatment were determined by counting ∼300 cells by two separate individuals.
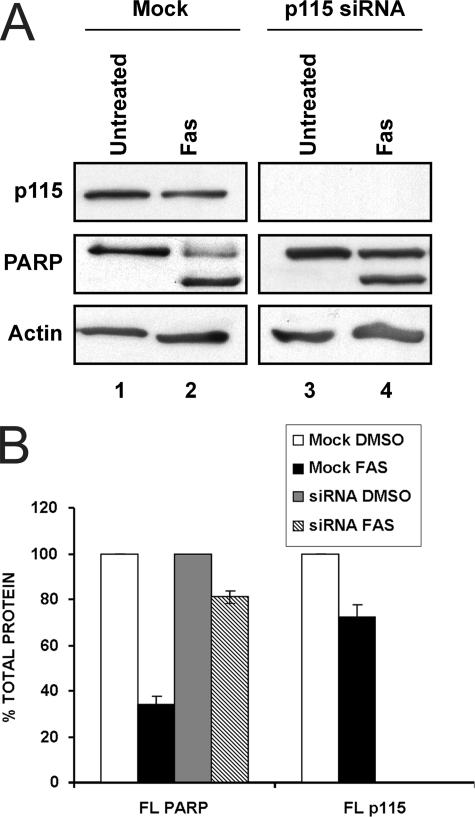
p115 Knockdown Delays Apoptosis—Our previous data are consistent with a model whereby caspase cleavage of p115 generates the CTF fragment that functions in enhancing or potentiating the apoptotic response. Consequently, depletion of p115 might transiently delay the onset of drug-induced apoptosis. To test this idea, cells depleted of p115 by using siRNA were incubated in the absence or presence of agonistic Fas antibody to induce apoptosis and the level of PARP, a marker of apoptosis was determined (Fig. 4). Fas treatment of cells transfected with mock siRNA led to efficient cleavage of full-length (∼110 kDa) PARP (∼66% of FL PARP was cleaved after 4 h) to release the p85 fragment (Fig. 4, panels A and B). In contrast, in cells with diminished p115 levels, ∼19% of full-length PARP was cleaved after 4 h compared with untreated cells (compare lanes 2 and 4 in Fig. 4, A and B) suggesting that the presence of p115 CTF enhanced apoptosis. Surprisingly, the apparent level of the p85 fragment did not appear significantly different in mock and p115 siRNA cells. This was likely due to differential recognition of FL PARP and the p85 fragment by the PARP antibody and to the possibility that the fragment was further cleaved at later stages of apoptosis, which would be delayed by the absence of p115 CTF. Similar results were obtained when cells were treated with anisomycin and apoptosis was induced via the intrinsic pathway (data not shown).
FIGURE 4.
p115 knockdown delays the induction of apoptosis. A, HeLa cells were transfected with mock siRNA (lanes 1 and 2) or p115 siRNA for 96 h (lanes 3 and 4), after which they were either treated with 0.5 μg/ml Fas antibody CH-11 in the presence of 10 μg/ml of cycloheximide for 4 h or with cycloheximide alone. Cell lysates were analyzed by Western blot using antibodies to p115, PARP, and actin. B, quantitation of PARP and p115 cleavage in response to Fas treatment was determined by comparing the levels of full-length PARP and p115, respectively, in response to the indicated treatments. Data are the average of three independent experiments.
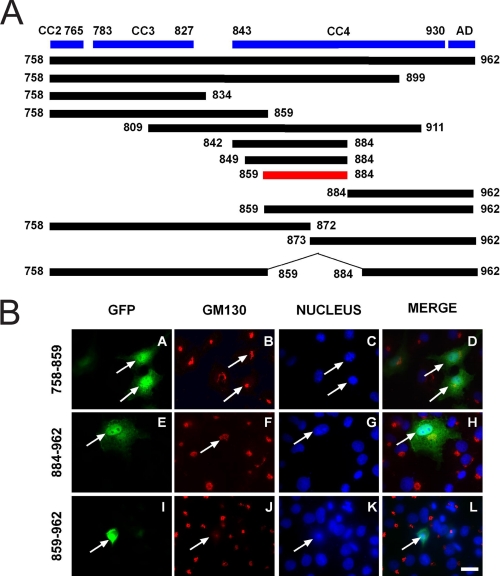
Identification of the p115 CTF Minimal Apoptotic Domain— The preceding data demonstrated that generation of the CTF and its nuclear import stimulated the apoptotic response. To identify the minimal domain within the CTF that could induce apoptosis, a series of N- and C-terminal truncation mutants were generated (Fig. 5, panel A). Representative micrographs of three such GFP-tagged constructs (residues 758–859, 884–962, and 859–962) are shown (Fig. 5B); FLAG-tagged CTF constructs gave similar results (data not shown). All three chimeras translocated to the nucleus; however, neither GFP-CTF-(758–859) nor GFP-CTF-(884–962) caused significant apoptosis. Most importantly, expression of the CTF-(859–962) truncation mutant induced apoptosis. These experiments suggested that amino acids 859–884 were required to generate the apoptotic phenotype. Indeed, inclusion of amino acids 859–884 in any of the GFP-CTF constructs resulted in both Golgi fragmentation and cell death, whereby cells became rounded, shrunken and in many cases the Golgi apparatus was not evident (supplement Fig. S2).
FIGURE 5.
Deletion mutagenesis reveals a key domain in the p115 CTF that confers the apoptotic phenotype. A, schematic representation of various GFP- and FLAG-tagged constructs of human p115 CTF. The positions of the coiled-coil (CC) domains 2, 3, and 4 are indicated as well as that of the C-terminal acidic GM130/giantin binding region. B, COS-7 cells were transfected with GFP-tagged CTF constructs: p115-(758–859) (panels A–D), p115-(884–962) (panels E–H), and p115-(859–962) (panels I–L). Cells were fixed after 18 h of transfection, and stained with a monoclonal antibody to GM130 (red) and Hoechst 33238 (blue). Transfected cells were identified by GFP fluorescence (green). Bar, 10 μm. Similar results were obtained using FLAG-tagged constructs (data not shown).
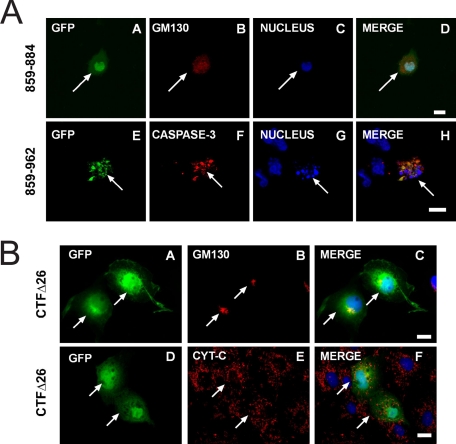
Confirmation that residues 859–884 were key in effecting apoptosis came from expression of a GFP construct comprising 859–884 alone (Fig. 6A, panels A–D). In cells expressing this construct, GM130 staining was diffuse and the normal tight Golgi morphology of COS-7 cells was absent. Additionally, cells expressing a construct encoding amino acids 859–962 fused to GFP were positive for staining with an antibody that recognizes only active caspase-3 and not the precursor procaspase-3 (Fig. 6A, panels E–H). Furthermore, cells transfected with this chimera also exhibited a markedly fragmented nuclei characteristic of apoptosis. These results suggested that amino acids 859–884 were sufficient to induce apoptosis, albeit less efficiently than the full-length CTF (Fig. 7). Thus, whereas ∼65% of cells expressing GFP-CTF-(758–962) were apoptotic and had a fragmented Golgi apparatus, this number was reduced to ∼45% in cells expressing the 859–884 CTF construct. Additional evidence that residues 859–884 were necessary for apoptosis was provided by a GFP-CTF construct lacking these amino acids (GFP-CTFΔ26) (Fig. 6B). In this case, cells expressing the CTFΔ26 mutant appeared to have normal Golgi apparatus (Fig. 6B, panels A–C) and the mitochondrial morphology was indistinguishable from control cells (Fig. 6B, panels D–F); additionally, its expression was only slightly more effective than GFP alone in mediating apoptosis (Fig. 7). Further dissection of this domain by dividing it into two separate halves, residues 758–872 and 873–962, showed that neither construct induced apoptosis (supplement Fig. S2). Quantitative analysis of all the various GFP-CTF truncation mutants confirmed that residues 859–884 constituted the most potent apoptotic domain of the p115 CTF in that expression of any construct that included this stretch of amino acids resulted in ∼50–60% of the cells exhibiting both an apoptotic phenotype and a fragmented Golgi apparatus (Fig. 7 and supplemental Fig. S3).
FIGURE 6.
Expression of p115 residues 859–884 is necessary and sufficient to induce apoptosis. A, COS-7 cells were transfected with either cDNA encoding GFP-tagged p115-(859–884) or GFP-tagged CTF-(859–962). At 18 h post-transfection, the cells were stained with an antibody to GM130 (panel B), an antibody that recognizes only active caspase-3 (panel F), and Hoechst to identify nuclei (blue). B, COS-7 cells were transfected with GFP-CTFΔ26 (panels A–F). Cells were fixed at 18 h post-transfection and stained with a monoclonal antibody to GM130 (panel B) or cytochrome c (panel E) and Hoechst (blue). Bar, 10 μm.
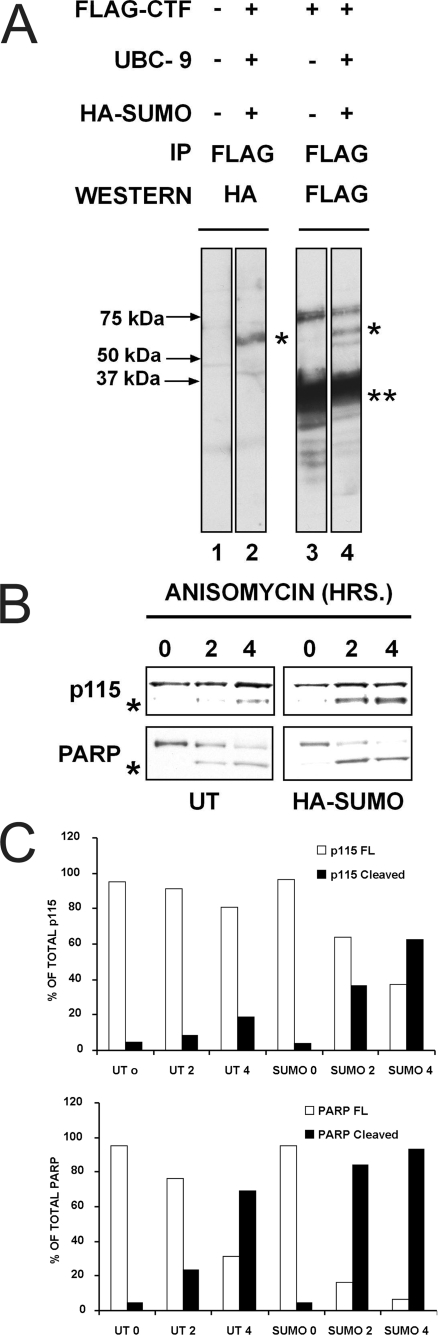
p115 CTF Is SUMOylated—Sequence analysis revealed that the CTF lacks a nuclear localization signal; consequently it was unclear how this polypeptide translocated into the nucleus. By exploiting a bioinformatics program, we predicted that residues 860–863 (Leu-Lys-Asp-Glu), 790–793 (Leu-Lys-Gln-Glu), and 902–905 (Ser-Lys-Lys-Glu) constituted putative SUMOylation sites. Significantly, it has been found in other contexts that SUMOylation is crucial for transcription, signal transduction, and most relevant to our studies, nuclear transport, stress response, and apoptosis (25, 26). To test the prediction that CTF might be SUMOylated in vivo, HeLa cells were transfected with cDNAs encoding either FLAG-tagged CTF alone, or FLAG-tagged CTF together with HA-tagged SUMO-1 and UBC9, a SUMO ligase. Cell lysates were then immunoprecipitated with a monoclonal anti-FLAG antibody, followed by Western blotting using either antibodies against FLAG or HA (Fig. 8). As expected, control untransfected cells did not show any SUMO-related polypeptides (Fig. 8A, lanes 1). Strikingly, cells transfected with all three constructs revealed that the CTF was SUMOylated in that a SUMO-immunoreactive polypeptide of ∼55 kDa was evident in these cells (Fig. 8A, lanes 2 and 4, asterisks). To determine whether SUMOylation affected the induction/or efficiency of apoptosis, cells were transfected with a plasmid encoding HA-SUMO and treated with anisomycin to induce apoptosis (Fig. 8B). When compared with control untransfected cells, those expressing HA-SUMO were more sensitive to anisomycin treatment as determined by p115 and PARP cleavage. In these cells, p115 cleavage to the 90-kDa fragment occurred earlier and more efficiently than in control cells; similarly, the efficiency of PARP processing to p85 was enhanced in HA-SUMO-transfected cells (Fig. 8C). Following 4 h treatment with anisomycin, in ∼63% of cells expressing HA-SUMO, p115 was cleaved compared with ∼20% in untransfected cells. Similarly, the kinetics of PARP processing were enhanced in SUMO expressing cells; thus following a 2-h anisomycin treatment ∼84% of cells had generated p85 PARP, whereas PARP processing was only ∼24% in untransfected, anisomycin-treated cells at this time (Fig. 8, B and C). Together, these data strongly suggest that SUMOylation of p115 CTF enhanced its proapoptotic activity.
FIGURE 8.
The p115 CTF is SUMOylated in vivo. A, HeLa cells were either untransfected (lane 1), transfected with 3×FLAG-tagged CTF (lane 3), or co-transfected with plasmids encoding 3×FLAG-tagged CTF, UBC9, and HA-tagged SUMO (lanes 2 and 4). At 18 h post-transfection, cell lysates were immunoprecipitated using a monoclonal antibody to the FLAG tag coupled to Sepharose beads. The immunoprecipitated material was resolved by SDS-PAGE and subjected to Western blot analysis using monoclonal antibodies to either HA (lanes 1 and 2) or FLAG (lanes 3 and 4). Single asterisks indicate SUMOylated FLAG-tagged CTF; double asterisks correspond to FLAG-tagged CTF. The band migrating above SUMOylated FLAG-CTF (∼75 kDa) likely corresponds to a dimeric form of CTF (lanes 3 and 4). B, HeLa cells were either untransfected or transfected with HA-SUMO and treated with 5 μg/ml anisomycin for 0, 2, and 4 h. Cell lysates were analyzed by Western blot using antibodies to p115 and PARP; asterisks correspond to the cleavage products of p115 (90 kDa) and PARP (p85), respectively. C, quantitation of PARP and p115 cleavage in response to anisomycin treatment in the presence or absence of HA-SUMO expression.
DISCUSSION
Nuclear Translocation of the p115 CTF Is Required for Apoptosis—p115 is a high molecular weight coiled-coil protein, which plays a key role in Golgi biogenesis and assembly (14, 27, 28). Its knockdown results in fragmentation of the Golgi apparatus (20). Different regions of the p115 N terminus have been implicated in binding to SNAREs, Rab1, and more recently COG2 (18, 29, 30) although how these interactions are integrated to mediate Golgi assembly is still not well understood. Additionally, the C terminus of p115 has been shown to bind GM130 and giantin via an ∼30 residue highly acidic domain and these interactions were proposed to mediate post-mitotic Golgi assembly (31, 32). Previous work from our laboratory has shown that during apoptosis p115 is cleaved by both upstream and executioner caspases, namely caspases-8 and -3, respectively, to generate an N terminus of 757 amino acids and a C-terminal fragment (CTF) of 205 residues; significantly, in cells activated by antibody cross-liking of the Fas receptor, this was demonstrated to be an early apoptotic event (13). Most significantly, expression of the p115 CTF led to its localization in the cell nucleus and the induction of apoptosis (10). However, it was unclear whether nuclear localization was a consequence of CTF overexpression, if the endogenous p115 CTF polypeptide would localize similarly and more importantly, if CTF nuclear translocation were a prerequisite for the induction of apoptosis. Furthermore, although the C-terminal acidic domain is not required for apoptosis (10), the actual CTF residues that confer apoptosis had not been determined.
To address these questions we used an antibody that specifically recognized the C-terminal domain of p115 and confirmed that in apoptotic cells endogenous immunoreactive-CTF was present in nuclei (Fig. 1). Consistent with our previous results, following 2 h Fas activation, the p115 CTF was present in nuclei prior to significant fragmentation of the Golgi apparatus or apoptosis. These data showed that the appearance of nuclear immunoreactive p115 was not a function of overexpression and that it was an early apoptotic event. p115 was also present in the Golgi apparatus at this time, suggesting that a fraction of the protein remained uncleaved. This observation is consistent with our previous data (13) that showed that although CTF was evident early during apoptosis, complete cleavage of p115 was not observed until later times. This would explain why at early times after induction of apoptosis the nuclear accumulation of CTF did not correlate with Golgi fragmentation (Fig. 1C). Our data also demonstrated that nuclear translocation of p115 CTF was essential to induce apoptosis because appending a nuclear export signal to the CTF suppressed its proapoptotic activity and furthermore, treatment of cells with leptomycin abrogated the NES effect (Fig. 2). This result suggested that CTF transport into the nucleus was necessary to induce and accelerate apoptosis triggered by external stimuli. In cells expressing the CTF-NES chimera, the fusion protein had both a perinuclear and reticular localization characteristic of the ER (Fig. 3). We had expected that it would accumulate in the cytoplasm and act as dominant-negative with respect to p115 interaction with GM130 and giantin resulting in Golgi fragmentation but not apoptosis. Although Golgi fragmentation was evident in some cells expressing the CTF-NES (data not shown), in most cells the Golgi morphology was normal. It was possible that the 11 amino acids of the HIV-1 Rev NES altered CTF structure such that it was unable to interact with either GM130 and/or giantin. However, this was unlikely because this sequence was located at the N terminus, upstream of the acidic domain (residues 933–962) required for GM130/giantin binding. In cells expressing CTF-NES its staining had a corona-like morphology (Fig. 3); based on this appearance, we speculate that the CTF may interact with proteins on the cytoplasmic face of the nuclear envelope and consequently, may be prevented from interacting with the Golgi apparatus. However, because in the presence of leptomycin this construct was retained in the nucleus and caused Golgi disruption as well as apoptosis, this suggested its action was via activation of apoptosis rather than due to a dominant negative effect on interactions between GM130, giantin, and p115. Furthermore, knockdown of p115 revealed that the absence of CTF led to a partial delay in apoptosis (Fig. 4). This observation was consistent with earlier studies showing that a caspase-resistant triple mutant of golgin-160 could delay the onset of apoptosis and strongly supports our idea that p115 cleavage plays a role in accelerating the apoptotic response.
A series of truncated CTF polypeptides were constructed that allowed us to define 26 amino acids (residues 859–884) that were essential for apoptosis (Figs. 5, 6, 7). Analysis of this “26-mer” domain revealed that it has a pI of 4.63 and is predicted to form an α-helix; it is also noteworthy that it lies within the fourth coiled-coil domain of p115 (Fig. 5). Given the lack of an obvious nuclear localization signal, we predicted that the CTF would translocate to the nucleus by “piggy-backing” on a protein(s) that normally shuttles to and from the nucleus. To identify such a protein, we used GST-CTF, His-tagged-CTF chimeras, and a TAP-tagged CTF for expression in HeLa cells to identify putative binding partners; however, other than GM130, we were unable to identify novel interacting proteins either in vitro or in vivo (data not shown).
p115 CTF Is SUMOylated—Inspection of the 26-amino acid sequence revealed that it contained three potential SUMOylation sites. Strikingly, co-expression of the CTF, with the SUMO-ligase UBC9 and HA-tagged SUMO confirmed that it was SUMOylated in vivo (Fig. 8). Most significantly, co-expression of SUMO and its ligase led to increased p115 cleavage and enhanced apoptosis, as measured by PARP processing, demonstrating that SUMO is important for stimulating the function of the p115 CTF (Fig. 8). Like ubiquitination, SUMO modification occurs on many proteins including those that regulate gene expression including transcription factors and co-factors, chromatin-modifying enzymes, histones, and proteins associated with the nuclear pore complex (33). However, our attempts to identify the endogenous SUMOylated CTF were unsuccessful; in part this was due of the transient nature of SUMOylation (25) and because of the rapid turnover of the CTF.3 SUMO modification of proteins has been implicated in protein targeting to the nuclear pore complex, e.g. RanGAP1 and regulating nucleocytoplasmic shuttling of transcription factors (34). Furthermore, during anisomycin-induced apoptosis SUMOylation was shown to mediate caspase-8 localization to nuclei (25). In light of these observations, we speculated that SUMOylation mediated efficient translocation of CTF to nuclei and tested this idea by generating CTF mutants in which Lys791, Lys861, and Lys902 were mutated to Ala. Surprisingly, mutation of each site individually or together did not perturb apoptosis and the CTF was nevertheless SUMOylated (data not shown). This suggested that either CTF may not be SUMOylated at the predicted sites or that other non-canonical SUMO sites served as substrates for SUMOylation by the promiscuous activity of UBC9. In this context, several studies including proteomic approaches have defined lysine modifications in non-canonical SUMO sequences (35, 36). Currently, experiments are in progress to map these sites and determine the role of SUMO in mediating CTF nuclear import.
Finally, our observations suggest that CTF could be part of a more general cellular stress program, the common feature of which is the nuclear import of a polypeptide derived from a cytoplasmic organelle, which alters transcription. For example, in response to the accumulation of misfolded proteins in the ER or in hepatocytes deprived of cholesterol, ATF6 and SREBP, respectively, are transported from the ER to the Golgi apparatus where they undergo S1P- and S2P-mediated proteolysis that releases a polypeptide that upon delivery to the nucleus alters transcription (reviewed in Refs. 37 and 38). Although there are clear differences between the cleavage of these proteins and p115, we speculate that similar to these proteins, the CTF acts as a co-factor to modify expression of apoptotic genes; currently, experiments are in progress to test this idea.
Supplementary Material
Acknowledgments
We thank Dr. Sara Salinas for suggesting that CTF may posses a SUMOylation site; Drs. Moshe Sadofsky and Vyacheslav Yurchenko for help and advice on the SUMOylation experiments as well as generous gifts of anti-SUMO and UBC9 antibodies. We thank Drs. Gerry Waters and Dr. Adam Linstedt for antibodies to p115 and Dr. Yukio Ikehara for the plasmid encoding p115. We also thank Dr. Christian Riebeling and Jenny Nachbar for helpful suggestions with the manuscript.
This paper is dedicated to the memory of Dr. Dennis Shields.
This work was supported, in whole or in part, by National Institutes of Health Grant DK21860 (to D. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PARP, poly(ADP-ribose) polymerase; HA, hemagglutinin; NES, nuclear export signal; siRNA, small interfering RNA; GFP, green fluorescent protein; CTF, C-terminal fragment; SUMO, small ubiquitin-like modifier.
S. Mukherjee and D. Shields, unpublished observations.
References
- 1.Shorter, J., and Warren, G. (2002) Annu. Rev. Cell Dev. Biol. 18 379-420 [DOI] [PubMed] [Google Scholar]
- 2.Warren, G., and Malhotra, V. (1998) Curr. Opin. Cell Biol. 10 493-498 [DOI] [PubMed] [Google Scholar]
- 3.Sutterlin, C., Hsu, P., Mallabiabarrena, A., and Malhotra, V. (2002) Cell 109 359-369 [DOI] [PubMed] [Google Scholar]
- 4.Maag, R. S., Hicks, S. W., and Machamer, C. E. (2003) Curr. Opin. Cell Biol. 15 456-461 [DOI] [PubMed] [Google Scholar]
- 5.Sesso, A., Fujiwara, D. T., Jaeger, M., Jaeger, R., Li, T. C., Monteiro, M. M., Correa, H., Ferreira, M. A., Schumacher, R. I., Belisario, J., Kachar, B., and Chen, E. J. (1999) Tissue Cell 31 357-371 [DOI] [PubMed] [Google Scholar]
- 6.Lowe, M., Lane, J. D., Woodman, P. G., and Allan, V. J. (2004) J. Cell Sci. 117 1139-1150 [DOI] [PubMed] [Google Scholar]
- 7.Barr, F. A., and Short, B. (2003) Curr. Opin. Cell Biol. 15 405-413 [DOI] [PubMed] [Google Scholar]
- 8.Lane, J. D., Lucocq, J., Pryde, J., Barr, F. A., Woodman, P. G., Allan, V. J., and Lowe, M. (2002) J. Cell Biol. 156 495-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancini, M., Machamer, C. E., Roy, S., Nicholson, D. W., Thornberry, N. A., Casciola-Rosen, L. A., and Rosen, A. (2000) J. Cell Biol. 149 603-612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, R., Novikov, L., Mukherjee, S., and Shields, D. (2002) J. Cell Biol. 159 637-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker, A., Ward, C., Sheldrake, T. A., Dransfield, I., Rossi, A. G., Pryde, J. G., and Haslett, C. (2004) Biochem. Biophys. Res. Commun. 316 6-11 [DOI] [PubMed] [Google Scholar]
- 12.Maag, R. S., Mancini, M., Rosen, A., and Machamer, C. E. (2005) Mol. Biol. Cell 16 3019-3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee, S., Chiu, R., Leung, S. M., and Shields, D. (2007) Traffic 8 369-378 [DOI] [PubMed] [Google Scholar]
- 14.Sapperstein, S. K., Walter, D. M., Grosvenor, A. R., Heuser, J. E., and Waters, M. G. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 522-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shorter, J., Beard, M. B., Seemann, J., Dirac-Svejstrup, A. B., and Warren, G. (2002) J. Cell Biol. 157 45-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shorter, J., and Warren, G. (1999) J. Cell Biol. 146 57-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez, C., Fujita, H., Hubbard, A., and Sztul, E. (1999) J. Cell Biol. 147 1205-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allan, B. B., Moyer, B. D., and Balch, W. E. (2000) Science 289 444-448 [DOI] [PubMed] [Google Scholar]
- 19.Puthenveedu, M. A., and Linstedt, A. D. (2001) J. Cell Biol. 155 227-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puthenveedu, M. A., and Linstedt, A. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1253-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puthenveedu, M. A., Bachert, C., Puri, S., Lanni, F., and Linstedt, A. D. (2006) Nat. Cell Biol. 8 238-248 [DOI] [PubMed] [Google Scholar]
- 22.Sohda, M., Misumi, Y., Yoshimura, S., Nakamura, N., Fusano, T., Sakisaka, S., Ogata, S., Fujimoto, J., Kiyokawa, N., and Ikehara, Y. (2005) Biochem. Biophys. Res. Commun. 338 1268-1274 [DOI] [PubMed] [Google Scholar]
- 23.Danial, N. N., and Korsmeyer, S. J. (2004) Cell 116 205-219 [DOI] [PubMed] [Google Scholar]
- 24.Haga, N., Fujita, N., and Tsuruo, T. (2003) Oncogene 22 5579-5585 [DOI] [PubMed] [Google Scholar]
- 25.Besnault-Mascard, L., Leprince, C., Auffredou, M. T., Meunier, B., Bourgeade, M. F., Camonis, J., Lorenzo, H. K., and Vazquez, A. (2005) Oncogene 24 3268-3273 [DOI] [PubMed] [Google Scholar]
- 26.Lin, D. Y., Huang, Y. S., Jeng, J. C., Kuo, H. Y., Chang, C. C., Chao, T. T., Ho, C. C., Chen, Y. C., Lin, T. P., Fang, H. I., Hung, C. C., Suen, C. S., Hwang, M. J., Chang, K. S., Maul, G. G., and Shih, H. M. (2006) Mol. Cell 24 341-354 [DOI] [PubMed] [Google Scholar]
- 27.Seemann, J., Jokitalo, E. J., and Warren, G. (2000) Mol. Biol. Cell 11 635-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, D. S., Alvarez, C., Gao, Y. S., Garcia-Mata, R., Fialkowski, E., and Sztul, E. (1998) J. Cell Biol. 143 319-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sohda, M., Misumi, Y., Yoshimura, S., Nakamura, N., Fusano, T., Ogata, S., Sakisaka, S., and Ikehara, Y. (2007) Traffic 8 270-284 [DOI] [PubMed] [Google Scholar]
- 30.Beard, M., Satoh, A., Shorter, J., and Warren, G. (2005) J. Biol. Chem. 280 25840-25848 [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, N., Lowe, M., Levine, T. P., Rabouille, C., and Warren, G. (1997) Cell 89 445-455 [DOI] [PubMed] [Google Scholar]
- 32.Sonnichsen, B., Lowe, M., Levine, T., Jamsa, E., Dirac-Svejstrup, A. B., and Warren, G. (1998) J. Cell Biol. 140 1013-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, E. S., and Blobel, G. (1999) J. Cell Biol. 147 981-994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salinas, S., Briancon-Marjollet, A., Bossis, G., Lopez, M. A., Piechaczyk, M., Jariel-Encontre, I., Debant, A., and Hipskind, R. A. (2004) J. Cell Biol. 165 767-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panse, V. G., Hardeland, U., Werner, T., Kuster, B., and Hurt, E. (2004) J. Biol. Chem. 279 41346-41351 [DOI] [PubMed] [Google Scholar]
- 36.Melchior, F., Schergaut, M., and Pichler, A. (2003) Trends Biochem. Sci. 28 612-618 [DOI] [PubMed] [Google Scholar]
- 37.Ron, D., and Walter, P. (2007) Nat. Rev. Mol. Cell. Biol. 8 519-529 [DOI] [PubMed] [Google Scholar]
- 38.Espenshade, P. J. (2006) J. Cell Sci. 119 973-976 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.