Abstract
Glycoprotein Ib α (GpIbα), a trans-membrane glycoprotein, is expressed on the surface of megakaryocytes and platelets, where, in association with glycoprotein Ib β, glycoprotein V, and glycoprotein IX, it normally forms the von Willebrand factor receptor (vWFR). A fully functional vWFR is necessary for platelet attachment, aggregation, and activation and has also been shown to regulate megakaryocyte ploidy. We have recently shown that the gene encoding GpIbα is a transcriptional target for the c-Myc oncoprotein and is more widely expressed than previously thought, with particularly high levels occurring in transformed cells. Indeed, GpIbα can substitute for c-Myc in promoting growth, transformation, and genomic instability. In the current work, we have demonstrated that, despite the promiscuous expression of GpIbα, other vWFR subunits remain largely restricted to megakaryocytes. We have characterized a panel of GpIbα mutants and shown that some regions of the protein essential for vWFR activity are not necessary for c-Myc-like functions. Specifically, the six C-terminal amino acids of the cytoplasmic domain, which mediate vWFR signaling, are entirely dispensible for the c-Myc-like functions of GpIbα. Instead, these require a more membrane-proximal filamin-binding domain. Also important is the GpIbα signal peptide, which, in the absence of other vWFR subunits, directs GpIbα to the endoplasmic reticulum rather than the membrane. Together, these results provide strong evidence that the domains of GpIbα mediating c-Myc-like functions are modular, genetically distinct, and independent of those involved in vWFR signaling.
Glycoprotein Ib α (GpIbα)2 is a Type I trans-membrane glycoprotein that is expressed on the surface of megakaryocytes and platelets, where it associates with three other trans-membrane proteins, glycoprotein Ibβ (GpIbβ), glycoprotein V (GpV), and glycoprotein IX (GpIX), to form the von Willebrand factor receptor (vWFR) (1–3). GpIbα is initially synthesized as a 627-amino acid precursor, with a 610-amino acid membrane-bound mature form resulting from cleavage of the N-terminal signal peptide. vWF is exposed on damaged vascular endothelium, which allows for the attachment, aggregation, and activation of vWFR-bearing platelets during the initial stages of hemostasis.
In addition to its role in hemostasis, GpIbα also regulates bone marrow megakaryocyte ploidy and proliferation (4–6). These functions, as well as those associated with platelet aggregation and activation, require the extreme C terminus of the GpIbα cytoplasmic domain, which interacts with signaling proteins, such as 14-3-3ζ, c-Src, and phosphatidylinositol 3-kinase (4–8). In addition, a more membrane-proximal cytoplasmic domain communicates indirectly with the actin cytoskeleton via its association with filamin. This so-called filamin-binding domain serves to stabilize the entire vWFR and facilitate its transport to the cell surface (9, 10).
Recently, we showed that GpIbα is more widely expressed than previously appreciated and participates in aspects of c-Myc oncoprotein function that are unrelated to and independent of its role in platelet physiology. Specifically, we showed that the GPIBA gene is a direct downstream target for c-Myc and that GpIbα is both necessary and sufficient to promote the genomic instability (GI) that typically accompanies c-Myc deregulation (11–16). Consistent with this finding, cancer cell lines that overexpress c-Myc tend to have extremely high levels of GpIbα, whereas untransformed cells or primary cells tend to have extremely low or undetectable levels (15). GpIbα overexpression also transforms established cells in vitro, promotes tumorigenesis in vivo, reduces growth factor requirements, and enhances survival in the face of growth factor deprivation (15). Moreover, enforced expression of GpIbα in primary cells leads to the induction of tetraploidy and double-stranded DNA breaks (DSBs) and to p53-dependent premature senescence reminiscent of that seen in response to many transforming oncogenes (15, 17–19). Taken together, these unanticipated oncoprotein-like aspects of GpIbα indicate that it plays an integral role in mediating a number of distinct c-Myc phenotypes.
These unexpected properties of GpIbα raise a number of important questions. For example, which regions of the molecule are necessary for these multiple behaviors, and are they genetically separable? How do these regions relate to those that execute the traditional vWFR-dependent functions of GpIbα? Finally, if GpIbα is expressed more widely than previously realized, are GpIbβ, GpV, and GpIX also similarly expressed, and, if so, is their association with GpIbα required for its oncoprotein-like properties?
In the current work, we have investigated these and other questions pertaining to the means by which GpIbα exerts its oncoprotein-like functions. Our findings provide evidence that cells other than megakaryocytes do not coordinately express all vWFR subunits, that GpIbα need not be expressed at the cell surface in order for it to carry out its c-Myc-like functions, that certain of these functions are genetically separable, and that these regions may be quite different from those that mediate vWFR functions. Our results also imply that, in nonmegakaryocytic cells, GpIbα plays roles that are distinct from those related to hemostatasis.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions—Rat1a fibroblasts, amphotropic Phoenix retroviral packaging cells, human foreskin fibroblasts, and BJ/TTR cells (14, 15) were grown in Dulbecco's modified Eagle's minimal essential medium containing 10% fetal calf serum and supplemented with 2 mm glutamine, 100 units/ml penicillin G, and 100 mg/ml streptomycin. HL60 promyelocytic cells, HeLa, MCF7 breast cancer cells, and CaLu1 lung cancer cells were grown as described previously (20, 21). The Mo7e megakaryocyte line was cultured in Iscove's modified Dulbecco's modified Eagle's minimal essential medium containing 10% fetal calf serum, the above cited amounts of penicillin G and streptomycin, and 10 ng/ml recombinant granulocyte-macrophage colony-stimulating factor or 50 ng/ml recombinant thrombopoietin, respectively. All cell culture reagents were obtained from MediaTech (Herndon, VA) or R&D Systems (Minneapolis, MN). Normal human prostate epithelium and keratinocytes were purchased from Cambrex (East Rutherford, NJ) and grown in the cell type-specific media recommended by the supplier. Soft agar clonogenicity assays of Rat1a cells were performed as described previously (11, 12).
Rat1a proliferation assays were performed as described previously (13). Cells were trypsinized and seeded into 6-well plates a density of 2 × 104 cells/well. The following day, fresh medium containing 1% serum was added. At regular intervals thereafter and until cultures become confluent, viable cell counts were performed on triplicate samples using trypan blue exclusion.
Apoptosis assays were performed as described previously (13). Briefly, cells were seeded into 6-well plates and allowed to achieve ∼50% confluence. Monolayers were then washed three times in serum-free medium and maintained in serum-free medium for the remainder of the experiment. Viable cell counts using trypan blue exclusion were then performed at regular intervals in triplicate.
Immunoblotting and Subcellular Fractionation—Immunoblotting following SDS-PAGE was performed essentially as described using polyvinylidene difluoride membranes (Millipore, Billerica, MA) (13). Antibodies consisted of a monoclonal antibody (mAb) directed against the c-Myc epitope tag present on the C termini of all GpIbα constructs (clone 9E10; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), a mAb directed against β-tubulin (clone D10; Santa Cruz Biotechnology), and a rabbit polyclonal antibody directed against calnexin (SPA-860; StressGen Biotechnologies, San Diego CA). A murine mAb against human GpIbα antibody (WM23) was a generous gift from Dr. Michael Berndt). Membranes were blocked for at least 1 h in PBS, 0.15% Tween (PBS-T) containing 5% nonfat dry milk, followed by the addition of primary antibodies at the dilutions recommended by the supplier. Primary antibody incubations were performed overnight at 4 °C with constant agitation, followed by extensive washing in PBS-T. The membranes were then incubated with 1:20,000 dilutions of horseradish peroxidase-labeled goat anti-mouse IgG (sc-2005; Santa Cruz Biotechnology) or donkey anti-rabbit IgG (sc-2313; Santa Cruz Biotechnology) for 2 h at room temperature in PBS-T-milk followed by repeated washing. Membranes were developed using an enhanced chemiluminescence kit (SuperSignal West Femto Maximum Sensitivity Substrate; Pierce). Isolation of the endoplasmic reticulum (ER) was performed with a Sigma endoplasmic reticulum isolation kit (catalog number ER0100 –1KT) according to the directions of the supplier.
Fluorescence and Confocal Microscopy—Cells were trypsinized and seeded into 12-well plates containing 18-mm glass coverslips (VWR, Inc., West Chester, PA). 1–2 days later, coverslips were washed three times with PBS and fixed at room temperature for 10 min with freshly prepared 4% paraform-aldehyde-PBS. After washing an additional 2–3 times with PBS, coverslips were either used immediately or stored in PBS for no longer than 24 h. Immunostaining was performed with the above described anti-c-Myc epitope tag mAb, with the above described WM23 anti-GpIbα mAb, with another GpIbα mAb (LJ-Ibα1 mAb; gift of Dr. Zaverio Ruggieri), or with an anti-γ-H2AX mAb (JBW301; Upstate, Temacula, CA). Secondary antibodies consisted of AlexaFluor 594-conjugated goat anti-mouse and rabbit IgGs (1:1500 dilution) (Invitrogen). To show the lack of localization of GpIbα to the surface, Rat1a cells expressing various forms of the protein were stained with the LJ-IBα1 mouse anti-GpIbα mAb. Secondary staining was with AlexaFluor 610-labeled goat anti-mouse IgG. The cells were also stained with a rabbit polyclonal antibody directed against the transforming growth factor-β2 receptor (Santa Cruz Biotechnology) and secondarily stained with AlexaFluor 488 goat anti-rabbit IgG. Final nuclear staining was with 4′,6-diamidino-2-phenylindole.
Fluorescence microscopy was performed with an Olympus Provis fluorescence microscope (Olympus, Inc., Center Valley, PA), as described previously (13, 22). Confocal images were taken with an Olympus Fluoview 1000 confocal microscope, and Adobe Photoshop CS2 (version 9.0) was employed for image analysis.
Quantitative Real Time PCR (qRT-PCR)—Total RNA was isolated as described previously (14, 23) and was treated with Turbo DNase (Ambion, Inc., Austin, TX) according to the directions of the supplier. qRT-PCR was used to quantify the expression of mRNAs encoding each subunit of the human VWFR. We used the Primer 3 program (available on the World Wide Web) to select optimal primers for the coding regions of the human mRNAs for GpIbα (GenBank™ accession number NM_000173), GpIbβ (NM_000407), GpV (L11238), and GpIX (NM_000174). Specific primers and their predicted products are listed in Table S1. Primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). For each qRT-PCR reaction, we used 50 ng of total input RNA in a SYBR Green-based assay (Quantitect; Qiagen, Inc., Chatsworth, CA) according to the directions of the supplier. Reactions were performed in triplicate on a Roche LightCycler 2 (Roche Applied Science) using the conditions recommended by the supplier. All analyses used LightCycler Relative Quantification Software 4. S.E. values on all samples were <2%. Cτ values for each reaction were normalized to those for glyceraldehyde-3-phosphate dehydrogenase reactions that were included in parallel for each set of qRT-PCR runs (13). The specificity of all primer sets was confirmed by electrophoresis of select amplified DNAs on 4% NuSieve-GTG low melt agarose gels (Cambrex Biosciences, Inc., Rockland, ME) in order to demonstrate the presence of a single amplified fragment of the predicted size (not shown).
Generation and Expression of Gp1bα Deletion Mutants—Most GpIbα deletion mutants were generated using a QuikChange mutagenesis kit (Stratagene). Oligonucleotides were synthesized by IDT, Inc. and were purified by polyacrylamide gel electrophoresis. Sequences of mutagenic primers are shown in Table S2. The starting template in all cases consisted of the pLXSN-EYFP vector containing the wild-type Gp1bα coding region tagged at its C terminus with a c-Myc epitope (14, 15, 24). All deletions were sequenced in their entirety for verification. Plasmid DNAs were transfected into Phoenix A cells using Superfect reagent (Qiagen) (24). Viral supernatants were harvested 48–72 h later and used to infect recipient cells in the presence of 8 μg/ml Sequabrene (Sigma). Stable transfectants were then isolated by cell sorting (24), pooled, and expanded for all further studies.
RESULTS
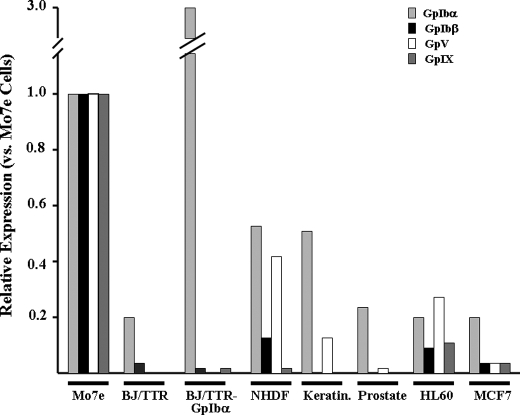
Widespread Expression of Endogenous GpIbα Occurs in the Absence of Other vWFR Subunits—We previously reported that GpIbα is expressed to varying degrees by many established and primary cells (14, 15). Elevated levels are seen in c-Myc-overexpressing cancer cell lines, particularly those with rearranged or amplified CMYC genes, such as Burkitt's lymphoma and HL60 promyelocytic leukemia (16, 17). However, the status of other vWFR subunits was not determined in those studies. To investigate this in more detail, we quantified transcript levels for GpIbα and each of the other three vWFR subunits in several nontransformed and transformed human cell lines and primary cell strains using qRT-PCR. As positive controls, we utilized the human megakaryoblastic cell line Mo7e, which expresses a functional vWFR (25), and a telomerase-immortalized line of fibroblasts (BJ/TTR cells) (14, 26) transduced with a GpIbα retroviral expression vector (BJ/TTR-GpIbα cells) (14). The latter cells express high levels of GpIbα, as determined by immunoblotting (16). As seen in Fig. 1, transcripts for GpIbα, GpIbβ, GpV, and GpIX were readily detected in Mo7e cells. In contrast BJ/TTR-GpIbα cells, although expressing GpIbα transcripts at levels 3-fold higher than those expressed by Mo7e cells, expressed barely detectable levels of GpIbβ, GpV, and GpIX transcripts. All other cell types tested expressed GpIbα transcripts at levels 20–50% of those measured in Mo7e cells but in most cases failed to express significant levels of GpIbβ, GpV, or GpIX transcripts. The only exception to this was seen in HL60 myeloid leukemia cells, which expressed low levels of all four transcripts. Although normal human diploid fibroblasts also expressed low levels of GpV and even lower levels of GpIbβ, they did not express significant levels of GpIX. These studies confirm our own earlier findings and those of others that GpIbα expression occurs in variety of cell types (14, 15, 27–29). They further show that, in most cases, this occurs independently of any significant or coordinated up-regulation of the other three vWFR subunit transcripts.
FIGURE 1.
Quantification of GpIbα, GpIbβ, GpV, and GpIX transcripts in established human cell lines and primary cells. Total RNAs from each of the indicated cell types were examined by qRT-PCR, as described under “Experimental Procedures.” All transcript levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase from the homologous RNA source. Mo7e megakaryocytes served as a positive control for all vWFR transcripts (27). Primary human cells examined included normal diploid fibroblasts (NHDF), keratinocytes (Keratin.), and normal prostate epithelium (Prostate). Established lines included telomerase-immortalized BJ/TTR fibroblasts, BJ/TTR-GpIbα fibroblasts (16, 17), HL60 promyelocytic leukemia cells, and MCF7 breast cancer cells (22, 23). In all cases, S.E. values of the triplicate samples were <2%.
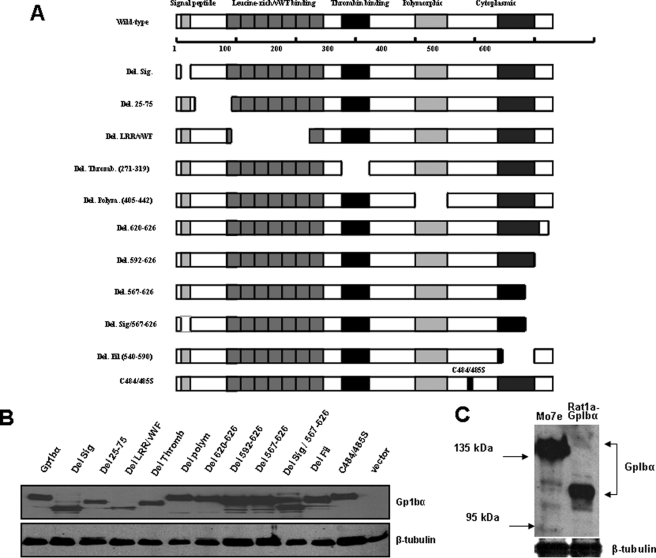
Expression of GpIbα Mutants—The paucity of transcripts for vWFR subunits other than GpIbα in most cell types (Fig. 1) suggested that the previously reported effects of GpIbα on transformation, proliferation, and genomic integrity (14, 15) were independent of its role in hemostasis. At the same time, it raised the question of how GpIbα performs these nontraditional functions and which regions of the molecule are involved. To address these issues, we generated a series of Myc epitope-tagged GpIbα mutants in the same retroviral backbone previously used to express the full-length protein (14, 15). These mutants consisted primarily of deletions in regions of the molecule involved in acknowledged vWFR-related functions (Fig. 2A and Table S3). For example, we generated mutants to test the need for GpIbα trafficking to the ER and cell surface (Del. Sig.), for vWF binding (Del.(25–75) and Del. LRR/vWF(75–210)), and for two distinct intracellular domains involved in 14-3-3ζ binding and phosphatidylinositol 3-kinase regulation (Del.(620–627)) and filamin binding (Del. Fil.(540–590)) in response to engagement of the receptor by vWF (5–7, 9, 10, 30). Another mutant (Del. Polym.) removed a 13-amino acid polymorphic region between residues Glu-396 and Thr-411 that is repeated up to three times in certain GpIbα alleles (31). This region has been proposed to be a site of O-glycosylation that can affect the susceptibility of platelets to shear-induced activation (31). Finally, a double point mutation (C384S/C385S) was generated to determine the consequences of abrogating the ability of GpIbα to be expressed on the cell surface in association with GpIbβ (3).
FIGURE 2.
Expression of human GpIbα mutants. A, schematic diagram depicting all of the mutants listed in Table S3. The top shows the full-length, 627-amino acid GpIbα protein and several of the domains known to be relevant for its function as a subunit of the vWFR. In platelets, the signal peptide is typically cleaved to yield a mature protein of 611 amino acids. However, the numbering of the protein here includes the signal peptide. Note that the cytoplasmic domain comprises two previously described functional elements; these comprise a membrane-proximal filamin-binding domain and a 6-amino acid C-terminal tail involved in vWRF-related signaling. All mutants were generated in the pLXSN-EYFP retroviral vector and were stably expressed as C-terminal c-Myc epitope-tagged proteins in Rat1a cells. B, immunoblotting of Rat1a cells stably expressing each of the mutants depicted in A. Comparable amounts of whole cell extracts were resolved by SDS-PAGE and processed for Western blotting, as described previously (16). The upper portion of the blot was probed with a mAb specific for the c-Myc epitope tag, and the bottom portion was probed with a mAb specific for β-tubulin. C, the molecular mass of GpIbα in transfected Rat1a cells was compared with that of endogenous GpIbα in Mo7e megakaryoyte cells, which express an intact vWFR (27). The blot was probed with the WM23 mAb that is specific for human GpIbα. Note that the latter protein possessed an apparent molecular mass significantly higher than that of the former (∼130–140 kDa versus ∼110–115 kDa) as a result of its more extensive post-translational glycosylation (29–32).
The above GpIbα mutants were stably expressed in Rat1a fibroblasts. As seen in Fig. 2B, most GpIbα proteins were expressed at similar levels. However, the wild-type protein, along with small deletion mutations or point mutations, migrated substantially faster (apparent molecular mass ∼115–120 kDa) than did endogenous GpIbα from the megakaryocytic cell line Mo7e (apparent molecular mass ∼135–140 kDa) (9, 32) (Fig. 2C). Several additional mutants, notably Del. Sig. and Del. Sig.(567–627), also migrated faster than predicted and/or were expressed at lower levels than the wild-type protein. Discrepancies in apparent molecular mass values probably reflect the failure to properly glycosylate the mature protein as a result of an impaired ability to traverse the ER → Golgi → cell surface pathway when other vWFR subunits are absent (9, 32, 33) (see below). This might be expected to activate the ER-associated degradation pathway, which has been shown to affect the half-life of an improperly processed or folded protein (32–34) (see below).
Subcellular Localization of GpIbα Mutants—Efficient cell surface localization of GpIbα requires that it be co-expressed with other subunits of the vWFR and covalently linked to GpIbβ via disulfide bonds (3, 30, 35). Because these subunits are not coordinately expressed in cells other than megakaryocytes (Fig. 1), we predicted that most of the free GpIbα would be inefficiently transported to the cell surface and thus remain confined to the ER. Indeed, initial imunohistochemical localization of ectopically expressed GpIbα in Rat11a cells and of endogenous human GpIbα in HeLa cells indicated that the protein was highly restricted to the cytoplasm (Fig. S1, A–D). These findings were further confirmed by confocal microscopy, which showed that GpIbα failed to co-localize at the cell surface with the transforming growth factor-β2 receptor (Fig. S2).
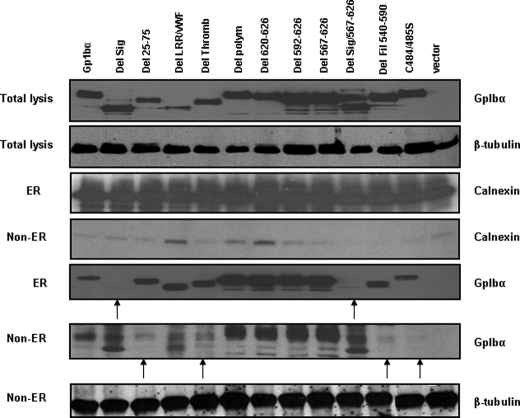
We next performed subcellular fractionation studies of Rat1a cells and examined the association of mutant GpIbα proteins with the ER. As controls for both protein loading and differential purification, the ER and non-ER cytoplasmic fractions were each probed for calnexin, an ER-associated protein, and for β-tubulin, a non-ER-associated protein. As shown in Fig. 3, all GpIbα proteins except those lacking the signal peptide were associated with the ER, whereas those proteins failing to localize to the ER were enriched in the ER-depleted cytoplasm. In four cases, namely those of Del. 25–75, Del. Thromb., Del. Fil., and C484/485S, the paucity of protein in the ER-depleted fraction suggested that the mutants were more tightly associated with the ER and/or were more rapidly degraded in the cytoplasm. We conclude that the proper transport of GpIbα to the ER requires its hydrophobic N-terminal signal peptide and that its subsequent proper glycosylation and transport to the cell surface requires the presence of and proper interaction with other vWFR subunits, as described previously (3, 30, 35).
FIGURE 3.
Subcellular localization of mutant GpIbα proteins. ER-enriched and ER-depleted fractions from Rat1a cells expressing the GpIbα proteins depicted in Fig. 2A were subjected to Western blotting. Equivalent amounts of lysate from each fraction were electrophoresed. As a control for ER enrichment, the blot was probed with an antibody directed against calnexin and β-tubulin, which are ER-associated and non-ER-associated, respectively.
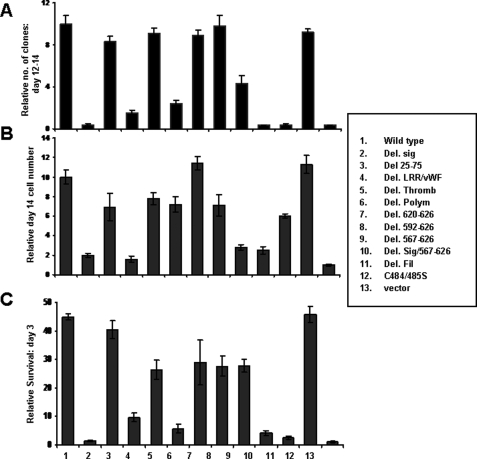
Transformation by GpIbα Mutants—We next asked how the mutations shown in Fig. 2A differed in their abilities to recapitulate various oncoprotein-like properties (14, 15). We first examined anchorage-independent growth. Pooled Rat1a cells expressing each of the mutants were plated in soft agar, as described previously (14, 15), and colonies were enumerated 12–14 days later. The results of these studies (Fig. 4A) showed that the regions of GpIbα most important for transformation included the signal peptide, the leucine-rich/vWF (LRR/vWF) binding domain, the polymorphic domain, and the filamin-binding domain. Mutants notable for a lack of effect on transformation included the thrombin binding domain, the C384S/C385S double point mutation, and progressively larger mutations affecting the 14-3-3ζ binding site (Del (620–627) and Del (592–627)). Indeed, even a larger deletion upstream of this latter site (Del (567–627)), which retained only 34 amino acids of the cytoplasmic domain and impinged on the filamin binding domain, still allowed for anchorage-independent growth at nearly half the efficiency of full-length GpIbα. Together with the results shown in Fig. 3, the collection of mutants described here allows us to conclude that neither efficient localization to the cell surface, GpIbα-GpIbβ association, nor signaling via 14-3-3ζ are necessary for the transforming activity of GpIbα. In contrast, localization to the ER and the retention of extracellular segments containing the LRR/vWF domain, the polymorphic domain, and the filamin binding domain are highly important for transformation by GpIbα.
FIGURE 4.
Identification of GpIbα domains responsible for imparting growth in soft agar and for promoting growth and survival. A, clonogenic assays. Rat1a cells expressing each of the mutants depicted in Fig. 2A were plated in triplicate in soft agar (4 × 103 cells/plate), as described previously, and enumerated 12–14 days later (25). B, growth of Rat1a cell lines in 1% serum. Each of the above described Rat1a cell lines was plated at low density in triplicate 6-well plates in standard growth medium containing 10% fetal calf serum. The following day, the medium was replaced with fresh medium containing 1% fetal calf serum. Viable cell counts were then performed periodically thereafter (17, 25, 26). Relative cell numbers at day 14 are shown here. C, survival of Rat1a cell lines following complete serum deprivation. Each of the indicated cell lines was grown to ∼50% confluence, washed three times in serum-free medium, and then maintained in serum-free medium. Viable cell counts were then performed after 3 days, as described previously (17).
Growth Promotion and Survival Mediated by GpIbα Mutants—Overexpression of GpIbα significantly reduces the in vitro growth factor requirements of both fibroblasts and myeloid cells, whereas suppression of endogenous GpIbα has the opposite effect (15).3 Specifically in the former case, Rat1a-GpIbα cells proliferate at a high rate in medium containing 1% serum, whereas Rat1a-vector cells are nearly quiescent. The mutants depicted in Fig. 2A thus provided an opportunity to define the specific regions of GpIbα responsible for this growth-promoting property and to compare and contrast them with transformation-specific domains. We found a strong but not absolute correlation among the regions of GpIbα needed to impart anchorage-independent growth in soft agar and those needed to allow for robust growth in low serum (Fig. 4B). The two notable differences involved the polymorphic region, which enhanced transformation but was largely dispensable for growth promotion, and the filamin binding domain, which was much more important for transformation than for promoting growth. From these experiments, we conclude that, whereas transformation and growth promotion by GpIbα require certain common domains, others, notably the polymorphic domain and the filamin binding domain, are distinctly different and genetically separable and independently influence these properties.
In addition to its effects on proliferation, GpIbα overexpression also prolongs survival in the absence of growth factors, whereas GpIbα depletion accelerates apoptotic death (15). Thus, we measured survival in each of the above cell lines following complete serum deprivation. As seen in Fig. 4C, the regions of GpIbα most critical to the survival of these cells were identical to those required for transformation. In contrast, proliferation and survival were distinguishable by virtue of the former being independent of the polymorphic region, whereas survival required the retention of this domain.
Three Levels of GI Mediated by GpIbα—In addition to its effects on transformation, proliferation, and survival, the deregulation of GpIbα also leads to at least three distinct types of profound GI (14, 15). First, in the presence of mitotic spindle poisons, GpIbα overexpression causes the rapid accumulation of tetraploid and aneuploid cells in a large proportion of the population. Second, GpIbα-overexpressing cells often acquire two or more nuclei, which are often of unequal size and shape and are frequently associated with chromosomal bridging and multiple micronuclei. In primary cells, these properties are particularly prominent following depletion of the p53 tumor suppressor (14, 15). Third, GpIbα overexpression leads to high levels of DSBs, the accumulation of which is largely p53-independent. The compromise of p53, however, allows such damaged cells to be rescued from eventual senescence (14, 15). The relationship among these various forms of GI has not been studied in detail, although the presence of aneuploidy does accelerate the in vivo tumorigenicity of Rat1a-GpIbα cells (15). Therefore, we next asked whether the above Rat1a cell lines could be used to map the domains of GpIbα responsible for imparting each of these distinctly different forms of GI.
We employed two methods to evaluate chromosomal and nuclear instability in Rat1a cells. First, cells were exposed to the mitotic spindle poison colcemid for 16 h, and their DNA content was then assessed by propidium iodide staining and flow cytometry (11, 12, 14, 15). Under these conditions, control Rat1a cells typically arrest in G2/M with a 4 n DNA content, whereas Rat1a-GpIbα cells undergo an aberrant round of DNA synthesis without an intervening mitosis and arrest in G2/M with a tetraploid (8 n) or pseudotetraploid genome (14, 15). In the second method, cells were exposed to the actin depolymerizing agent dihydrocytochalasin B (DCB), stained with 4′,6-diamidino-2-phenylindole, and examined at the single cell level by fluorescence microscopy. As a result of cytokinesis failure, control Rat1a cells become binucleate at a high frequency, whereas Rat1a-GpIbα cells acquire more than two nuclei, which are often dysmorphic and show evidence of chromosomal bridging (14). In order to monitor double-stranded DNA breaks, we used immunostaining with an antibody specific for Ser-139-phosphorylated histone H2A (γ-H2AX), which tends to accumulate focally at the sites of these lesions (36).
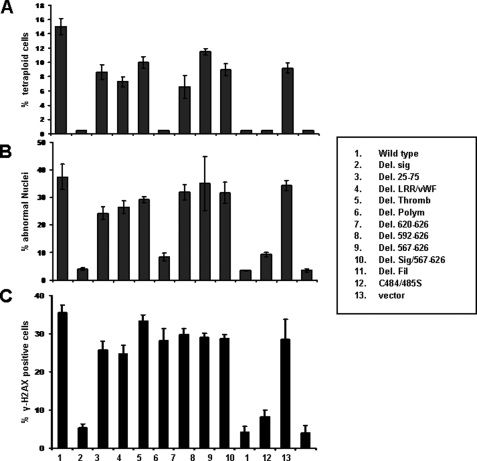
The results of our first method of evaluation are shown in Fig. 5A and Fig. S3. As predicted, control Rat1a-vector cells efficiently arrested in G2/M following exposure to colcemid, whereas Rat1a-GpIbα cells developed tetraploidy at a high frequency. In contrast, GpIbα mutants with deletions of the signal peptide, the polymorphic region, and the filamin-binding domain were unable to promote tetraploidy, whereas the remaining deletions demonstrated at least a partial retention of this activity.
FIGURE 5.
Identification of GpIbα domains responsible for promoting GI. A, logarithmically growing Rat1a cells expressing the GpIbα proteins depicted in Fig. 2A were exposed to colcemid, and their DNA content was assessed by flow cytometry of propidium iodide-stained nuclei, as described previously (14, 16, 17). Tetraploid/aneuploid populations are included in the “8 n” population. Note that Rat1a-vector control cells demonstrate little, if any, evidence of tetraploidy/aneuploidy. Numbers are the average of triplicate determinations, and bars indicate ±1 S.E. See Fig. S3 for examples of actual propidium iodide profiles. B, quantification of nuclear abnormalities in Rat1a cells following DCB treatment. Rat1a cell lines were exposed to DCB for 24 h and were then stained with 4′,6-diamidino-2-phenylindole. The percentage of cells containing a single nucleus, two nuclei, or “abnormal nuclei” was then determined. In the last case, this group included cells with >2 nuclei, nuclei of unequal size and shape, micronuclei, or other abnormalities, such as chromosomal bridging. All determinations were based on counting at least 200 cells from randomly chosen fields. See Fig. S4 for examples of the various abnormalities seen in cells expressing full-length GpIbα and its active mutants. C, γ-H2AX staining. Logarithmically growing Rat1a cells were fixed in paraformaldehyde and stained for γ-H2Ax using an antibody against Ser-139 of histone H2A, as described previously (17). The percentage of positively staining cells was then determined based on counting >200 cells/sample from triplicate cultures. See Fig. S5 for examples of images from cell lines with detectable levels of γ-H2AX.
Nuclear morphologies of individual Rat1a cells following DCB treatment are shown in Fig. 5B and Fig. S4. Control cells showed the expected binucleation following DCB treatment, whereas very few such cells were observed in non-DCB-treated cultures. Moreover, nuclei in the former cells were invariably of similar size and shape. In marked contrast, DCB-treated Rat1a-GpIbα cells often displayed multiple and dysmorphic nuclei along with micronuclei and a high frequency of chromosomal bridging. Based on the available mutants, we conclude that the GpIbα domains necessary to generate the above described nuclear abnormalities are identical to those needed to impart tetraploidy.
Finally, we examined each of the Rat1a cell groups for evidence of DSBs, as revealed by the presence of γ-H2Ax foci (36). As seen in Fig. 5C and Fig. S5, logarithmically growing control cells showed little evidence for DSBs, whereas Rat1a-GpIbα cells showed high levels of on-going DNA damage, as described previously (15). Interestingly, although the signal peptide and filamin-binding domain were necessary for the generation of ongoing DSBs, the polymorphic region appeared to be entirely dispensible. From these studies, we conclude that at least three distinct domains of GpIbα are responsible for the different forms of GI. These include the signal peptide, the polymorphic region, and the filamin binding domain. Whereas all three of these regions are necessary to promote the aneuploidy and large scale nuclear abnormalities, the polymorphic region is dispensable for the induction of DSBs.
DISCUSSION
Our prior observation that ectopic expression of GpIbα promotes transformation, proliferation, survival, and various forms of GI (14, 15) raised a number of questions that we address in the current work. Among these was whether these oncoprotein functions require cell surface localization of GpIbα and/or the participation of other vWFR subunits (35, 37). Our conclusion that they do not is supported by several lines of evidence. First is the demonstration that many primary and established cells express GpIbα (14, 15) but that only megakaryocytes express the full complement of subunits needed to generate a functional and properly glycosylated vWFR. In the few exceptions where transcripts for other subunits were detected (Fig. 1), they were observed at low levels and in the absence of coordinate expression of the remaining transcripts. The actual extent of GpIbα expression in other cells remains to be established, although it has been previously reported by others in breast cancers, normal endothelium, and dermal dendrocytes (27–29) and by us in a variety of transformed and nontransformed cell types (14, 15). Second, we showed that cysteines 384 and 385 of GpIbα, which are critical for vWFR formation (3), are entirely dispensable for its oncoprotein-like functions. Third, virtually none of the ectopically expressed or endogenous GpIbα in cells other than megakaryocytes localizes to the cell surface (Fig. 3 and Figs. S1 and S2). Rather, most of the wild-type protein accumulates within the ER (Fig. 3). This observation is consistent with the notion that efficient and correct transport and localization of GpIbα occurs only in the context of other vWFR subunits, as demonstrated previously (3, 7, 10, 30, 38). Retention by the ER appears to be essential for all of the oncoprotein-like function of GpIbα, as evidenced by the lack of any observable activity of the Del. Sig. mutant, which is otherwise intact. However, these functions are not merely a nonspecific consequence of perturbed ER homeostasis resulting from the accumulation of GpIbα in this compartment, as evidenced by our finding that some GpIbα mutants associate with the ER as well as the wild-type protein but appear to be biologically inactive (e.g. Del.(25–75) and Del. Fil.). In addition, Rat1a cells treated with brefeldin A, which causes a generalized accumulation of glycoproteins in the ER due to inhibition of ER → Golgi transport (39), showed no evidence for any of the behaviors typically associated with GpIbα ectopic expression (not shown). Thus, although ER sequestration is necessary for the oncoprotein properties of GpIbα, it is be no means sufficient and appears to be related to other functions of GpIbα that can be mapped to specific domains.
Another question is whether the regions of GpIbα required for vWFR signaling are identical to those controlling its oncoprotein-like properties. Because the C-terminal 6 amino acids of GpIbα play such a critical role in the former set of functions (6–8, 40, 41), it was surprising to find that they were entirely dispensable for the latter ones. Indeed, even the deletion of more than 50 additional amino acids upstream of the C terminus (Del.(567–627)) only marginally affected any of the oncoprotein activities of GpIbα. Collectively, our observations strongly support the idea that the vWFR-related and oncoprotein-related domains of GpIbα are functionally distinct and genetically separable.
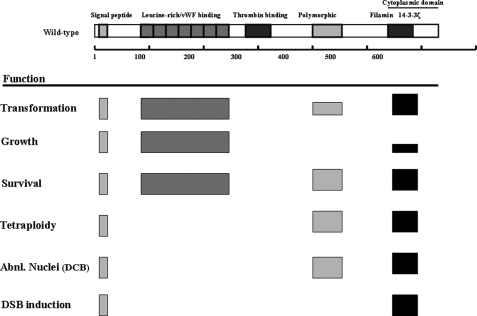
The above observations raise another important question, namely which regions of GpIbα are required for its oncoprotein-like properties? Our analyses point to four critical domains, which also serve to emphasize the functionally modular nature of the full-length protein (Fig. 6). The first is the signal peptide, which was necessary for every function. Because this domain is required for the localization of GpIbα to the ER, our results provide yet additional evidence that the intracellular signaling pathways associated with oncoprotein functions are initiated from this specific subcellular location. The second important region was the filamin-binding domain, which, in the classical vWFR context, probably serves to indirectly anchor GpIbα to the cytoplasmic actin cytoskeleton and ensure its stable retention at the cell surface (9, 10). Filamin binding by GpIbα has also been reported to be essential for proper platelet adhesion, activation, and morphologic integrity (10). By virtue of their ability to interact with over 30 different membrane receptors and intracellular signaling molecules, filamins have been proposed to regulate cell shape and mobility, possibly by integrating and propagating environmental cues and by organizing the actin cytoskeleton into a dynamic three-dimensional structure (42, 43).
FIGURE 6.
Summary of the regions of GpIbα involved in mediating c-Myc-like properties. Functional domains of the protein that were deleted are shown at the top of the diagram and are taken from Fig. 2A. Boxes in the lower portion of the figure indicate those regions that, when deleted, resulted in loss of the indicated function. The heights of the boxes are meant to represent the approximate degree of resulting impairment. For example, deletion of the filamin binding domain resulted in the loss of all functions except for growth promotion, which was only partially affected.
In addition to the signal peptide and the filamin-binding region, the LRR/vWF domain and the polymorphic regions were also required, albeit for distinct and more limited subsets of activities. For example, the LRR/vWF domain was necessary for transformation, the promotion of growth, and survival but was otherwise dispensable for all genome-destabilizing functions. By contrast, the polymorphic region was required for transformation, survival, the induction of aneuploidy, and the abnormal nuclear behavior in response to actin depolymerization. However, this region was not required for DSB generation, thus suggesting that this form of GI is not only mediated by a different mechanism but is neither required for nor a precursor of the other two types of GI. In the normal context of the vWFR, the polymorphic region, which consists of 1–3 repeats of the sequence SEPAPSPTTPEPT, serves as a site for O-glycosylation and has been proposed to regulate the degree to which platelets can be activated in response to shear stress (31, 32). The particular “wild-type” clone that was used in these studies contained one copy of this region. It will be of interest in future work to determine whether variable numbers of repeats of this segment influence any of the GpIbα functions described here. Several additional lines of evidence argue for the specificity of the oncoprotein-like properties of GpIbα (14, 15). First, the majority of tumor cell lines examined express extremely high levels of GpIbα, which correlate well with c-Myc levels (15). Second, we have shown that the knockdown of endogenous GpIbα leads to a loss of c-Myc-mediated GI (14), thus implicating the protein in a process that is believed to lie at the heart of tumor cell pathogenesis and evolution (44). Finally, recent work from our laboratory has demonstrated that the stable knockdown of endogenous GpIbα in several human cancer cell lines is associated with a profound loss of soft agar clonogenicity and the ability to grow as tumor xenografts (not shown). Taken together, these results are consistent with the idea that the oncoprotein-like properties of GpIbα reflect true alternate functions rather than aberrant ones lacking true physiologic correlates.
In summary, the results presented here address the means by which GpIbα affects transformation, proliferation, survival, and three distinct forms of GI. These non-vWFR-related properties map to specific domains with functions previously defined solely in the context of the vWFR. Furthermore, unlike the transmembrane signaling of the vWFR, these oncoprotein functions require neither the intact vWFR complex, cell surface expression of GpIbα, nor signaling by its C-terminal segment. Rather, association with the ER and subsequent intracellular signaling through the LRR/vWF binding domain, the polymorphic domain, and the filamin-binding domain appear to be essential for this novel alternate pathway. It will clearly be important to define further the nature of the specific mechanisms involved in these signaling events.
Supplementary Material
Acknowledgments
We thank Allan Gewirtz for graciously providing the Mo7e megakaryocyte cell line; Michael Berndt and Zaverio Ruggieri for supplying mAbs WM23 and LJ-Ibα1, respectively; Massimo Trucco for use of the real time PCR equipment; and Bill Saunders for valuable suggestions, discussions, and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA105033 and CA078259 (to E. V. P.). This work was also supported by a postdoctoral fellowship award from the Children's Hospital of Pittsburgh Research Advisory Committee (to Y. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
Footnotes
The abbreviations used are: GbIbα, glycoprotein Ib α; GpIbβ, glycoprotein Ib β; GpV, glycoprotein V; GpIX, glycoprotein IX; ER, endoplasmic reticulum; vWF, von Willebrand factor; vWFR, von Willebrand factor receptor; GI, genomic instability; DSB, double-stranded DNA break; mAb, monoclonal antibody; PBS, phosphate-buffered saline; qRT, quantitative reverse transcription; LRR, leucine-rich region; DCB, dihydrocytochalasin B.
Y. J. Li and E. V. Prochownik, unpublished observations.
References
- 1.Lopez, J. A. (1994) Blood Coagul. Fibrinolysis 5 97–119 [PubMed] [Google Scholar]
- 2.Berndt, M. C., Shen, Y., Dopheide, S. M., Gardiner, E. E., and Andrews, R. K. (2001) Thromb. Haemostasis 86 178–188 [PubMed] [Google Scholar]
- 3.Luo, S. Z., Mo, X., Afshar-Kharghan, V., Srinivasan, S., Lopez, J. A., and Li, R. (2007) Blood 109 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng, S., Christodoulides, N., and Kroll, M. H. (1999) Blood 93 4256–4263 [PubMed] [Google Scholar]
- 5.Wu, Y., Asazuma, N., Satoh, K., Yatomi, Y., Takafuta, T., Berndt, M. C., and Ozaki, Y. (2003) Blood 101 3469–3476 [DOI] [PubMed] [Google Scholar]
- 6.Kanaji, T., Russell, S., Cunningham, J., Izuhara, K., Fox, J. E., and Ware, J. (2004) Blood 104 3161–3168 [DOI] [PubMed] [Google Scholar]
- 7.Canobbio, I., Balduini, C., and Torti, M. (2004) Cell. Signal. 16 1329–1344 [DOI] [PubMed] [Google Scholar]
- 8.Yin, H., Stojanovic, A., Hay, N., and Du, X. (2008) Blood 111 658–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, S., Lu, X., and Kroll, M. H. (2005) J. Biol. Chem. 280 6709–6715 [DOI] [PubMed] [Google Scholar]
- 10.Nakamura, F., Pudas, R., Heikkinen, O., Permi, P., Kilpeläinenm, I., Munday, A. D., Hartwig, J. H., Stossel, T. P., and Ylänne, J. (2006) Blood 107 1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin, X. Y., Grove, L., Datta, N. S., Long, M. W., and Prochownik, E. V. (1999) Oncogene 18 1177–1184 [DOI] [PubMed] [Google Scholar]
- 12.Yin, X. Y., Grove, L., Datta, N. S., Katula, K., Long, M. W., and Prochownik, E. V. (2001) Cancer Res. 61 6487–6493 [PubMed] [Google Scholar]
- 13.Rogulski, K. R., Cohen, D. E., Corcoran, D. L., Benos, P. V., and Prochownik, E. V. (2005) Proc. Natl. Acad. Sci. U. S. A 102 18968–18973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y., Lu, J., and Prochownik, E. V. (2007) Proc. Natl. Acad. Sci. U. S. A 104 3490–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Y., Lu, J., Cohen, D., and Prochownik, E. V. (2007) Oncogene 27 1599–1609 [DOI] [PubMed] [Google Scholar]
- 16.Prochownik, E. V., and Li, Y. (2007) Cell Cycle 6 1024–1029 [DOI] [PubMed] [Google Scholar]
- 17.Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D., and Lowe, S. W. (1997) Cell 88 593–602 [DOI] [PubMed] [Google Scholar]
- 18.Bartkova, J., Rezaei, N., Liontos, M., Liontos, M., Karakaidos, P., Kletsas, D., Issaeva, N., Vassiliou, L. V., Kolettas, E., Niforou, K., Zoumpourlis, V. C., Takaoka, M., Nakagawa, H., Tort, F., Fugger, K., Johansson, F., Sehested, M., Andersen, C. L., Dyrskjot, L., Ørntoft, T., Lukas, J., Kittas, C., Helleday, T., Halazonetis, T. D., Bartek, J., and Gorgoulis, V. G. (2006) Nature 444 633–637 [DOI] [PubMed] [Google Scholar]
- 19.Braig, M., Lee, S., Loddenkemper, C., Rudolph, C., Peters, A. H., Schlegelberger, B., Stein, H., Dörken, B., Jenuwein, T., and Schmitt, C. A. (2005) Nature 436 660–665 [DOI] [PubMed] [Google Scholar]
- 20.Wang, H., Hammoudeh, D. I., Follis, A. V., Reese, B. E., Lazo, J. S., Metallo, S. J., and Prochownik, E. V. (2007) Mol. Cancer Ther. 6 2399–2408 [DOI] [PubMed] [Google Scholar]
- 21.Wang, H., Mannava, S., Grachtchouk, V., Zhuang, D., Soengas, M. S., Gudkov, A. V., Prochownik, E. V., and Nikiforov, M. A. (2007) Oncogene 27 1905–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin, X., Landay, M. F., Han, W., Levitan, E. S., Watkins, S. C., Levenson, R. M., Farkas, D. L., and Prochownik, E. V. (2001) Oncogene 20 4650–4664 [DOI] [PubMed] [Google Scholar]
- 23.Yin, X., Grove, L., Rogulski, K., and Prochownik, E. V. (2002) J. Biol. Chem. 277 19998–20010 [DOI] [PubMed] [Google Scholar]
- 24.Rothermund, K., Rogulski, K., Fernandes, E., Whiting, A., Sedivy, J., Pu, L., and Prochownik, E. V. (2005) Cancer Res. 65 2097–2107 [DOI] [PubMed] [Google Scholar]
- 25.Miyazawa, K., Nishimaki, J., Katagiri, T., Yaguchi, M., Iwase, O., Gotoh, A., Tauchi, T., Kawanishi, Y., Toyama, K., Ohyashiki, K., Ishibashi, T., and Broxmeyer, H. E. (2000) Hematology 5 233–246 [DOI] [PubMed] [Google Scholar]
- 26.Hahn, W. C., Counter, C. M., Lundberg, A. S., Beijersbergen, R. L., Brooks, M. W., and Weinberg, R. A. (1999) Nature 400 464–468 [DOI] [PubMed] [Google Scholar]
- 27.Konkle, B. A., Shapiro, S. S., Asch, A. S., and Nachman R. L. (1990) J. Biol. Chem. 265 19833–19838 [PubMed] [Google Scholar]
- 28.Oleksowicz, L., Dutcher, J. P., DeLeon-Fernandez, M., and Etkind, P. (1997) Exp. Cell Res. 237 110–117 [DOI] [PubMed] [Google Scholar]
- 29.Monteiro, M. R., Shapiro, S. S., Takafuta, T., Menezes, D. W., and Murphy, G. F. (1999) J. Invest. Dermatol. 113 272–276 [DOI] [PubMed] [Google Scholar]
- 30.Lopez, J. A., Leung, B., Reynolds, C. C., Li, C. Q., and Fox, J. E. (1992) J. Biol. Chem. 267 12851–12859 [PubMed] [Google Scholar]
- 31.Lopez, J. A., Ludwig, E. H., and McCarthy, B. J. (1992) J. Biol. Chem. 267 10055–10061 [PubMed] [Google Scholar]
- 32.Ulsemer, P., Strassel, C., Baas, M.-J., Salamero, J., Chasserot-Golaz, S., Cazenave, J. P., De La Salle, C., and Lanza, F. (2001) Biochim. J. 358 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegde, R. S., and Bernstein, H. D. (2006) Trends Biochem. Sci. 31 563–571 [DOI] [PubMed] [Google Scholar]
- 34.Schroder, M., and Kaufman, R. J. (2005) Annu. Rev. Biochem. 74 739–789 [DOI] [PubMed] [Google Scholar]
- 35.Mo, X., Lu, N., Padilla, A., Lopez, J. A., and Li, R. (2006) J. Biol. Chem. 281 23050–23059 [DOI] [PubMed] [Google Scholar]
- 36.Sedelnikova, O. A., Horikawa, I., Zimonjic, D. B., Popescu, N. C., Bonner, W. M., and Barrett, J. C. (2004) Nat. Cell Biol. 6 168–170 [DOI] [PubMed] [Google Scholar]
- 37.Dong, J. F., Gao, S., and Lopez, J. A. (1998) J. Biol. Chem. 273 31449–31454 [DOI] [PubMed] [Google Scholar]
- 38.Strassel, C., Pasquet, J. M., Alessi, M. C., Juhan-Vague, I., Chambost, H., Combrié, R., Nurden, P., Bas, M. J., De La Salle, C., Cazenave, J. P., Lanza, F., and Nurden, A. T. (2003) Biochemistry 42 4452–4462 [DOI] [PubMed] [Google Scholar]
- 39.Hunziker, W., Whitney, J. A., and Mellman, I. (1992) FEBS Lett. 307 93–96 [DOI] [PubMed] [Google Scholar]
- 40.Geddis, A. E., Fox, N. E., and Kaushansky, K. (2001) J. Biol. Chem. 276 34473–34479 [DOI] [PubMed] [Google Scholar]
- 41.Dai, K., Bodnar, R., Berndt, M. C., and Du, X. (2005) Blood 106 1975–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stossel, T. P., Condeelis, J., Cooley, L., Hartwig, J. H., Noegel, A., Schleicher, M., and Shapiro, S. S. (2001) Nat. Rev. Mol. Cell. Biol. 2 138–145 [DOI] [PubMed] [Google Scholar]
- 43.Feng, Y., and Walsh, C. A. (2004) Nat. Cell Biol. 6 1034–1038 [DOI] [PubMed] [Google Scholar]
- 44.Beckman, R. A., and Loeb, L. A. (2005) Semin. Cancer Biol. 15 423–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.