Abstract
Cytoplasmic folate-mediated one carbon (1C) metabolism functions to carry and activate single carbons for the de novo synthesis of purines, thymidylate, and for the remethylation of homocysteine to methionine. C1 tetrahydrofolate (THF) synthase, encoded by Mthfd1, is an entry point of 1Cs into folate metabolism through its formyl-THF synthetase (FTHFS) activity that catalyzes the ATP-dependent conversion of formate and THF to 10-formyl-THF. Disruption of FTHFS activity by the insertion of a gene trap vector into the Mthfd1 gene results in embryonic lethality in mice. Mthfd1gt/+ mice demonstrated lower hepatic adenosylmethionine levels, which is consistent with formate serving as a source of 1Cs for cellular methylation reactions. Surprisingly, Mthfd1gt/+ mice exhibited decreased levels of uracil in nuclear DNA, indicating enhanced de novo thymidylate synthesis, and suggesting that serine hydroxymethyltransferase and FTHFS compete for a limiting pool of unsubstituted THF. This study demonstrates the essentiality of the Mthfd1 gene and indicates that formate-derived 1Cs are utilized for de novo purine synthesis and the remethylation of homocysteine in liver. Further, the depletion of cytoplasmic FTHFS activity enhances thymidylate synthesis, affirming the competition between thymidylate synthesis and homocysteine remethylation for THF cofactors.
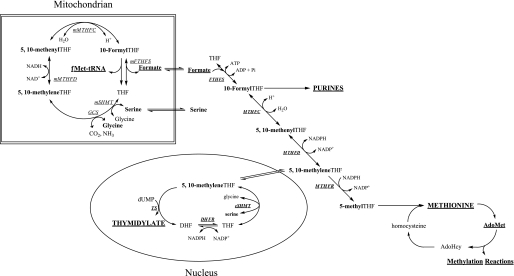
Folate-mediated one-carbon (1C)3 metabolism is compartmentalized in the cytoplasm, mitochondria, and nucleus of mammalian cells (1). In the cytoplasm, 1C metabolism functions to carry and chemically activate single carbons for the de novo synthesis of purines, thymidylate, and for the remethylation of homocysteine to methionine (2) (see Fig. 1). Methionine can be adenosylated to form S-adenosylmethionine (AdoMet), the major cellular methyl group donor required for the methylation of DNA, RNA, histones, small molecules, and lipids. Nuclear 1C metabolism functions to synthesize thymidylate from dUMP and serine during S phase through the small ubiquitin-like modifier-dependent translocation of cytoplasmic serine hydroxymethyltransferase (cSHMT), dihydrofolate reductase, and thymidylate synthase into the nucleus (3).
FIGURE 1.
Folate-mediated one-carbon metabolism occurs in the mitochondria, nucleus, and cytoplasm. Mitochondrial-derived formate traverses to the cytoplasm where it is incorporated into the folate-activated one-carbon pool through the activity of FTHFS and utilized in the synthesis of purines, thymidylate, and the methylation of homocysteine to methionine. Methionine can be converted to a methyl donor through its adenosylation to AdoMet. Thymidylate biosynthesis occurs in the cytoplasm and nucleus. The one-carbon unit is labeled in bold. GCS, glycine cleavage system; mSHMT, mitochondrial serine hydroxymethyltransferase; mMTHFD, mitochondrial methylenetetrahydrofolate dehydrogenase; mMTHFC, mitochondrial methenyltetrahydrofolate cyclohydrolase; mFTHFS, mitochondrial formyltetrahydrofolate synthetase; MTHFD, methylenetetrahydrofolate dehydrogenase; MTHFC, methenyltetrahydrofolate cyclohydrolase; FTHFS, formyltetrahydrofolate synthetase; MTHFR, methylenetetrahydrofolate reductase; TS, thymidylate synthase; DHFR, dihydrofolate reductase; and cSHMT, cytoplasmic serine hydroxymethyltransferase.
Serine, through its conversion to glycine by SHMT, is a primary source of 1Cs for nucleotide and methionine synthesis (4). SHMT generates 1Cs in the cytoplasm, mitochondria, and nucleus, although the generation of 1Cs through SHMT activity in the cytoplasm is not essential in mice, indicating the essentiality of mitochondria-derived 1Cs for cytoplasmic 1C metabolism (5). In mitochondria, the hydroxymethyl group of serine and the C2 carbon of glycine are transferred to tetrahydrofolate (THF) to generate 5,10-methylene-THF by the mitochondrial isozyme of SHMT and the glycine cleavage system, respectively (6). The 1C carried by methylene-THF is oxidized and hydrolyzed to generate formate by the NAD-dependent methylene-THF dehydrogenase (MTHFD) and methenyl-THF cyclohydrolase (MTHFC) activities encoded by a single gene, Mthfd2 (7), and 10-formyl-THF synthetase (FTHFS) activity, encoded by Mthfd1L (8) (see Fig. 1).
In the cytoplasm, the product of the Mthfd1 gene, C1THF synthase, is a trifunctional enzyme that contains NADP-dependent MTHFD and MTHFC activities on the N-terminal domain of the protein, and FTHFS activity on the C-terminal domain (9). These three activities collectively catalyze the interconversion of THF, 10-formyl-THF, 5,10-methenyl-THF, and 5,10-methylene-THF (10) (Fig. 1). The ATP-dependent FTHFS activity of C1THF synthase condenses mitochondria-derived formate with THF to form 10-formyl-THF, which is required for the de novo synthesis of purines (9). The MTHFC and MTHFD activities convert 10-formyl-THF to methylene-THF (11). Methylene-THF is utilized in the de novo synthesis of thymidylate or, alternatively, can be irreversibly reduced by methylene-THF reductase to 5-methyl-THF, which is used in the remethylation of homocysteine to methionine (12).
Impairments in 1C metabolism, due to insufficient folate cofactors and/or single nucleotide polymorphisms in genes that encode folate-dependent enzymes, are associated with numerous pathologies and developmental anomalies, including cancers, cardiovascular disease, and neural tube defects. The causal mechanisms underlying the folate-pathology relationship(s) remains to be established. However, a number of hypotheses have been proposed related to the role of 1C metabolism in genome stability and gene expression. Decreased thymidylate synthesis results in increased uracil misincorporation into DNA and decreased rates of cell division, causing double strand breaks in DNA and genomic instability (13). Decreased AdoMet synthesis alters methylation patterns in CpG islands in DNA and can result in histone hypomethylation, which can alter gene expression (2). Proliferating cells also require the de novo synthesis of purines to maintain rates of DNA synthesis (14).
It has been shown that the gene product of Mthfd2, mitochondrial MTHFC/MTHFD is essential in mice, and Mthfd2 deficiency results in embryonic lethality (15). This protein is required for the generation of formate from serine in the mitochondria of embryonic cells. Here, we have investigated the essentiality of the Mthfd1 gene in mice and the effect of altered Mthfd1 gene expression on biomarkers of cytoplasmic 1C metabolism. Our data demonstrate that Mthfd1 is an essential gene in mice and that Mthfd1-deficient mice are a model for the study of folate-associated pathologies.
EXPERIMENTAL PROCEDURES
Generation of Mthfd1gt/+ Mice—All study protocols were approved by the Institutional Animal Care and Use Committee of Cornell University and conform to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Mouse embryonic stem cells (E14Tg2a.4) containing a gene trap vector insertion between exons 26 and 27 of one allele of the Mthfd1 gene (cell line XB175) were obtained from Bay-Genomics (San Francisco, CA). The gene trap vector pGT0pfs contains the engrailed 2 (En2) intron located 5′ to a promoterless βgeo cassette (16). The integration of the gene trap vector within the Mthfd1 gene was verified by 5′-rapid amplification of cDNA ends PCR at BayGenomics. XB175 embryonic stem cells were expanded and injected into C57Bl/6 mouse blastocysts at the Cornell University Transgenic Mouse Core Facility (Ithaca, NY). Germ line transmission of the Mthfd1gt(pGT0pfs)Stov (Mthfd1gt/t) allele was confirmed by PCR using purified tail nuclear DNA. Mice were backcrossed onto the 129P2/OlaHsd (N>10) and the C57Bl/6 (N6) backgrounds. All mice were maintained under specific-pathogen free conditions.
Genotyping of Mthfd1gt/+ Mice—Genotyping was carried out by PCR using nuclear DNA isolated from tail tissue using a DNeasy DNA purification kit (Qiagen). A duplex PCR reaction was used to detect the wild-type Mthfd1 and Mthfd1gt alleles. The primers were as follows: Mthfd1 forward, 5′-tttggcttgaagagggacatgagg-3′; Mthfd1 reverse, 5′-aggaccttagaggactagcagggt-3′; and En2 reverse, 5′-gtcctacaacacacactccaacct-3′ (priming sites indicated in Fig. 2A). The PCR conditions were as follows: 94 °C for 20 s, 59 °C for 20 s, and 72 °C for 60 s.
FIGURE 2.
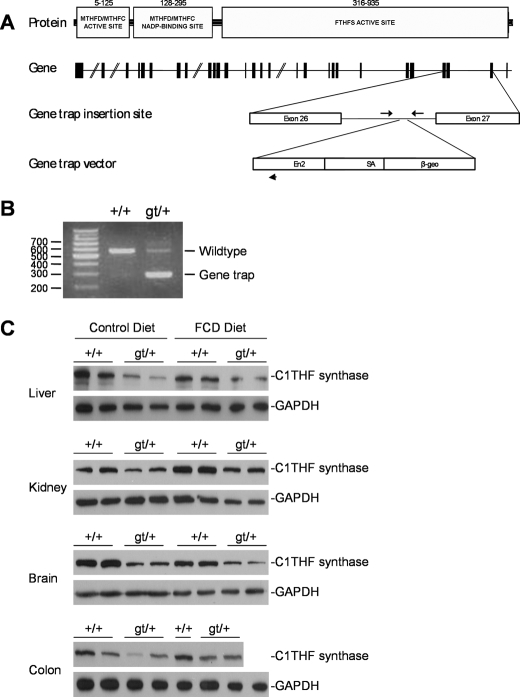
Generation of Mthfd1gt mice. A, C1THF synthase consists of three domains: the MTHFD/MTHFC catalytic domain, the MTHFD/MTHFC NADP-binding domain, and the FTHFS catalytic domain. The amino acid residues forming each domain are indicated. The Mthfd1 gene consists of 28 exons. The gene trap vector inserted into intron 26–27 of the Mthfd1 gene. PCR priming sites are indicated by arrows. B, the gene trap vector cassette in the Mthfd1 gene is detected as a 320-bp PCR product, whereas the wild-type allele is detected as a 553-bp PCR product. C, Western blot analysis of liver, kidney, brain, and colon from Mthfd1gt mice. Tissue lysates were probed with polyclonal sheep anti-mouse C1THF synthase antibody and mouse anti-human glyceraldehyde-3-phosphate dehydrogenase antibody, which served as a loading control. FTHFS, formyltetrahydrofolate synthetase; MTHFC, methenyltetrahydrofolate cyclohydrolase; MTHFD, methylenetetrahydrofolate dehydrogenase; THF, tetrahydrofolate.
C1THF Synthase Purification from Liver—Freshly isolated liver from an Mthfd1gt/+ mouse was homogenized in 50 mm potassium phosphate buffer, pH 7.2, and the suspension was clarified by centrifugation (10,000 × g). Ammonium sulfate was added to 15%, and precipitated proteins were pelleted by centrifugation, resuspended in 10 mm potassium phosphate buffer, pH 7.2, and dialyzed against the same buffer. The protein extract was loaded onto a DEAE-Sephacel column and eluted with a KCl gradient (0–300 mm and 100 ml). Fractions were collected and assayed for C1THF synthase-β-galactosidase-neomycin fusion protein (250 kDa) and C1THF synthase protein (105 kDa) by immunoblotting using rat anti-rabbit C1THF synthase antibodies generously provided by Dean Appling (University of Texas at Austin, see “Immunoblotting”). Fractions 20–25 contained the C1THF synthase-β-galactosidase-neomycin fusion protein without contaminating C1THF synthase protein; fractions 40–47 contained C1THF synthase protein without contaminating C1THF synthase-β-galactosidase-neomycin fusion protein.
Enzyme Activity Assays—FTHFS and MTHFD activities were assayed as described by Cheek and Appling (17).
Diets—Breeding mice were fed a standard rodent chow diet (Harlan Teklad LM-485). For the diet study, 3-week-old male mice were randomly weaned onto AIN-93G (control (C) diet, Dyets, Inc., Bethlehem, PA), which contained 2 mg/kg folic acid and 2.5 g/kg choline bitartrate or modified AIN-93G, which lacked folic acid and choline bitartrate (FCD diet, Dyets, Inc.). Mice were maintained on the diet for 5 weeks.
Diet Study Tissue Harvest—The animal feeding cycle was synchronized prior to tissue harvest to ensure AdoMet levels reflected homocysteine remethylation capacity with minimal contributions from dietary methionine. Food was removed 24 h prior to killing the animals. After 12 h, each animal was given one food pellet, and the animals were killed 12 h later. Blood was collected in heparin-coated tubes by cardiac puncture. Plasma was isolated by centrifugation. Plasma and tissues were flash-frozen in liquid nitrogen and stored at –80 °C.
Immunoblotting—Total protein was extracted and quantified from tissue (18). Immunoblotting was performed as previously described (5). Two different antibodies were used to detect C1THF synthase protein. Sheep anti-mouse C1THF synthase antibody was generated from the peptide NYVPDDTKPNGRKVVG (amino acid residues 239–254) and affinity purified using the same biotin-conjugated peptide. This antibody was diluted 1:10,000, and horseradish peroxidase-conjugated rabbit anti-sheep IgG secondary antibody (Pierce) was diluted 1:20,000. Rat anti-rabbit C1THF synthase antibodies were the generous gift of Dean Appling (University of Texas at Austin); immunoblots generated with this antibody were performed as described elsewhere (17). For glyceraldehyde-3-phosphate dehydrogenase detection, mouse anti-human glyceraldehyde-3-phosphate dehydrogenase antibody (Novus) was diluted 1:1,000,000 and the secondary antibody goat anti-mouse IgG conjugated to horseradish peroxidase (Pierce) was diluted 1:20,000.
Determination of AdoMet and AdoHcy Concentrations—Frozen tissues were sonicated in 500 μl of 0.1 m sodium acetate buffer (pH 6), and protein was precipitated by adding 312 μl of 10% perchloric acid to each sample. After vortexing, samples were centrifuged at 2,000 × g for 10 min at 4 °C. AdoMet and AdoHcy were determined as described previously (19). AdoMet and AdoHcy values were normalized to cellular protein content (18).
Plasma and Tissue Folate Concentration—Folate concentration of plasma and tissues was quantified using the Lactobacillus casei microbiological assay as described previously (20).
Uracil Content in Nuclear DNA—Nuclear DNA was extracted from 25–50 mg of tissue using DNeasy Tissue and Blood Kit (Qiagen), including an incubation with RNase A (Sigma) and RNase T1 (Ambion) for 30 min at 37 °C. 10 μg of DNA was treated with 1 unit of uracil DNA glycosylase (Epicenter) for 1 h at 37 °C. Immediately following incubation, 10 pg of [15N2]Uracil (Cambridge Isotopes) was added to each sample as an internal standard, and the sample was dried completely in a speed vacuum. 50 μl of acetonitrile, 10 μl of triethylamine, and 1 μl of 3,5-bis(trifluoromethyl)benzyl bromide were added to each sample and incubated for 25 min at 30 °C with shaking at 500 rpm. 50 μl of water followed by 100 μl of isooctane were added to each sample. Samples were vortexed and centrifuged. Organic extraction of derived uracil was completed by the removal of the aqueous phase and analysis of the organic phase. Uracil content in nuclear DNA was analyzed by gas chromatography mass spectrometry, as previously described (5).
Metabolite Profile from Plasma—Total homocysteine, cystathionine, total cysteine, methionine, glycine, serine, α-aminobutyric acid, N,N-dimethylglycine, and N-methylglycine were assayed in mouse plasma by stable isotope dilution capillary gas chromatography mass spectrometry as previously described (21, 22).
Statistical Analyses—Differences in genotype distribution were analyzed by the Chi square test. When comparing two groups, differences were determined by Student's t test analysis. Diet x genotype effects were analyzed by two-way ANOVA and Tukey's HSD post hoc test. Groups were considered significantly different when p ≤ 0.05. Data were normalized by log transformation. Data are presented as mean ± S.E. All statistics were performed using JMP IN software, release 5.1.2 (SAS Institute Inc.).
RESULTS
The Gene Trap Insertion in the Mthfd1 Gene Inactivates 10-Formyl-THF Synthetase Activity—The insertion of the gene trap vector into the Mthfd1 gene was identified by 5′ rapid amplification of cDNA ends at BayGenomics. The gene trap vector insertion site is located within the FTHFS domain of the Mthfd1 gene. We confirmed the presence of the gene trap vector by designing forward and reverse PCR primers specific to intron 26–27 that flank the gene trap insertion site, as well as a reverse primer that was specific to the En2 intron of the gene trap vector (Fig. 2A). The Mthfd1gt allele generated a 320-bp PCR product, whereas the wild-type allele resulted in a 553-bp PCR product (Fig. 2B).
The C1THF synthase-β-galactosidase-neomycin fusion protein and C1THF synthase protein were partially purified from the liver of an Mthfd1gt/+ mouse using DEAE-Sephacel and FTHFS and MTHFD activities were assayed. Fractions containing wild-type C1THF synthase, as determined by immunoblotting using rat anti-rabbit C1THF synthase antibodies (not shown), contained 0.8 μmol·min–1·mg–1 FTHFS activity and 0.40 μmol·min–1·mg–1 MTHFD activity. Fractions containing the C1THF synthase fusion protein exhibited comparable MTHFD activity (0.36 μmol·min–1·mg–1). These fractions contained essentially no FTHFS activity (0.001 μmol·min–1·mg–1). These data indicate that the gene trap insertion disrupted the FTHFS activity of C1THF synthase.
Mthfd1 Is Essential in Mice—To determine the viability of Mthfd1gt/gt mice, heterozygous B6.129P2-Mthfd1gt(N6) mice were intercrossed and the genotype distribution was determined (Table 1). A total of 139 pups from 26 litters were examined. The average litter size was 5.3 pups. We found that the Mthfd1 genotypes were not distributed as expected for Mendelian inheritance of the Mthfd1gt allele (Chi square analysis, p = 2.7 × 10–11). The ratio of Mthfd1+/+ to Mthfd1gt/+ to Mthfd1gt/gt mice was 39:100:0, demonstrating that homozygosity for the Mthfd1gt allele is embryonic lethal. When it was assumed that Mthfd1gt/gt was lethal, the Mthfd1+/+ and Mthfd1gt/+ genotypes were observed in the expected distribution. Both sexes were found at the expected frequency. Mthfd1gt/+ mice appear healthy and breed normally. The data indicate that in dams fed a standard rodent chow diet, homozygosity for the Mthfd1gt allele is embryonic lethal.
TABLE 1.
Mthfd1 null mice are not viable
Mthfd1gt/+ mice were intercrossed and their progeny genotyped. The expected genotype distribution was calculated based on a Mendelian distribution. Differences between observed and expected genotype and sex distributions were analyzed by Chi square analysis. p values ≤ 0.05 were considered significantly different.
|
Genotype
|
Observed genotype distribution
|
Expected genotype distribution
|
||||
|---|---|---|---|---|---|---|
| Male | Female | Total | Male | Female | Total | |
| Mthfd1+/+ | 15 | 24 | 39 | 17.5 | 17.5 | 35 |
| Mthfd1gt/+ | 46 | 54 | 100 | 34.5 | 34.5 | 69 |
| Mthfd1gt/- | 0 | 0 | 0 | 17.5 | 17.5 | 35 |
| Total | 61 | 78 | 139 | 69.5 | 69.5 | 139 |
| Number of litters observed | 26 | |||||
| Mean litter size (mean ± S.E.) | 5.3 ± 0.4 | |||||
| p value, observed versus expected genotype distribution (1:2:1) | 2.7 × 10-11 | |||||
| p value, observed versus expected genotype distribution assuming homozygous lethality (1:2:0) | nsa | |||||
| p value, observed versus expected sex distribution | ns | |||||
ns, not significant.
C1THF Synthase Protein Levels in Mthfd1gt/+ Mice—Heterozygous 129P2-Mthfd1gt male mice were crossed to C57Bl/6 females to produce B6129P2F1 Mthfd1+/+ and Mthfd1gt/+ progeny. The mice were weaned onto either the control diet or the FCD diet and maintained for 5 weeks. The Mthfd1gt/+ mice consistently exhibited an approximate 50% decrease in wild-type C1THF synthase protein in kidney, liver, colon, and brain, as detected by immunoblotting using sheep anti-mouse C1THF synthase antibodies (Fig. 2C). C1THF synthase protein levels were not influenced by the folate/choline-deficient diet in these mice.
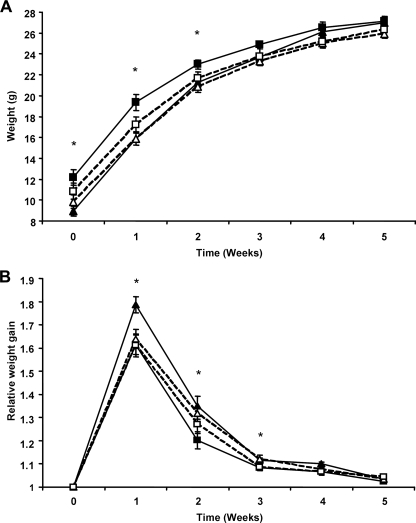
Mthfd1gt/+ Mice Have a Larger Body Weight at Weaning—Mthfd1gt/+ mice had a significantly larger body weight independent of diet than their wild-type siblings at weaning and for the first 2 weeks of the study (Fig. 3A, Student's t test, p ≤ 0.01). Relative weight gain was significantly increased in wild-type mice in comparison with Mthfd1gt/+ mice for the first 3 weeks, which by week 3 had compensated for the initial differences in body weight (Fig. 3B, Student's t test, p ≤ 0.03). No difference in body weight was observed between wild-type and Mthfd1gt/+ mice from weeks 3–5.
FIGURE 3.
Weight gain in wild-type and Mthfd1gt/+ mice fed a folate/choline sufficient or-deficient diet. Weight gain (A) and relative week (B) over week weight gain. In both panels, a solid line represents control diet fed mice and a dashed line represents folate/choline-deficient diet-fed mice. Full and empty triangles represent wild-type mice, and full and empty squares represent Mthfd1gt/+ mice. *, a significant difference, p ≤ 0.03 as determined by Student's t test, between wild-type and Mthfd1gt/+ mice.
After 5 weeks, the folate/choline-deficient diet resulted in a significant decrease in plasma folate concentrations in comparison with the control diet (20.4 versus 40.4 ng/ml, Student's t test, p = 0.006, Table 2). The diet had no significant effect on body weight (Fig. 3A). The Mthfd1 genotype did not influence plasma folate concentration (Table 2).
TABLE 2.
Plasma folate in Mthfd1gt/+ mice after 5 weeks on diet
Differences between genotypes and diets were analyzed by Student's t-test. Genotype × diet effects were analyzed by two-way ANOVA using Tukey's HSD post-hoc analysis. Data are presented as the mean ± S.E. values. p values ≤ 0.05 were considered significantly different (n = 3 per group).
| Mthfd1 genotype | Diet | Plasma |
|---|---|---|
| ng/ml | ||
| Mthfd1+/+ | AIN-93G (C) | 38.2 ± 2.57 |
| AIN-93G (FCD) | 24.1 ± 8.90 | |
| Mthfd1gt/+ | AIN-93G (C) | 42.7 ± 1.65 |
| AIN-93G (FCD) | 16.9 ± 5.11 | |
| p value, diet effect | 0.006 | |
| p value, genotype effect | nsa | |
| p value, diet × genotype effect | ns |
ns, not significant.
C1THF Synthase Alters Liver AdoMet Concentrations—Wild-type Mthfd1 mice fed the FCD diet tended to have lower liver AdoMet than wild-type mice fed the control diet (0.3 versus 0.5 pmol/μg of protein, Student's t test, p = 0.06), but overall the FCD diet did not significantly reduce AdoMet concentrations in liver after 5 weeks (Table 3). However, liver AdoMet was significantly reduced by 40% in Mthfd1gt/+ mice in comparison to Mthfd1+/+ mice fed the control diet (0.3 versus 0.5 pmol of AdoMet/μg of protein; two-way ANOVA and Tukey's HSD post-hoc test, p = 0.05, Table 3). The FCD diet resulted in more than a 50% decrease in liver AdoHcy (0.4 versus 0.9, Student's t test, p < 0.0001) and an increase in the AdoMet/AdoHcy ratio (0.9 versus 0.5, Student's t test, p = 0.005) in comparison to mice fed the control diet.
TABLE 3.
Liver AdoMet, AdoHcy, AdoMet/AdoHcy, and uracil in nuclear DNA in Mthfd1gt/+ mice after 5 weeks on diet
Differences between genotypes and diets were analyzed by Student's t-test. Genotype × diet effects were analyzed by two-way ANOVA using Tukey's HSD post-hoc analysis. Data are presented as the mean ± S.E. values. p values ≤ 0.05 were considered significantly different. n = 7-11 per group.
| Mthfd1 Genotype | Diet | AdoMet | AdoHcy | AdoMet/AdoHcy | Uracil DNA content | |
|---|---|---|---|---|---|---|
| pmol/μg protein | pg/μg DNA | |||||
| Mthfd1+/+ | AIN-93G (C) | 0.5 ± 0.07 | 1.0 ± 0.11 | 0.6 ± 0.12 | 0.4 ± 0.08 | |
| AIN93G (FCD) | 0.3 ± 0.05 | 0.4 ± 0.03 | 0.9 ± 0.16 | 1.0 ± 0.37 | ||
| Mthfd1gt/+ | AIN-93G (C) | 0.3 ± 0.07 | 0.8 ± 0.07 | 0.5 ± 0.14 | 0.7 ± 0.13 | |
| AIN93G (FCD) | 0.4 ± 0.05 | 0.4 ± 0.05 | 0.9 ± 0.11 | 0.7 ± 0.11 | ||
| p value, diet effect | nsa | <0.0001 | 0.005 | ns | ||
| p value, genotype effect | ns | ns | ns | ns | ||
| p value, diet × genotype effect | 0.05b | ns | ns | ns | ||
ns, not significant.
Control Mthfd1+/+ is significantly different from control Mthfd1gt/+, p < 0.05, as analyzed by two-way ANOVA and Tukey's HSD post-hoc test for genotype × diet effect.
In the colon, diet, but not genotype mediated changes in AdoMet, AdoHcy, and the AdoMet/AdoHcy ratio (Table 4). The FCD diet significantly increased AdoMet by ∼25% (0.2 versus 0.15 pmol of AdoMet/μg of protein, Student's t test, p = 0.02), decreased AdoHcy by 25% (0.3 versus 0.4 pmol of AdoHcy/μg protein, p = 0.0005) and increased the AdoMet/AdoHcy ratio by almost 2-fold (0.7 versus 0.4, p ≤ 0.0001).
TABLE 4.
Colon AdoMet, AdoHcy, AdoMet/AdoHcy and uracil in nuclear DNA in Mthfd1gt/+ mice after 5 weeks on diet
Differences between genotypes and diets were analyzed by Student's t-test. Genotype × diet effects were analyzed by two-way ANOVA using Tukey's HSD post-hoc analysis. Data are presented as the mean ± S.E. values. p values ≤ 0.05 were considered significantly different (n = 6-10 per group).
| Mthfd1 genotype | Diet | AdoMet | AdoHcy | AdoMet/AdoHcy | Uracil DNA content | |
|---|---|---|---|---|---|---|
| pmol/μg protein | pg/μg DNA | |||||
| Mthfd1+/+ | AIN-93G (C) | 0.2 ± 0.06 | 0.4 ± 0.04 | 0.4 ± 0.07 | 0.2 ± 0.02 | |
| AIN93G (FCD) | 0.2 ± 0.01 | 0.3 ± 0.02 | 0.7 ± 0.06 | 0.5 ± 0.10 | ||
| Mthfd1gt/+ | AIN-93G (C) | 0.1 ± 0.02 | 0.4 ± 0.03 | 0.3 ± 0.04 | 0.1 ± 0.02 | |
| AIN93G (FCD) | 0.2 ± 0.02 | 0.3 ± 0.03 | 0.7 ± 0.08 | 0.2 ± 0.02 | ||
| p value, diet effect | 0.02 | <0.0005 | <0.0001 | <0.0001 | ||
| p value, genotype effect | nsa | ns | ns | <0.0001 | ||
| p value, diet × genotype effect | ns | ns | ns | ns | ||
ns, not significant.
Mthfd1gt/+ Mice Are Less Susceptible to Folate-mediated Uracil Misincorporation in Colonic DNA—After 5 weeks, liver uracil tended to be increased in Mthfd1+/+ mice fed the FCD diet in comparison to those fed the control diet (0.4 versus 1.0 pg/μg DNA, Student's t test, p = 0.10). However, when considering both wild-type and Mthfd1gt/+ mice, it was found that the FCD diet did not significantly affect uracil incorporation into liver nuclear DNA (Table 3).
The FCD diet was associated with a significant 1.5-fold increase in uracil content in DNA in the colon (Table 4, 0.3 versus 0.2 pg/μg of DNA, p < 0.0001). Overall, Mthfd1gt/+ mice had significantly less uracil content in nuclear DNA from colon in comparison with their wild-type counterparts independent of diet (0.2 versus 0.3 pg of uracil/μg of DNA, Student's t test, p < 0.0001).
Mthfd1- and Diet-mediated Changes in the Trans-sulfuration Pathway—The FCD diet was associated with changes in markers of homocysteine metabolism (Table 5). Mice fed the FCD diet for 5 weeks demonstrated a 25% increase in plasma homocysteine (6.5 versus 5.2 μm, Student's t test, p = 0.02) and a 28% increase in plasma α-aminobutyric acid (5.9 versus 4.6 μm, p = 0.03). Plasma glycine was significantly reduced in mice fed the FCD diet (286 versus 325 μm, p = 0.04).
TABLE 5.
Metabolic profile of plasma in Mthfd1gt/+ mice after 5 weeks on diet
Differences between diets and genotypes were analyzed by Student's t-test. Genotype × diet effects were analyzed by two-way ANOVA using Tukey's HSD post-hoc analysis. Data are presented as the mean ± S.E. values. p values ≤ 0.05 were considered significantly different (n = 6 per group).
|
Metabolite
|
Mthfd1+/+
|
Mthfd1gt/+
|
p value of model effect
|
||||
|---|---|---|---|---|---|---|---|
| AIN-93G (C) | AIN-93G (FCD) | AIN-93G (C) | AIN-93G (FCD) | Diet | Genotype | Diet × genotype | |
| Homocysteine (μm) | 5.0 ± 0.5 | 5.8 ± 0.8 | 5.3 ± 0.3 | 7.1 ± 0.3 | 0.02 | nsa | ns |
| Cystathionine (nm) | 1159 ± 103 | 956 ± 118 | 789 ± 58 | 1042 ± 67 | ns | ns | 0.02b |
| Cysteine (μm) | 200 ± 16 | 219 ± 17 | 227 ± 9 | 222 ± 16 | ns | ns | ns |
| α-Aminobutyric acid (μm) | 3.6 ± 0.3 | 5.9 ± 0.9 | 5.6 ± 0.3 | 5.8 ± 0.3 | 0.03 | ns | 0.07c |
| Methionine (μm) | 40.3 ± 4.3 | 36.6 ± 3.6 | 39.9 ± 3.9 | 44.7 ± 1.8 | ns | ns | ns |
| Glycine (μm) | 341 ± 24 | 275 ± 19 | 308 ± 18 | 297 ± 7 | 0.04 | ns | ns |
| Serine (μm) | 150 ± 14 | 138 ± 10 | 151 ± 15 | 142 ± 3 | ns | ns | ns |
| Dimethylglycine (μm) | 8.49 ± 0.55 | 7.43 ± 0.50 | 7.76 ± 0.64 | 7.50 ± 0.72 | ns | ns | ns |
| Methylglycine (μm) | 1.79 ± 0.07 | 1.41 ± 0.13 | 1.46 ± 0.07 | 1.44 ± 0.15 | ns | ns | ns |
ns, not significant.
Control Mthfd1+/+ is significantly different than control Mthfd1gt/+, p < 0.05, as analyzed by two-way ANOVA and Tukey's HSD post-hoc test for diet × genotype effect.
Control Mthfd1+/+ is significantly different than folate/choline-deficient Mthfd1+/+ and folate/choline-deficient Mthfd1gt/+, p < 0.05, as analyzed by two-way ANOVA and Tukey's HSD post-hoc test for diet × genotype effect.
While the Mthfd1 genotype alone did not affect markers of homocysteine metabolism, a significant diet x genotype effect was observed for plasma cystathionine and α-aminobutyric acid. Mthfd1gt/+ mice fed the control diet, but not those fed the FCD diet, had decreased cystathionine concentrations in comparison with wild-type mice fed the control diet (789 versus 1159 nm, two-way ANOVA and Tukey's HSD post-hoc test, p < 0.05). The FCD diet was associated with an increase in plasma α-aminobutyric acid in wild-type mice, but not in Mthfd1gt/+ mice (3.6 versus 5.9 μm). The Mthfd1gt/+ mice on both diets maintained an overall higher concentration of α-aminobutyric acid than wild-type mice.
DISCUSSION
There are two major entry points of 1Cs into folate metabolism in the cytoplasm; the synthesis of 10-formyl-THF from formate catalyzed by FTHFS, and the formation of methylene-THF from serine catalyzed by cSHMT. Previously, we have demonstrated that cSHMT, encoded by the Shmt1 gene, is not essential in mice (5). This study demonstrates that the Mthfd1 gene is essential in mice and affirms the fundamental role of mitochondria in the production of formate for 1C metabolism in the cytoplasm. Although the gene trap insertion targeted and impaired the activity of the FTHFS domain of the C1THF synthase enzyme, the fusion protein did not accumulate to a significant level in most tissues (<10% wild-type levels), as determined by Western blotting, indicating that the gene trap insertion reduced all three activities of C1THF synthase by reducing total protein levels. Primary mouse fibroblasts derived from cultured embryonic stem cells lacking Mthfd1 expression were shown to be purine auxotrophs (8), therefore, it is likely that the Mthfd1gt/gt mice are not viable due to inadequate purine synthesis to sustain growth and development.
The mitochondrial NAD-dependent MTHFD/MTHFC (encoded by Mthfd2) has also been shown to be essential in mice (15). This enzyme is responsible for the synthesis of 10-formyl-THF in mitochondria (23). Mitochondrial 10-formyl-THF can be used in the generation of formate through FTHFS activity encoded by Mthfd1L (8, 24) or used to formylate the initiator methionyl-tRNA. Changes in mitochondrial protein synthesis were not associated with in utero lethality in Mthfd2-deficient embryos (15). Rather, it was suggested that the decrease in 10-formyl-THF available for purine synthesis compromised embryonic development, which is what we propose occurs in Mthfd1-deficient embryos.
Although the Mthfd1gt/+ mice appear healthy and breed well, the analysis of biomarkers indicates folate-mediated 1C metabolism is impaired. Mthfd1gt/+ mice fed the control diet had decreased levels of AdoMet in liver compared with wild-type mice, indicating that formate-derived 1Cs are utilized for the remethylation of homocysteine in liver, which is consistent with previous isotope tracer metabolic studies in cell cultures (19). Liver AdoMet concentrations in wild-type Mthfd1 mice also tended to decrease in response to the FCD diet, whereas Mthfd1gt/+ mice were refractory to a further diet-mediated decrease in liver AdoMet. Other studies have demonstrated that AdoMet levels are tightly buffered and that compensatory mechanisms exist to spare AdoMet levels when methionine synthesis is inhibited (25). Unlike that observed in liver, the Mthfd1 genotype had no effect on AdoMet or AdoHcy in the colon. The differences in AdoMet and AdoHcy levels across tissues may be due to tissue-specific differences in gene expression, enzyme activity, or responses to altered 1C pools. Homocysteine metabolism was also altered in Mthfd1gt/+ mice. Mthfd1gt/+ mice demonstrated decreased plasma cystathionine and increased plasma α-aminobutyric acid, suggesting that homocysteine metabolism via the trans-sulfuration pathway was influenced by changes in the amount of 1Cs available for homocysteine remethylation.
Uracil misincorporation into nuclear DNA was also influenced by Mthfd1 status, but only significantly in colon. The Mthfd1gt/+ mice exhibited lower uracil levels in colonic DNA compared with wild-type mice, and this effect was independent of diet, suggesting that the 1Cs utilized in thymidylate synthesis are not solely derived from formate. This supports previous studies that demonstrate cSHMT provides an independent pool of 1Cs for thymidylate biosynthesis through nuclear folate-mediated 1C metabolism (5, 19), and that maintenance of cSHMT levels is important to prevent uracil accumulation in DNA (5). The decrease in uracil content in DNA associated with FTHFS disruption indicates that FTHFS and SHMT compete for a limiting pool of THF; other studies have demonstrated that the concentration of folate-dependent enzymes exceeds the cellular concentration folate cofactors (19). Depletion of FTHFS activity may make THF more available to cSHMT and the thymidylate synthesis cycle and prevent uracil misincorporation into DNA (3, 26).
Similar to the Mthfd1gt/+ mice, a common single nucleotide polymorphism exists in the formyl-THF synthetase domain of the human Mthfd1 gene and results inaGtoA substitution at position 1958 (27). The single nucleotide polymorphism has been associated with a number of folate-mediated pathologies. In some populations, G1958A homozygosity has been shown to be a maternal risk factor for late pregnancy loss (28), severe abruptio placentae (29), neural tube defects (30–32), and congenital heart defects (33). The mechanisms underlying Mthfd1-associated reproductive and birth defects in humans are not understood; however, it has been suggested that it is a deficiency in de novo purine synthesis due to decreased formyl-THF synthetase activity that promotes pathogenesis (31, 33). Our data demonstrate that the Mthfd1gt mouse is a good model to study the effect of altered folate metabolism in C1THF synthase-associated embryonic defects and loss, as well as other folate-mediated pathologies. Current studies in our laboratory are focusing on clarifying the metabolic role played by C1THF synthase in embryonic development and cancer.
Acknowledgments
We thank Sylvia Allen, Rachel Slater, Anna Beaudin, and Martha Field for technical assistance. We appreciate the generous gift of rat anti-rabbit C1THF synthase antibodies from Dean Appling (University of Texas at Austin).
This work was supported, in whole or in part, by National Institutes of Health Grants CA105440 (to P. J. S. and B. S.) and DK42033 (to B. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: 1C, one carbon; AdoMet, S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine; C, control; cSHMT, cytoplasmic serine hydroxymethyltransferase; FCD, folate/choline-deficient; FTHFS, formyltetrahydrofolate synthetase; MTHFC, methenyltetrahydrofolate cyclohydrolase; MTHFD, methylenetetrahydrofolate dehydrogenase; SHMT, serine hydroxymethyltransferase; THF, tetrahydrofolate; ANOVA, analysis of variance.
References
- 1.Appling, D. R. (1991) FASEB J. 5 2645–2651 [DOI] [PubMed] [Google Scholar]
- 2.Stover, P. J. (2004) Nutr. Rev. 62 S3–S12; discussion S13 [DOI] [PubMed] [Google Scholar]
- 3.Woeller, C. F., Anderson, D. D., Szebenyi, D. M., and Stover, P. J. (2007) J. Biol. Chem. 282 17623–17631 [DOI] [PubMed] [Google Scholar]
- 4.Davis, S. R., Stacpoole, P. W., Williamson, J., Kick, L. S., Quinlivan, E. P., Coats, B. S., Shane, B., Bailey, L. B., and Gregory, J. F., 3rd. (2004) Am. J. Physiol. 286 E272–E279 [DOI] [PubMed] [Google Scholar]
- 5.MacFarlane, A. J., Liu, X., Perry, C. A., Flodby, P., Allen, R. H., Stabler, S. P., and Stover, P. J. (2008) J. Biol. Chem. 283 25846–25853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motokawa, Y., and Kikuchi, G. (1971) Arch. Biochem. Biophys. 146 461–464 [DOI] [PubMed] [Google Scholar]
- 7.Mejia, N. R., and MacKenzie, R. E. (1988) Biochem. Biophys. Res. Commun. 155 1–6 [DOI] [PubMed] [Google Scholar]
- 8.Christensen, K. E., Patel, H., Kuzmanov, U., Mejia, N. R., and MacKenzie, R. E. (2005) J. Biol. Chem. 280 7597–7602 [DOI] [PubMed] [Google Scholar]
- 9.Hum, D. W., Bell, A. W., Rozen, R., and MacKenzie, R. E. (1988) J. Biol. Chem. 263 15946–15950 [PubMed] [Google Scholar]
- 10.Tan, L. U., Drury, E. J., and MacKenzie, R. E. (1977) J. Biol. Chem. 252 1117–1122 [PubMed] [Google Scholar]
- 11.Pelletier, J. N., and MacKenzie, R. E. (1995) Biochemistry 34 12673–12680 [DOI] [PubMed] [Google Scholar]
- 12.Vanoni, M. A., and Matthews, R. G. (1984) Biochemistry 23 5272–5279 [DOI] [PubMed] [Google Scholar]
- 13.Duthie, S. J., and Hawdon, A. (1998) FASEB J. 12 1491–1497 [PubMed] [Google Scholar]
- 14.Kondo, M., Yamaoka, T., Honda, S., Miwa, Y., Katashima, R., Moritani, M., Yoshimoto, K., Hayashi, Y., and Itakura, M. (2000) J. Biochem. 128 57–64 [DOI] [PubMed] [Google Scholar]
- 15.Di Pietro, E., Sirois, J., Tremblay, M. L., and MacKenzie, R. E. (2002) Mol. Cell. Biol. 22 4158–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skarnes, W. C. (2000) Methods Enzymol. 328 592–615 [DOI] [PubMed] [Google Scholar]
- 17.Cheek, W. D., and Appling, D. R. (1989) Arch. Biochem. Biophys 270 504–512 [DOI] [PubMed] [Google Scholar]
- 18.Bensadoun, A., and Weinstein, D. (1976) Anal. Biochem. 70 241–250 [DOI] [PubMed] [Google Scholar]
- 19.Herbig, K., Chiang, E. P., Lee, L. R., Hills, J., Shane, B., and Stover, P. J. (2002) J. Biol. Chem. 277 38381–38389 [DOI] [PubMed] [Google Scholar]
- 20.Suh, J. R., Oppenheim, E. W., Girgis, S., and Stover, P. J. (2000) J. Biol. Chem. 275 35646–35655 [DOI] [PubMed] [Google Scholar]
- 21.Stabler, S. P., Lindenbaum, J., Savage, D. G., and Allen, R. H. (1993) Blood 81 3404–3413 [PubMed] [Google Scholar]
- 22.Allen, R. H., Stabler, S. P., and Lindenbaum, J. (1993) Metabolism 42 1448–1460 [DOI] [PubMed] [Google Scholar]
- 23.Mejia, N. R., and MacKenzie, R. E. (1985) J. Biol. Chem. 260 14616–14620 [PubMed] [Google Scholar]
- 24.Prasannan, P., Pike, S., Peng, K., Shane, B., and Appling, D. R. (2003) J. Biol. Chem. 278 43178–43187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmore, C. L., Wu, X., Leclerc, D., Watson, E. D., Bottiglieri, T., Krupenko, N. I., Krupenko, S. A., Cross, J. C., Rozen, R., Gravel, R. A., and Matthews, R. G. (2007) Mol. Genet. Metab. 91 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson, D. D., Woeller, C. F., and Stover, P. J. (2007) Clin. Chem. Lab. Med. 45 1760–1763 [DOI] [PubMed] [Google Scholar]
- 27.Hol, F. A., van der Put, N. M., Geurds, M. P., Heil, S. G., Trijbels, F. J., Hamel, B. C., Mariman, E. C., and Blom, H. J. (1998) Clin. Genet. 53 119–125 [DOI] [PubMed] [Google Scholar]
- 28.Parle-McDermott, A., Pangilinan, F., Mills, J. L., Signore, C. C., Molloy, A. M., Cotter, A., Conley, M., Cox, C., Kirke, P. N., Scott, J. M., and Brody, L. C. (2005) Mol. Hum. Reprod. 11 477–480 [DOI] [PubMed] [Google Scholar]
- 29.Parle-McDermott, A., Mills, J. L., Kirke, P. N., Cox, C., Signore, C. C., Kirke, S., Molloy, A. M., O'Leary, V. B., Pangilinan, F. J., O'Herlihy, C., Brody, L. C., and Scott, J. M. (2005) Am. J. Med. Genet. A 132 365–368 [DOI] [PubMed] [Google Scholar]
- 30.De Marco, P., Merello, E., Calevo, M. G., Mascelli, S., Raso, A., Cama, A., and Capra, V. (2006) J. Hum. Genet. 51 98–103 [DOI] [PubMed] [Google Scholar]
- 31.Brody, L. C., Conley, M., Cox, C., Kirke, P. N., McKeever, M. P., Mills, J. L., Molloy, A. M., O'Leary, V. B., Parle-McDermott, A., Scott, J. M., and Swanson, D. A. (2002) Am. J. Hum. Genet. 71 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parle-McDermott, A., Kirke, P. N., Mills, J. L., Molloy, A. M., Cox, C., O'Leary, V. B., Pangilinan, F., Conley, M., Cleary, L., Brody, L. C., and Scott, J. M. (2006) Eur. J. Hum. Genet. 14 768–772 [DOI] [PubMed] [Google Scholar]
- 33.Christensen, K. E., Rohlicek, C. V., Andelfinger, G. U., Michaud, J., Bigras, J. L., Richter, A., Mackenzie, R. E., and Rozen, R. (2008) Hum. Mutat., in press [DOI] [PubMed]