Abstract
Protein kinase A (PKA)-dependent phosphorylation is regulated by targeting of PKA to its substrate as a result of binding of regulatory subunit, R, to A-kinase-anchoring proteins (AKAPs). We investigated the effects of disrupting PKA targeting to AKAPs in the heart by expressing the 24-amino acid regulatory subunit RII-binding peptide, Ht31, its inactive analog, Ht31P, or enhanced green fluorescent protein by adenoviral gene transfer into rat hearts in vivo. Ht31 expression resulted in loss of the striated staining pattern of type II PKA (RII), indicating loss of PKA from binding sites on endogenous AKAPs. In the absence of isoproterenol stimulation, Ht31-expressing hearts had decreased +dP/dtmax and -dP/dtmin but no change in left ventricular ejection fraction or stroke volume and decreased end diastolic pressure versus controls. This suggests that cardiac output is unchanged despite decreased +dP/dt and -dP/dt. There was also no difference in PKA phosphorylation of cardiac troponin I (cTnI), phospholamban, or ryanodine receptor (RyR2). Upon isoproterenol infusion, +dP/dtmax and -dP/dtmin did not differ between Ht31 hearts and controls. At higher doses of isoproterenol, left ventricular ejection fraction and stroke volume increased versus isoproterenol-stimulated controls. This occurred in the context of decreased PKA phosphorylation of cTnI, RyR2, and phospholamban versus controls. We previously showed that expression of N-terminal-cleaved cTnI (cTnI-ND) in transgenic mice improves cardiac function. Increased cTnI N-terminal truncation was also observed in Ht31-expressing hearts versus controls. Increased cTnI-ND may help compensate for reduced PKA phosphorylation as occurs in heart failure.
Specificity of PKA2 signaling is mediated in large part by binding of PKA to AKAPs. AKAP-bound PKA is targeted at subcellular locations adjacent to PKA substrates and activated by local pools of cAMP (1, 2). AKAPs bind dimers of regulatory subunits of PKA (mainly RII) (3, 4). We investigated the effect of disrupting PKA targeting to AKAPs to determine the effect of AKAP targeting of PKA on regulation of cardiac function in vivo. Because the inotropic response to β-adrenergic receptor (β-AR) stimulation is dependent upon PKA protein phosphorylation, we predicted an impaired response to β-AR stimulation upon disruption of PKA targeting to substrates in the heart.
In addition to regulation by PKA-dependent phosphorylation, cardiac TnI (cTnI) (5), is subject to proteolysis. Truncation of 28–31 amino acids from the cardiac-specific N-terminal of cTnI, most likely by calpain-dependent proteolysis (6–8), eliminates the PKA-dependent phosphorylation sites serines 23 and 24 but preserves regions of cTnI homologous to skeletal muscle TnI, which contain the principal binding sites for other thin filament proteins. Thus, the inhibitory function of cTnI is not affected by N-terminal cleavage (6, 7). Consistent with this idea, we previously showed that transgenic mice expressing low (16–30% of total cTnI) or high (41–82%) levels of cTnI-ND showed beneficial effects on cardiac function (Table 2 and Figs. 4 and 6 in Ref. 7) that is, notably improved myocardial relaxation including decreased EDP and increased LV filling (7), thus mimicking the effects of PKA phosphorylation of N-terminal serines.
Recapitulation of effects of PKA phosphorylation of cTnI by N-terminal truncation may be due to the fact that phosphorylation of serines 23 and 24 results in folding of the N-terminal away from the long axis of cTnI such that it no longer binds to the C terminus of TnC (9–11). Proteolytic cleavage of the N terminus of cTnI, which eliminates interaction with cTnC, mimics this loss of binding. In addition, in vitro studies have shown that cleavage of the N terminus of cTnI occurs to a greater extent under conditions of decreased PKA phosphorylation of serines 23 and 24 (8, 12, 13). As indicated above, conformational changes in cTnI occur upon PKA phosphorylation. Thus, it is likely that when N-terminal serines are unphosphorylated, changes in the shape of cTnI alter accessibility of the N-terminal cleavage site to the protease.
PKA phosphorylation of several PKA substrates is reduced in heart failure; thus, the observation of increased N-terminal cleavage of cTnI upon decreased PKA phosphorylation is of great clinical interest. We propose that N-terminal proteolysis of cTnI compensates for decreased activity of the β-adrenergic signaling pathway and avoids potentially deleterious effects of prolonged cAMP elevation.
In this study we first tested whether increased PKA phosphorylation of cTnI in rat cardiac myocytes recapitulates PKA phosphorylation-dependent differences in proteolysis of cTnI observed in vitro. We predicted that PKA-dependent cTnI phosphorylation would be decreased by disruption of PKA targeting to AKAPs in the heart in vivo. If this hypothesis proved to be correct, we hypothesized that decreased PKA-dependent cTnI phosphorylation would then result in increased cleavage of the N terminus of cTnI.
Cardiac function is improved with overexpression of cTnI-ND in transgenic mice (7). Furthermore, impaired cardiac dysfunction in Gsα-deficient transgenic mice is partially restored by crossing Gsα mice with cTnI-ND-overexpressing mice (14). We, therefore, hypothesize that disruption of PKA targeting to endogenous AKAPs in rat hearts in vivo has salutary effects on contractility, compensating for reduced PKA phosphorylation of other PKA substrates.
We tested this hypothesis by introducing the 24 amino acid, RII-binding peptide, Ht31, by adenoviral gene transfer into rat hearts in vivo. This 24-amino acid peptide has been widely used to disrupt PKA/AKAP interaction; for example, Ht31 inhibits aquaporin-2 phosphorylation by PKA in renal cells (15), Ht31 suppresses PKA-dependent KvLQT1/Isk channel activity in Cos-7 cells (16), Ht31 blocks PKA phosphorylation of β-adrenergic receptors in A431 cells (17), and Ht31 diminishes forskolin-dependent activation of the cystic fibrosis transmembrane conductance regulator Cl- channel in myocytes (18).
EXPERIMENTAL PROCEDURES
Adenovirus Construction—Recombinant adenovirus encoding HA-tagged Ht31 peptide (Ad-HA-Ht31493–515; Ht31) or HA-tagged Ht31P (proline-substituted derivative) (Ad-HA-Ht31P493–515; Ht31P) were generated (19). Ht31P, with a proline replacing an isoleucine in the RII binding domain, disrupts the amphipathic helix required for RII binding; thus, Ht31P expression does not block PKA/AKAP interaction and serves as a negative control.
Using the AdEasy™ XL adenoviral vector system, the cDNA encoding either Ht31493–515 (residues 493–515) or Ht31P493–515 (residues 493–515; I507P) (kindly supplied by Dr. John Scott, Vollum Institute, Oregon Health Science University, Portland, OR) were cloned into the pShuttle-CMV vector. Adenoviral stock solutions (stored at -80 °C) were diluted with phosphate-buffered saline (1:1), thawed, and maintained at 37 °C until the time of injection. In all cases a viral solution was injected into rat hearts containing 250 μl of buffer containing 1.2 × 1012 particles/ml adenovirus or buffer alone, as described (20, 21). In addition to Ht31P, adenovirus encoding enhanced green fluorescence protein (Ad-eGFP) served as control. No significant differences were observed in cardiac function, protein phosphorylation, or cTnI proteolytic cleavage between Ht31P or eGFP-expressing hearts; thus, Ht31P and eGFP were together reported as controls. Adenoviral preparations (Ht31, Ht31P, eGFP) were obtained from the NIH/NHLBI-sponsored Preclinical Vector Core Facility, University of Pittsburgh.
In Vivo Adenoviral Gene Transfer into Rat Hearts—Male Sprague-Dawley rats (250–300 g) were used for gene transfer into rat hearts in vivo. Experiments were performed under the supervision of a full-time veterinarian. Veterinary care was maintained in Assessment and Accreditation of Laboratory Animal Care-approved facilities. The Animal Research Committee at the Cleveland Clinic approved all protocols and the methods of anesthesia, surgery, and euthanasia.
Gene transfer of Ht31, Ht31P, or eGFP into rat hearts was performed as described (20, 21). In brief, rats were anesthetized with 50 mg/kg sodium pentobarbital, placed in a supine position, intubated, and ventilated at 80 breaths per min. Mid-sternotomy was performed, the pericardium was opened, and a 7-0 suture was placed at the apex of the LV to secure the heart for catheter injection. A 24-gauge catheter containing 250 μl of adenovirus was advanced from the apex of the LV to the aortic root. Aorta and pulmonary arteries were clamped distal to the catheter, and then the adenoviral solution was injected. The clamp was maintained for 60 s. This allowed for perfusion at low pressure, and, thus, efficient adenoviral infection of the endocardium. Within 60 s of clamp release, heart rate and ventricular pressure quickly recovered to base line. The sternum and skin were then closed by creating a negative pressure gradient using a 10-ml syringe placed in the closed chest to reduce any residual pneumothorax. Once completed, the animals were ventilated for an additional 10–15 min, extubated, and transferred back to their cages.
Cardiac Function—Transthoracic two-dimensional and M-mode echocardiography were performed using a 15-mHz linear-array transducer coupled with an Acuson Sequoia C256 (Siemens Medical Solutions, Erlangen, Germany) (22, 23). Animals were lightly sedated with ketamine (50 mg/kg; intraperitoneal injection) for basal echocardiographic measurements. For physiological evaluation during isoproterenol stimulation, animals were sedated in a chamber using 5% isoflurane/95% oxygen mixture, intubated, artificially ventilated with a pressure cycled rodent ventilator (Kent Scientific Corp., RSP1002) at 80 breaths per min and maintained under anesthesia (2.5% isoproterenol flurane, 97.5% O2). Cardiac dimensions were measured between the intraventricular septum wall thickness and posterior wall thickness from the short-axis view at the level of papillary muscles from at least three cardiac cycles. A second catheter was inserted into the left jugular vein to deliver incremental doses of isoproterenol, at 0.1, 0.5 and 1.0 μg/kg of body weight/min. Base-line measurements were obtained before isoproterenol infusion, and then the effect of isoproterenol stimulation was determined 2 min after the start of isoproterenol infusion. Percent LV ejection fraction (LVEF) was calculated as (EDV - ESV)/EDV × 100, where EDV = end diastolic volume, and ESV = end systolic volume; stroke volume = EDV - ESV. Heart rate = 1/(R - R interval), expressed as beats/min.
LV systolic and diastolic properties were measured in vivo (23, 24). Rats were intubated and artificially ventilated. A miniaturized impedance/micro-manometer microtip pressure-volume catheter (Millar Instruments) was inserted into the right carotid vein and advanced into the LV under pressure control. After evaluation of cardiac function, hearts were harvested for biochemical measurements, immediately stored in liquid N2, or placed in Cyro-O.C.T. compound (Tissue-Tek) and stored at -80 °C.
Western Blot Analysis—PKA-dependent protein phosphorylation was measured 7-days after in vivo gene transfer of Ht31, Ht31P, or eGFP in LV homogenates (25, 26). Tissue was obtained immediately after maximal isoproterenol infusion or, in a subset of animals, under base-line conditions. For Western blot analysis of protein phosphorylation in homogenates of rat heart LV, proteins were separated by SDS-PAGE: Tris-glycine 4–12% gradient gels for cTnI and PLB, Tris acetate 3–8% gradient NUPAGE gels for RyR2, Tris-glycine 12% gels for cTnI-ND/cTnI, and Tris-glycine 16% gels for lysates obtained from adult rat cardiac myocytes.
Extent of PKA-dependent protein phosphorylation was assessed using antibodies recognizing total and phosphorylated PKA substrates (cTnI, RyR2, and PLB). The ratio of phosphorylated to total protein was then determined. Restore stripping buffer (Pierce) was used to remove the first primary antibody from the immunoblots (anti-phospho-TnI, anti-phospho-RyR2, and anti-phospho-PLB, respectively). After stripping, blots were reprobed with the new primary antibody (anti-TnI, anti-RyR2, and anti-PLB).
Antibody description and dilution are as follows. (i) Phospho-TnI (cardiac) (Ser-23/24) rabbit polyclonal primary antibody (1:2500 dilution; Cell Signaling) recognizes cTnI phosphorylated at serines 23/24; cTnI rabbit polyclonal antibody (1:2500 dilution; Cell Signaling) recognizes total cTnI. Phospho-TnI and total-TnI antibodies were probed with anti-rabbit IgG, horseradish peroxidase-linked secondary antibody (1:2500 dilution; Cell Signaling). (ii) RyR2 phosphorylated at serine 2809 was measured with phospho-RyR2 (Ser-2809) rabbit monoclonal antibody (1:5000 dilution; Badrilla) and RyR2 mouse polyclonal antibody (1:2500 dilution; Affinity BioReagents; clone 34C). Phospho-RyR2 and total RyR2 antibodies were probed with anti-rabbit IgG horseradish peroxidase (HRP)-linked secondary antibody (1:2500 dilution; Cell Signaling) and anti-mouse IgG, HRP-linked secondary antibody (1:2500 dilution; Cell Signaling), respectively. (iii) PLB phosphorylated by PKA at serine 16 was determined with anti-phospho-PLB (Ser-16) rabbit polyclonal antibody (1:5000 dilution; Upstate) and PLB mouse monoclonal antibody to PLB (1:5000 dilution; Upstate). Phospho-PLB and total-PLB primary antibodies were then probed using the anti-rabbit IgG and anti-mouse IgG horseradish peroxidase-linked secondary antibody (1:5000 dilution; Cell Signaling), respectively. Staining was detected with the SuperSignal West Pico Chemiluminescent Substrate (Pierce). Antibody-labeled proteins were quantified by densitometry using a Kodak Gel Logic 100 Imaging System.
The amount of truncated cTnI was assessed in LV homogenates isolated 7 days after in vivo gene transfer and after isoproterenol infusion. Western blot analysis was performed using anti-TnI-1 monoclonal antibody (1:2000 dilution) (25, 26). Anti-TnI-1 antibody recognizes an epitope at the C terminus of cTnI and, thus, identifies N-terminal cTnI cleavage products (25, 26). Blots were probed using the anti-mouse IgG, alkaline phosphatase-linked secondary antibody (1:3000 dilution; Sigma) and subsequently detected by 5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium substrate reaction. Non-infected mouse heart control and cloned recombinant mouse cTnI-ND protein (∼21.5 kDa) serve as alignment or molecular weight marker for the expression of the N-terminal cTnI cleavage products on the Western blot. The ratio of truncated cTnI/total cTnI was determined in Ht31 (n = 4)- versus Ht31P (n = 4)-expressing hearts.
Western blot analysis of N-terminal deleted cardiac TnI products was carried out using the anti-4H6 monoclonal antibody (25, 26), which recognizes an epitope specific to cTnI and, thus, identifies whether the TnI observed is a re-expression of skeletal muscle TnI isoforms or cardiac TnI cleavage products. Skeletal muscle was obtained from rat extensor digitorum longus muscle (expressing fast skeletal TnI) and rat soleus muscle (expressing slow skeletal TnI). Thus, the anti-4H6 antibody allowed us to determine that the N-terminal cTnI products were cardiac cleavage products and not skeletal isoforms.
Morphology and Immunohistochemistry—Seven days after in vivo gene transfer and after functional evaluation in vivo, rats were sacrificed, and hearts were excised, rinsed in phosphate-buffered saline, and weighed. Cross-sections (5–6 μm) of Cyro-O.C.T. compound (Tissue-Tek)-embedded LV were transferred to glass slides (2) and stained with hematoxylin and eosin or fixed and immunostained with anti-CD45 antibody (1:50; BD Pharmingen clone OX-1) followed by visualization with polyclonal biotin conjugated anti-mouse IgG and Vectastain elite ABC reagent together with the DAB detection system. Rat liver served as a positive control. Sections were also fixed and immunostained with anti-RII antibody (1:500 dilution; Upstate) followed by the secondary Alexa 568 (red) antibody (1:500 dilution; Molecular Probes) or immunostained with fluorescein-conjugated (green) anti-HA antibody (1:500 dilution; Roche clone 12CA5), which recognizes the HA epitope-tagged Ht31 and Ht31P.
Hearts infected with adenovirus and probed with secondary antibody (negative control) or hearts not infected with adenovirus and probed with either the fluorescein-conjugated or the secondary Alexa 568 antibodies (negative control) showed background fluorescence only (data not shown). Images were obtained using a TE 2000 Nikon inverted microscope equipped with a 10×, 40×, 60× objective, fluorescent filters, and a Diagnostic Instruments, Inc. SPOT camera (model 7.0.1).
Statistical Analysis—All data are expressed as the means ± S.E. Statistical significance was determined using unpaired Student's t test to evaluate the null hypothesis. Analysis was also performed using two-way repeated measures analysis of variance to compare means of multiple groups. A value of p < 0.05 was considered statistically significant.
RESULTS
Cardiac Morphology and Immunohistochemistry—No significant differences in heart weight-to-body weight ratio (Table 1) or LV mass (Table 2) were observed between hearts expressing Ht31 and control hearts. After gene transfer, there was no evidence of an adverse inflammatory response (infiltration of inflammatory cells, indicated by a lack of anti-CD45 staining) (Fig. 1). Positive staining for anti-CD45 was verified using rat liver (Fig. 1, panel C). There was also no evidence of histological abnormalities (e.g. myocyte hypertrophy or myofibrillar disarray) in experimental hearts (Ht31) versus controls (Fig. 1, panels D–G). The thickness of the intraventricular septum and LV posterior wall was not significantly different (Table 1).
TABLE 1.
Characteristics of Sprague-Dawley rats The echocardiographic parameters interventricular septum (IVS) thickness and posterior wall (PW) thickness were measured under base-line conditions in the absence of isoproterenol stimulation. NS, not significant.
| Control (n = 8) | Ht31 (n = 8) | p valuea | |
|---|---|---|---|
| mean ± S.E. | mean ± S.E. | ||
| Body weight (g) | 359.4 ± 9.1 | 344.4 ± 9.9 | NS |
| Heart weight (mg) | 852.5 ± 71.2 | 866.3 ± 37.1 | NS |
| Heart weight/body weight (mg/g × 100) | 236.0 ± 15.6 | 251.7 ± 9.0 | NS |
| IVS-diastole (mm) | 0.19 ± 0.01 | 0.20 ± 0.01 | NS |
| IVS-systole (mm) | 0.29 ± 0.02 | 0.30 ± 0.02 | NS |
| PW-diastole (mm) | 0.19 ± 0.01 | 0.18 ± 0.02 | NS |
| PW-systole (mm) | 0.30 ± 0.01 | 0.27 ± 0.02 | NS |
Compared by Student's t test
TABLE 2.
In vivo cardiac function of Ht31-expressing hearts versus controls Controls represent both gene transfer of Ht31P (inactive proline-substituted derivative of Ht31) and eGFP. dp/dtmax, maximum pressure derivative; dp/dtmin, minimum pressure derivative; ESP, end-systolic pressure; EDV, end-diastolic volume; ESV, end-systolic volume; tau, time constant of left ventricular isovolumetric relaxation. The dose of isoproterenol was 1.0 mg/kg of body weight/min × 2 min. NS, not significant.
| Control (n = 16) (base-line) | Control (n = 14) (isoproterenol) | p value | |
|---|---|---|---|
| mean ± S.E. | mean ± S.E. | ||
| LV mass (g) | 1.10 ± 0.07 | 1.16 ± 0.07 | NS |
| LV dp/dt (max) (mm Hg/s) | 4491 ± 427 | 8363 ± 467 | <0.05 |
| LV dp/dt (min) (mm Hg/s) | –4384 ± 457 | –5129 ± 475 | NS |
| LV EDP (mm Hg) | 11.02 ± 1.44 | 11.90 ± 1.00 | NS |
| LV ESP (mm Hg) | 83.39 ± 4.86 | 76.30 ± 4.93 | NS |
| LV EDV (ml) | 0.42 ± 0.04 | 0.43 ± 0.04 | NS |
| LV ESV (ml) | 0.20 ± 0.02 | 0.10 ± 0.02 | <0.05 |
| tau (ms) | 12.50 ± 0.71 | 8.71 ± 0.52 | <0.05 |
| Ht31 (n = 14) (base-line) | Ht31 (n = 5) (isoproterenol) | p value | |
| mean ± S.E. | mean ± S.E. | ||
| LV mass (g) | 1.20 ± 0.12 | 1.01 ± 0.16 | NS |
| LV dp/dt (max) (mm Hg/s) | 3069 ± 328 | 7713 ± 662 | <0.05 |
| LV dp/dt (min) (mm Hg/s) | –2787 ± 341 | –4128 ± 295 | <0.05 |
| LV EDP (mm Hg) | 5.12 ± 1.46 | 11.11 ± 2.36 | <0.05 |
| LV ESP (mm Hg) | 76.45 ± 4.64 | 68.73 ± 4.38 | NS |
| LV EDV (ml) | 0.48 ± 0.04 | 0.46 ± 0.03 | NS |
| LV ESV (ml) | 0.24 ± 0.03 | 0.06 ± 0.01 | <0.05 |
| tau (ms) | 14.56 ± 0.80 | 8.31 ± 0.71 | <0.05 |
FIGURE 1.
Morphology and immunohistochemistry of hearts expressing Ht31 and Ht31P controls. Immunohistochemistry shows no evidence of an adverse inflammatory response in Ht31 (A) or Ht31P controls (B), as indicated by lack of anti-CD45 staining. Positive staining for anti-CD45 was verified using rat liver, as indicated by brown labeling of cell membranes (C). Hematoxylin and eosin-stained sections of excised hearts 7 days after gene transfer of Ht31 (D and E) versus Ht31P control (F and G) show no evidence of histological abnormalities in response to gene transfer. Panels E and G (scale bar = 10 μm) are high magnification of panels D and F (scale bar = 100 μm).
Ht31 and Ht31P Expression in Rat Heart in Vivo—Immunostained cryostat sections of LV (Fig. 2; panels A–D) show expression of HA-Ht31 and HA-Ht31P. Ht31 or Ht31P expression is indicated by green immunolabeling of anti-HA (Fig. 2; panels A, A′, B, and B′). AKAP-Lbc, the parent protein of Ht31 and Ht31P, as well as other AKAPs expressed in the heart are localized to specific and unique subcellular locations (3). In contrast, Ht31 and Ht31P are short soluble 24 amino acid peptides lacking targeting domains and are, thus, free to diffuse throughout the cytoplasm as indicated in Fig. 2, panels panels A, A′, B, and B′.
FIGURE 2.
Ht31 expression disrupts RII localization in rat heart in vivo. Fluorescent microscopy of cryostat sections shows the effect of disruption of PKA/AKAP targeting by Ht31 in rat hearts in vivo. Panels A and B shows expression of the anti-HA epitope-labeled Ht31P peptide (Ad-HA-Ht31P) control (A and A′) and the Ht31 peptide (Ad-HA-Ht31) (B and B′) using fluorescein-conjugated (green) anti-HA antibody. Panels D and D′, Ht31 expression results in a significantly decreased cross-striation pattern of RII labeling as compared with Ht31P control (C and C′) (yellow arrows). Red, anti-RII; green, anti-HA-Ht31 or anti-HA-Ht31P control peptide. Panels A–D, scale bar = 30 μm. Panels A′–D′, 3-fold magnification of images in white boxed region in panels A–D.
We previously showed co-localization of RII and mAKAP (previously called AKAP100) and co-localization of mAKAP andα-actinin in rat cardiac myocytes, as indicated by periodic cross-striated staining (2). In the current study, 7 days after expression of the control peptide, Ht31P, in vivo RII immunostaining was observed as periodic cross-striated staining (yellow arrows, Fig. 2, panels C and C′), demonstrating that binding of RII to endogenous AKAPs is not altered by introduction of the control Ht31P peptide by gene transfer in vivo. In contrast, 7 days post-infection with the active, RII binding peptide, Ht31, the striated pattern of RII was lost, indicating that Ht31 competes for RII bound to endogenous AKAPs in the myocardium (Fig. 2; panels D and D′). These results show that Ht31 and Ht31P can be successfully expressed in LV of rat hearts in vivo by adenoviral gene transfer and that Ht31 (but not Ht31P) expression disrupts RII localization.
Cardiac Function in Hearts Expressing Ht31—Cardiac function was assessed 7 days after in vivo gene transfer (Figs. 3 and 4). Echocardiographic and hemodynamic measurements were obtained at base line and after dose-dependent increases in isoproterenol infusion. At base line, disruption of PKA/AKAP interaction by Ht31 resulted in a significant decrease in dP/dtmax and dP/dtmin but also a significant decrease in LV EDP. LV EDP is frequently elevated under conditions of impaired cardiac function (28); thus, decreased LV EDP, as observed in our study, is indicative of increased efficiency of relaxation. There was no difference in end-systolic pressure (Fig. 4), LVEF, or stroke volume (SV) (Fig. 3) in Ht31-treated hearts versus controls. Taken together, we conclude that cardiac function at base line is not impaired after Ht31 expression.
FIGURE 3.
Cardiac function in hearts expressing Ht31. A, % LVEF at base line (0.0) and with increasing doses of infused isoproterenol in Ht31 and control hearts. B, LV SV at baseline (0.0) and with increasing doses of infused isoproterenol in hearts expressing Ht31 versus controls. Measurements were recorded 2 min after the start of isoproterenol infusion. Data are expressed as the mean ± S.E.; #, p < 0.05 (Ht31 versus control), as determined by two-way repeated measures analysis of variance; *, p < 0.05 Ht31 versus control.
FIGURE 4.
Hemodynamics in hearts expressing Ht31. Shown are LV pressure and LV dP/dt (A and C), control (B and D), Ht31-expressing hearts (A and B), and base-line conditions (C and D) after maximal isoproterenol stimulation. Pressure-volume loop recordings are shown in panels E–H; Ht31P control-expressing hearts are shown at base line (E) and upon maximal isoproterenol stimulation (F); Ht31 expressing hearts are shown in base line (G) and upon isoproterenol stimulation (H). The bar graphs show LV dP/dtmax (I), LV dP/dtmin (J), LV EDP (K), and LV end-systolic pressure (L). Measurements were recorded 2 min after the start of maximal isoproterenol infusion. Data are expressed as mean ± S.E.; *, p < 0.05 Ht31 versus control.
Upon infusion of isoproterenol, differences in dP/dtmax or dP/dtmin as well as EDP between Ht31- and Ht31P-expressing hearts were no longer observed; however, LVEF (at 0.5 and 1.0 μg/kg of body weight/min isoproterenol infusion) and SV (at 0.5 μg/kg of body weight/min of isoproterenol infusion) were significantly greater in Ht31-expressing hearts versus controls (Fig. 3; panels A and B), indicative of increased pumping ability. This is also illustrated in the wider pressure-volume loop in Fig. 4, panel F, versus panel H.
PKA Protein Phosphorylation— When expressed in rat hearts in vivo, Ht31 competes for PKA with endogenous AKAPs, such as mAKAP, AKAP15/18 (Fig. 2). Therefore, we predicted that PKA protein phosphorylation in Ht31-expressing hearts would be decreased versus controls. We performed Western blot analysis of LV homogenates of rat hearts expressing Ht31 or Ht31P immunoblotted with antibodies that recognize PKA-specific sites on cTnI, RyR2, and PLB and normalized the amount of PKA-phosphorylated protein to the total amount of the same protein (cTnI, RyR2 or PLB) on the blot.
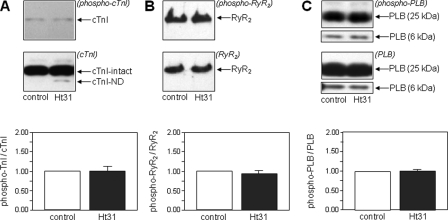
At base line there was no significant difference in PKA phosphorylation of cTnI, PLB, or RyR2 between Ht31 and control hearts (Fig. 5). In contrast, maximal isoproterenol stimulation of Ht31-expressing hearts was associated with significantly decreased PKA phosphorylation of cTnI, RyR2, and PLB versus isoproterenol-stimulated Ht31P controls (Fig. 6) (Western blot analysis of PKA phosphorylation at base line was performed at a different time than Western blot analysis of isoproterenol-stimulated hearts, accounting for different intensities of the bands in Figs. 5 and 6). Therefore, increased LVEF and SV during isoproterenol stimulation in Ht31-expressing hearts versus controls occurs under conditions of decreased PKA-dependent protein phosphorylation.
FIGURE 5.
Phosphorylation of PKA substrates under base-line conditions. PKA-dependent phosphorylation of cTnI (A), RyR2 (B), and PLB (C) in hearts expressing Ht31 versus controls at base line (unstimulated conditions). For each PKA substrate, the upper panel shows a Western blot with anti-phosphoprotein antibody, and the lower panel shows a Western blot with antibody to total protein with the latter antibody recognizing both phosphorylated and unphosphorylated species. The bar graphs show the ratio of phosphorylated to total protein. Data are expressed as mean ± S.E.; Ht31 versus control was not significantly different.
FIGURE 6.
Phosphorylation of PKA substrates after β-adrenergic stimulation. PKA-dependent phosphorylation after isoproterenol stimulation of cTnI (A), RyR2 (B), and PLB (C) in hearts expressing Ht31 versus controls. For each PKA substrate, the upper panel shows a Western blot with anti-phosphoprotein antibody, and the lower panel shows the Western blot with antibody to total protein, with the antibody recognizing both phosphorylated and unphosphorylated species. The bar graphs show the ratio of phosphorylated to total protein. Data are expressed as the mean ± S.E.; *, p < 0.05 Ht31 versus control.
Proteolytic Cleavage of cTnI—We previously showed that transgenic expression of cTnI-ND in mice results in improved cardiac function (7). N-terminal truncation of cTnI to produce cTnI-ND is observed in hearts under physiological conditions and in response to increased cardiac stress (6, 7). In vitro studies have also shown that N-terminal truncation of cTnI occurs to a greater extent when PKA phosphorylation is reduced (8, 12, 29, 30).
Because N-terminal truncation of cTnI occurs under physiological conditions (7), we predicted that we would observe N-terminal truncation in cardiac myocytes isolated from normal rat hearts. We, thus, investigated whether N-terminal truncation of cTnI is greater in isolated adult rat myocytes when PKA phosphorylation of cTnI is reduced. We compared the extent of cTnI truncation in isolated adult rat cardiac myocytes with and without β-adrenergic stimulation. As expected, there was significantly more PKA phosphorylation of N-terminal serines of cTnI in rat cardiac myocytes stimulated for 10 min by 10 μm forskolin plus 100 μm IBMX versus unstimulated myocytes (Fig. 7, panels C and D). Fig. 7, panels B and E, confirm that N-terminal truncation of cTnI occurs in isolated myocytes but also show that the amount of cTnI cleavage (cTnI-ND/intact cTnI) is significantly less (4 ± 1%) in cells stimulated with forskolin plus IBMX than in unstimulated myocytes (15 ± 2%) (p < 0.001). Both the recombinant cTnI-ND (Fig. 7, panel B, rm-TnI-ND) and truncated cTnI products from rat hearts in vivo (Fig. 7, panel B, adult rat myocyte) were identified using the anti-cTnI antibody but not with the antibody to phosphorylated N-terminal serines 23 and 24 (Fig. 7, panel C). Therefore, we can conclude that the truncated cTnI in these cells lacks the N terminus, containing the PKA sites.
FIGURE 7.
PKA phosphorylation and proteolytic cleavage of cTnI in adult rat cardiomyocytes. A, SDS-PAGE of non-stimulated and forskolin (forsk) + IBMX-stimulated isolated adult rat cardiomyocytes. Western blot analysis using anti-total-cTnI antibody (B) and anti-phospho-cTnI antibody (C) in non-stimulated myocytes and in myocytes stimulated with forskolin + IBMX. Also shown is the cloned recombinant mouse cTnI-ND (rm-TnI-ND), serving as a Mr marker for expression of the cTnI-ND cleavage product (see Fig. 8). Shown is quantification of PKA-dependent cTnI phosphorylation (D) and cTnI cleavage in forskolin + IBMX-stimulated (E) versus non-stimulated controls (n = 4 myocyte preps). Data are expressed as the mean ± S.E.; *, p < 0.001 forskolin + IBMX-stimulated myocytes versus non-stimulated control by unpaired t tests.
Our results show that in intact myocytes, when PKA phosphorylation of N-terminal serines of cTnI is increased, there is decreased N-terminal truncation of cTnI. Conversely, decreased PKA phosphorylation of cTnI should result in increased N-terminal truncation. We can test this idea in Ht31 versus Ht31P-expressing rat hearts, as we already have shown after isoproterenol stimulation a significant decrease in PKA phosphorylation of cTnI, RyR2, and PLB when PKA targeting to endogenous AKAPs is abrogated by Ht31 expression (Fig. 6, panel A). We next determined whether N-terminal cleavage of cTnI occurs in hearts of rats expressing Ht31 and Ht31P and, if so, whether N-terminal cleavage of cTnI is greater in Ht31-expressing hearts versus controls.
Fig. 8 shows Western blots using a TnI-1 monoclonal antibody (31) which recognizes the C-terminal epitope (amino acids 193–211) of cTnI (molecular mass, 24 kDa). A Western blot using the TnI-1 anti-cTnI C terminus antibody shows the cTnI cleavage products together with TnI in control homogenates from rat heart, rat skeletal muscle (expressing slow skeletal and fast skeletal TnI), and recombinant mouse cTnI-ND29–211 protein (Mr 21.5 kDa) (Fig. 8, panel B). Fig. 8C shows a Western blot using the cardiac specific 4H6 antibody,3 which recognizes rat cTnI isoforms including cTnI proteolytic products but not skeletal muscle TnI. Cardiac TnI and lower Mr bands are recognized by TnI-1 and 4H6 antibodies, indicating that we are observing N-terminal cleavage of cTnI. These results eliminate the possibility that the lower Mr bands in Ht31 or Ht31P expressing rat hearts are re-expressed skeletal muscle TnI isoforms (32).
FIGURE 8.
Proteolytic cleavage of cTnI in hearts expressing Ht31. A, SDS-PAGE (top) and Western blot analysis (bottom) of LV homogenates from Ht31 and Ht31P expressing hearts 7 days after in vivo gene transfer. Also shown are non-infected mouse heart control and cloned recombinant mouse cTnI-ND protein (rm-cTnI-ND) control. The Western blot was probed using the anti-cTnI C terminus monoclonal antibody TnI-1. The cloned recombinant cTnI-ND serves as a molecular weight marker for expression of these cleavage products. B, Western blot analysis using the TnI-1 monoclonal antibody of LV homogenate from a Ht31-expressing heart and controls isolated from a rat extensor digitorum longus (EDL) muscle (expressing fast skeletal TnI (fs-TnI)), rat soleus muscle (expressing slow skeletal TnI (ss-TnI)) and recombinant mouse cTnI-ND. Cloned recombinant mouse cTnI-ND protein serves as a protein marker for the cTnI cleavage products identified by the TnI-1 monoclonal antibody. C, Western blots analysis using the cardiac-specific 4H6 monoclonal antibody of LV homogenates from a Ht31 expressing heart and a rat skeletal extensor digitorum longus muscle. D, quantification of increased cleavage of Ht31 (n = 4) versus control adenoviral (n = 4)-expressing hearts. Data are expressed as the mean ± S.E.; *, p < 0.05 Ht31 versus control as determined by unpaired t test analysis.
Based upon recognition of these cTnI cleavage products by the TnI-1 monoclonal antibody, the apparent molecular weights of these cleavage products on SDS gels, differing mobility from fast and slow skeletal TnI (Fig. 8, panel B), and previous studies (7, 31), we conclude that the lower Mr bands are truncated cTnI lacking the N-terminal amino acid extension.
Previous studies have shown that calpain-dependent cleavage may produce more than one N-terminal cleavage product of cTnI (6, 7), representing different calpain-dependent cleavage sites of cTnI (se Fig. 1 in Ref. 7). All of these truncated forms of cTnI share similar properties, they lack the cardiac-specific N-terminal domain containing PKA phosphorylation sites but retain domains containing the core inhibitory function of cTnI (i.e. regions of cTnI homologous to skeletal muscle TnI). Thus, the primary binding sites for other thin filament proteins are preserved. Consistent with these previous findings, we also observe two bands of N-terminal-truncated cTnI in rat hearts expressing Ht31 or Ht31P. Based upon the previous work, we predict that these represent two different cleavage products.
Consistent with prior observations that N-terminal cTnI truncation can occur under physiological conditions (6) and with our findings in isolated rat cardiac myocytes (this study) (Fig. 7), we observe N-terminal-truncated cTnI in both Ht31- and Ht31P-expressing rat hearts. Furthermore, consistent with results in Fig. 7, we observe significantly more N-terminal-truncated cTnI in hearts expressing Ht31 versus Ht31P. (Fig. 8, panels A and D).
To summarize, the increased LVEF and SV observed upon stimulation of Ht31-expressing versus Ht31P-expressing hearts with high dose isoproterenol is accompanied by decreased PKA phosphorylation of cTnI, PLB, and RyR2 and increased truncation of the N terminus of cTnI.
DISCUSSION
We have been able to disrupt subcellular targeting of AKAP-targeted PKA by in vivo gene transfer of a competing RII-binding peptide, Ht31. We elicited redistribution of PKA away from its substrates and inhibition of phosphorylation of PKA substrates. Thus, we have shown that in vivo, maximal phosphorylation of PKA substrates upon β-adrenergic stimulation requires a local pool of PKA in close proximity to its protein substrates.
Each of the proteins showing decreased PKA phosphorylation after Ht31 expression in vivo is likely regulated by a local pool of AKAP-tethered PKA plus other complementary signaling molecules. For example, PKA, PDE4D3, and PP2A are targeted to RyR2 by mAKAP (2, 33–35); PKA associates with PLB via AKAP15/18δ (36). AKAP targeting of PKA to myofilaments is less well delineated. We have shown that the intermediate filament protein, synemin, localized at the Z-discs, is an AKAP and may participate in regulation of PKA-dependent myofilament protein phosphorylation (37). PKA also reportedly binds TnT, positioning PKA in an ideal location for cTnI phosphorylation (38). Our results in Ht31-expressing rat hearts indicate that the binding of PKA by Ht31 displaces PKA from binding sites on these (and other) AKAPs, decreasing the local pool of PKA near PKA substrates, resulting in decreased substrate phosphorylation.
Another major finding in this study is that we can elicit a robust inotropic response to β-adrenergic stimulation in rat hearts in vivo under conditions (Ht31 expression) where PKA phosphorylation of RyR2, PLB, and cTnI is reduced compared with isoproterenol-stimulated controls. This represents the first evidence to our knowledge of an increased inotropic response in vivo under conditions of decreased PKA substrate phosphorylation.
We explored the mechanism behind this observation. We already know from previous in vitro studies that proteolytic cleavage of the N terminus of cTnI occurs to a greater extent when PKA phosphorylation of serines 23 and 24 of cTnI is decreased (8). We have now shown that upon activation of the β-adrenergic pathway in isolated adult myocytes, significantly less truncated cTnI is produced versus unstimulated myocytes. From this observation, we conclude that the extent of N-terminal proteolysis of cTnI is regulated by PKA activity. In other words, in addition to eliciting an increase in PKA-dependent protein phosphorylation, β-adrenergic stimulation can affect protein function by regulating extent of proteolysis.
Our observations of a reciprocal dependence of N-terminal cTnI truncation on PKA phosphorylation is most likely representative of a more general post-translational mechanism. Indeed, regulation of proteolysis by phosphorylation has been reported for several proteins in addition to cTnI; for example, proteolytic cleavage of microtubule-associated protein, tau, and cleavage of filamin are decreased when each of these substrates is phosphorylated by PKA (39–41). Very recently, a similar observation has been obtained by Sadayappan and Robbins for myosin-binding protein C.4
Consistent with our observations of decreased N-terminal cleavage of cTnI in cardiac myocytes upon increased PKA phosphorylation of cTnI, we observed decreased N-terminal cleavage of cTnI in rat hearts in vivo expressing Ht31 versus isoproterenol-stimulated rat heart controls. Taken together with our results from isolated myocytes, we conclude that the decrease in cTnI proteolysis in rat hearts in vivo expressing Ht31 versus controls results from differences in PKA phosphorylation of cTnI.
As discussed previously, expression of N-terminal-truncated cTnI in transgenic mice results in increased myocardial relaxation, decreased EDP, and increased LV filling (7). Overexpression of cTnI-ND in the Gsα-deficient mouse model of heart failure helps restore cardiac function (14). Consistent with structural studies of PKA phosphorylated cTnI (9, 10, 11), several of the functional changes observed in these mice indicate that N-terminal-truncated cTnI mimics the PKA-phosphorylated protein, for example, demonstrating a rightward shift in the force/Ca2+ curve versus intact cTnI.
In our prior study of transgenic mice overexpressing cTnI-ND (7), even expression of relatively low levels of cTnI-ND/total cTnI resulted in beneficial effects on cardiac contractility, including a significant decrease in EDP (also observed at base line in this study), mimicking functional effect of PKA phosphorylation of N-terminal serines. Similarly in the current study the significant increase in cTnI expression in Ht31 (Fig. 8)- versus Ht31P-expressing hearts is also likely to mimic effects of PKA phosphorylation of the N-terminal serines. Given the fact that increased expression of cTnI-ND partially restores function in Gsα-deficient mice (14), we conclude that the increased expression of N-terminal-truncated cTnI in Ht31-expressing rat hearts contributes to restoration of cardiac function upon isoproterenol stimulation versus Ht31P controls.
In summary, disruption of PKA/AKAP interactions in the heart in vivo displaces PKA from endogenous AKAPs, decreases PKA dependent substrate phosphorylation, increases cardiac contractility upon β-adrenergic stimulation, and results in an increase in N-terminal truncation of cTnI.
Therefore, under conditions of decreased PKA-dependent protein phosphorylation, such as occurs during heart failure (27, 42), N-terminal proteolysis of cTnI may represent a beneficial post-translational mechanism to compensate for decreased PKA phosphorylation and reduced inotropic response to β-adrenergic stimulation.
Acknowledgments
We thank Stacey McCulle for research support and Shawn Robinson (Dept. of Medicine, Division of Cardiology, University of Maryland School of Medicine) and Joseph Mauban and Sabrina Manni (Dept. of Physiology, University of Maryland School of Medicine) for careful review of this manuscript. We also thank Stephen Chong for research support.
This work was supported, in whole or in part, by National Institutes of Health Grants HL085487 (to B. M.), HL56256 and AG16613 (to M. B.), HL074400 and HL76491 (to M. P.), and HL078773 (to J.-P. Jin). This work was also supported by National Aeronautics and Space Administration Grant NNA04CK26G (to J.-P. Jin) and by the American Heart Association, mid-Atlantic affiliate, Beginning Grant-in-aid 0465470U (to B. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PKA, protein kinase A; AKAP, A-kinase-anchoring protein; TnI, troponin I; cTnI, cardiac TnI; HA, hemagglutinin; EDP, decreased end diastolic pressure; LV, left ventricular; LVEF, LV ejection fraction; eGFP, enhanced green fluorescence protein; PLB, phospholamban; RyR2, ryanodine receptor; SV, stroke volume; RII, type II; IBMX, isobutylmethylxanthine.
J.-P. Jin, unpublished results.
S. Sadayappan and J. Robbins, personal communication.
References
- 1.Colledge, M., and Scott, J. D. (1999) Trends Cell Biol. 9 216-221 [DOI] [PubMed] [Google Scholar]
- 2.Yang, J., Drazba, J. A., Ferguson, D. G., and Bond, M. (1998) J. Cell Biol. 142 511-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruehr, M. L., Russell, M. A., and Bond, M. (2004) J. Mol. Cell. Cardiol. 37 653-665 [DOI] [PubMed] [Google Scholar]
- 4.Carr, D. W., Stofko-Hahn, R. E., Fraser, I. D. C., Bishop, S. M., Acott, T. S., Brennan, R. G., and Scott, J. D. (1991) J. Biol. Chem. 266 14188-14192 [PubMed] [Google Scholar]
- 5.Kentish, J. C., McCloskey, D. T., Layland, J., Palmer, S., Leiden, J. M., Martin, A. F., and Solaro, R. J. (2001) Circ. Res. 88 1059-1065 [DOI] [PubMed] [Google Scholar]
- 6.Yu, Z. B., Zhang, L. F., and Jin, J.-P. (2001) J. Biol. Chem. 276 15753-15760 [DOI] [PubMed] [Google Scholar]
- 7.Barbato, J. C., Huang, Q. Q., Hossain, M. M., Bond, M., and Jin, J.-P. (2005) J. Biol. Chem. 280 6602-6609 [DOI] [PubMed] [Google Scholar]
- 8.Di Lisa, F., De Tullio, R., Salamino, F., Barbato, R., Melloni, E., Siliprandi, N., Schiaffino, S., and Pontremoli, S. (1995) Biochem. J. 308 57-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, W.-J., Chandra, M., Xing, J., Solaro, R. J., and Cheung, H. C. (1997) Biochemistry 36 6745-6753 [DOI] [PubMed] [Google Scholar]
- 10.Dong, W.-J., Chandra, M., Xing, J., She, M., Solaro, J., and Cheung, H. C. (1997) Biochemistry 36 6754-6761 [DOI] [PubMed] [Google Scholar]
- 11.Heller, W. T., Finley, N. L., Dong, W.-J., Timmins, P., Cheung, H. C., Rosevar, P. R., and Trewhella, J. (2003) Biochemistry 42 7790-7800 [DOI] [PubMed] [Google Scholar]
- 12.Feng, J., Schaus, B. J., Fallavollita, J. A., Lee, T. C., and Canty, J. M., Jr. (2001) Circulation 103 2035-2037 [DOI] [PubMed] [Google Scholar]
- 13.Barta, J., Tóth, A., Jaquet, K., Redlich, A., Édes, I., and Papp, Z. (2003) Mol. Cell. Biochem. 251 83-88 [PubMed] [Google Scholar]
- 14.Feng, H., Chen, M., Weinstein, L. S., and Jin, J.-P. (2008) J. Biol. Chem. 283 33384-33393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klussmann, E., Maric, K., Wiesner, B., Beyermann, M., and Rosenthal, W. (1999) J. Biol. Chem. 274 4934-4938 [DOI] [PubMed] [Google Scholar]
- 16.Potet, F., Scott, J. D., Mohammad-Panah, R., Escande, D., and Baro, I. (2001) Am. J. Physiol. 280 H2038-H2045 [DOI] [PubMed] [Google Scholar]
- 17.Shih, M., Lin, F., Scott, J. D., Wang, H. Y., and Malbon, C. C. (1999) J. Biol. Chem. 274 1588-1595 [DOI] [PubMed] [Google Scholar]
- 18.Kockskamper, J., Sendhoff, K., Erlenkamp, S., Bordusa, F., Cerovsky, V., and Glitsch, H. G. (2001) Pflüegers Arch. Eur. J. Physiol. 441 807-815 [DOI] [PubMed] [Google Scholar]
- 19.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509-2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsui, T., Tao, J., del Monte, F., Lee, K. H., Li, L., Picard, M., Force, T. L., Franke, T. F., Hajjar, R. J., and Rosenzweig, A. (2001) Circulation 104 330-335 [DOI] [PubMed] [Google Scholar]
- 21.del Monte, F., and Hajjar, R. J. (2003) Methods Mol. Biol. 219 179-193 [DOI] [PubMed] [Google Scholar]
- 22.Askari, A., Unzek, S., Goldman, C. K., Ellis, S. G., Thomas, J. D., DiCorleto, P. E., Topol, E. J., and Penn, M. S. (2004) J. Am. Coll. Cardiol. 43 1908-1914 [DOI] [PubMed] [Google Scholar]
- 23.Popovic, Z. B., Richards, K. E., Greenberg, N. L., Rovner, A., Drinko, J., Cheng, Y., Penn, M. S., Fukamachi, K., Mal, N., Levine, B. D., Garcia, M. J., and Thomas, J. D. (2006) Am. J. Physiol. 291 H762-H769 [DOI] [PubMed] [Google Scholar]
- 24.McConnell, B. K., Jones, K. A., Fatkin, D., Arroyo, L. H., Lee, R. T., Aristizabal, O., Turnbull, D. H., Georgakopoulos, D., Kass, D., Bond, M., Niimura, H., Schoen, F. J., Conner, D., Fischman, D. A., Seidman, C. E., and Seidman, J. G. (1999) J. Clin. Investig. 104 1235-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConnell, B. K., Moravec, C. S., Morano, I., and Bond, M. (1997) Am. J. Physiol. 273 H1440-H1451 [DOI] [PubMed] [Google Scholar]
- 26.McConnell, B. K., Moravec, C. S., and Bond, M. (1998) Am. J. Physiol. 274 H385-H396 [DOI] [PubMed] [Google Scholar]
- 27.Zakhary, D. R., Moravec, C. S., and Bond, M. (2000) Circulation 101 1459-1464 [DOI] [PubMed] [Google Scholar]
- 28.Gaasch, W. H., Zile, M. R., Hoshino, P. K., Weinberg, E. O., Rhodes, D. R., and Apstein, C. S. (1990) Circulation 81 1644-1653 [DOI] [PubMed] [Google Scholar]
- 29.Murphy, A. M., Kogler, H., Georgakopoulos, D., McDonough, J. L., Kass, D. A., Van Eyk, J. E., and Marban, E. (2000) Science 287 488-491 [DOI] [PubMed] [Google Scholar]
- 30.McDonough, J. L., Arrell, D. K., and Van Eyk, J. E. (1999) Circ. Res. 84 9-20 [DOI] [PubMed] [Google Scholar]
- 31.Jin, J. P., Yang, F. W., Yu, Z. B., Ruse, C. I., Bond, M., and Chen, A. (2001) Biochemistry 40 2623-2631 [DOI] [PubMed] [Google Scholar]
- 32.Thijssen, V. L., Ausma, J., Gorza, L., van der Velden, H. M., Allessie, M. A., Van Gelder, I. C., Borgers, M., and van Eys, G. J. (2004) Circulation. 110 770-775 [DOI] [PubMed] [Google Scholar]
- 33.Dodge-Kafka, K. L., Soughayer, J., Pare, G. C., Carlisle Michel, J. J., Langeberg, L. K., Kapiloff, M. S., and Scott, J. D. (2005) Nature 437 574-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapiloff, M. S., Jackson, N., and Airhart, N. (2001) J. Cell Sci. 114 3167-3176 [DOI] [PubMed] [Google Scholar]
- 35.Lehnart, S. E., Wehrens, X. H., Reiken, S., Warrier, S., Belevych, A. E., Harvey, R. D., Richter, W., Jin, S. L., Conti, M., and Marks, A. R. (2005) Cell 123 25-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lygren, B., Carlson, C. R., Santamaria, K., Lissandron, V., McSorley, T., Litzenberg, J., Lorenz, D., Wiesner, B., Rosenthal, W., Zaccolo, M., Taskén, K., and Klussmann, E. (2007) EMBO Rep. 8 1061-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, M. A., Lund, L. M., Haber, R., McKeegan, K., Cianciola, N., and Bond, M. (2006) Arch. Biochem. Biophys. 456 204-215 [DOI] [PubMed] [Google Scholar]
- 38.Sumandea, C. A., GarcÃa-CazarÃn, M. L., Bozio, C. H., Balke, C. W., Solaro, R. J., and Sumandea, M. P. (2007) Circulation 116 II_189 Abstract 957
- 39.Elvira, M., Díez, J. A., Wang, K. K., and Villalobo, A. (1993) J. Biol. Chem. 268 14294-14300 [PubMed] [Google Scholar]
- 40.Litersky, J. M., and Johnson, G. V. (1992) J. Biol. Chem. 267 1563-1568 [PubMed] [Google Scholar]
- 41.Goll, D. E., Thompson, V. F., Li, H., Wei, W., and Cong, J. (2003) Physiol. Rev. 83 731-801 [DOI] [PubMed] [Google Scholar]
- 42.Zakhary, D. R., Moravec, C. S., Stewart, R. W., and Bond, M. (1999) Circulation 99 505-510 [DOI] [PubMed] [Google Scholar]