Abstract
The Hermansky-Pudlak syndrome is a disorder affecting endosome sorting. Disease is triggered by defects in any of 15 mouse gene products, which are part of five distinct cytosolic molecular complexes: AP-3, homotypic fusion and vacuole protein sorting, and BLOC-1, -2, and -3. To identify molecular associations of these complexes, we used in vivo cross-linking followed by purification of cross-linked AP-3 complexes and mass spectrometric identification of associated proteins. AP-3 was co-isolated with BLOC-1, BLOC-2, and homotypic fusion and vacuole protein sorting complex subunits; clathrin; and phosphatidylinositol-4-kinase type II α (PI4KIIα). We previously reported that this membrane-anchored enzyme is a regulator of AP-3 recruitment to membranes and a cargo of AP-3 (Craige, B., Salazar, G., and Faundez, V. (2008) Mol. Biol. Cell 19,1415 -1426). Using cells deficient in different Hermansky-Pudlak syndrome complexes, we identified that BLOC-1, but not BLOC-2 or BLOC-3, deficiencies affect PI4KIIα inclusion into AP-3 complexes. BLOC-1, PI4KIIα, and AP-3 belong to a tripartite complex, and down-regulation of either PI4KIIα, BLOC-1, or AP-3 complexes led to similar LAMP1 phenotypes. Our analysis indicates that BLOC-1 complex modulates the association of PI4KIIα with AP-3. These results suggest that AP-3 and BLOC-1 act, either in concert or sequentially, to specify sorting of PI4KIIα along the endocytic route.
Membranous organelles along the exocytic and endocytic pathways are each defined by unique lipid and protein composition. Vesicle carriers communicate and maintain the composition of these organelles (2). Consequently defining the machineries that specify vesicle formation, composition, and delivery are central to understanding membrane protein traffic. Generally vesicle biogenesis uses multiprotein cytosolic machineries to select membrane components for inclusion in nascent vesicles (2, 3). Heterotetrameric adaptor complexes (AP-1 to AP-4) are critical to generate vesicles of specific composition from the different organelles constituting the exocytic and endocytic routes (2-4).
The best understood vesicle formation machinery in mammalian cells is the one organized around the adaptor complex AP-2 (5). This complex generates vesicles from the plasma membrane using clathrin. Our present detailed understanding of AP-2 vesicle biogenesis mechanisms and interactions emerged from a combination of organellar and in vitro binding proteomics analyses together with the study of binary interactions in cell-free systems (5-9). In contrast, the vesicle biogenesis pathways controlled by AP-3 are far less understood. AP-3 functions to produce vesicles that traffic selected membrane proteins from endosomes to lysosomes, lysosome-related organelles, or synaptic vesicles (10-13). AP-3 is one of the protein complexes affected in the Hermansky-Pudlak syndrome (HPS;3 Online Mendelian Inheritance in Man (OMIM) 203300). So far, mutations in any of 15 mouse or eight human genes trigger a common syndrome. This syndrome encompasses defects that include pigment dilution, platelet dysfunction, pulmonary fibrosis, and occasionally neurological phenotypes (14, 15). All forms of HPS show defective vesicular biogenesis or trafficking that affects lysosomes, lysosome-related organelles (for example melanosomes and platelet dense granules), and, in some of them, synaptic vesicles (11-13). Most of the 15 HPS loci encode polypeptides that assemble into five distinct molecular complexes: the adaptor complex AP-3, HOPS, and the BLOC complexes 1, 2, and 3 (14). Recently binary interactions between AP-3 and BLOC-1 or BLOC-1 and BLOC-2 suggested that arrangements of these complexes could regulate membrane protein targeting (16). Despite the abundance of genetic deficiencies leading to HPS and genetic evidence that HPS complexes may act on the same pathway in defined cell types (17), we have only a partial picture of protein interactions organizing these complexes and how they might control membrane protein targeting.
In this study, we took advantage of cell-permeant and reversible cross-linking of HPS complexes followed by their immunoaffinity purification to identify novel molecular interactions. Cross-linked AP-3 co-purified with BLOC-1, BLOC-2, HOPS, clathrin, and the membrane protein PI4KIIα. We previously identified PI4KIIα as a cargo and regulator of AP-3 recruitment to endosomes (1, 18). Using mutant cells deficient in either individual HPS complexes or a combination of them, we found that BLOC-1 facilitates the interaction of AP-3 and PI4KIIα. Our studies demonstrate that subunits of four of the five HPS complexes co-isolate with AP-3. Moreover BLOC-1, PI4KIIα, and AP-3 form a tripartite complex as demonstrated by sequential co-immunoprecipitations as well as by similar LAMP1 distribution phenotypes induced by down-regulation of components of this tripartite complex. Our findings indicate that BLOC-1 complex modulates the recognition of PI4KIIα by AP-3. These data suggest that AP-3, either in concert or sequentially with BLOC-1, participates in the sorting of common membrane proteins along the endocytic route.
EXPERIMENTAL PROCEDURES
Antibodies—Antibodies used in these studies are listed in Table 1 except for the vps33b antibodies. An antiserum was prepared against rat vps33b peptide DTLTAVENKVSKLVTDKAAGKITDAFSSL (amino acids 450-478 of the NCBI record NP_071622.1). This peptide is conserved in human and mouse vps33b. Peptide synthesis and rabbit immunization were performed by Alpha Diagnostic International (San Antonio, TX).
TABLE 1.
Antibodies used
IB, immunoblot; HC, heavy chain; DSHB, Developmental Studies Hybridoma Bank.
| Antibody | Source | Catalog no. | Type | IB dilution |
|---|---|---|---|---|
| Annexin II | BD Biosciences | 610068 | Monoclonal | 1:1000 |
| β-Actin | Sigma | A5451 | Monoclonal | 1:1000 |
| γ-Adaptin | BD Biosciences | A36120 | Monoclonal | 1:500 |
| α-Adaptin | Sigma | A4325 | Monoclonal | 1:500 |
| δ-Adaptin | DSHB | SA4 | Monoclonal | 1:500 |
| β3-Adaptin | V. Faundez | Polyclonal | 1:500 | |
| σ3-Adaptin | V. Faundez | Polyclonal | 1:1000 | |
| μ3-Adaptin | V. Faundez | Polyclonal | 1:200 | |
| Clathrin HC | BD Biosciences | C43820 | Monoclonal | 1:1000 |
| Dysbindin | E. Dell'Angelica | Polyclonal | 1:1000 | |
| Pallidin | Proteintech Group | 10891-1-AP | Polyclonal | 1:1000 |
| Pallidin | E. Dell'Angelica | Polyclonal | 1:500 | |
| Pallidin | E. Dell'Angelica | Monoclonal | 1:500 | |
| Muted | E. Dell'Angelica | Polyclonal | 1:1000 | |
| Hps6 ru | R. Swank | Polyclonal | 1:500 | |
| Hsp90 | BD Biosciences | 610418 | Monoclonal | 1:1000 |
| Hsc70 | Stressgen | SPA-815 | Monoclonal | 1:1000 |
| Myc epitope | Bethyl Laboratories | A190-105A | Polyclonal | 1:1000 |
| LAMP1 | DSHB Iowa | H4A3 | Monoclonal | 1:1000 |
| PI4KIIα | P. de Camilli | Polyclonal | 1:1000 | |
| Synaptophysin | Chemicon | MAB5258 | Monoclonal | 1:2000 |
| SYTL4 | Atlas Antibodies | HPA001475 | Polyclonal | 1:1000 |
| Transferrin receptor | Zymed Laboratories Inc. | 12-6800 | Monoclonal | 1:2000 |
| Tubulin | Sigma | T6199 | Monoclonal | 1:2000 |
| ZnT3 | V. Faundez | Polyclonal | 1:1000 |
Plasmids—Plasmid encoding mouse vps39-GFP in pEGFP-C1 was a generous gift of Dr. Robert Piper (19). Human vps41 (SC111791) was obtained from the TrueClone collection (Origene, Rockville, MD). vps41 was Myc-tagged by PCR at the carboxyl terminus and subcloned into pcDNA3.1D/V5-HIS-TOPO expression vector. The full open reading frame sequence was confirmed by sequencing. HA-tagged human PI4KIIα and the dileucine sorting mutant PI4KIIαL61A/L62A were described previously (1). Transfections were performed with Lipofectamine 2000 following the manufacturer's recommendations.
Cell Lines—Cells used are listed in Table 2. Mouse fibroblast and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Cellgro, Herndon, VA; 4.5 g/liter glucose) containing 10% fetal bovine serum (Hyclone, Logan, UT), 100 units/ml penicillin, and 100 mg/ml streptomycin. Hygromycin (200 μg/ml) was used for cells carrying retroviruses. ZnT3-transfected PC12 cell lines were cultured as described previously (20). PC12 cells were metabolically labeled with 50 μCi/ml 35[S]methionine (>1000 Ci (37.0 TBq)/mmol, PerkinElmer Life Sciences) for 12 h.
TABLE 2.
Cells used
| Cell type | Defective complex | Genotype | Source |
|---|---|---|---|
| Fibroblast | AP-3 | Ap3dmh/mh empty retrovirus | A. Peden |
| Fibroblast | None | Ap3dmh/mh δ retrovirus | A. Peden |
| Fibroblast | AP-3 | Ap3dmh/mh | V. Faundez |
| Fibroblast | AP-3 | Ap3b1pe/pe | E. Dell'Angelica |
| Fibroblast | BLOC-1 | Pldnpa/pa | E. Dell'Angelica |
| Fibroblast | BLOC-2 | Hps3coa/coa | E. Dell'Angelica |
| Fibroblast | BLOC-3 | Hps1ep/ep | E. Dell'Angelica |
| Fibroblast | BLOC-1 | Pldnpa/pa | R. Swank |
| Fibroblast | BLOC-1, -2 | Pldnpa/pa, Hps6ru/ru | R. Swank |
| Fibroblast | BLOC-2, -3 | Hps3coa/coa, Hps4le/le | R. Swank |
| Fibroblast | BLOC-1, -2, -3 | Pldnpa/pa, Hps5ru2j/ru2j, Hps1ep/ep | R. Swank |
| Fibroblast | None | C57B | E. Dell'Angelica |
| Fibroblast | None | C57B | R. Swank |
| PC12 | None | V. Faundez | |
| HEK293T | None | V. Faundez |
Cross-linking and Immunoprecipitation—Detailed procedures have been described previously (1). Briefly cells were washed twice in cold PBS plus CaCl2 and MgCl2 (PBS-CM) and incubated with 1 mm dithiobis(succinimidyl propionate)(DSP) (Pierce) or DMSO alone in PBS-CM for 2 h on ice. The reaction was stopped by adding 25 mm Tris, pH 7.4, for 15 min on ice and washing twice in PBS. Cells were lysed for 30 min on ice in buffer A (150 mm NaCl, 10 mm HEPES, 1 mm EGTA, and 0.1 mm MgCl2, pH 7.4), 0.5% Triton X-100, and Complete™ antiprotease mixture (Roche Applied Science). Homogenates were clarified by sedimentation at 16,100 × g for 10 min. 500-800 μg of supernatant were immunoprecipitated using Dynal magnetic beads M450 (Invitrogen) covered with monoclonal antibodies against AP-3 δ subunit, γ-adaptin (AP-1), or transferrin receptor for 3 h at 4 °C. Beads were washed six times in buffer A plus 0.1% Triton X-100 for 5 min each and incubated with SDS-PAGE sample buffer for 5 min at 75 °C. Samples were loaded on 4-20% PAGE-SDS Criterion precast gels (Bio-Rad) and analyzed by immunoblot.
Sucrose Sedimentation and Mass Spectrometry—Clarified Triton-soluble supernatants from PC12 cells either treated with DSP or DMSO alone were sedimented in a 5-20% sucrose gradient prepared in buffer A plus 0.5% Triton X-100 during 13 h at 187,000 × g in an SW55 rotor (1.5 mg per gradient) (20). 20 samples were collected from the bottom (250 μl each) and analyzed by Western blots. The following standards were used to calibrate the gradients: horse spleen apoferritin (443 kDa, 16.5 S), bovine serum albumin (66 kDa, 4.6 S), sweet potato β-amylase (200 kDa, 9.4 S), and bovine erythrocytes carbonic anhydrase (29 kDa, 2.9 S).
A total of ∼58 mg of clarified Triton-soluble supernatant from DSP-treated PC12 cells was fractionated by sucrose gradient sedimentation (∼20 gradients per preparation). AP-3 migration in the gradient was determined by immunoblot. Two preparations were performed. In one preparation sucrose fractions 1-8 (from the bottom) were pooled. In a second preparation, fractions 3-5 were pooled and further analyzed. Pooled fractions containing cross-linked AP-3 complexes were diluted in buffer A, and samples were subjected to immunoaffinity chromatography with AP-3 δ antibodies bound to Dynal M450 magnetic beads. Binding was performed for 3 h at 4 °C. After six washes in buffer A plus Triton X-100, AP-3 cross-linked complexes were eluted with a 50 μm concentration of a peptide corresponding to the epitope recognized by the anti-δ SA4 monoclonal antibody (AQQVDIVTEEMPENALPSDEDDKDPNDPYRA) corresponding to the amino acids 680-710 of human δ-adaptin (NCBI:AAD03777; gi:1923266). Elution was performed for 2 h at 0 °C. Peptide eluted material was precipitated on ice with 10% trichloroacetic acid for 30 min, and protein pellets were washed twice with 1:1 ethanol/ether (-20 °C), dried, and resuspended in 0.1 n NaOH. Solubilized protein pellets were incubated in Laemmli sample buffer (Bio-Rad) at 75 °C for 5 min, and proteins were resolved in a single lane of a 4-20% PAGE-SDS Criterion gel (Bio-Rad). Proteins were stained with SYPRO Ruby (Bio-Rad), and the lane was divided into 18 fractions. Each fraction was subjected to in-gel digestion with sequencing grade trypsin (Promega, Madison, WI) at 37 °C overnight, and the peptides were extracted as described previously (21). Protein identification was performed with nano-high performance liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS) analysis (for details, see Ref. 22); a QSTAR XL (Applied Biosystems, Foster City, CA) hybrid quadrupole tandem mass spectrometer interfaced with an Ultimate nano-HPLC system (LC Packings, Sunnyvale, CA) was used. The mass spectrometer was operated in positive ion mode using information-dependent acquisition to acquire a single MS scan (m/z 400-1900 scan range) followed by up to two MS/MS scans (m/z 50-1900 scan range). A rolling collision energy was used for the MS/MS scans. The data were analyzed using the ProID (Applied Biosystems) and Mascot (Matrix Science) search algorithms. We defined the following criteria to consider probable positive protein identification. Proteins were included for analysis 1) if represented by at least one peptide with a Mascot score >45 unless the proteins was already known to interact with AP-3 and/or 2) if represented in at least two of three spectrometry analyses. Peptides identified in a single MS run were also included if the corresponding proteins were part of a complex in which other subunits were identified in a distinct MS run.
Membrane Preparation—HEK293T cells, either treated or not with DSP, were washed in PBS and homogenized by 18 passages in a cell cracker in buffer A containing Complete antiprotease mixture. Homogenate was sedimented at 27,000 × g for 40 min to generate a P1 membrane fraction and an S1 supernatant. Cytosol free of membranes was obtained by centrifugation of S1 at 210,000 × g for 30 min in a Beckman Coulter TLA120.2 rotor. Membrane fractions or cytosol was solubilized in buffer A plus 0.5% Triton X-100 for 30 min on ice. Triton extracts were clarified by centrifugation at 16,100 × g for 10 min, and supernatants were immunoprecipitated followed by Western blot analysis.
Clathrin-coated Vesicle (CCV) Isolation—Clathrin-coated vesicles were prepared according to Girard et al. (23). Briefly PC12 cells were homogenized in buffer B (100 mm MES, pH 6.8, 0.5 mm EGTA, and 1 mm MgCl2) plus Complete antiprotease mixture. Homogenates were centrifuged at 17,000 × g for 20 min in a Sorvall SS-34 fixed angle rotor to obtain a supernatant (S1) and a pellet (P1). S1 was centrifuged at 56,000 × g for 1 h in a Beckman Type 40 ultracentrifuge fixed angle rotor to generate P2 and S2 fractions. P2 pellets were resuspended in buffer B plus antiprotease mixture and laid over a layer of 8% (w/v) sucrose in buffer B prepared in 50% (v/v) deuterium oxide. After 2 h of centrifugation at 116,000 × g in a Beckman SW55 rotor, the pellet of CCVs was resuspended in buffer B and kept at -80 °C.
siRNA Treatment—siRNA oligos for PI4KIIα and its control have been described previously (1, 24). Dharmacon ON-TARGETplus SMARTpool was used to silence expression of human AP-3 δ subunit (NCBI:L-016014-00-0005, NM_003938) at 50 nm and the BLOC1 subunit pallidin (NCBI:L-009637-01-0005, NM_012388) at 100 nm. The siCONTROL non-targeting siRNA 2 (5′-UAAGGCUAUGAAGAGAUAC-3′) was used as a control. Oligonucleotide sequences for δ were the following: Sense Sequence 1, CUACAGGGCUCUGGAUAUUUU; Antisense Sequence 1, 5′-PAAUAUCCAGAGCCCUGUAGUU; Sense Sequence 2, GGACGAGGCAAAAUACAUAUU; Antisense Sequence 2, 5′-PUAUGUAUUUUGCCUCGUCCUU; Sense Sequence 3, GAAGGACGUUCCCAUGGUAUU; Antisense Sequence 3, 5′-PUACCAUGGGAACGUCCUUCUU; Sense Sequence 4, CAAAGUCGAUGGCAUUCGGUU; Antisense Sequence 4, 5′-PCCGAAUGCCAUCGACUUUGUU. Oligonucleotide sequences for pallidin were the following: Sense Sequence 1, CAAAAGAGGCAGCGUGAUAUU; Antisense Sequence 1, 5′-PUAUCACGCUGCCUCUUUUGUU; Sense Sequence 2, CAAAGGUAGUACAGCGUUUUU; Antisense Sequence 2, 5′-PAAACGCUGUACUACCUUUGUU; Sense Sequence 3, CAAACAAACAAACAACGUAUU; Antisense Sequence 3, 5′-PUACGUUGUUUGUUUGUUUGUU; Sense Sequence 4, GAACCAAGUUGUAUUGUUAU; Antisense Sequence 4, 5′-PUAACAAUACAACUUGGUUCUU.
HEK293T cells were transfected twice on alternate days with siRNA-CONTROL 1 or 2 oligos or with siRNA oligos to silence the human PI4KIIα, the BLOC1 subunit pallidin, or the AP-3 δ subunit. Twenty-four hours after the second transfection cells were reseeded in 12-well plates for Western blots or on coverslips for immunofluorescence analysis. Samples were processed the next day.
RESULTS
Identification of Proteins Co-purifying with Cross-linked AP-3 Complexes—We used chemical cross-linking to stabilize labile protein interactions followed by purification of cross-linked protein complexes containing HPS gene products. We chose DSP, a homobifunctional cell-permeable cross-linker with a 12-Å spacer arm. This reagent contains a disulfide bond that allows cleavage of cross-linked products (25-27). Previously we demonstrated that the interaction of AP-3 with its cargo/regulator PI4KIIα on membranes could be structurally and functionally explored in vivo using DSP whole-cell cross-linking (1). Here we focused on the adaptor complex AP-3, which is affected in HPS2 syndrome (28), because it shows the most severe systemic and neurological phenotype of all HPS affected loci (29).
We first sought non-saturating in vivo DSP incubation conditions that could stabilize a small fraction of known AP-3-interacting proteins into high molecular weight complexes, “in vivo controlled cross-linking.” Triton-soluble extracts of control and DSP-treated PC12 cells were fractionated by sucrose sedimentation. We determined the sedimentation of AP-3 and its membrane protein cargoes, ZnT3 and PI4KIIα (1, 18, 20). After DSP treatment, a fraction of AP-3 exhibited a slightly increased sedimentation coefficient above 9.4 S (Fig. 1A). Similarly a discrete pool of ZnT3 and PI4KIIα was incorporated into complexes with a sedimentation coefficient between 9.4 and 16.5 S (Fig. 1A, lanes 1-7). The sedimentation of a non-AP-3 cargo synaptic vesicle protein, synaptophysin (20), remained unaffected by DSP treatment. Moreover the total protein sedimentation pattern between control and DSP-treated samples remained identical (Fig. 1B). These results show that a limited set of protein-protein interactions are stabilized in vivo by DSP treatment. We independently confirmed these findings by immunoprecipitation of AP-3 complexes from [S35]methionine-labeled whole-cell Triton extracts from control and DSP-treated PC12 cells. Because the majority of AP-3 remains uncross-linked in whole cell extracts of DSP-treated cells, we predicted that major radiolabeled polypeptides precipitated with AP-3 antibodies should be similar between DSP-treated and untreated cells. In fact, radiolabeled bands immunoprecipitated with AP-3 δ antibodies were identical irrespective of whether cells were treated or not treated with DSP (Fig. 1C, compare lanes 5 and 6). Moreover these bands differed from those immunoprecipitated with transferrin receptor (TrfR) antibodies (Fig. 1C, lanes 3 and 4). These experiments show that AP-3 complexes are selectively immunoisolated from cross-linked cells without background due to excessive cross-linking or nonspecific binding.
FIGURE 1.
Strategy to isolate AP-3 interactors from in vivo cross-linked cells. A, Triton-soluble extracts of PC12 cells incubated in the absence or presence of DSP were fractionated by sucrose sedimentation. Fractions were probed with antibodies against AP-3 β3-adaptin, the AP-3 cargoes ZnT3 and PI4KIIα, and synaptophysin (Sphysin). B, the graph depicts total protein distribution of the gradients in A. Closed circles, DSP-treated. Open circles, vehicle-treated. C, PC12 cells were metabolically labeled overnight with [35S]methionine and treated with (lanes 2, 4, and 6) and without DSP (lanes 1, 3, and 5). Clarified Triton (Tx100)-soluble extracts were immunoprecipitated (IP) with empty beads (lanes 1 and 2) and beads coated with monoclonal antibodies (Ab) against TrfR (lanes 3 and 4) or the δ subunit of AP-3 (lanes 5 and 6). D, diagram depicting the strategy to isolate putative AP-3 interactors and their identification by nano-liquid chromatography (nLC)-MS/MS. IB, immunoblot.
AP-3 bound to putative interactors was isolated from sucrose-fractionated Triton extracts of in vivo controlled cross-linked PC12 cells (Fig. 1D). Triton-soluble extracts were clarified by sedimentation, and supernatants were fractionated by sucrose sedimentation to resolve cross-linked AP-3 complexes (Fig. 1A, fractions 3-5) from the majority of non-cross-linked material (migrating under 9.4 S; Fig. 1A, fractions 6-21). AP-3 complexes sedimenting between 9.4 and 16.5 S were further purified by affinity chromatography with a monoclonal antibody against the AP-3 δ subunit. Bead-bound cross-linked AP-3 complexes were selectively eluted with a 31-mer δ antigenic peptide (Fig. 1D). Two independent purifications generated ∼5-6 μg of peptide eluate each from ∼58 mg of cell homogenate (Fig. 2A, lanes 2-5). The identity of the proteins co-purifying with cross-linked AP-3 complexes was determined by nano-liquid chromatography-MS/MS. Three independent mass spectrometry analyses identified 51 proteins (Fig. 2A and Table 3). Prominently represented were neuronal and non-neuronal AP-3 subunits (see Table 3, entries 1-7), clathrin chains (entries 8 and 9), BLOC-1 complex subunits (entries 10-13), molecular chaperones of the Hsc70 and Hsp90 families (entries 14-19), and diverse membrane proteins, including ZnT3 and PI4KIIα (entries 20-28). Half of all proteins identified by us were also found in other relevant vesicle proteomes including those from AP-3-derived vesicles (18), ARF1-GTPγS-recruited AP-3 to liposomes (30), neuronal and non-neuronal clathrin-coated vesicles (7, 8), and synaptic vesicles (31).
FIGURE 2.
Identification of components present in cross-linked AP-3-complexes. A, samples of total DSP-treated PC12 cell homogenate (Homog; lane 2), Triton-soluble supernatant (Sup; lane 3), pooled sucrose fractions 3-5 (Suc Fx 3-5; lane 4), and peptide (Pep.) eluate (lane 5) of cross-linked AP-3 complexes from δ antibody-coated magnetic beads were resolved by SDS-PAGE and silver-stained (inverted image is depicted). Enlarged lane 5 is shown together with some of the most prominent proteins identified by MS/MS and listed in Table 3. Lane 1 correspond to molecular weight markers. B, Triton-soluble supernatants of PC12 cells treated with (lanes 2, 4, and 6) and without DSP (lanes 1, 3, and 5) were immunoprecipitated with empty beads (lanes 1 and 2), beads coated with transferrin receptor (lanes 3 and 4), or δ AP-3 antibodies (lanes 5 and 6). Protein complexes were resolved by SDS-PAGE and analyzed by immunoblot (IB) with the indicated antibodies. Control immunoblots were performed with antibodies against synaptophysin (Sphysin) and β-actin. Lanes 7 and 8 correspond to 5% inputs. Note the arrow in Hsp90 blots to denote Hps90-specific bands. C, PC12 cells cross-linked with and without DSP were lysed in buffer A plus 0.1% Triton either in the absence (lanes 1 and 2) or presence of 0.05% SDS (lane 3 and 4). Samples were immunoprecipitated (IP) with AP-3 δ antibodies. Beads were washed six times, resuspended in sample buffer, and resolved in a 4-20% Criterion SDS-PAGE gel. AP-3 immunocomplexes were tested for the presence of clathrin, PI4KIIα, BLOC1, ZnT3-Myc, and granuphilin/SYTL4. A significant reduction in ZnT3-Myc and granuphilin associated with AP-3 was observed in the presence of SDS (lane 3 and 4). Inputs, 1.6% (lanes 5-8). D, wild type (lanes 1, 2, 5, 6, 9, and 10) and AP-3-null mouse skin fibroblasts (mocha mh/mh; lanes 3, 4, 7, 8, 11, and 12) were treated in the absence (odd lanes) or presence of DSP (even lanes). Detergent-soluble extracts were immunoprecipitated with antibodies against AP-3 δ-adaptin or AP-1 γ-adaptin. Immunocomplexes were resolved by SDS-PAGE and analyzed by immunoblot. Inputs equal 5%.
TABLE 3.
Proteins co-purifying with cross-linked AP-3 complexes
The table depicts proteins identified by AP-3 immunoaffinity chromatography. ∑ peptides and ∑ Mascot scores corresponds to the sum of all peptides identified and their individual Mascot scores in three MS/MS analyses from two independent purifications. Clathrin binding motifs were identified using gi-defined primary sequences fed into the Eukaryotic Linear Motif (ELM) search engine. Relevant previous proteomes correspond to those of 1) AP-3 microvesicles (18), 2) brain clathrin-coated vesicles (7), 3) HeLa cell clathrin-coated vesicles (8), 4) synaptic vesicles (31), and 5) GTP γS-dependent recruitment of AP-3 to proteoliposomes (30). Clathrin box corresponds to the consensus sequence L(I/V/L/M/F)X(I/V/L/M/F)(D/E).
| Protein | gi no. | ∑ peptides | ∑ Mascot scores | Clathrin box | Clathrin binding motif WXXW | Previous proteomes | |
|---|---|---|---|---|---|---|---|
| 1 | Ap3d1 protein | 34862288 | 303 | 16,302 | 1, 4, 5 | ||
| 2 | Ap3b1 protein | 157817716 | 276 | 12,454 | 387LVDVE391 | 1, 5 | |
| 974LLDLD978 | |||||||
| 1213LVTVE1217 | |||||||
| 3 | Ap3b2 protein | 52317148 | 247 | 11,592 | 228LIDVE232 | 1, 5 | |
| 808LLDLE812 | |||||||
| 1046LLTLD1050 | |||||||
| 4 | Ap3m1 protein | 1703027 | 48 | 2,081 | 77LFVIE81 | 3, 5 | |
| 5 | Ap3m2 protein | 5803000 | 23 | 1,058 | 77LFVIE81 | 4, 5 | |
| 6 | Ap3s1 protein | 4502861 | 35 | 1,659 | 1, 3, 5 | ||
| 7 | Ap3s2 protein | 109462206 | 12 | 583 | 4, 5 | ||
| 8 | Clathrin heavy chain 1 | 51491845 | 27 | 1,357 | 2, 3, 5 | ||
| 9 | Clathrin light chain a | 4502901 | 2 | 91 | 2,3 | ||
| 10 | Distrobrevin-binding protein 1 | 83415179 | 7 | 358 | 1, 3 | ||
| 11 | Reduced pigmentation protein | 109458315 | 9 | 482 | 3 | ||
| 12 | Cappuccino protein | 34878563 | 5 | 274 | 3 | ||
| 13 | Muted protein | 157823988 | 5 | 194 | 1, 3 | ||
| 14 | dnaK-type chaperone hsp72/uncoating ATPase | 347019 | 67 | 3,460 | 391LLLLD395 | 1, 2, 4 | |
| 15 | Heat shock protein 1, α/Hsp90 | 28467005 | 24 | 1,270 | 4 | ||
| 16 | Heat shock protein 1, βHsp90 | 40556608 | 55 | 2,741 | |||
| 17 | dnaK-type molecular chaperone hst70 | 92355 | 13 | 645 | 394LLLLD398 | ||
| 18 | heat shock 70-kDa protein 5 | 25742763 | 6 | 371 | 234LLTID238 | 4 | |
| 414LVLLD418 | |||||||
| 19 | Heat shock protein 70 | 396270 | 5 | 254 | 391LLLLD395 | ||
| 20 | Protocadherin 9 isoform 1 | 62661787 | 2 | 66 | |||
| 21 | Protocadherin 15 | 109509982 | 6 | 225 | |||
| 22 | Phosphatidylinositol 4-kinase type 2 α | 16758554 | 1 | 46 | 290LLQFE294 | 165WTKW168 | 1, 3, 4 |
| 296LVVLD300 | |||||||
| 23 | ZnT3 | 8134847 | 2 | 86 | 1, 4 | ||
| 24 | Neurotransmitter transporter NTT5 | 157820661 | 2 | 58 | |||
| 25 | CKAP4/p63 | 157823877 | 6 | 390 | |||
| 26 | Olfactory receptor 598 | 58801352 | 2 | 40 | |||
| 27 | Olfactory receptor Olr205 | 47576839 | 2 | 88 | |||
| 28 | Annexin II | 2143593 | 6 | 334 | 2 | ||
| 29 | Tyrosine hydroxylase | 6981652 | 4 | 176 | |||
| 30 | Granuphilin/SYTL4 | 17385946 | 1 | 41 | 492WTGW495 | ||
| 31 | Septin-7 | 9789726 | 3 | 114 | 2, 4, 5 | ||
| 32 | Cytoplasmic dynein intermediate chain 2B | 1151093 | 2 | 54 | |||
| 33 | Dynein, cytoplasmic, light intermediate chain 1 | 21955134 | 3 | 125 | |||
| 34 | Dynactin 2 | 51948450 | 1 | 54 | |||
| 35 | Dynein, cytoplasmic, heavy polypeptide 1 | 148491097 | 33 | 1,388 | 2543WSPW2546 | 2, 4 | |
| 36 | Kinesin family member 21B | 157821213 | 16 | 262 | |||
| 37 | Myosin, heavy polypeptide 9/mFLJ00279 protein | 6981236 | 22 | 819 | 825WQWW828 | ||
| 38 | Myosin regulatory light chain | 228542 | 6 | 318 | |||
| 39 | Microtubule-associated protein 1B (MAP 1B) | 19856246 | 10 | 543 | |||
| 40 | Intermediate filament protein nestin | 2209202 | 15 | 527 | |||
| 41 | Similar to A-kinase anchor protein 13 isoform 2 | 157823521 | 8 | 184 | 1435LLCLE1439 | ||
| 42 | Casein kinase II, α 1 polypeptide | 16758674 | 8 | 292 | 2 | ||
| 43 | Calcium/calmodulin-dependent protein kinase II, δ | 6978595 | 4 | 172 | |||
| 44 | Lactate dehydrogenase A | 8393706 | 5 | 255 | 2, 4 | ||
| 45 | Carbamoyl-phosphate synthetase 2 | 158534057 | 10 | 726 | |||
| 46 | Hemoflavoprotein b5/b5r | 12007117 | 7 | 255 | |||
| 47 | Development and differentiation-enhancing factor 2 | 157823109 | 7 | 236 | |||
| 48 | Rho guanine nucleotide exchange factor 7 | 18202066 | 6 | 218 | |||
| 49 | C9orf86/rbel1/F08G12.1 | 157819403 | 23 | 1,185 | |||
| 50 | Hypothetical protein HEAT repeat containing 4 | 109478489 | 8 | 372 | 687WNDW690 | ||
| 51 | E3 ubiquitin-protein ligase NEDD4 | 32469607 | 8 | 392 |
We confirmed whether proteins found by mass spectrometry within cross-linked AP-3 complexes selectively co-precipitated from control (Fig. 2B, lane 5) and/or DSP-treated PC12 Triton-soluble cellular extracts (Fig. 2B, lane 6). We used magnetic beads alone (Fig. 2B, lanes 1 and 2) or beads decorated with antibodies against TrfR as controls (Fig. 2B, lanes 3 and 4). This receptor does not interact with AP-3 (28). Intact AP-3 complexes were immunoprecipitated by δ-adaptin antibodies from in vivo cross-linked Triton cell extracts as determined by the presence of δ-, β3-, σ3-, and μ3-adaptins (Figs. 2B, lanes 5 and 6, and 6A, lanes 1-2′). In contrast, none of the AP-3 subunits were detected in transferrin receptor immunocomplexes or empty beads (Fig. 2B, lanes 1-4). Furthermore cross-linked AP-3 immunocomplexes were free of several abundant membrane and cytosolic proteins such as transferrin receptor, synaptophysin, and β-actin. AP-3-coated beads preferentially or selectively precipitated clathrin heavy chain (Fig. 2B), BLOC-1 subunits (Fig. 2B for pallidin; muted and dysbindin not shown), membrane proteins (Fig. 2B; ZnT3, PI4KIIα, and annexin II), molecular chaperones (Fig. 2B; Hsc70 and Hsp90), and granuphilin. We tested whether these interactions were resistant to more stringent conditions by preparing PC12 cell extracts and performing immunoprecipitations in the presence of 0.1% Triton X-100 either in the absence (Fig. 2C, lanes 1 and 2) or presence of 0.05% SDS (Fig. 2C, lanes 3 and 4). Clathrin, PI4KIIα, the BLOC-1 subunit pallidin, and ZnT3 co-precipitated with AP-3 from cross-linked cell extracts irrespective of whether Triton or SDS was used (Fig. 2C, compare lanes 2 and 4). In contrast, the specific association of ZnT3 and granuphilin detected in the absence of DSP was either reduced (granuphilin) or abolished (ZnT3) (Fig. 2, B (lane 5) and C (compare lanes 1 and 3)). However, the association of these proteins with AP-3 was still evident in the presence of DSP.
FIGURE 6.
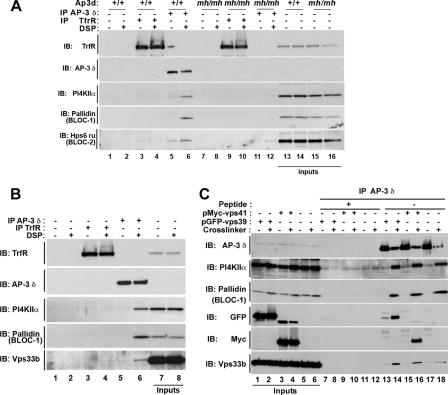
HPS gene deficiencies selectively affect the composition of cross-linked AP-3 complexes. Skin mouse fibroblast isolated from wild type C57B and HPS gene-deficient mice were incubated in the absence (n lanes) or presence (n′ lanes) of DSP. Triton-soluble supernatants were immunoprecipitated (IP) with δ AP-3 antibodies. Immunocomplexes were resolved by SDS-PAGE and analyzed by immunoblot (IB) with antibodies indicated to the left of the figure. Two different cells lines of wild type and HPS deficiencies were used in A and B, and they are numbered at the bottom of the figure. A depicts experiments performed with BLOC-1- (Pldnpa/pa; lanes 3-4′) and AP-3 (Ap3b1pe/pe; lanes 5-6′)-deficient cells. Note that Ap3b1pe/pe corresponds to an AP-3 hypomorph phenotype. B depicts deficiencies in BLOC-2 (Hps3coa/coa; lanes 3-4′) or BLOC-3 (Hps1ep/ep; lanes 5-6′). C depicts BLOC-1 (Pldnpa/pa; lanes 2 and 2′), BLOC-1 and -2 (Pldnpa/pa, Hps6ru/ru; lanes 3 and 3′), BLOC-1, -2, and -3 (Pldnpa/pa, Hps5ru2J/ru2J, Hps1ep/ep lanes 4 and 4′), or BLOC-2 and -3 (Hps4le/le, Hps3coa/coa; lanes 5 and 5′) deficiencies.
We further examined the specificity of these AP-3 interactions in mouse fibroblasts from wild type (Ap3d+/+) and AP-3-deficient mocha mice (Ap3dmh/mh). Immunoisolation of AP-3 complexes from DSP-treated wild type fibroblasts precipitated clathrin, PI4KIIα, and the BLOC-1 subunit pallidin (Fig. 2D, lane 6). Importantly these proteins were not observed in immunoprecipitations from AP-3-null mocha (mh/mh) cross-linked cell extracts, indicating that proteins present in AP-3 cross-linked complexes were not due to spurious binding to bead-antibody complexes (Fig. 2D, compare lanes 6 and 8). Finally we analyzed whether some of these AP-3 interactors were shared with the adaptor complex AP-1 (Fig. 2D). Clathrin was present in a complex with either AP-1 or AP-3. However, the interactions of BLOC-1 and PI4KIIα with cross-linked AP-3 complexes were restricted to this adaptor complex (Fig. 2D, compare lanes 2 and 6). Importantly PI4KIIα interaction with AP-1 was negligible in wild type cells, yet it increased beyond background levels in mocha cells. These results are consistent with our findings that AP-1 and PI4KIIα colocalize only in AP-3-null cells (18).
To assess whether proteins identified by mass spectrometry associate into a complex, we asked whether clathrin, BLOC-1 (detected by its subunit pallidin), and PI4KIIα sedimented together with AP-3 by sucrose sedimentation analysis (Fig. 3A). Triton-soluble extracts from control and DSP-treated PC12 cells were fractionated by sucrose sedimentation. Sucrose fractions were immunoisolated with AP-3 δ antibodies followed by immunoblot analysis of immunocomplexes (Fig. 3A). Clathrin, BLOC-1, and PI4KIIα co-precipitated with AP-3 as a heterogeneous complex whose sedimentation ranged from 9.4 to 16.5 S. Such a complex was not observed in uncross-linked PC12 Triton-soluble extracts (Fig. 3A, lanes 1-8). AP-3, BLOC-1, and PI4KIIα could form a tripartite complex. We tested this hypothesis performing sequential immunoprecipitations with antibodies against AP-3 δ and an HA epitope engineered into either PI4KIIα or PI4KIIαL61A/L62A. PI4KIIαL61A/L62A is a mutant that lacks a dileucine sorting motif necessary to bind AP-3, thus providing a stringent control for specificity (Fig. 3B) (1). HEK293 cells transfected with HA-tagged forms of wild type and PI4KIIαL61A/L62A were treated with DSP, and cross-linked complexes were precipitated with AP-3 δ antibodies. Cross-linked complexes bound to AP-3 beads were efficiently eluted with the δ epitope peptide (Fig. 3B, compare lanes 3 and 4 with lanes 5 and 6). Eluates underwent a second round of immunoprecipitation with anti-HA antibodies (Fig. 3B, lanes 7 and 8). PI4KIIα-HA co-precipitated AP-3 and BLOC-1 subunits detected by immunoblot, yet this tripartite complex failed to be isolated from cells expressing PI4KIIαL61A/L62A-HA. These data indicate that BLOC-1, PI4KIIα, and AP-3 associate into a high molecular weight complex. Collectively our results demonstrate that in vivo controlled cross-linking is a viable approach to identify proteins associated into labile macromolecular complexes. In the case of AP-3, the macromolecular complex possesses as a minimum BLOC-1 and PI4KIIα.
FIGURE 3.
AP-3, BLOC-1, and PI4KIIα form a tripartite molecular complex. A, clarified detergent-soluble extracts from DSP-treated PC12 cells were resolved by sucrose sedimentation as described in Fig. 1D. Fractions 1-13 were immunoprecipitated (IP) with AP-3 δ antibodies, and immunocomplexes were analyzed by immunoblot (IB) with the indicted antibodies. Note that the bands in pallidin blots seen in the absence and presence of DSP correspond to IgG light chains. Both immunoprecipitating and immunoblotting antibodies were raised in mice. B, HEK293T cells transfected with PI4KIIα-HA wild type or the mutant PI4KIIαL61A/L62A-HA were incubated with DSP and lysed, and supernatants were immunoprecipitated with AP-3 δ antibodies. AP-3 immunocomplexes were eluted with 50 μm δ antigenic peptide during 3 h at 4 °C (Eluate; lanes 3 and 4). After peptide elution, beads were heated at 75 °C for 10 min to release remaining AP-3 complexes (Beads; lanes 5 and 6). δ antigenic peptide eluate (lanes 3 and 4) or heat eluted material remaining in beads (lanes 5 and 6) were immunoprecipitated again with rabbit antibodies against HA epitope to isolate complexes containing HA-tagged PI4KIIα (lanes 5-8). Immunocomplexes were resolved by SDS-PAGE and analyzed by Western blot. AP-3 isolates a ternary complex containing PI4KIIα-HA and the BLOC1 complex (lane 8). PI4KIIαL61A/L62A-HA mutant severely reduced BLOC-1 and AP-3 associated to the kinase (lane 7). Φ denotes that the procedure was not performed. Lanes 1 and 2 represent the initial input in the experiment (10%). The arrow indicates IgG chains.
AP-3, BLOC-1, and PI4KIIα Concentrate in Clathrin-coated Vesicles—Adaptors and clathrin cycle between cytosolic and membrane-bound states. Once on membranes, adaptors and clathrin are concentrated along with membrane proteins into CCVs (3, 4). We analyzed the subcellular distribution of AP-3, PI4KIIα, BLOC-1, and clathrin by fractionation of DSP-cross-linked PC12 cells into soluble and total membrane fractions (Fig. 4A, lanes 1-6 and 7-12, respectively). Cytosol and membranes were solubilized with detergent, and cross-linked AP-3 complexes were isolated with AP-3 δ-adaptin antibodies (Fig. 4A, lanes 5 and 6 and lanes 11 and 12). Cytosolic fractions were free of membranes as evidenced by the negligible content of PI4KIIα (Fig. 4A, compare lanes 13 and 14 with lanes 15 and 16). AP-3 complexes isolated from cross-linked cytosolic fractions contained AP-3 and the BLOC-1 subunit pallidin but were devoid of other cytosolic proteins found in AP-3 cross-linked complexes obtained from whole-cell extracts such as clathrin (Fig. 4A, lane 6). In contrast, AP-3 complexes isolated from cross-linked membrane fractions were enriched in AP-3 and its membrane protein cargo/regulator PI4KIIα, clathrin, and the BLOC-1 subunit pallidin (Fig. 4A, compare lanes 6 and 12). Binding of these proteins to AP-3 was not due to cytosolic contaminants in the membrane fraction as indicated by the absence of tubulin (Fig. 4A, lane 12). Furthermore background binding of all these proteins was negligible as determined by precipitations using beads lacking antibodies or decorated with transferrin receptor antibodies (Fig. 4A, lanes 1-4 and 7-10). We confirmed the association of AP-3, clathrin, and PI4KIIα in membranes immunoisolating cross-linked clathrin complexes from DSP-treated PC12 cells (Fig. 4B). Detergent-soluble extracts from vehicle- and DSP-treated cells were immunoisolated with the clathrin monoclonal antibody X22. AP-3 and PI4KIIα co-isolated with clathrin yet only in the presence of DSP (Fig. 4B, compare lanes 9 and 10). Similarly immunoisolation of cross-linked AP-3 complexes yielded clathrin and PI4KIIα in DSP-treated extracts (Fig. 4B, compare lanes 7 and 8). In contrast; AP-3, clathrin, and PI4KIIα levels were negligible in control immunoisolations performed with empty beads (Fig. 3B, lanes 5 and 6) or transferrin receptor antibody-containing beads (Fig. 4B, lanes 3 and 4).
FIGURE 4.
AP-3, BLOC-1, and PI4KIIα are enriched in membrane compartments and in clathrin-coated vesicles. A, cytosol (Cyt; lanes 1-6) and membrane (Mem; lanes 7-12) fractions of PC12 cells treated in the absence (odd lanes) or presence (even lanes) of DSP were immunoprecipitated with empty beads (lanes 1, 2, 7, and 8), beads coated with TrfR antibodies (lanes 3, 4, 9, and 10) or δ AP-3 (lanes 5, 6, 11, and 12) antibodies. Immunocomplexes were resolved by SDS-PAGE and analyzed by immunoblot (IB) with antibodies indicated to the right of the figure. B, Triton-soluble extracts of PC12 cells incubated in the absence or presence of DSP were immunoprecipitated (IP) with beads decorated with TrfR antibodies (lanes 3 and 4), no antibodies (lanes 5 and 6), AP-3 δ antibodies (lanes 7 and 8), and clathrin X22 antibodies (lanes 9 and 10). Immunocomplexes were resolved by SDS-PAGE and analyzed by immunoblot. C, CCV-enriched fractions (lane 6) prepared as described by Girard et al. (23) were resolved by SDS-PAGE and analyzed by immunoblot. Compare CCV with total homogenate (H) and fractions P1, S1, P2, and S2 (see “Experimental Procedures”). All lanes contain 1 μg of protein.
We next tested whether membrane-bound AP-3, BLOC-1, and PI4KIIα present on membrane fractions were concentrated in clathrin-coated vesicles. We prepared clathrin-coated vesicle-enriched fractions from non-cross-linked PC12 cells. CCV enrichment with respect to total homogenate was monitored by the content of clathrin and clathrin-binding adaptors (Fig. 4C, AP-1 γ and AP-2 α, compare lanes 1 and 6). Similar to clathrin, AP-3 subunits (Fig. 4C, β3- and μ3-adaptin) as well as BLOC-1 complex subunits (Fig. 4C, pallidin and dysbindin) were enriched in clathrin-coated vesicle fractions (Fig. 4C). Concomitantly membrane proteins that bind AP-3, such as PI4KIIα and ZnT3, co-purified and were enriched in CCV (Fig. 4C). These results demonstrate that the AP-3 adaptor complex forms a complex with BLOC-1, clathrin, and PI4KIIα on membranes. The association of PI4KIIα and BLOC-1 with AP-3 and clathrin into a complex likely determines PI4KIIα and BLOC-1 concentration in CCVs.
AP-3, BLOC-1, and PI4KIIα Co-precipitate with the HPS Complexes BLOC-2 and HOPS—The co-isolation of two HPS complexes, AP-3 and BLOC-1, with PI4KIIα prompted us to examine whether other HPS complexes not detected by mass-spectrometry in PC12 cells could co-precipitate with cross-linked AP-3 complexes. Our rationale was founded in the detection of the HOPS subunit vps33b in PI4KIIα-containing microvesicles (18) and the binary interactions between BLOC-1 and BLOC-2 described by Di Pietro et al. (16). To test for the presence of BLOC-2 and HOPS complex subunits in cross-linked AP-3 complexes, we used mouse skin fibroblasts and human HEK293 cells. In contrast with PC12 cells, endogenous BLOC-2 and HOPS complex subunits were readily detectable in these cell types (Figs. 5 and 6). Cross-linked AP-3 complexes were immunoisolated from detergent-soluble extracts from wild type mouse fibroblasts incubated in the absence and presence of DSP (Fig. 5A, lanes 1-6). Similar to our findings in PC12 cells, AP-3 cross-linked complexes were co-isolated with clathrin, BLOC-1, and PI4KIIα from DSP-treated wild type fibroblasts (Fig. 5A, lane 6). In addition, the BLOC-2 subunit Hps6 ruby was also detected in these AP-3 cross-linked complexes containing PI4KIIα (Fig. 5A, lane 6). The specificity of a BLOC-2 interaction with AP-3 cross-linked complexes was confirmed by the absence of AP-3, PI4KIIα, BLOC-1, and the BLOC-2 subunit Hps6 ruby from transferrin receptor immunoisolations (Fig. 5A, lane 4). We further tested the association of BLOC-2 subunits with AP-3 cross-linked complexes using mouse fibroblasts carrying genetic deficiencies in HPS genes. First AP-3 δ antibody immunoisolations performed with δ-null mocha fibroblasts abrogated the detection of the BLOC-2 subunit Hsp6 ruby (Fig. 5A, mh/mh, lane 12). Finally deficiencies in the BLOC-1 subunit pallidin decreased the content of Hps6 ruby in AP-3 cross-linked complexes (Fig. 6C, Pldnpa/pa, compare lanes 1′ and 2′).
FIGURE 5.
BLOC-2 and HOPS subunits associate with cross-linked AP-3 complexes. DSP cross-linking was performed in skin fibroblast from mocha mice transduced with an empty retrovirus (mh/mh) or with a retrovirus containing the δ subunit of AP-3 (AP-3+/+). A, Triton-soluble supernatant of cells treated in the absence (odd lanes) or presence of DSP (even lanes) were immunoprecipitated (IP) with empty beads (lanes 1, 2, 7, and 8), with beads coated with TrfR (lanes 3, 4, 9, and 10), or with δ antibodies (lanes 5, 6, 11, and 12). Immunocomplexes were resolved by SDS-PAGE and analyzed by immunoblot with antibodies against AP-3 and subunits of BLOC-1 (pallidin) and BLOC-2 (Hsp6 ru). B, HEK293 Triton-soluble cell supernatants treated with (even lanes) and without DSP (odd lanes) were immunoprecipitated with empty beads (lanes 1 and 2) and beads coated with monoclonal antibodies against TrfR (lanes 3 and 4) or the δ subunit of AP-3 (lanes 5 and 6). Immunocomplexes were resolved by SDS-PAGE and analyzed by immunoblot with the antibodies specified to the left. C, Triton-soluble supernatants of untransfected HEK293 cells or cells transfected with Myc-tagged vps41 or GFP-tagged vps39 treated with (even lanes) and without DSP (odd lanes) were immunoprecipitated with beads decorated with δ AP-3 antibodies (lanes 7 and 18). Controls were performed by competition with 10 μm δ peptide (lanes 7-12). Protein complexes were resolved by SDS-PAGE and analyzed by immunoblot (IB) with the indicated antibodies. All inputs correspond to 5%.
We focused on HEK293 cells to test whether HOPS complex subunits could be co-isolated with AP-3 cross-linked complexes. These cells endogenously express high levels of the HOPS subunit vps33b (Fig. 5B, lanes 7 and 8). Much like PC12 cells and mouse skin fibroblasts, AP-3 complexes isolated from DSP-treated HEK293 cell extracts co-precipitated with PI4KIIα and the BLOC-1 subunit pallidin. However, in addition to these proteins, the HOPS complex subunit vps33b was readily detectable in cross-linked AP-3 complexes (Fig. 5B, lane 6). The presence of vps33b in AP-3 complexes was selective because Vps33b was absent from control immunoisolations with transferrin receptor antibody-coated beads (Fig. 5B, lane 4) and from AP-3 δ antibody immunoisolations out-competed with a δ antigenic peptide (Fig. 5C, compare lanes 12 and 18). The interaction of HOPS complex subunits with AP-3 cross-linked complexes was not limited just to endogenous vps33b. In fact, exogenous expressed vps39-GFP or vps41-Myc also associated selectively with cross-linked AP-3 complexes isolated from HEK293 cells (Fig. 5C, lanes 14 and 16, respectively). Similar to vps33b, PI4KIIα, and BLOC-1 these proteins were eliminated by competing AP-3 δ antibody with a δ antigenic peptide. These results indicate that subunits of three HPS complexes, BLOC-1, BLOC-2, and HOPS, co-isolate with AP-3.
HPS Gene Deficiencies Selectively Affect the Composition of the AP-3-BLOC-1-PI4KIIα Protein Complexes—PI4KIIα, BLOC-1, BLOC-2, and HOPS complex subunits are present in cross-linked AP-3 complexes. We hypothesized that these interactions occurred to facilitate the targeting of PI4KIIα. To explore this hypothesis, we analyzed the effects of HPS mutants either null or hypomorphs for AP-3, BLOC-1 to -3, or combinations thereof in the inclusion of PI4KIIα into AP-3 cross-linked complexes. These studies used primary skin fibroblasts from wild type C57BL mice and mutants either in 1) the AP-3 complex (Ap3b1pe/pe, hypomorph mutant with an almost complete absence of β3-μ3 subunits yet still possessing δ-σ3), 2) BLOC-1 complex (Pldnpa/pa), 3) BLOC-2 complex (Hps3coa/coa, Hps5ru-2J/ru-2J, or Hps6ru/ru), or 4) BLOC-3 complex (Hps1ep/ep or Hps4le/le) (17). Cross-linked AP-3 complexes isolated from Ap3b1pe/pe cells contained δ-σ3 subunits but showed a severe reduction of PI4KIIα and BLOC-1 subunit pallidin (Fig. 6A, compare lanes 1-2′ with lanes 5-6′). Surprisingly despite the severe reduction of β3 chains in Ap3b1pe/pe, there was clathrin associated with the remaining δ-σ3 AP-3 cross-linked subunits. In contrast with the Ap3b1pe/pe mutant phenotypes, the absence of BLOC-1 in Pldnpa/pa cells decreased the content of PI4KIIα present in AP-3 cross-linked complexes without affecting the levels of clathrin (Fig. 6A, compare lanes 1-2′ with lanes 3-4′). None of the PI4KIIα and clathrin phenotypes found in Ap3b1pe/pe and Pldnpa/pa cells were observed in cells either lacking the BLOC-2 or BLOC-3 complexes (Fig. 6B, BLOC-2 deficient, Hps3coa/coa, lanes 3-4′; BLOC-3-deficient, Hps1ep/ep, lanes 5-6′).
We explored the effects of single, double, and triple BLOC complex deficiencies to test whether combinations of BLOC complexes could regulate the content of PI4KIIα present in AP-3 cross-linked complexes. Complexes isolated from cells lacking BLOC-1 possessed reduced levels of PI4KIIα and the BLOC-2 subunit Hps6 ru without affecting clathrin content (Fig. 6C, Pldnpa/pa, lanes 2 and 2′). The reduced PI4KIIα level phenotype observed in BLOC-1-deficient Pldnpa/pa cells remained unchanged irrespective of whether BLOC-1 deficiency was combined with a BLOC-2 defect (Fig. 6C, Pldnpa/pa, Hps6ru/ru cells, lanes 3 and 3′) or was part of a triple BLOC-1, -2, and -3 deficiency (Fig. 6C, Pldnpa/pa, Hps5ru-2J/ru-2J, Hps4le/le cells, lanes 4 and 4′). Predictably the combined absence of BLOC-2 and BLOC-3 did not affect PI4KIIα or clathrin levels present in AP-3 cross-linked complexes (Fig. 6C, Hps3coa/coa, Hps4le/le cells, lanes 5 and 5′).
These results indicate that AP-3 forms a complex(es) with clathrin, PI4KIIα, BLOC-1, BLOC-2, and HOPS. A tripartite complex between AP-3, BLOC-1, and PI4KIIα is at the core of these interactions where BLOC-1 selectively regulates the abundance of PI4KIIα that associates with AP-3.
AP-3 and BLOC-1 Down-regulation Phenocopies PI4KIIα Knockdown—Association between AP-3, BLOC-1, and PI4KIIα suggests that deficiencies in components of this complex should alter traffic of membrane proteins to lysosomes in a similar fashion. We tested this prediction analyzing the appearance of LAMP1 enlarged endosomes after down-regulation of PI4KIIα, AP-3, or BLOC-1. LAMP1 is a lysosomal membrane protein that appears in enlarged endosomes induced by acute down-regulation of PI4KIIα (1). Rescue of this phenotype requires the presence of an AP-3 dileucine sorting motif as well as the kinase activity in PI4KIIα (1). siRNA-mediated down-regulation of PI4KIIα (Fig. 7A, lane 2) reduced the content of this enzyme without affecting the levels of either components of the tripartite complex (AP-3 or BLOC-1) or proteins that co-precipitate with AP-3 and PI4KIIα, such as clathrin and the HOPS subunit vps33b. Similarly down-regulation of pallidin or AP-3 δ selectively decreased the levels of subunits found in each one of these individual protein complexes (Fig. 7A, lanes 4 and 5). Reduction in the content of multiple AP-3 or BLOC-1 subunits by siRNA knockdown of only one subunit recapitulates phenotypes observed in single gene deficiencies affecting AP-3 and BLOC-1 in mice (32, 33). Acute siRNA-mediated down-regulation of PI4KIIα induced the appearance of LAMP1-positive enlarged endosomes in 25% of the cells analyzed. In contrast ∼5% in control siRNA-treated cells displayed this phenotype (Fig. 7, B and C, gray bars). Similarly acute down-regulation of AP-3 or BLOC-1 subunits induced LAMP1 enlarged organelles with the same penetrance as PI4KIIα siRNA (Fig. 7C, gray bars). In addition to LAMP1 enlarged endosomes, either BLOC-1 pallidin knockdown (Fig. 7B), PI4KIIα, or AP-3 δ siRNA-treated cells (data not shown) possessed “doughnut”-shaped LAMP1-positive organelles clustered in the periphery of cells (Fig. 7B). Control cells exhibited this phenotype in less than 2% of cells in contrast with 30-50% of either BLOC-1-, AP-3-, or PI4KIIα-down-regulated cells (Fig. 7C, black bars). These data provide functional evidence supporting a sorting role of a BLOC-1-PI4KIIα-AP-3 tripartite complex.
FIGURE 7.
AP-3, BLOC-1, and PI4KIIα knockdown alters the subcellular distribution of a lysosomal membrane protein, LAMP1. HEK293T cells were transfected either with controls, human PI4KIIα-, BLOC1 subunit pallidin-, or AP-3 δ subunit-targeting siRNA oligonucleotides. Twenty-four hours after the second siRNA transfection cells were split and reseeded in 12-well plates for Western blots (A) or on coverslips for immunofluorescence (B) analysis. A depicts immunoblot (IB) analysis of control and knockdown cells. No changes in expression levels were observed for LAMP1, clathrin, the HOPS subunit vps33b, TrfR, or β-actin. B, indirect immunofluorescence detection of human LAMP1. Experimental knockdowns resulted in redistribution of the lysosomal protein LAMP1 either toward enlarged endosomes (gray bars; C) or as doughnut-shaped organelles redistributed toward cell periphery (black bars; C). Quantification of both phenotypes is presented in C. Numbers in parentheses represent total number of cells counted. Bar, 10 μm.
DISCUSSION
Here we used an in vivo chemical cross-linking approach coupled with immunoaffinity chromatography to identify and explore the role of molecules co-purifying with AP-3 (Fig. 1). This strategy yielded 51 molecules identified by mass spectrometry in PC12 cells (Table 3 and Fig. 8). Prominently represented among these proteins were the membrane-anchored proteins PI4KIIα and ZnT3 and cytosolic factors such as BLOC-1 subunits and clathrin chains. Moreover a targeted search for subunits from other HPS complexes identified BLOC-2 and HOPS in non-neuronal cells (Figs. 5 and 6). These results provide novel insight into the molecular organization of HPS protein complexes indicating that AP-3 forms complexes with clathrin and cargoes, such as PI4KIIα, and three other HPS complexes, BLOC-1, -2, and HOPS. The relevance of these associations is illustrated by the effect that genetic deficiencies in HPS complexes exert on the composition of AP-3 cross-linked complexes. For example, an AP-3 hypomorph allele (Ap3b1pe/pe) predictably decreased the association of PI4KIIα, BLOC-1, and clathrin to cross-linked AP-3 complexes (Fig. 6). However, a BLOC-1-null background (Plndpa/pa) significantly decreased the association of PI4KIIα and BLOC-2 with AP-3 cross-linked complexes without affecting clathrin (Fig. 6). These data are consistent with a model where BLOC-1 modulates the association of PI4KIIα to AP-3. Furthermore our present findings in BLOC-1-deficient cells provide a molecular mechanism to understand our previous observation that PI4KIIα and AP-3 colocalization is reduced in BLOC-1-deficient Pldnpa/pa cells (34). In vivo chemical cross-linking uniquely allows the isolation of multiprotein complexes containing coats, adaptors, enzymatic regulators of vesicle biogenesis, and specific membrane protein cargoes recognized by adaptors. In the present case, PI4KIIα and ZnT3, two well established AP-3 cargoes, associate with AP-3 but not transferrin receptor. Our data provide biochemical, genetic, and functional evidence for complexes containing a minimum of AP-3, BLOC-1, and PI4KIIα (Figs. 3B, 6, and 7, respectively).
FIGURE 8.
Architecture of the AP-3-HPS protein interaction network. A describes the subunit composition of the AP-3, BLOC-1, BLOC-2, and HOPS complexes depicted in B. Subunits highlighted in red are those identified in this study by immunoblot and/or MS/MS. B, proposed diagram of interactions among components identified. Proteins pairs linked by lines highlighted with a single asterisk indicate interactions curated in BioGRID. Double asterisks denote interactions documented here and by others (16, 40). Annx II, annexin II; HC, heavy chain; LC, light chain.
Components co-purifying with AP-3 isolated from DSP-treated cells could correspond to molecules that either directly interact with AP-3 or lay in proximity to the adaptor. To minimize those molecules that could spuriously co-purify with cross-linked AP-3, we used diverse criteria to filter protein identification by mass spectrometry (22), immunoprecipitation specificity with AP-3 but neither transferrin receptor nor AP-1 antibodies, and the absence of AP-3-immunoprecipitated proteins in AP-3-null Ap3dmh/mh cells (Fig. 2). Using these criteria 50% of those components identified in our studies were also represented in other relevant analyses such as those performed in clathrin-coated vesicles by us and others (7, 8), synaptic vesicles (31), AP-3 microvesicles (18), and AP-3 complexes assembled into liposomes (30). Our analysis does not represent an exhaustive identification of all components of an AP-3 interaction “network.” In vivo controlled cross-linking possesses inherent limitations. First, the nature of the molecules retained in cross-linked complexes is strictly dependent on the cross-linker chemistry. Second, antibodies used to isolate cross-linked components select for complexes where the epitope is exposed. This is evident with antibodies against δ-adaptin that recognize an AP-3 epitope in the proximity of where VAMP7-TI binds AP-3 (35). This could explain our inability to detect this v-(R)-SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) in cross-linked AP-3 complexes. Third, a component attached to detergent-insoluble membranes or the cytoskeleton (36) would be poorly represented. Finally we cannot assess the in vivo stoichiometry of components present in immunoprecipitated cross-linked complexes. This idea is based on the subsaturating DSP conditions used to stabilize supramolecular complexes yet still maintain low background of spurious interactions. This is an important consideration because either HPS complexes could function together in an array of several HPS complexes, or individual HPS complexes could constitute functional units of their own. This could be the case for BLOC-1 in melanocytes where it controls targeting of TYRP1 and ATP7A independently of AP-3 (37, 38). Similarly Saccharomyces cerevisiae HOPS regulates in vitro homotypic fusion of vacuoles independently of AP-3 (39).
A striking observation was the presence of clathrin in mutant AP-3 cross-linked complexes isolated from Ap3b1pe/pe cells (Fig. 5). These mutant cells completely lack β3-adaptin but retain their δ-σ3 subcomplexes (33). β3-Adaptin possesses a clathrin binding motif of the L(I/V/L/M/F)X(I/V/L/M/F)(D/E) type. This motif, called clathrin box, is necessary for clathrin and AP-3 β3 interaction in vitro (40). However, it appears to be dispensable in vivo (33). These published data suggest either that clathrin is not required for AP-3-dependent vesicle biogenesis or that other molecular determinants in addition to the clathrin box in β3 are required for clathrin engagement in AP-3-dependent vesicle biogenesis. Computational analysis identified clathrin binding motifs of the L(I/V/L/M/F)X(I/V/L/M/F)(D/E) and WXXW type in one of every four molecules identified by mass spectrometry or immunoblot (see Table 3) (41, 42). This suggests that multiple components present in cross-linked AP-3 complexes could mediate clathrin binding. Notoriously PI4KIIα possesses three putative clathrin binding motifs. Consistent with an association between clathrin and PI4KIIα, this enzyme is concentrated in CCVs, clathrin antibodies immunoprecipitated PI4KIIα (Fig. 3), and siRNA-mediated knockdown of clathrin selectively decreased the total cellular levels of PI4KIIα (data not shown).
Interactions between some HPS complexes have been described previously. These include AP-3 and BLOC-1, BLOC-1 and BLOC-2 (16, 40), BLOC-2 and clathrin (43), and AP-3 and clathrin. The significance of the latter remains controversial (33, 40). Here we describe novel and DSP-sensitive molecular associations among HPS complexes. These include AP-3 and HOPS (Fig. 5, B and C); AP-3, BLOC-1, and PI4KIIα (see above); clathrin, AP-3, and PI4KIIα (Fig. 4); and AP-3 and BLOC-2 by mechanisms sensitive to AP-3 alleles (Ap3dmh/mh and Ap3b1pe/pe) as well as the BLOC-1 Pldnpa/pa allele (Figs. 5A and 6). The absence of BLOC-1 (Plndpa/pa) significantly decreased the association of BLOC-2 and PI4KIIα with AP-3 (Fig. 6). These findings suggest that BLOC-1 regulates the association of PI4KIIα and BLOC-2 with AP-3 complexes. We previously established that PI4KIIα sorting from endosomes requires its interaction with AP-3 by means of a dileucine sorting signal (1). Ablation of this dileucine signal completely prevents PI4KIIα interaction with AP-3. However, PI4KIIα interaction with AP-3 is reduced in the absence of BLOC-1. How can these observations be reconciled? Although speculative, BLOC-1 could act as an AP-3-specific accessory sorting factor capable of modulating the recognition by AP-3 of the dileucine sorting motif present in PI4KIIα (1, 44-46). Alternatively BLOC-1 or a BLOC-1-interacting factor could activate PI4KIIα kinase activity and increase the association of AP-3 to membrane domains enriched in phosphatidylinositol 4-phosphate. Elucidation of these mechanisms would require the identification of direct protein interactions between PI4KIIα with either BLOC-1 subunits or unknown BLOC-1-interacting factors.
Acknowledgments
We are indebted to members of the Faundez laboratory and Drs. A. Kowalczyk and S. L'Hernault for comments.
This work was supported, in whole or in part, by National Institutes of Health Grants NS42599 and GM 077569 (to V. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HPS, Hermansky-Pudlak syndrome; PI4KIIα, phosphatidylinositol-4-kinase type II α; GFP, green fluorescent protein; HA, hemagglutinin; PBS, phosphate-buffered saline; DSP, dithiobis(succinimidyl propionate); HPLC, high performance liquid chromatography; MS, mass spectrometry; MS/MS, tandem mass spectrometry; CCV, clathrin-coated vesicle; MES, 4-morpholineethanesulfonic acid; siRNA, small interfering RNA; TrfR, transferrin receptor; GTPγS, guanosine 5′-O-(thiotriphosphate); HOPS, homotypic fusion and vacuole protein sorting; BLOC, biogenesis of liposome-related organelles complex.
References
- 1.Craige, B., Salazar, G., and Faundez, V. (2008) Mol. Biol. Cell 191415 -1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino, J. S., and Glick, B. S. (2004) Cell 116153 -166 [DOI] [PubMed] [Google Scholar]
- 3.Robinson, M. S. (2004) Trends Cell Biol. 14167 -174 [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino, J. S., and Traub, L. M. (2003) Annu. Rev. Biochem. 72395 -447 [DOI] [PubMed] [Google Scholar]
- 5.Schmid, E. M., and McMahon, H. T. (2007) Nature 448883 -888 [DOI] [PubMed] [Google Scholar]
- 6.Conner, S. D., and Schmid, S. L. (2003) Nature 42237 -44 [DOI] [PubMed] [Google Scholar]
- 7.Blondeau, F., Ritter, B., Allaire, P. D., Wasiak, S., Girard, M., Hussain, N. K., Angers, A., Legendre-Guillemin, V., Roy, L., Boismenu, D., Kearney, R. E., Bell, A. W., Bergeron, J. J., and McPherson, P. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 1013833 -3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borner, G. H., Harbour, M., Hester, S., Lilley, K. S., and Robinson, M. S. (2006) J. Cell Biol. 175571 -578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid, E. M., Ford, M. G., Burtey, A., Praefcke, G. J., Peak-Chew, S. Y., Mills, I. G., Benmerah, A., and McMahon, H. T. (2006) PLoS Biol. 4e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danglot, L., and Galli, T. (2007) Biol. Cell 99349 -361 [DOI] [PubMed] [Google Scholar]
- 11.Newell-Litwa, K., Seong, E., Burmeister, M., and Faundez, V. (2007) J. Cell Sci. 120531 -541 [DOI] [PubMed] [Google Scholar]
- 12.Raposo, G., and Marks, M. S. (2007) Nat. Rev. Mol. Cell Biol. 8786 -797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raposo, G., Marks, M. S., and Cutler, D. F. (2007) Curr. Opin. Cell Biol. 19 394-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Pietro, S. M., and Dell'Angelica, E. C. (2005) Traffic 6525 -533 [DOI] [PubMed] [Google Scholar]
- 15.Wei, M. L. (2006) Pigm. Cell Res. 1919 -42 [DOI] [PubMed] [Google Scholar]
- 16.Di Pietro, S. M., Falcon-Perez, J. M., Tenza, D., Setty, S. R., Marks, M. S., Raposo, G., and Dell'Angelica, E. C. (2006) Mol. Biol. Cell 174027 -4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautam, R., Novak, E. K., Tan, J., Wakamatsu, K., Itoh, S., and Swank, R. T. (2006) Traffic 7 779-792 [DOI] [PubMed] [Google Scholar]
- 18.Salazar, G., Craige, B., Wainer, B. H., Guo, J., De Camilli, P., and Faundez, V. (2005) Mol. Biol. Cell 163692 -3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson, S. C., Winistorfer, S. C., Poupon, V., Luzio, J. P., and Piper, R. C. (2004) Mol. Biol. Cell 151197 -1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar, G., Love, R., Werner, E., Doucette, M. M., Cheng, S., Levey, A., and Faundez, V. (2004) Mol. Biol. Cell 15575 -587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, G., Waltham, M., Anderson, N. L., Unsworth, E., Treston, A., and Weinstein, J. N. (1997) Electrophoresis 18391 -402 [DOI] [PubMed] [Google Scholar]
- 22.Khwaja, F. W., Reed, M. S., Olson, J. J., Schmotzer, B. J., Gillespie, G. Y., Guha, A., Groves, M. D., Kesari, S., Pohl, J., and Meir, E. G. (2007) J. Proteome Res. 6 559-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girard, M., Allaire, P. D., Blondeau, F., and McPherson, P. S. (2004) in Current Protocols in Cell Biology (Bonifacino, J. S., Dasso, M., Harford, J., Lippincott-Schwartz, J., and Yamada, K., eds) pp.3.13.1 -3.13.31, John Wiley & Sons, New York18228454
- 24.Balla, A., Tuymetova, G., Barshishat, M., Geiszt, M., and Balla, T. (2002) J. Biol. Chem. 27720041 -20050 [DOI] [PubMed] [Google Scholar]
- 25.Alloza, I., Martens, E., Hawthorne, S., and Vandenbroeck, K. (2004) Anal. Biochem. 324137 -142 [DOI] [PubMed] [Google Scholar]
- 26.Lomant, A. J., and Fairbanks, G. (1976) J. Mol. Biol. 104243 -261 [DOI] [PubMed] [Google Scholar]
- 27.Xiang, C. C., Mezey, E., Chen, M., Key, S., Ma, L., and Brownstein, M. J. (2004) Nucleic Acids Res. 32 e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dell'Angelica, E. C., Shotelersuk, V., Aguilar, R. C., Gahl, W. A., and Bonifacino, J. S. (1999) Mol. Cell 3 11-21 [DOI] [PubMed] [Google Scholar]
- 29.Kantheti, P., Qiao, X., Diaz, M. E., Peden, A. A., Meyer, G. E., Carskadon, S. L., Kapfhamer, D., Sufalko, D., Robinson, M. S., Noebels, J. L., and Burmeister, M. (1998) Neuron 21 111-122 [DOI] [PubMed] [Google Scholar]
- 30.Anitei, M., Baust, T., Czupalla, C., Parshyna, I., Bourel, L., Thiele, C., Krause, E., and Hoflack, B. (2008) Mol. Biol. Cell 191942 -1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takamori, S., Holt, M., Stenius, K., Lemke, E. A., Gronborg, M., Riedel, D., Urlaub, H., Schenck, S., Brugger, B., Ringler, P., Muller, S. A., Rammner, B., Grater, F., Hub, J. S., De Groot, B. L., Mieskes, G., Moriyama, Y., Klingauf, J., Grubmuller, H., Heuser, J., Wieland, F., and Jahn, R. (2006) Cell 127831 -846 [DOI] [PubMed] [Google Scholar]
- 32.Li, W., Zhang, Q., Oiso, N., Novak, E. K., Gautam, R., O'Brien, E. P., Tinsley, C. L., Blake, D. J., Spritz, R. A., Copeland, N. G., Jenkins, N. A., Amato, D., Roe, B. A., Starcevic, M., Dell'Angelica, E. C., Elliott, R. W., Mishra, V., Kingsmore, S. F., Paylor, R. E., and Swank, R. T. (2003) Nat. Genet. 35 84-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peden, A. A., Rudge, R. E., Lui, W. W., and Robinson, M. S. (2002) J. Cell Biol. 156327 -336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salazar, G., Craige, B., Styers, M. L., Newell-Litwa, K. A., Doucette, M. M., Wainer, B. H., Falcon-Perez, J. M., Dell'angelica, E. C., Peden, A. A., Werner, E., and Faundez, V. (2006) Mol. Biol. Cell 174014 -4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Arca, S., Rudge, R., Vacca, M., Raposo, G., Camonis, J., Proux-Gillardeaux, V., Daviet, L., Formstecher, E., Hamburger, A., Filippini, F., D'Esposito, M., and Galli, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 1009011 -9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Styers, M. L., Salazar, G., Love, R., Peden, A. A., Kowalczyk, A. P., and Faundez, V. (2004) Mol. Biol. Cell 155369 -5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Setty, S. R., Tenza, D., Sviderskaya, E. V., Bennett, D. C., Raposo, G., and Marks, M. S. (2008) Nature 4541142 -1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setty, S. R., Tenza, D., Truschel, S. T., Chou, E., Sviderskaya, E. V., Theos, A. C., Lamoreux, M. L., Di Pietro, S. M., Starcevic, M., Bennett, D. C., Dell'Angelica, E. C., Raposo, G., and Marks, M. S. (2007) Mol. Biol. Cell 18768 -780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickner, W. (2002) EMBO J. 211241 -1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dell'Angelica, E. C., Klumperman, J., Stoorvogel, W., and Bonifacino, J. S. (1998) Science 280431 -434 [DOI] [PubMed] [Google Scholar]
- 41.Dell'Angelica, E. C. (2001) Trends Cell Biol. 11315 -318 [DOI] [PubMed] [Google Scholar]
- 42.Miele, A. E., Watson, P. J., Evans, P. R., Traub, L. M., and Owen, D. J. (2004) Nat. Struct. Mol. Biol. 11 242-248 [DOI] [PubMed] [Google Scholar]
- 43.Helip-Wooley, A., Westbroek, W., Dorward, H., Mommaas, M., Boissy, R. E., Gahl, W. A., and Huizing, M. (2005) BMC Cell Biol. 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blagoveshchenskaya, A. D., Hewitt, E. W., and Cutler, D. F. (1999) Mol. Biol. Cell 103979 -3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honing, S., Sandoval, I. V., and von Figura, K. (1998) EMBO J. 171304 -1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theos, A. C., Tenza, D., Martina, J. A., Hurbain, I., Peden, A. A., Sviderskaya, E. V., Stewart, A., Robinson, M. S., Bennett, D. C., Cutler, D. F., Bonifacino, J. S., Marks, M. S., and Raposo, G. (2005) Mol. Biol. Cell 165356 -5372 [DOI] [PMC free article] [PubMed] [Google Scholar]