Abstract
The N-end rule pathway is a ubiquitin-dependent system where E3 ligases called N-recognins, including UBR1 and UBR2, recognize type-1 (basic) and type-2 (bulky hydrophobic) N-terminal residues as part of N-degrons. We have recently reported an E3 family (termed UBR1 through UBR7) characterized by the 70-residue UBR box, among which UBR1, UBR2, UBR4, and UBR5 were captured during affinity-based proteomics with synthetic degrons. Here we characterized substrate binding specificity and recognition domains of UBR proteins. Pull-down assays with recombinant UBR proteins suggest that 570-kDa UBR4 and 300-kDa UBR5 bind N-degron, whereas UBR3, UBR6, and UBR7 do not. Binding assays with 24 UBR1 deletion mutants and 31 site-directed UBR1 mutations narrow down the degron-binding activity to a 72-residue UBR box-only fragment that recognizes type-1 but not type-2 residues. A surface plasmon resonance assay shows that the UBR box binds to the type-1 substrate Arg-peptide with Kd of ∼3.4 μm. Downstream from the UBR box, we identify a second substrate recognition domain, termed the N-domain, required for type-2 substrate recognition. The ∼80-residue N-domain shows structural and functional similarity to 106-residue Escherichia coli ClpS, a bacterial N-recognin. We propose a model where the 70-residue UBR box functions as a common structural element essential for binding to all known destabilizing N-terminal residues, whereas specific residues localized in the UBR box (for type 1) or the N-domain (for type 2) provide substrate selectivity through interaction with the side group of an N-terminal amino acid. Our work provides new insights into substrate recognition in the N-end rule pathway.
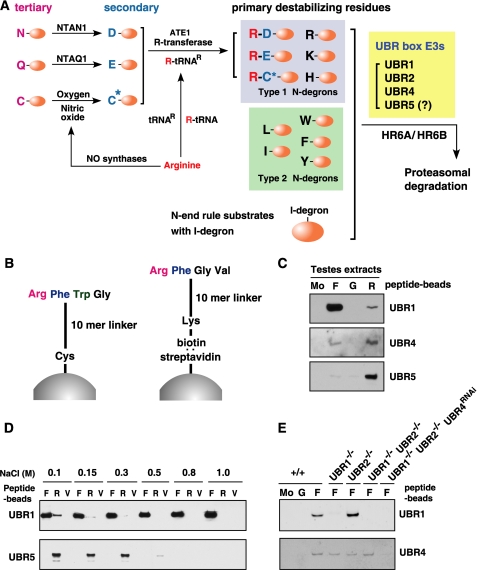
The N-end rule pathway is a ubiquitin (Ub)2-dependent proteolytic system in which N-terminal residues of short-lived proteins function as an essential component of degradation signals (degrons) called N-degrons (Fig. 1A) (1-15). An N-degron can be created from a pre-N-degron through specific N-terminal modifications (12). Specifically, in mammals, N-terminal Asn and Gln are tertiary destabilizing residues that function through their deamidation by N-terminal amidohydrolases into the secondary destabilizing N-terminal residues Asp and Glu, respectively (6, 16) (Fig. 1A). N-terminal Asp and Glu are secondary destabilizing residues that function through their arginylation by ATE1 R-transferase, which creates the primary destabilizing residue Arg at the N terminus (4, 8) (Fig. 1A). N-terminal Cys can also function as a tertiary destabilizing residue through its oxidation in a manner depending on nitric oxide and oxygen (O2); the oxidized Cys residue is subsequently arginylated by ATE1 (8, 13, 17).
FIGURE 1.
A, the mammalian N-end rule pathway. N-terminal residues are indicated by single-letter abbreviations for amino acids. Yellow ovals denote the rest of a protein substrate. C* denotes oxidized N-terminal Cys, either Cys-sulfinic acid [CysO2(H)] or Cys-sulfonic acid [CysO3(H)]. The Cys oxidation requires nitric oxide and oxygen (O2) or its derivatives. The oxidized Cys is arginylated by ATE1 Arg-tRNA-protein transferase (R-transferase). N-recognins also recognize internal (non-N-terminal) degrons in other substrates of the N-end rule pathway. B, the X-peptide pull-down assay. Left, a 12-mer peptide bearing N-terminal Arg (type 1), Phe (type 2), Trp (type 2), or Gly (stabilizing control) residue was cross-linked through its C-terminal Cys residue to Ultralink Iodoacetyl beads. Right, the otherwise identical 12-mer peptide, bearing C-terminal biotinylated Lys instead of Cys, was conjugated, via biotin, to the streptavidin-Sepharose beads. C, the X-peptide pull-down assay of endogenous UBR proteins using testes extracts. Extracts from mouse testes were mixed with bead-conjugated X-peptides bearing N-terminal Phe (F), Gly (G), or Arg (R). After centrifugation, captured proteins were separated and subjected to anti-UBR immunoblotting. Mo, a pull-down reaction with mock beads. D, the X-peptide pull-down assays using rat testis extracts were performed in the presence of varying concentrations of NaCl. After incubation and washing, bound proteins were eluted by 10 mm Tyr-Ala for Phe-peptide, 10 mm Arg-Ala for Arg-peptide, and 5 mm Tyr-Ala and 5 mm Arg-Ala for Val-peptide. Eluted proteins were subjected to immunoblotting for UBR1 and UBR5. E, cytoplasmic fractions of wild-type (+/+), Ubr1-/-, Ubr2-/-, Ubr1-/-Ubr2-/-, and Ubr1-/-Ubr2-/-Ubr4RNAi MEFs were subjected to X-peptide pull-down assay. Precipitated proteins were separated and analyzed by immunoblotting for UBR1 and UBR4.
N-terminal Arg together with other primary destabilizing N-terminal residues are directly bound by specific E3 Ub ligases called N-recognins (3, 7, 9). Destabilizing N-terminal residues can be created through the removal of N-terminal Met or the endoproteolytic cleavage of a protein, which exposes a new amino acid at the N terminus (12, 13). N-terminal degradation signals can be divided into type-1 (basic; Arg, Lys, and His) and type-2 (bulky hydrophobic; Phe, Leu, Trp, Tyr, and Ile) destabilizing residues (2, 12). In addition to a destabilizing N-terminal residue, a functional N-degron requires at least one internal Lys residue (the site of a poly-Ub chain formation) and a conformational feature required for optimal ubiquitylation (1, 2, 18). UBR1 and UBR2 are functionally overlapping N-recognins (3, 7, 9). Our proteomic approach using synthetic peptides bearing destabilizing N-terminal residues captured a set of proteins (200-kDa UBR1, 200-kDa UBR2, 570-kDa UBR4, and 300-kDa UBR5/EDD) characterized by a 70-residue zinc finger-like domain termed the UBR box (10-12). UBR5 is a HECT E3 ligase known as EDD (E3 identified by differential display) (19) and a homolog of Drosophila hyperplastic discs (20). The mammalian genome encodes at least seven UBR box-containing proteins, termed UBR1 through UBR7 (10). UBR box proteins are generally heterogeneous in size and sequence but contain, with the exception of UBR4, specific signatures unique to E3s or a substrate recognition subunit of the E3 complex: the RING domain in UBR1, UBR2, and UBR3; the HECT domain in UBR5; the F-box in UBR6 and the plant homeodomain domain in UBR7 (Fig. 2B). The biochemical properties of more recently identified UBR box proteins, such as UBR3 through UBR7, are largely unknown.
FIGURE 2.
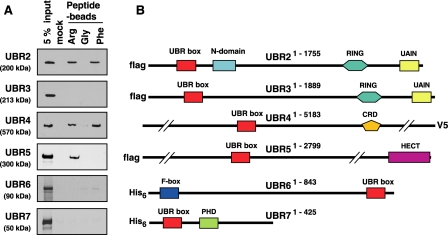
The binding properties of the UBR box family members to type-1 and type-2 destabilizing N-terminal residues. A, the X-peptide pull-down assay with overexpressed, full-length UBR proteins: UBR2, UBR3 (in S. cerevisiae cells), UBR4, UBR5 (in COS7 cells), and UBR6 and UBR7 (in the wheat germ lysates). Precipitates were analyzed by immunoblotting (for UBR2, UBR3, UBR4, and UBR5) with tag-specific antibodies as indicated in B or autoradiography (for UBR6 and UBR7). B, the structures of UBR box proteins. Shown are locations of the UBR box, the N-domain, and other E3-related domains. UBR, UBR box; RING, RING finger; UAIN, UBR-specific autoinhibitory domain; CRD, cysteine-rich domain; PHD, plant homeodomain; HECT, HECT domain.
Studies using knock-out mice implicated the N-end rule pathway in cardiac development and signaling, angiogenesis (8, 15), meiosis (9), DNA repair (21), neurogenesis (15), pancreatic functions (22), learning and memory (23, 24), female development (9), muscle atrophy (25), and olfaction (11). Mutations in human UBR1 is a cause of Johanson-Blizzard syndrome (22), an autosomal recessive disorder with multiple developmental abnormalities (26). Other functions of the pathway include: (i) a nitric oxide and oxygen (O2) sensor controlling the proteolysis of RGS4, RGS5, and RGS16 (8, 13, 17), (ii) a heme sensor through hemin-dependent inhibition of ATE1 function (27), (iii) the regulation of short peptide import through the peptide-modulated degradation of the repressor of the import (28, 29), (iv) the control of chromosome segregation through the degradation of a separate produced cohesin fragment (30), (v) the regulation of apoptosis through the degradation of a caspase-processed inhibitor of apoptosis (31, 32), (vi) the control of the human immunodeficiency virus replication cycle through the degradation of human immunodeficiency virus integrase (10, 33), and (vii) the regulation of leaf senescence in plants (34).
In the present study we characterized substrate binding specificities and recognition domains of UBR proteins. In our binding assays, UBR1, UBR2, UBR4, and UBR5 were captured by N-terminal degradation determinants, whereas UBR3, UBR6, and UBR7 were not. We also report that in contrast to other E3 systems that usually recognize substrates through protein-protein interface, UBR1 and UBR2 have a general substrate recognition domain termed the UBR box. Remarkably, a 72-residue UBR box-only fragment fully retains its structural integrity and thereby the ability to recognize type-1 N-end rule substrates. We also report that the N-domain, structurally and functionally related with bacterial N-recognins, is required for recognizing type-2 N-end rule substrates. We discuss the evolutionary relationship between eukaryotic and prokaryotic N-recognins.
EXPERIMENTAL PROCEDURES
Overexpression of UBR Proteins in Mammalian Cells—The plasmid pcDNA3flagUBR2 (9) was used to express N-terminal FLAG-tagged mouse UBR2 in COS7 cells from the PCMV promoter. The human UBR4 cDNA was excised from plasmid 7124A (a gift from Dr. Scott Vande Pol, University of Virginia) using SalI and NotI, and was subcloned into the plasmid pENTR3C (Invitrogen) to yield the entry vector pENTR3ChUBR4. By using the Gateway system (Invitrogen), the 15.9-kb UBR4 open reading frame was transferred from the pENTR3ChUBR4 to the destination vector pcDNA6.2/clumio-DEST (Invitrogen), yielding plasmid pcDNA6.2/cluvhUBR4 that expresses C-terminal lumio-V5-tagged UBR4 with a size of 570 kDa in COS7 cells from the PCMV promoter. Plasmids S503 and S3 (gifts from Drs. Michelle Henderson and CKW Watts, Garvan Institute of Medical Research) were used to express N-terminal FLAG-tagged full-length human UBR5 and its truncated derivative UBR5-(889-1877) in mammalian cells from PSV40 promoter. Cells were harvested 48 h after transfection, and cytosolic extracts were prepared as described (10). Ubr1-/-, Ubr2-/-, Ubr1-/-Ubr2-/-, and Ubr1-/-Ubr2-/-Ubr4RNAi mouse embryonic fibroblasts (MEFs) have been previously established (7, 9, 10).
Protein Expression Using a Continuous-exchange Cell-free (CECF) System—Full-length UBR proteins or their fragments were expressed in vitro and labeled with [35S]methionine using the RTS 100 Wheat Germ CECF system (Roche, Germany) according to the manufacturer's protocol. Briefly, the PT7 promoter-based linear DNA templates were generated by using two-step PCR. DNA templates for the first PCR were m-Ubr1 (7), m-Ubr2 (9), m-Ubr3 (11), h-UBR4 (7124A), h-UBR5 (S503), m-Ubr6 (cDNA clone IMAGE 4237432), and m-Ubr7 (cDNA clone IMAGE 6812389). Escherichia coli clps gene was amplified using genomic DNA from the strain DH5α cells (Invitrogen). The wheat germ lysate containing amino acids, RNA polymerases, DNA templates, and [35S]methionine (Amersham Bioscience) was incubated for 24 h at 25 °C in the CECF unit. Glycerol (50 μl) was added to the reaction mixture (50 μl) to stabilize expressed proteins. To evaluate the level of synthesized proteins, the incorporated 35S was counted using trichloroacetic acid precipitation. Synthesized proteins were stored at -20 °C and used for assays within 3 days. Proteins were also expressed using the transcription-translation-coupled TnT system (Promega, Madison, WI) as described (10).
The X-peptide Pull-down Assay—For the X-peptide pull-down assay, a set of 12-mer peptides (X-peptides) bearing N-terminal Arg (type 1), Phe (type 2), Trp (type 2), or Gly (stabilizing control) residues were cross-linked through the C-terminal Cys residue to Ultralink Iodoacetyl beads (Pierce) as described previously (10) (Fig. 1B, left). The ratio of peptides to beads was ∼1 μmol of peptides per 1-ml beads. Alternatively, the otherwise identical 12-mer peptides, bearing C-terminal biotinylated Lys instead of Cys, were conjugated, via biotin, to streptavidin-Sepharose beads (Amersham Bioscience) to a ratio of 1-1.5 μmol of peptides per 1 ml of beads (Fig. 1B, right). RTS wheat germ lysates (50 μl) expressing 35S-labeled proteins were diluted 2-fold and centrifuged to remove precipitates. An aliquot (10-20 μl) of soluble extract, containing 50-100 μg of total protein, was diluted in 250 μl of binding buffer A (0.1% Nonidet P-40, 10% glycerol, 0.15 m KCl, and 20 mm HEPES, pH 7.9) and mixed with X-peptide beads (7.5-μl packed volume for cross-linked peptide beads or 10-μl packed volume for biotinylated peptide beads). The mixtures were incubated at 4 °C for 4 h with a gentle rotation in the presence or absence of dipeptides. The beads were pelleted by centrifugation at 2,400 × g for 30 s, washed three times with 0.25 ml of binding buffer A, resuspended in 15 μl of SDS-PAGE sample buffer, and heated at 55 °C for 30 min. To analyze the binding property of endogenous UBR proteins, extracts from mouse testes were prepared and subjected to the X-peptide pull-down assay essentially as described (10). COS7 cell extracts expressing UBR proteins were diluted to ∼1.0 mg/ml in binding buffer A for pull-down assays. Denatured proteins in SDS-PAGE sample buffer were separated in 10% BisTris acrylamide gel with MES buffer, followed by fixing with solution A (50% methanol and 10% acetic acid in water) for 20 min and subsequently with solution B (25% methanol and 10% acetic acid in water) for 10 min. The 35S-labeled proteins were detected using autoradiography or, for quantitation, using a PhosphorImager (Bio-Rad).
Site-directed Mutagenesis—Site-directed mutagenesis was employed to express 31 mutant 50-kDa UBR1-(1-453) proteins, each of which contained a mutation to alanine (Ala), by using overlap extension PCR (35). Two first round PCRs, with primers containing mismatch nucleotides, generated two DNA fragments with overlap extension of 15-24 bp. PCR primer sequences are available upon request. The second round PCR, using two DNA fragments from the first round PCRs as templates, yielded a single chimeric DNA fragment containing a mutation to Ala. The resulting PCR fragments were used as templates in the third PCR to generate the PT7 promoter-based linear DNA templates for the RTS protein expression system. The final mutant DNA fragments were evaluated by sequencing.
Bacterial Expression and Purification of MBP-UBR1-(91-191)-His6 and MBP-His6—A cDNA fragment encoding C-terminal His6-tagged UBR1-(91-191) was subcloned into a bacterial expression vector pMAL-c4X (New England Biolabs, Boston, MA) that expresses maltose-binding protein (MBP). The resulting plasmid pMalUBRbox expresses MBP-UBR1-(91-191)-His6. MBP-UBR1-(91-191)-His6 and its control, MBP-His6, were overexpressed in E. coli BL21(DE3) cells and subjected to two-step purification using amylose-resin column chromatography (New England Biolabs) and cobalt-Sepharose column chromatography (Pierce). Purified proteins (5 μg) were subjected to the X-peptide pull-down assay for 1 h in the binding buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.05% Tween 20).
Surface Plasmon Resonance (Biacore) Assay—The direct surface plasmon resonance was measured using a Biacore T100 biosensor (GE Healthcare). Prior to peptide loading, the surface of the SA sensor chip was conditioned by 4-5 washes of 1 m NaCl and 50 mm NaOH at a flow rate of 30 μl/min for 30 s to wash off nonspecifically or poorly bound streptavidin. Biotinylated Arg-peptide, Phe-peptide, and Gly-peptide, adjusted to 20 nm in the binding buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, and 0.05% P20 surfactant), were immobilized on the surface of the SA sensor chip at a flow rate of 20 μl/min to reach ∼200 response units. Purified MBP-UBR1-(91-191)-His6 and MBP-His6, adjusted to 5 μm in the binding buffer, were injected at a flow rate of 40 μl/min. After dissociation for 60 s, the surface was regenerated back to bound peptides by a 30-s injection of 50 mm EDTA at a flow rate of 10 μl/min. Kinetics experiments were performed using MBP-UBR1-(91-191)-His6 at varying concentrations (0.01, 0.02, 0.04, 0.08, 0.16, 0.32, 0.64, 1.25, 2.5, and 3.13 μm). A duplicate concentration at 3.13 μm was used to determine cycle to cycle variability, and all data were double-referenced to a buffer-only control and a reference flow cell without biotinylated peptides. The data were fit using a 1:1 interaction model where A + B = AB, including terms for mass transport, using Biacore T100 Evaluation Software version 1.1.1.
RESULTS
Binding Properties of UBR Proteins to Destabilizing N-terminal Residues—We employed X-peptide pull-down assays to characterize the binding specificities of UBR proteins for destabilizing N-terminal residues. X-peptide beads (X = Arg (type 1), Phe (type 2), or Gly (stabilizing control)) (Fig. 1B) were mixed with mouse testes extracts and precipitated by centrifugation, followed by immunoblotting to detect the presence of UBR proteins in precipitates. Arg-peptide captured endogenous UBR1, UBR2, UBR4, and UBR5 from testes extracts, whereas Phe-peptide brought down UBR1, UBR2, and UBR4, but not UBR5 (Fig. 1C, data not shown). None of these UBR proteins were detected in precipitates prepared by Gly-peptide. Thus, we confirmed that endogenous UBR1, UBR2, UBR4, and UBR5 can be captured by destabilizing N-terminal residues using a different experimental setting. (Because either method for peptide-bead conjugation, through C-terminal Cys or biotin, gave essentially the same results, the conjugation method is not specified in subsequent experiments.)
The binding of UBR1 (Fig. 1D), UBR2, and UBR4 (data not shown) to the type-2 substrate Phe was not inhibited by increasing salt (NaCl) concentrations up to 1.0 m. This result indicates that type-2 substrate recognition involves hydrophobic interaction, consistent with type-2 N termini (Phe, Leu, Trp, Tyr, and Ile) being bulky “hydrophobic.” In contrast, the binding of UBR1 and UBR5 to the type-1 substrate Arg was significantly affected by salt concentrations at a range of 0.15-0.5 m (Fig. 1D), in agreement with type-1 substrates (Arg, Lys, and His) being “basic.” The level of UBR4 precipitated by Phe-peptide from MEFs was diminished in UBR1-/-UBR2-/- UBR4RNAi (retrovirus-mediated knockdown of UBR4 in Ubr1-/-Ubr2-/- cells) but not in Ubr1-/-, Ubr2-/-, and Ubr1-/-Ubr2-/- MEFs (Fig. 1E), suggesting that UBR4 binding to destabilizing N-terminal residues is independent from UBR1 and UBR2.
We next examined the interaction of recombinant UBR proteins to destabilizing N-terminal residues. As expected, Arg-peptide and Phe-peptide, but not Gly-peptide, precipitated recombinant UBR2, which were expressed in Saccharomyces cerevisiae cells (Fig. 2A) or in the CECF-based wheat germ lysate (data not shown). However, under the same conditions, recombinant UBR3 did not bind to these N termini (Fig. 2A). We constructed a mammalian plasmid (pcDNA6.2/cluvhUBR4) expressing 570-kDa human UBR4 with a C-terminal lumio-V5 tag (Fig. 2B). Recombinant full-length UBR4, expressed in COS7 cells, bound to Phe-peptide and Arg-peptide, but not to Gly-peptide (Fig. 2A). The 300-kDa HECT E3 ligase UBR5, expressed in COS7 cells, bound to Arg-peptide but not to Phe-peptide or Gly-peptide. In contrast, 90-kDa UBR6 and 50-kDa UBR7 expressed in the wheat germ lysate did not bind to X-peptides (Fig. 2A). These results together suggest that UBR1, UBR2, UBR4, and UBR5 are operationally N-recognins, whereas the full-length UBR3, UBR6, and UBR7 proteins do not bind to destabilizing N-terminal residues under experimental conditions used in this study.
The UBR Box Is the Substrate Recognition Domain of UBR1 and UBR2—To determine whether there is a distinct domain that recognizes destabilizing N-terminal residues, we developed an efficient in vitro binding assay using X-peptides and protein production in the RTS Wheat Germ CECF system (see “Experimental Procedures”). The RTS system uses a semi-permeable membrane that creates a reaction compartment and a feeding compartment into which inhibitory byproducts diffuse and from which substrates and energy components are continuously supplied (36). The production yield of UBR1 fragments in the RTS system was markedly higher than traditional in vitro expression technologies, making it possible to efficiently and reproducibly determine the binding specificity of a test protein to various N-terminal residues of synthetic peptides.
Because the 70-residue UBR box, a Cys/His-based zinc finger-like domain, is the only domain conserved in UBR1, UBR2, UBR4, and UBR5 (Figs. 2B and 3, A and B), we first tested whether the 50-kDa UBR1 fragment (His6-UBR1-(1-453)) containing the UBR box has the ability to bind to destabilizing N-terminal residues. The 50-kDa fragment (1 in Fig. 3C), expressed in the CECF-based wheat germ lysate, was pulled down by both Arg-peptide and Phe-peptide but not by Gly-peptide (Fig. 3C). In contrast, 82-kDa UBR1-(308-1010) (5 in Fig. 4A), lacking the UBR box, did not bind to destabilizing N termini. A corresponding UBR2 fragment (50-kDa UBR2-(1-452)) also bound to Arg-peptide and Phe-peptide but not Gly-peptide (Fig. 4B). We conclude that the 50-kDa N-terminal region in both UBR1 and UBR2 contains the activity for binding to type-1 and type-2 N-end rule substrates.
FIGURE 3.
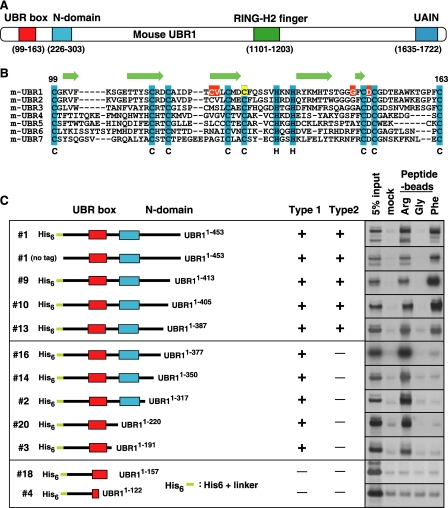
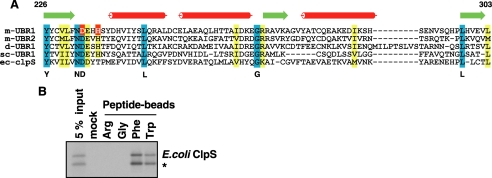
The UBR box is the substrate recognition domain of UBR1. A, locations of the UBR box, the N-domain, the RING domain, and the UAIN domain in mouse UBR1. B, sequence alignment of UBR boxes in mouse UBR proteins, in which conserved Cys and His residues are highlighted (cyan). Indicated by yellow highlight is the Cys residue of mouse UBR1 deduced from the Cys residue of Arabidopsis BIG/UBR4 whose missence mutation perturbs auxin transport (42). Indicated by red highlight are the residues of mouse UBR1 deduced from those of S. cerevisiae UBR1 that were identified to be essential for degradation of type-1 N-end rule substrates (41). Predicted secondary structure elements of the UBR box of mouse UBR1 (arrow, β-sheet) are shown above the sequence alignment. C, the X-peptide pull-down assay with C-terminal deleted UBR1 fragments expressed in the CECF-based wheat germ extracts. The identification numbers for UBR1 fragments are shown to the left. Most UBR1 fragments are N-terminal His6-tagged. The binding activity of each fragment was recorded as either positive (+) or negative (-). The autoradiography for the X-peptide pull-down assay is shown to the right. UAIN, UBR-specific autoinhibitory domain.
FIGURE 4.
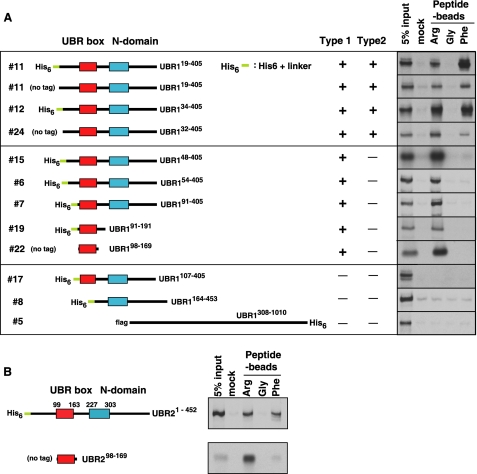
The X-peptide pull-down assay of deletion mutants of UBR1 and UBR2. A, the X-peptide pull-down assay with N-terminal deleted and other related UBR1 fragments that were expressed in the CECF-based wheat germ extracts. The identification numbers for UBR1 fragments are shown to the left. Most UBR1 fragments are N-terminal His6-tagged. The binding activity of each fragment was recorded as either positive (+) or negative (-). The autoradiography for the X-peptide pull-down assay is shown to the right. B, the X-peptide pull-down assay with UBR2 fragments expressed in the wheat germ extracts.
To further dissect the substrate recognition domain, we expressed 23 deletion mutants of 50 kDa UBR1-(1-453) (1 in Fig. 3C) using the CECF-based wheat germ extracts and examined their binding specificities to various N termini using the X-peptide pull-down assay. Unexpectedly, serial C-terminal deletion of UBR1-(1-453) revealed the presence of two distinct substrate recognition domains of the N-end rule pathway. For example, UBR1-(1-387) (13 in Fig. 3C) bound to both type-1 and type-2 N termini. However, UBR1-(1-377) (16 in Fig. 3C), with a C-terminal deletion of 10 amino acids from UBR1-(1-387), had an intact activity to type-1 substrate but was almost completely abolished in type-2 substrate binding, indicating that sequence elements recognizing type-1 and type-2 N-end rule substrates may be mechanistically distinct. UBR1-(1-191) (3 in Fig. 3C), containing the UBR box (residues 99-163), still retained the ability to bind to the type-1 substrate, whereas UBR1-(1-157) (18 in Fig. 3C), with a C-terminal deletion of six amino acids from the UBR box, showed no detectible activity toward N-degrons. The 11-kDa UBR1-(91-191) fragment (19 in Fig. 4A), with a deletion of N-terminal 90 residues from UBR1-(1-191), retained a significant activity. However, UBR1-(107-405) (17 in Fig. 4A), with an N-terminal deletion of eight residues (residue 99-106) from the UBR box, abolished its binding to the type-1 substrate. Remarkably, UBR1-(98-169) (22 in Fig. 4A), the 72-residue UBR box-alone fragment (residue 98-169), retained an almost complete activity for the type-1 substrate. We conclude that the 70-residue UBR box of UBR1 is the substrate recognition domain for type-1 substrates.
We next determined the binding properties of isolated UBR box fragments from other UBR proteins to the type-1 N terminus. Approximately 70-residue UBR box-only fragments from UBR2 through UBR7 were expressed in the RTS Wheat Germ CECF system. Among these, only UBR2, UBR3, and UBR4 fragments were expressed in levels sufficient for the X-peptide pull-down assay. Consistent with the finding with UBR1, the UBR box-only fragment of UBR2 exhibited a strong affinity to the type-1 substrate Arg but not to the type-2 substrate Phe or the control N terminus Gly (Fig. 4B). In contrast, the corresponding UBR3 and UBR4 fragments did not show significant and selective binding to any of these N termini (data not shown). We then determined whether a larger UBR4 fragment, containing the UBR box, can bind to destabilizing N-terminal residues; the human 80-kDa UBR4-(1524-2254) fragment, expressed in the wheat germ lysate, did not bind to tested N termini (data not shown). Likewise, UBR5-(889-1877), containing the UBR box, did not bind to Arg-peptide (data not shown). Thus, the binding activity of the UBR box to the N terminus requires additional components, including specific residues in the context of appropriate conformation.
The N-domain Is Required for Recognition of Type-2 N-end Rule Substrates—UBR1-(1-387) (13 in Fig. 3C) binds to both type-1 and type-2 N termini, whereas UBR1-(1-377) (16 in Fig. 3C) binds to only type-1 N terminus. Given that the UBR box spans residues 99-163, the ∼200-residue region (residues 164-387) immediately downstream of the UBR box (residues 99-163) is essential for recognizing type-2 substrates but not type-1 substrates (Fig. 5A). It has been noticed that a 78-residue domain (residues 226-303), termed here the N-domain, exhibits weak, but significant, similarity in primary and secondary sequences to E. coli ClpS with a size of 106 residues (37). ClpS has been recently implicated as an N-recognin that recognizes bacterial N-end rule substrates (38). Based on these results, we reasoned that the N-domain is a distinct domain required for recognition of type-2 N-end rule substrates, which is further supported by site-directed mutagenesis analysis (see below). We then tested whether the N-domain is sufficient for type-2 substrate recognition; UBR1-(164-453) (8 in Fig. 4A), containing the N-domain but not the UBR box, did not bind to the type-2 substrate. This suggests that the N-domain itself is not sufficient for substrate binding, but its specific residues participate in the recognition of type-2 substrates. Whereas UBR1-(34-405) (12 in Fig. 4A), with N-terminal 33-residue deletion, bound both type-1 and type-2 N termini, UBR1-(48-405) (15 in Fig. 4A), with N-terminal 47-residue deletion, bound only type-1 N terminus. Thus, the N-terminal region is also required for optimal recognition of type-2 substrates, but not for type-1 substrates, perhaps through its interaction with the N-domain.
FIGURE 5.
The binding properties of E. coli ClpS to mammalian N-end rule substrates. A, the sequence alignment of the ∼80-residue N-domain of eukaryotic N-recognins with E. coli ClpS with a size of 106 amino acids. Indicated by red highlight are the residues of mouse UBR1 deduced from those of S. cerevisiae UBR1 that were identified to be essential for degradation of type-2 N-end rule substrates (41). Predicted secondary structure elements of the N-domain of mouse UBR1 (arrow, β-sheet; cylinder, α-helix) are shown above the sequence alignment. B, the X-peptide pull-down assay with E. coli ClpS expressed in the wheat germ extracts. E. coli ClpS binds to type-2, but not type-1, N termini in the context of a mammalian N-end rule substrate. *, a putative cleavage product of His6-ClpS.
The proteolytic system that creates and recognizes N-degrons is present both in prokaryotes and eukaryotes, even though prokaryotes lack the Ub-proteasome system (2, 39). In the bacterium E. coli, the primary destabilizing N-terminal residues (Phe, Leu, Trp, and Tyr) and the secondary destabilizing residues (Arg and Lys) function as an essential component of N-degrons (2). Prokaryotic proteasome enzymes, ClpA and ClpP, cooperate to degrade N-end rule substrates. E. coli ClpS, the ClpA adaptor protein, has been identified as a substrate recognition protein for the bacterial N-end rule pathway (38). The role of ClpS in the bacterial N-end rule degradation was complicated by the finding that it suppressed the degradation rate when the substrate level was high (40). To address the functional relationship between eukaryotic and prokaryotic N-recognins, we tested whether E. coli ClpS can bind to destabilizing N termini in the context of mammalian N-end rule substrates. The X-peptide pull-down assay using E. coli ClpS, expressed in the wheat germ lysate, demonstrated that ClpS can be brought down by Phe-peptide and Trp-peptide but not by Arg-peptide or Gly-peptide (Fig. 5B). In addition to confirming that ClpS is an N-recognin, these results suggest that the machinery required for recognition of type-2 substrates is evolutionarily conserved, at least in part, in eukaryotes and prokaryotes.
We observed the presence of the N-domain in UBR1, UBR2, and their (putative) homologs in other species (type-1/2 N-recognins), but not in UBR5 (type-1 N-recognin) (Fig. 2B) or their (putative) homologs. Mouse UBR4 (type-1/2 N-recognin) also contains a region with weak homology to the N-domain, whose function is to be tested (data not shown). Thus, the presence of the N-domain is largely correlated with the ability to recognize type-2 N termini.
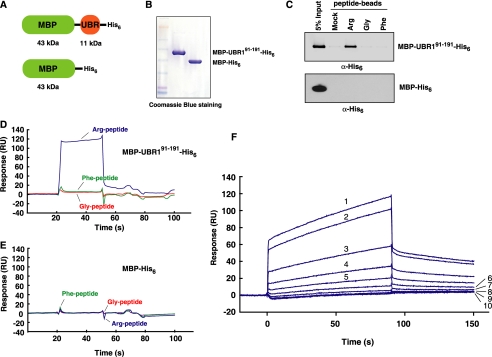
Surface Plasmon Resonance (Biacore) Assay to Determine the Affinity of the UBR Box to N-terminal Amino Acids—We determined the interaction between the UBR box and various N-terminal amino acids using surface plasmon resonance (Biacore) assay. X-peptide pull-down assays indicate that 11-kDa UBR1-(91-191) (22 in Fig. 4A), containing only UBR box, binds to the type-1 substrate as efficient as UBR1-(1-453) (1 in Fig. 3C), containing both the UBR box and the N-domain. To determine the binding properties of the UBR box, we expressed and purified MBP-UBR1-(91-191)-His6 and MBP-His6 to near homogeneity (Fig. 6, A and B). MBP-UBR1-(91-191)-His6 bound to the type-1 substrate but not the type-2 substrate (Fig. 6C). (We were unable to purify MBP-UBR1-(1-453) due to its instability in bacteria.) Biotinylated X-peptides were immobilized on the streptavidin-coated sensor chip, and MBP-UBR1-(91-191)-His6 or MBP-His6 was injected over the immobilized peptides. MBP-UBR1-(91-191)-His6 showed strong binding to Arg-peptide but not to Phe-peptide or Gly-peptide (Fig. 6D). The control protein MBP-His6 showed no detectible binding to these substrates (Fig. 6E). Kinetic analysis with serially diluted MBP-UBR1-(91-191)-His6 indicated that its affinity to Arg-peptide is ∼3.4 μm (Fig. 6F). In addition to confirming that the UBR box is a substrate binding domain, these results suggest that the selective binding of the UBR box to destabilizing N-terminal residues is a critical biochemical event in the ubiquitylation of N-end rule substrates.
FIGURE 6.
Surface plasmon resonance (Biacore) analysis for the interaction between MBP-UBR1-(91-191)-His6 and X-peptides (X = Arg, Phe, or Gly). Biotinylated X-peptides, adjusted to 20 nm in the binding buffer (see “Experimental Procedures”), were immobilized to a sensor chip. A, the structures of MBP-UBR1-(91-191)-His6 and MBP-His6. B, purified MBP-UBR1-(91-191)-His6 and MBP-His6 were separated on SDS-PAGE and stained with Coomassie Brilliant Blue R-250 (Bio-Rad). C, the X-peptide pull-down assay. Purified MBP-UBR1-(91-191)-His6 or MBP-His6 was mixed with X-peptide beads, followed by anti-His6 immunoblotting of precipitated proteins. D, Biacore sensorgram illustrating the binding of MBP-UBR1-(91-191)-His6 to Arg-peptide but not to Phe-peptide or Gly-peptide. MBP-UBR1-(91-191)-His6, adjusted to 5 μm in the binding buffer, was injected over immobilized X-peptides at a flow rate of 40 μl/min. E, Biacore sensorgram with MBP-His6 showing no detectible affinity to X-peptides. F, kinetic analysis of the affinity of MBP-UBR1-(91-191)-His6 to Arg-peptide. MBP-UBR1-(91-191)-His6 was serially diluted in the binding buffer and subsequently injected over immobilized Arg-peptide at a flow rate of 40 μl/min. Numbers (1-10) represent, respectively, the concentrations (3.13, 2.5, 1.25, 0.63, 0.31, 0.16, 0.08, 0.04, 0.02, and 0.01 μm) of injected MBP-UBR1-(91-191)-His6.
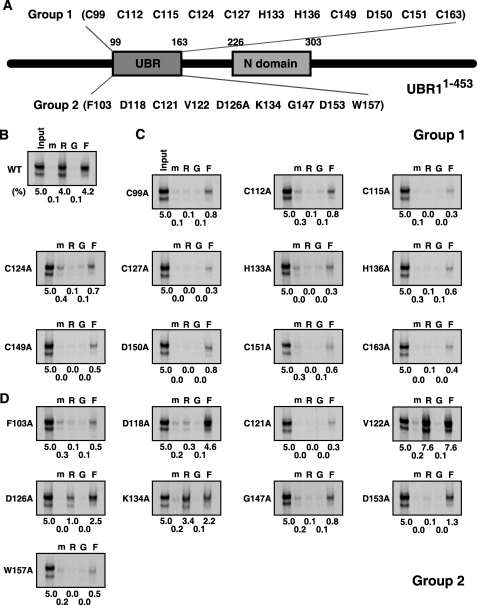
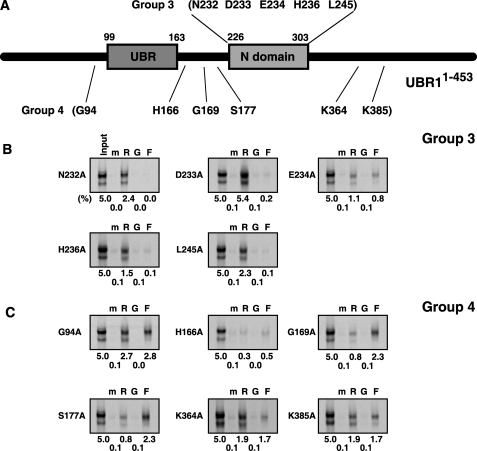
Site-directed Mutagenesis Analysis of the Substrate Recognition Domains of the N-end rule Pathway—To further dissect substrate recognition domains of mouse UBR1, we employed site-directed mutagenesis to generate 31 mutants of UBR1-(1-453), each of which contained a mutation to alanine (Ala), and determined the effect of individual mutations using the X-peptide pull-down assay (Figs. 7 and 8). The mutants can be categorized into four groups. Group-1 residues (8 Cys, 2 His, and 1 Asp), localized in the 70-residue UBR box, are conserved in all known UBR proteins (Fig. 7A). The mutation of any of these residues almost completely abolished the ability to bind to the type-1 substrate and less severely to the type-2 substrate (Fig. 7C), compared with wild-type UBR1-(1-453) (Fig. 7B). This indicates that these Cys and His residues provide a structural element for the recognition of both type-1 and type-2 substrates, perhaps by forming a zinc finger-like structure. In addition to these Cys/His residues, the mutation of the conserved Asp-150 residue also exhibited a similar effect, suggesting that this residue may be a substrate binding site (see “Discussion”) (Fig. 7C). In contrast to well conserved residues, the mutation of less conserved residues (Group 2) within the UBR box yielded variable effects on the binding specificities (Fig. 7, A and C). Group 3 consists of conserved residues within the N-domain (Fig. 8A). Remarkably, the mutation of Group-3 residues showed a clear tendency to specifically eliminate binding to the type-2 substrate but not to the type-1 substrate (Fig. 8B), suggesting that specific residues within the N-domain participate in the type-2 substrate recognition. Mutations of residues localized outside the UBR box and the N-domain (Group 4) exhibited variable effects on the binding properties (Fig. 8, A and C). Taken together, we propose a model where the UBR box provides a structural element for the recognition of both type-1 and type-2 N-end rule substrates, whereas specific residues within the UBR box and the N-domain, respectively, participate in the interaction with type-1 and type-2 substrates.
FIGURE 7.
Site-directed mutagenesis analysis of the UBR box of mouse UBR1. A, diagram of the UBR1-(1-453) fragment showing residues that are mutated into alanine. Group 1 is 11 residues (8 Cys, 2 His, and 1 Asp), which are localized within the 70-residue UBR box and conserved in all known UBR proteins. Group 2 is 9 residues that are incompletely conserved within the UBR box. B, the X-peptide pull-down assay with wild-type UBR1-(1-453), expressed in the wheat germ lysate, and bead-conjugated X-peptides bearing N-terminal Arg (R), Gly (G), or Phe (F). The levels of signals compared with 5% input signal are shown at the bottom. The lane m represents a pull-down reaction with mock beads. C and D, the X-peptide pull-down assays with Group-1 (C) and Group-2 (D) UBR1-(1-453) mutants. Note that Group-1 mutations show clear tendency to completely abolish the type-1 substrate binding activity.
FIGURE 8.
Site-directed mutagenesis analysis of the N-domain. A, the diagram of UBR1-(1-453) showing residues that are mutated into Ala. Group 3 is 5 residues within the N-domain, and Group 4 is 6 residues localized outside the UBR box and the N-domain. B and C, the X-peptide pull-down assay with Group 3 (B) and Group 4 (C) UBR1-(1-453) mutants and bead-conjugated X-peptides bearing N-terminal Arg (R), Gly (G), or Phe (F). The levels of signals compared with 5% input signal are shown to the bottom. Lane m represents a pull-down reaction with mock beads.
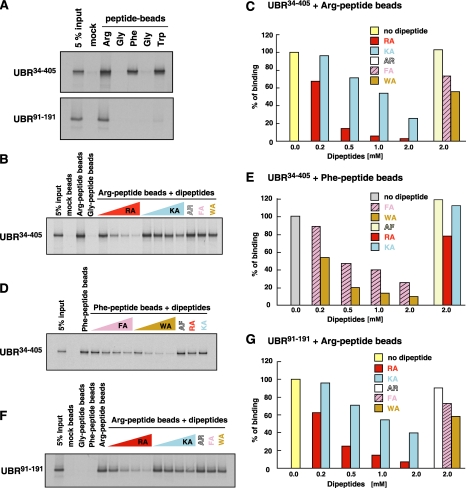
Selective Inhibition of the Interaction between the UBR Box and N-end Rule Substrates by Dipeptides—Dipeptides bearing destabilizing N-terminal residues have been used to inhibit the N-end rule pathway in vivo and in vitro (2, 14). To further characterize the binding of the UBR box to N-degrons, we performed the X-peptide pull-down assay in the presence of varying concentrations of dipeptides (Fig. 9). UBR1-(34-405), containing both the UBR box and the N-domain, bound to X-peptides bearing N-terminal Arg (type 1), Phe (type 2), or Trp (type 2), but not Gly (stabilizing) (Fig. 9A). The binding of Arg-peptide to UBR1-(34-405) was selectively inhibited by the type-1 dipeptide Arg-Ala with an approximate half-maximal inhibitory concentration (IC50) of 0.26 mm, but not significantly by 2 mm of the dipeptides Phe-Ala, Trp-Ala (type 2), or Ala-Arg (stabilizing) (Fig. 9, B and C). In contrast to Arg-Ala, the type-1 dipeptide Lys-Ala showed notably weak inhibitory efficacy (IC50 ∼1.1 mm) (Fig. 9, B and C). Likewise, the binding of Phe-peptide to UBR1-(34-405) was selectively inhibited by the type-2 dipeptides Phe-Ala (IC50 ∼ 0.44 mm) and Trp-Ala (IC50 ∼ 0.24 mm), but not significantly by the dipeptides Arg-Ala, Lys-Ala, or Ala-Phe (Fig. 9, D and E). In agreement with the above results, 11-kDa UBR1-(91-191) bound to Arg-peptide but not to Phe-peptide, Trp-peptide, or Gly-peptide (Fig. 9A). The binding of Arg-peptide to UBR1-(91-191) was inhibited by Arg-Ala (IC50 ∼0.28 mm) and Lys-Ala (IC50 ∼1.3 mm), but not by Phe-Ala, Trp-Ala, or Ala-Arg (Fig. 9, F and G). These results together suggest that the UBR box is a common structural element responsible for binding to all known N-terminal degradation determinants and that, whereas the affinity in substrate recognition is mainly determined by the identity of N-terminal residues, it is also affected by the length and sequence of the N-terminal region of the protein.
FIGURE 9.
Selective inhibition of the interaction between the UBR box and the X-peptide by dipeptides. A, the X-peptide pull-down assay with UBR1-(34-405) (top) and UBR1-(91-191) (bottom), expressed in the CECF-based wheat germ extracts, using X-peptides that bear N-terminal Arg, Phe, Trp, or Gly. B, inhibition of the interaction between UBR1-(34-405) and Arg-peptide by dipeptides bearing various N-terminal residues. The X-peptide pull-down assay was performed in the presence of varying concentrations of dipeptides. RA, Arg-Ala; KA, Lys-Ala; AR, Ala-Arg; FA, Phe-Ala; WA, Trp-Ala. C, quantitation of B. D, inhibition of the interaction between UBR1-(34-405) and Phe-peptide by dipeptides bearing various N-terminal residues. E, quantitation of D. F, inhibition of the interaction between UBR1-(91-191) and Arg-peptide by dipeptides bearing various N-terminal residues. G, quantitation of F.
DISCUSSION
Although the N-end rule pathway is well characterized in its hierarchical structure, physiological functions, degrons, and inhibitors, it has remained elusive whether N-degron is recognized by a specific domain that is conserved in primary, secondary, and/or tertiary structure. Our results demonstrate that the 70-residue UBR box is a general substrate recognition domain in the N-end rule pathway. Given that the UBR box is a zinc finger-like domain where conserved Cys and His residues provide structural integrity, it is parsimonious to speculate that it provides an essential structural element on which active site residues, within or outside the UBR box, interact with the N terminus of a short-lived protein. Its strong structural integrity is manifested by the data that the 72-residue UBR box-only fragment (out of 1,757 amino acids) retains virtually intact activity for binding to the type-1 substrate (Fig. 4A), when compared with larger recombinant UBR1 fragments (Figs. 3C and 4A) and endogenous UBR proteins (10) (Fig. 2A, data not shown). This indicates that all the active site residues required for type-1 degron recognition reside within the 70-residue region. One such type-1 residue may be the well conserved Asp-150, whose mutation completely abolishes substrate binding (Fig. 7C). The recognition of type-2 substrates requires an additional domain, termed the N-domain (Fig. 3A), which is localized downstream of the UBR box. In contrast to the UBR box, however, the N-domain alone is not sufficient for substrate binding (8 in Fig. 4A), indicating that specific residues within the N-domain are localized in the three-dimensional proximity of the UBR box, where they interact with N-degrons. By using surface plasmon resonance (Biacore) assay, we demonstrate that UBR1-(91-191) binds to Arg-peptide, with Kd of ∼3.4 μm (Fig. 6, D and F), but shows no detectible affinity to Phe-peptide and Gly-peptide (Fig. 6D). These results suggest that the strong difference in affinity to N-terminal residues is the molecular basis of substrate selectivity in the N-end rule pathway. Perhaps, the combination of moderate affinity (Kd of ∼3.4 μm) and strong selectivity (difference in Kd between destabilizing and stabilizing residues) makes it possible to achieve an appropriate balance between substrate selectivity and enzymatic processivity, ensuring both “selective binding” to a substrate and “rapid dissociation” from the N terminus to transfer Ub to an internal Lys residue.
Our findings are consistent with a genetic screening with S. cerevisiae ubr1, which identified specific mutations impairing the degradation of type-1 or type-2 N-end rule substrates (41). A sequence comparison indicates that type-1-specific residues (Cys-121/Val-122, Gly-147, and Asp-150) of mouse UBR1, deduced from the yeast UBR1 residues, are all localized inside the UBR box (Fig. 3B, red highlight). It is also notable that among type 2-specific residues (Asp-233, His-236, and Glu-407 in mouse UBR1), Asp-233 and His-236 are localized within the N-domain (Fig. 5A, red highlight). In agreement with these observations, our pull-down assays demonstrate that mutations of type-1 residues (Cys-121, Gly-147, and Asp-150) have clear tendencies to completely abolish the type-1 activity of mouse UBR1 (Fig. 7), whereas mutations of type-2 residues, Asp-233 and His-236, disrupt the type-2 activity (Fig. 8B). Based on these findings, we propose a model where the UBR box of UBR1 and UBR2 provides a common structural element essential for binding to all known destabilizing N-terminal residues, whereas a set of specific residues inside (for type 1) or outside (for type 2) the UBR box interact with their side groups.
We tested whether isolated UBR box fragments from other UBR proteins are capable of binding to the type-1 substrate Arg. In contrast to UBR1 and UBR2, the UBR box of other N-recognins, 600-kDa UBR4 and 300-kDa UBR5, did not show detectible affinity to the N terminus (data not shown). This suggests that the UBR box of UBR4 and UBR5 may have a minimal role in substrate recognition. However, it is more likely that the UBR box of these two proteins do act as a substrate recognition domain. One parsimonious interpretation would be that, whereas Cys/His residues of the UBR box provide a structural element (i.e. the thermodynamic free energy in conformation), the binding of isolated UBR box fragments requires specific residues in a manner depending on appropriate conformation. This conjecture is supported by the finding that a missense mutation of a conserved Cys residue (Fig. 3B, yellow highlight) within the UBR box impairs the function of Arabidopsis UBR4/BIG in auxin transport and growth (42).
The N-end rule pathway mediates non-Ub proteolysis in prokaryotes that do not have the Ub-proteasome system. In contrast to eukaryotes where the UBR box is a signature of N-recognins (Refs. 10 and 11 and this study), the identity of prokaryotic N-recognins has been elusive until recently. E. coli ClpS, an adaptor protein of the ClpA/ClpP proteolytic complex, has been implicated as a bacterial N-recognin (38). The binding of a bacterial N-recognin to mammalian type-2 N-end rule substrates (Fig. 5B) indicates that bacterial N-recognins could be evolutionarily and functionally related with the type-2 N-end rule pathway in eukaryotes. Our data show that mouse UBR1 has a distinct domain, termed the N-domain, with homology to the bacterial N-recognin in primary and secondary sequences (Fig. 5A) and participates in the recognition of type-2 mammalian substrates (Figs. 3 and 4). The mechanistic similarity in substrate recognition between eukaryotic and prokaryotic N-end rule pathways, despite fundamental differences in downstream proteolytic processes (the proteasome complex versus the ClpA/ClpP complex), indicates the ancient origin of the N-end rule pathway. How do mammalian N-recognins have a signature of bacterial N-recognin? One intriguing possibility is that a type-2 N-recognin in bacteria has been recruited, sometime during evolution, to a eukaryotic N-recognin that originally recognized only type-1 substrates, resulting in a chimeric N-recognin that binds to both type-1 and type-2 substrates. Because of the presence of the UBR box whose zinc finger-like structure based on Cys/His residues is stronger compared with the N-domain, the structural integrity of the N-domain may have been gradually diminished, leaving a few residues that directly interact with type-2 substrates. The finding that the UBR box alone has no detectible affinity to the type-2 substrate Phe (Fig. 6D) further supports the model that the N-end rule pathway in ancient eukaryotes originally evolved to recognize only type-1 substrates.
Substrate recognition by Ub ligases is usually based on protein-protein interface, making it difficult to define a general substrate recognition domain. For example, F-box proteins in the Skp1-Cullin-F-box E3 complex have an F-box motif that binds to SKP1 for assembly into the SKP1·CUL1 complex but do not have a conserved substrate recognition domain (43). In contrast to other E3 systems, the N-end rule pathway recognizes a single amino acid at the N terminus as a primary degradation signal, making it possible to employ a general domain that recognizes the universal structure of the N terminus of the protein. Although the UBR box is conserved in all known N-recognins (UBR1, UBR2, UBR4, and UBR5) in mammals, it is also found in other UBR proteins (UBR3, UBR6, and UBR7) (Fig. 2). It remains to be investigated which residues in the UBR box discriminate N-recognins from non-N-recognins. It has been reported that Arabidopsis thaliana has an N-recognin called PRT1 that recognizes aromatic N-terminal residues (Phe, Tyr, and Trp), a subset of type-2 N-end rule substrates (44, 45). The 45-kDa RING finger protein with two ZZ domains shows no significant sequence homology to any of the UBR proteins nor does it contain a canonical UBR box or the N-domain, suggesting that the UBR box is not needed to recognize aromatic N-terminal residues in certain conditions. Perhaps, there may be a set of eukaryotic N-recognins that do not have the UBR box.
Acknowledgments
We are grateful to Jai Wha Seo for genotyping and maintaining mouse strains, Yong Li for critical suggestions, to the staff of the animal facility at the University of Pittsburgh for the care and maintenance of mice, and Kwon lab members for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM69482, GM074000, HL083365 (to Y. T. K.), and CA078039 (to J. S. L.). This work was also supported by grants from the American Heart Association (to Y. T. K.) and the Hartwell Foundation (to J. S. L. and D. D. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Ub, ubiquitin; degron, degradation signal; N-degron, N-terminal degron; MEF, mouse embryonic fibroblast; CECF, continuous-exchange cell-free; MBP, maltose-binding protein; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MES, 4-morpholineethanesulfonic acid.
References
- 1.Bachmair, A., and Varshavsky, A. (1989) Cell 56 1019-1032 [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12142-12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon, Y. T., Reiss, Y., Fried, V. A., Hershko, A., Yoon, J. K., Gonda, D. K., Sangan, P., Copeland, N. G., Jenkins, N. A., and Varshavsky, A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7898-7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon, Y. T., Kashina, A. S., and Varshavsky, A. (1999) Mol. Cell. Biol. 19 182-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon, Y. T., Lévy, F., and Varshavsky, A. (1999) J. Biol. Chem. 274 18135-18139 [DOI] [PubMed] [Google Scholar]
- 6.Kwon, Y. T., Balogh, S. A., Davydov, I. V., Kashina, A. S., Yoon, J. K., Xie, Y., Gaur, A., Hyde, L., Denenberg, V. H., and Varshavsky, A. (2000) Mol. Cell. Biol. 20 4135-4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon, Y. T., Xia, Z., Davydov, I. V., Lecker, S. H., and Varshavsky, A. (2001) Mol. Cell. Biol. 21 8007-8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon, Y. T., Kashina, A. S., Davydov, I. V., Hu, R. G., An, J. Y., Seo, J. W., Du, F., and Varshavsky, A. (2002) Science 297 96-99 [DOI] [PubMed] [Google Scholar]
- 9.Kwon, Y. T., Xia, Z., An, J. Y., Tasaki, T., Davydov, I. V., Seo, J. W., Sheng, J., Xie, Y., and Varshavsky, A. (2003) Mol. Cell. Biol. 23 8255-8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasaki, T., Mulder, L. C., Iwamatsu, A., Lee, M. J., Davydov, I. V., Varshavsky, A., Muesing, M., and Kwon, Y. T. (2005) Mol. Cell. Biol. 25 7120-7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tasaki, T., Sohr, R., Xia, Z., Hellweg, R., Hortnagl, H., Varshavsky, A., and Kwon, Y. T. (2007) J. Biol. Chem. 282 18510-18520 [DOI] [PubMed] [Google Scholar]
- 12.Tasaki, T., and Kwon, Y. T. (2007) Trends Biochem. Sci. 32 520-528 [DOI] [PubMed] [Google Scholar]
- 13.Lee, M. J., Tasaki, T., Moroi, K., An, J. Y., Kimura, S., Davydov, I. V., and Kwon, Y. T. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15030-15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, M. J., Pal, K., Tasaki, T., Roy, S., Jiang, Y., An, J. Y., Banerjee, R., and Kwon, Y. T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 100-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An, J. Y., Seo, J. W., Tasaki, T., Lee, M. J., Varshavsky, A., and Kwon, Y. T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6212-6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigoryev, S., Stewart, A. E., Kwon, Y. T., Arfin, S. M., Bradshaw, R. A., Jenkins, N. A., Copeland, N. G., and Varshavsky, A. (1996) J. Biol. Chem. 271 28521-28532 [DOI] [PubMed] [Google Scholar]
- 17.Hu, R. G., Sheng, J., Qi, X., Xu, Z., Takahashi, T. T., and Varshavsky, A. (2005) Nature 437 981-986 [DOI] [PubMed] [Google Scholar]
- 18.Prakash, S., Tian, L., Ratliff, K. S., Lehotzky, R. E., and Matouschek, A. (2004) Nat. Struct. Mol. Biol. 11 830-837 [DOI] [PubMed] [Google Scholar]
- 19.Callaghan, M. J., Russell, A. J., Woollatt, E., Sutherland, G. R., Sutherland, R. L., and Watts, C. K. (1998) Oncogene 17 3479-3491 [DOI] [PubMed] [Google Scholar]
- 20.Mansfield, E., Hersperger, E., Biggs, J., and Shearn, A. (1994) Dev. Biol. 165 507-526 [DOI] [PubMed] [Google Scholar]
- 21.Ouyang, Y., Kwon, Y. T., An, J. Y., Eller, D., Tsai, S. C., Diaz-Perez, S., Troke, J. J., Teitell, M. A., and Marahrens, Y. (2006) Mut. Res. 596 64-75 [DOI] [PubMed] [Google Scholar]
- 22.Zenker, M., Mayerle, J., Lerch, M. M., Tagariello, A., Zerres, K., Durie, P. R., Beier, M., Hülskamp, G., Guzman, C., Rehder, H., Beemer, F. A., Hamel, B., Vanlieferinghen, P., Gershoni-Baruch, R., Vieira, M. W., Dumic, M., Auslender, R., Gil-da-Silva-Lopes, V. L., Steinlicht, S., Rauh, M., Shalev, S. A., Thiel, C., Ekici, A. B., Winterpacht, A., Kwon, Y. T., Varshavsky, A., and Reis, A. (2005) Nat. Genet. 37 1345-1350 [DOI] [PubMed] [Google Scholar]
- 23.Balogh, S. A., Kwon, Y. T., and Denenberg, V. H. (2000) Learn. Mem. 7 279-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balogh, S. A., McDowell, C. S., Kwon, Y. T., and Denenberg, V. H. (2001) Brain Res. 892 336-343 [DOI] [PubMed] [Google Scholar]
- 25.Lecker, S. H., Solomon, V., Price, S. R., Kwon, Y. T., Mitch, W. E., and Goldberg, A. L. (1999) J. Clin. Investig. 104 1411-1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johanson, A., and Blizzard, R. (1971) J. Pediatr. 79 982-987 [DOI] [PubMed] [Google Scholar]
- 27.Hu, R. G., Wang, H., Xia, Z., and Varshavsky, A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 76-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du, F., Navarro-Garcia, F., Xia, Z., Tasaki, T., and Varshavsky, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14110-14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner, G. C., Du, F., and Varshavsky, A. (2000) Nature 405 579-583 [DOI] [PubMed] [Google Scholar]
- 30.Rao, H., Uhlmann, F., Nasmyth, K., and Varshavsky, A. (2001) Nature 410 955-959 [DOI] [PubMed] [Google Scholar]
- 31.Ditzel, M., Wilson, R., Tenev, T., Zachariou, A., Paul, A., Deas, E., and Meier, P. (2003) Nat. Cell Biol. 5 467-473 [DOI] [PubMed] [Google Scholar]
- 32.Varshavsky, A. (2003) Nat. Cell Biol. 5 373-376 [DOI] [PubMed] [Google Scholar]
- 33.Lloyd, A. G., Ng, Y. S., Muesing, M. A., Simon, V., and Mulder, L. C. (2007) Virology 360 129-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, S., Ito, M., Gallis, J., Nishida, I., and Watanabe, A. (2002) Plant J. 32 129-137 [DOI] [PubMed] [Google Scholar]
- 35.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989) Gene (Amst.) 77 51-59 [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann, M., Nemetz, C., Madin, K., and Buchberger, B. (2004) Biotechnol. Annu. Rev. 10 1-30 [DOI] [PubMed] [Google Scholar]
- 37.Lupas, A. N., and Koretke, K. K. (2003) J. Struct. Biol. 141 77-83 [DOI] [PubMed] [Google Scholar]
- 38.Erbse, A., Schmidt, R., Bornemann, T., Schneider-Mergener, J., Mogk, A., Zahn, R., Dougan, D. A., and Bukau, B. (2006) Nature 439 753-756 [DOI] [PubMed] [Google Scholar]
- 39.Tobias, J. W., Shrader, T. E., Rocap, G., and Varshavsky, A. (1991) Science 254 1374-1377 [DOI] [PubMed] [Google Scholar]
- 40.Wang, K. H., Sauer, R. T., and Baker, T. A. (2007) Genes Dev. 21 403-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia, Z., Webster, A., Du, F., Piatkov, K., Ghislain, M., and Varshavsky, A. (2008) J. Biol. Chem. 283 24011-24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K. C., Putterill, J., Palme, K., Estelle, M., and Chory, J. (2001) Genes Dev. 15 1985-1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W., and Elledge, S. J. (1996) Cell 86 263-274 [DOI] [PubMed] [Google Scholar]
- 44.Potuschak, T., Stary, S., Schlogelhofer, P., Becker, F., Nejinskaia, V., and Bachmair, A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7904-7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stary, S., Yin, X. J., Potuschak, T., Schlögelhofer, P., Nizhynska, V., and Bachmair, A. (2003) Plant Physiol. 133 1360-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]