Abstract
Detection of foreign RNA by the innate immune system can trigger the induction of type I interferon (IFN) and apoptosis. Important antiviral defense pathways that result in type I IFN production following the recognition of foreign double-stranded RNA (dsRNA) include the RIG-I family helicases and IPS-1 adaptor cytosolic pathway and the Toll-like receptor 3 and TIR domain-containing adaptor-inducing IFN-β (TRIF) adaptor membrane-associated pathway, both of which activate IFN regulatory factor 3 (IRF3). In addition to triggering an innate immune response, dsRNAs are widely used to mediate gene-selective silencing in mammalian cells by the RNA interference pathway. We investigated the ability of short interfering RNAs, including T7 phage polymerase-synthesized RNA (PRNA), which like some viral RNAs contains a 5′-triphosphate, to selectively silence gene expression and to cause induction of IFN-β and apoptosis. We found that PRNA-mediated gene silencing and associated nonspecific pro-apoptotic and IFN-inducing effects were dependent on the cell line and RNA length. Double-stranded PRNAs 50 nucleotides long as well as polyinosinic-polycytidylic acid activated the RNA-dependent protein kinase (PKR) and induced significant levels of IFN-β and apoptosis, whereas shorter PRNAs and chemically synthesized dsRNAs did not. Effector caspase activation and apoptosis following RNA transfection was enhanced by pretreatment with IFN, and removal of the 5′-phosphate from PRNAs decreased induction of both IFN-β and apoptosis. PKR, in addition to IPS-1 and IRF3 but not TRIF, was required for maximal type I IFN-β induction and the induction of apoptosis by both transfected PRNAs and polyinosinic-polycytidylic acid.
The mammalian innate immune system recognizes and responds to a diverse array of pathogen-associated molecular patterns, one of which is foreign RNA (1–6). Characterization of viral RNAs that activate the innate immune response identified the retinoic acid-inducible protein (RIG-I)2-like receptor (RLR) and Toll-like receptor (TLR) signaling networks that sense and respond to different viruses and RNAs in a selective manner (6–12). The RLR- and TLR-mediated innate immune responses include activation of the type I interferon (IFN) system leading to the production of proteins with antiviral activities as well as the activation of pathways leading to apoptosis (3, 6). Although much has been learned about the biochemical mechanism of IFN production following viral infection (11, 12), the possible interplay between the IFN induction and apoptosis induction processes in response to foreign RNA including relatively short RNAs used for RNA interference (RNAi)-mediated gene silencing (13, 14) is unclear.
RLR and TLR nucleic acid sensors localize differently within cells and recognize specific characteristics of foreign RNA (4, 6, 11, 12, 15). The TLRs localize to the host cell surface or within endosomes (2, 12). TLR3 recognizes double-stranded RNA (dsRNA) and certain long single-stranded RNAs (ssRNAs) and uses the Toll/IL-1 receptor (TIR) domain-containing adaptor-inducing IFN-β (TRIF) protein to mediate downstream signaling (16, 17). TLR7 and 8 detect ssRNAs in a sequence-dependent manner and signal through the adaptor protein myeloid differentiation factor-88 (4, 6, 12). Following RNA ligand recognition, these TLR proteins mediate activation of type I IFN production (12). The RLR class of RNA sensors are cytoplasmic sensors that selectively recognize specific RNAs (7, 8, 11). For example, RIG-I detects rhabdoviridae, flaviviridae, orthomyxoviridae, and paramyxoviridae RNAs and RNAs possessing a 5′-triphosphate, whereas melanoma differentiation-associated gene 5 (MDA5) RLR responds to picornaviridae and synthetic polyinosinic-polycytidylic acid (pIpC) RNAs (7, 8, 10, 18–20). Recently, the RLRs have been shown to exhibit size-dependent activity, with RIG-I activated by shorter dsRNA and MDA5 by longer dsRNA molecules including pIpC (21). Both RIG-I and MDA5 possess caspase activation and recruitment domains. Following RNA-mediated activation, they signal through the IFN-β promoter stimulator-1 (IPS-1, also known as MAVS/VISA/Cardif) adaptor localized on the outer mitochondrial membrane (9, 22–24) to activate the NF-κB and IFN regulatory factor 3 (IRF3) factors that transcriptionally induce type I IFN production (11). An additional cytoplasmic RNA sensor, the RNA-activated protein kinase (PKR), also binds dsRNA as well as structured ssRNAs that then mediate autophosphorylation and activation of PKR (3, 25, 26). The best characterized substrate of activated PKR remains eukaryotic translation initiation factor eIF-2α (27), which when phosphorylated on serine 51 subsequently inhibits cellular translation initiation and plays an important role in apoptosis (3, 28–30).
Although long dsRNA molecules are well established and potent molecular triggers for the modulation of gene expression and function and the innate immune response (1, 3, 31), within both plants and animals dsRNA also mediates the RNAi process that permits sequence selective suppression of gene expression (32–34). Either short dsRNA molecules or plasmids that express short dsRNAs as hairpins with homology to a target messenger RNA can selectively silence expression of a targeted gene when introduced into mammalian cells by lipid-based transfection (32–35). Although RNAi represents a powerful functional genomics tool and potential therapeutic strategy including for viral infections, nonspecific effects have been described even when short dsRNAs were used to achieve RNAi (13, 14, 36). Such nonspecific responses have the potential to cause misinterpretation of silencing data (13). An important reason that short (∼20 bp) dsRNAs are used to mediate RNAi in mammalian cells is to minimize the activation of the IFN system (3, 33, 35). However, experimental evidence directly linking IFN system components to the nonspecific effects sometimes observed with RNAi technology is limited.
Our initial observation of significant nonspecific gene silencing caused by T7 phage polymerase-synthesized siRNAs (PRNAs) in both human (HeLa) and mouse (MEF) cells led us to investigate the potential causes of this effect. We examined PRNA-mediated induction of type I IFN-β and apoptotic responses and the relationship between these processes, together with the specific PRNA characteristics required to induce these effects. We found that PRNAs with sufficient structure to activate PKR, including 50-bp and 50-nucleotide (nt) PRNAs, induced IFN-β and triggered apoptosis, whereas shorter PRNAs and chemically synthesized RNAs did not. The 5′-triphosphate of PRNAs, although required for maximal induction of IFN-β and apoptosis, was not required for PKR activation. Using transient and stable knockdown strategies, we established that in addition to IPS-1 and IRF3, PKR is a key mediator of both type I IFN induction and the induction of apoptosis by both polymerase-synthesized RNAs and pIpC. These results implicate PKR as an important component of the cytosolic sensor-mediated response to dsRNA.
EXPERIMENTAL PROCEDURES
Cell Maintenance, Transfection, and Dual Luciferase Assay— HeLa (PKR+), HeLa stably knocked down for PKR expression (PKRkd), MEF, and Chinese hamster ovary (CHO) cells were cultured as previously described (30, 37). For luciferase construct transfection, cells were seeded into 24-well plates and transfected with 190 ng of pGL3 firefly (Promega) and 10 ng of phRL Renilla (Promega) luciferase plasmid complexed with Lipofectamine 2000 (Invitrogen) in Opti-MEM I (Invitrogen) per well. Following transfection with the reporter constructs, transfection complexes with 100 ng of RNA (CsiLUC, PLUC, or pIpC) and Lipofectamine 2000 in Opti-MEM I were added as indicated. The cells were harvested after 24 h for dual luciferase assay as described by the manufacturer (Promega) using a Victor3 V plate reader model 1420 with injectors (PerkinElmer Life Sciences). The values were normalized to four vector control wells (no silencing RNA) distributed throughout each plate (38).
RNA Preparation and Transfection—The chemically synthesized siRNA targeting firefly luciferase (35), CsiLUC20 with the sequence ACUUACGCUGAGUACUUCGA and a UU 3′ overhang, was purchased from Dharmacon. The negative control siRNA, CsiNEG1, was purchased from Ambion. T7 RNA polymerase synthesized siRNAs were prepared using the Silencer® siRNA construction kit (Ambion) according to the manufacturer's protocol. Single-stranded RNAs and dsRNA were not treated with RNase T1 unless otherwise noted as treated (+T1). For phosphatase treatment, 2 units CIP (New England Biolabs) was used per μg RNA; incubation was at 37 °C for 1 h. Gel purification of siRNAs synthesized with T7 polymerase was as described by Ambion. The sequence of the PsiLUC20 was the same as the chemically synthesized CsiLUC20 20-bp siRNA; the sense sequence of the T7 transcribed 50-bp PsiLUC50 was UUACAGAUGCACAUAUCGAGGUGGACAUCACUUACGCUGAGUACUUCGAA.
IFN-β Expression—Expression of IFN-β was measured by real time PCR. The cells were seeded in 24-well plates and transfected with 280 ng (560 ng for stable or transient knockdown experiments) of RNA complexed with Lipofectamine 2000 in Opti-MEM I. RNA was purified at 5 h using an RNeasy mini kit (Qiagen) following the manufacturer's protocol. Random-primed reverse transcription was carried out using 500 ng of RNA and SuperScript II (Invitrogen) according to the manufacturer's protocol. The following primer pairs were used: glyceraldehyde-3-phosphate dehydrogenase, forward primer GCCTTCCGTGTCCCCACTG and reverse primer CGCCTGCTTCACCACCTTC; IFN-β, forward primer ACGCCGCATTGACCATCTATG and reverse primer CGGAGGTAACCTGTAAGTCTGT. Real time PCR analyses were performed in duplicate or triplicate with each reverse transcription product, using IQ SYBR Green Supermix (Bio-Rad) and a Bio-Rad MyIQ real time real time PCR machine (3-min hot start followed by 30 s at 95 °C, 45 s at 58 °C, and 45 s at 72 °C, repeated 40 times). IFN-β values were normalized to glyceraldehyde-3-phosphate dehydrogenase values.
Caspase 3/7 Activation Assays—Effector caspase activation was measured using the Caspase-Glo 3/7 assay (Promega) following the manufacturer's recommendations. The cells in 96-well plates were analyzed 8 h following RNA transfection as described above using a Victor3 V plate reader (PerkinElmer Life Sciences) to measure luciferase reporter activity as described (30).
IPS-1, TRIF, IRF3, and PKR Knockdown—The target sequences of the CsiRNAs prepared by Dharmacon with dTdT overhangs for human cell studies were: IPS-1, UAGUUGAUCUCGCGGACGA; TRIF, GACCAGACGCCACUCCAAC; IRF3, CCGCAAAGAAGGGUUGCGU; and PKR, GCAGGGAGUAGUACUUAAAUA. For transient knockdown experiments, HeLa cells in 60-mm dishes at ∼60% confluency were transfected with 50 nm of siRNA with Lipofectamine 2000 on days 1 and 3. On day 2 the cells were reseeded in 60-mm dishes for the second siRNA transfection, and on day 4 the cells were seeded in 24-well plates for RNA transfection on day 5.
Western Blot Analysis—Whole cell extracts were prepared with extract buffer containing protease inhibitors (37); for eIF-2α and PKR phosphorylation, 50 nm NaF and 2 nm Na2VO3 were also present (30). Protein fractionation and immunoblot analysis was as described (30, 37) using the following primary antibodies: PKR (Santa Crux Biotechnology); IRF3 (Santa Cruz Biotechnology); MAVS/VISA/IPS-1 (Bethyl Laboratories); eIF-2α (Cell Signaling Technology); phospho-eIF-2α Ser51 (Cell Signaling Technology); phospho-PKR Thr446 (Santa Cruz Biotechnology), PARP (BD Pharmingen); TRIF (Alexis Biochemicals); IκB-α (Santa Cruz Biotechnology); and α-tubulin (Sigma). The blots were visualized using a Li-Cor Odyssey infrared imager system (Li-Cor Biosciences).
Immunofluorescence—Cells grown on coverslips in 12-well plates were transfected with 560 ng of RNA complexed with Lipofectamine 2000, fixed at the indicated time following transfection with Accustain 10% formalin solution (Sigma) for 1 h at room temperature. Following PBS washes, the cells were blocked and permeabilized with 3% bovine serum albumin containing 0.1% Triton X-100 in PBS for 1 h at room temperature, washed with PBS, and incubated with IRF3 primary antibody (generously provided by M. David, University of California, San Diego) at 4 °C overnight. The cells were then washed and incubated with Alexa 488-conjugated anti-rabbit IgG (Invitrogen) in 3% bovine serum albumin in PBS for 1 h with Prolong Gold antifade reagent with 4′,6′-diamino-2-phenylindole (Invitrogen). The images were captured using an Olympus IX71 microscope with Q-Capture PRO software (QImaging).
Electrophoretic Mobility Shift Assay—HeLa cells, PKR+ or PKRkd, were transfected as indicated and then harvested after 4 h. Nuclear extracts were prepared as previously described (39). Aliquots of 5 μg of nuclear extracts were incubated in 20 μl of total reaction volume containing 10 mm Tris-HCl, 1 mm dithiothreitol, 1 mm EDTA, 10% glycerol, 0.1% Triton X-100, 1 μg of poly(dI-dC), 0.1 mg/ml bovine serum albumin, and 50 mm KCl for 5 min. The 32P-labeled oligonucleotide probe with the sense strand sequence AGTTGAGGCGACTTTCCCAGGC was added to the reaction mixture and then incubated for 20 min. The reaction products were analyzed by electrophoresis in a 5% polyacrylamide gel with 0.5× TBE buffer. The gels were dried and analyzed by autoradiography.
RESULTS
Polymerase-synthesized siRNAs Induce Nonspecific Gene Silencing—Because long dsRNAs are known to induce IFN signaling and activate PKR (1, 3, 11, 13), we tested the effect on gene silencing selectivity of increasing the length of an siRNA from the commonly utilized 20 bp to 50 bp. A T7 RNA polymerase-based strategy was used to synthesize RNAs including a 20-bp siRNA (PsiLUC20) possessing the same nucleotide sequence as a chemically synthesized 20-bp siRNA (CsiLUC20) and also a 50-bp siRNA (PsiLUC50) that included the 20-bp siRNA sequence along with additional target sequence. The siRNAs targeted the message of a firefly luciferase reporter, whereas Renilla luciferase was used as an internal measure of nonspecific gene silencing.
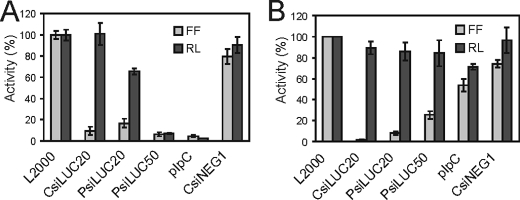
The CsiLUC20 targeting firefly luciferase showed high selectivity and no significant nonspecific gene silencing when tested at 36 nm (100 ng) of siRNA in human HeLa cells (Fig. 1A), hamster CHO cells (Fig. 1B), and mouse (MEF) cells (data not shown). A negative control siRNA, CsiNEG1, did not display any detectable gene-specific silencing and like CsiLUC20 only showed nonspecific gene silencing at very high concentrations, 180 nm (data not shown). In contrast to the selectivity seen with 100 ng of CsiLUC20, surprisingly high nonspecific gene silencing was caused by the same amount (100 ng) of PsiRNA when tested in either HeLa cells (Fig. 1A) or MEFs (data not shown). Furthermore, the 50-bp PsiLUC50 siRNA caused even greater nonspecific gene silencing in these cells than the 20-bp PsiLUC20 siRNA, comparable with pIpC. CHO cells, a rodent cell line where selective RNAi can be mediated by long dsRNA (40), showed sequence specific silencing for all of the siRNAs tested including PsiLUC50 (Fig. 1B).
FIGURE 1.
Nonspecific gene silencing caused by transfection of PRNAs. A, HeLa; B, CHO. Cells were transfected with reporter plasmids expressing firefly (FF) and Renilla (RL) luciferase and either chemically synthesized (C) siRNA, T7 polymerase-synthesized (P) siRNA 20 or 50 or pIpC. Normalized firefly and Renilla luciferase values are shown along with the standard deviation determined from three to four independent experiments.
Polymerase-synthesized Single and Double-stranded RNAs Induce IFN-β—To further explore the cause of nonspecific gene silencing by T7 RNA polymerase-synthesized RNAs (PRNAs), ssRNAs in addition to the PsiLUC dsRNAs were synthesized. The structural integrity of the PRNAs was verified by denaturing gel electrophoresis. For the dsRNAs that were not treated with T1 RNase prior to gel analysis to remove the 5′-terminal single-stranded sequence and triphosphate moiety, one major RNA population was observed. T1 nuclease treatment, while creating a population of shorter PsiLUC20 and PsiLUC50 RNAs, typically appeared incomplete, especially for PsiLUC50 (data not shown).
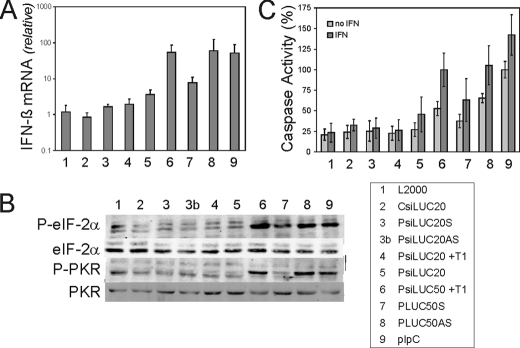
Because PsiRNAs, but not chemically synthesized CsiRNAs, caused pronounced nonspecific gene silencing in HeLa cells (Fig. 1A), and because dsRNA is a known inducer of type I IFN, we assessed the ability of these transfected RNAs to induce IFN-β mRNA production. The IFN-β transcript level seen following transfection with the PLUC20 sense (S) or antisense (AS) ssRNAs or the PsiLUC20 dsRNA remained low and comparable with that of the liposome (L2000) vehicle alone (Fig. 2A). However, the PLUC50AS ssRNAs and PsiLUC50 dsRNA induced high levels of IFN-β mRNA production when transfected, similar to that observed with pIpC, whereas the PLUC50S ssRNA was a weak inducer of IFN-β (Fig. 2A). By contrast, the PsiLUC50 and pIpC RNAs added directly to cells without transfection reagent were inactive (data not shown).
FIGURE 2.
Interferon induction, PKR, and eIF-2α phosphorylation, and caspase activity in transfected cells. HeLa cells were transfected with CsiRNA; PRNA sense (S) or antisense (AS) ssRNA 20- or 50-nt; or PsiRNA dsRNA 20- or 50-bp treated with T1 (+T1) or not; or pIpC. A, real time PCR measurement of human IFN-β induction relative to glyceraldehyde-3-phosphate dehydrogenase following RNA transfection. Total RNA was isolated at 5 h post-transfection. The means and standard deviations determined from three independent experiments are shown. B, Western blot analysis showing the phosphorylation of PKR and eIF-2α in HeLa cells at 8 h following transfection. C, caspase 3/7 activity at 8 h post-transfection, with and without IFN pretreatment for 20 h. The results are normalized to control pIpC transfection as 100%; the mean caspase 3/7 activity and standard deviations were determined from three independent experiments.
Activation of eIF-2α Phosphorylation and Global Translation Attenuation by PRNAs—As shown in Fig. 2B, transfection of HeLa cells with the 50-nt PLUC50AS ssRNA or T1-treated PsiLUC50 50-bp dsRNA led to an increase in the phosphorylation of eIF-2α similar to that seen with pIpC. By contrast, transfection of the PLUC50S ssRNA resulted in only a slight increase in eIF-2α phosphorylation, and the shorter 20-nt PLUC20 and 20-bp PsiLUC20 RNAs did not display any detectable effect. The phosphorylation of eIF-2α at residue Ser51 is one of the best characterized mechanisms of translational control in animal cells (41). Among the multiple eIF-2α kinases including PKR, HRI, PERK, and GCN2 known in eukaryotes, the PKR kinase from human cells is activated by dsRNA by a mechanism that involves autophosphorylation at threonine 446 (3). When Thr446 phosphorylation was measured, we observed a similar pattern for activation of PKR as was seen for eIF-2α phosphorylation (Fig. 2B). As an indicator of the effect of RNA transfection on overall cellular protein synthesis activity, we examined global translational activity by pulse labeling with 35S-labeled amino acids and measuring hot trichloroacetic acid insoluble incorporation of radioactivity. The PsiLUC50 RNA caused significant inhibition of global translation at 6 h after transfection, comparable with that seen with pIpC, whereas the shorter PsiLUC20 RNA and the chemically synthesized CsiLUC20 and CsiNEG1 siRNAs only slightly reduced activity from that of L2000 lipid alone (data not shown).
Induction of Apoptosis by PRNAs—Because of the nonspecific inhibition of both the Renilla luciferase reporter and overall cellular protein synthesis seen in cells transfected with 50-bp PRNA, we considered the possibility that the decrease in translation might be associated with an induction of apoptosis. As a measure of the ability of the transfected RNAs to induce apoptosis, we first examined effector caspase 3 and 7 (3/7) activation. Transfection of HeLa cells with PLUC50AS ssRNA, PsiLUC50 dsRNA, or pIpC caused the activation of caspases 3/7 (Fig. 2C). By contrast, neither the shorter 20-nt ss or the 20-bp ds PRNAs nor CsiLUC20 significantly affected caspase activity in transfected HeLa cells. Pretreatment of cells with IFN-α enhanced effector caspase activity in cells subsequently transfected with 50-nt PRNAs, suggesting the involvement of proapoptotic IFN system components. The RNA-dependent activation of poly(ADP-ribose) polymerase (PARP) cleavage, an independent indicator of apoptosis, correlated with the activation of effector caspase activity by RNA (data not shown).
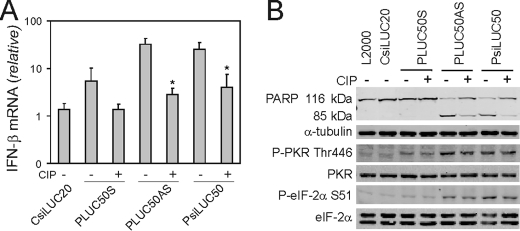
PRNA 5′-Phosphate Moiety Contributes to Induction of IFN-β and Activation of Apoptosis—To assess the role of 5′-terminal phosphate moieties in the induction of IFN-β and the apoptotic response in HeLa cells, the inducing capacities of PRNAs were analyzed with and without calf intestinal phosphatase (CIP) treatment. Transfection of the PLUC50S RNA induced low levels of IFN-β, which decreased following CIP treatment of the RNA (Fig. 3A) and did not result in significant levels of PARP cleavage even without CIP treatment (Fig. 3B, top panel). CIP treatment of PLUC50AS and PsiLUC50 decreased the IFN-β mRNA inducing capacity of these RNAs (Fig. 3A). Likewise, CIP treatment of the 50-nt PLUC50AS and 50-bp PsiLUC50 PRNAs reduced apoptosis as measured by PARP cleavage (Fig. 3B, top panel). By contrast, CIP treatment of these PRNAs did not affect either eIF-2α or PKR phosphorylation (Fig. 3B, bottom panel). These results suggest that the 5′-triphosphate moiety of transfected RNA enhances both the induction of IFN-β and the activation of the apoptotic response and that PKR activation is insufficient to maximally activate these effects.
FIGURE 3.
5′-Phosphate contributes to IFN-β induction and apoptosis caused by PRNAs. Cells were transfected with 50-nt PLUC50S, AS, or 50-bp PsiLUC50 PRNAs or CsiLUC20. A, effect of CIP phosphatase treatment compared with buffer alone on IFN-β induction at 5 h as measured by real time PCR. The mean and standard deviations from three independent experiments are shown. *, p < 0.05. B, Western blot analysis showing PARP cleavage at 24 h and phosphorylation of PKR and eIF-2α at 8 h following transfection of CIP-treated or untreated RNAs.
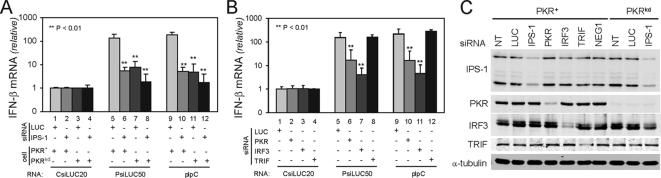
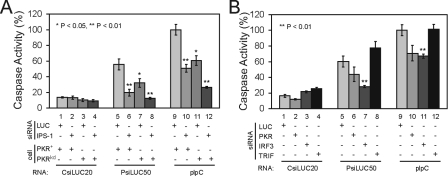
PKR in Addition to IPS-1 and IRF3 but Not TRIF Is a Mediator of the Induction of IFN-β—In an attempt to identify mediators of IFN-β induction by PRNAs, we first measured IFN-β induction in PKR-sufficient parental HeLa cells (PKR+) compared with PKR-deficient HeLa cells (PKRkd), which stably express a short hairpin RNA that reduces PKR protein levels to <5% of parental cells (30). These HeLa cells were transfected with either a chemically synthesized LUC siRNA as a control or a chemically synthesized siRNA targeting the IPS-1 adaptor protein, prior to subsequent transfection with the CsiLUC, PRNAs, or pIpC RNAs. The PKR+ cells showed ∼100-fold increase in IFN-β RNA after transfection with PsiLUC50 or pIpC compared with CsiLUC20 (Fig. 4A, lanes 5 and 9). The PKRkd cells showed a significant reduction in RNA-mediated IFN-β induction compared with PKR+ HeLa cells (Fig. 4A, lanes 7 and 11). IPS-1 knockdown in PKR-sufficient parental cells (lanes 6 and 10) significantly reduced RNA-mediated IFN-β induction as anticipated (11). The IPS-1 siRNA further decreased the already low IFN-β production seen in the stably deficient PKRkd cells (lanes 8 and 12).
FIGURE 4.
Mediators of IFN-β induction by transfected PsiRNA and pIpC. A, IFN-β induction following the knockdown of IPS-1 in parental HeLa cells (PKR+) and PKR stable knockdown HeLa cells (PKRkd). B, IFN-β induction in parental HeLa cells treated with CsiRNAs to transiently knockdown PKR, IRF3, and TRIF. The cells were transfected with CsiLUC20, PsiLUC50, or pIpC and IFN-β transcript levels were determined by real time PCR at 5 h post-transfection. For A and B, the mean inductions of IFN-β mRNA and standard deviations from four independent experiments are shown. **, p < 0.01. C, representative Western blot showing the knockdown of IPS-1, PKR, IRF3, and TRIF in parental and PKRkd HeLa cells.
We then assessed IFN-β induction in the parental HeLa cells in which either PKR, IRF3, or TRIF had been transiently knocked down. IRF3 knockdown reduced IFN-β induction over 30-fold (Fig. 4B, lanes 7 and 11) to the similar low level seen following IPS-1 knockdown (Fig. 4A). By contrast, TRIF knockdown (Fig. 4B, lanes 8 and 12) had no effect on IFN-β induction mediated by PsiLUC50 or pIpC transfection. This was anticipated because the RNA without transfection reagent did not induce IFN-β, suggesting that cytoplasmic delivery was required for induction. Furthermore, transient knockdown of PKR gave the same phenotype as stable knockdown of PKR; HeLa cells deficient in PKR protein either stably (Fig. 4A, lanes 7 and 11) or transiently (Fig. 4B, lanes 6 and 10) showed a >90% decrease in IFN-β induction following PsiLUC50 or pIpC transfection. The same effects were seen with the PLUC50AS (data not shown). Finally, the siRNA-mediated knockdown of the IPS-1, PKR, IRF3, and TRIF proteins in parental PKR+ and PKRkd HeLa cells was verified by Western immunoblot analysis (Fig. 4C); the transient protein knockdowns were both significant and selective with each of the siRNAs.
PKR, IPS-1, and IRF3 but Not TRIF Are Mediators of the Induction of Apoptosis by PRNAs—To gain further insight into the RNA sensor pathways activated by PRNAs, we next examined the effect of knockdown of the IPS-1, TRIF, IRF3, and PKR proteins on effector caspase activity. As was seen for IFN-β induction (Fig. 4A), the PKR-deficient PKRkd cells showed a greatly reduced caspase 3/7 activity compared with parental PKR-sufficient parental cells (Fig. 5A, lanes 7 and 11). Knockdown of IPS-1 in PKR+ parental cells reduced caspase activation to a similar low level as the PKRkd cells (Fig. 5A, lanes 6 and 10), whereas knockdown of IPS-1 in the stable PKRkd cells caused an even further reduction, which was most striking following pIpC transfection (Fig. 5A, lanes 8 and 12). In addition, PKR knockdown and IRF3 knockdown both reduced caspase 3/7 activity for the 50-bp PsiLUC50 pRNAs and pIpC, whereas the TRIF knockdown had no effect (Fig. 5B). Although the low level of PKR remaining after transient knockdown varied somewhat between several independent experiments, quantitation revealed that the lower the level of residual PKR protein, the lower the corresponding caspase activation that was observed following RNA transfection (data not shown).
FIGURE 5.
Mediators of apoptosis induction by PRNAs and pIpC. A, caspase 3/7 activity measured at 8 h after transfection in parental PKR+ and stable PKRkd HeLa cells pretreated with IPS-1 siRNA or control LUC siRNA prior to transfection with the indicated RNA. B, caspase 3/7 activity at 8 h in parental HeLa cells treated with either PKR, IRF3, or TRIF siRNA prior to RNA transfection. The results are normalized to control pIpC transfection as 100%; the mean caspase 3/7 activities and standard deviations from three to four independent experiments is shown. *, p < 0.05; **, p < 0.01.
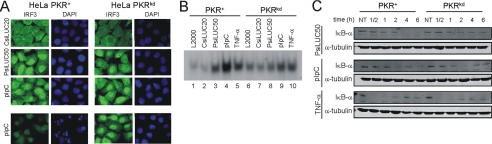
PKR Is Required for Maximal NF-κB Activation but Not for IRF3 Nuclear Localization Following dsRNA Transfection—As one approach to assess whether PKR contributes to the induction of IFN-β and apoptosis through the same pathway as IPS-1 mediated activation of IRF3, we used immunofluorescence to monitor IRF3 nuclear localization following transfection of either PKR+ or PKRkd cells with dsRNA. Although chemically synthesized CsiLUC20 dsRNA did not lead to IRF3 nuclear localization in either PKR+ or PKRkd cells, transfection with either PsiLUC50 dsRNA or pIpC caused an enhanced nuclear localization of IRF3 in both PKR-sufficient and -deficient cells (Fig. 6A). In the parental PKR+ cells, significant nuclear fragmentation and apoptosis was seen by 8 h, whereas this was minimal in the PKRkd cells (Fig. 6A, bottom panel). The similarity in IRF3 nuclear localization observed between PKR+ and PKRkd cells suggests that PKR enhances proapoptotic responses by affecting a step other than IRF3 nuclear localization.
FIGURE 6.
PKR is required for maximal NF-κB activation but not IRF3 nuclear localization. A, immunofluorescence images showing IRF3 (green) and 4′,6′-diamino-2-phenylindole (DAPI, blue) staining following RNA transfection with CsiLUC20, PsiLUC50 or pIpC. The cells were fixed at 4 h (top three rows) and 8 h (bottom row) post-transfection. B, electrophoretic mobility shift analysis using nuclear extracts from treated HeLa cells, parental PKR+, or stable PKRkd, incubated with a NF-κB oligonucleotide probe. C, Western blot analysis of IκB-α and phosphorylated eIF-2α using PsiLUC50, pIpC, or TNF-α transfected parental HeLa and PKRkd cells. Extracts were prepared at the indicated time following treatment. Tubulin was used as a loading control.
In addition to IRF-binding sites in the IFN-β enhancer, there is an NF-κB site present in positive regulatory domain II (42, 43), and dsRNA is known to activate NF-κB signaling in human cells (44). To test whether PKR is important for NF-κB activation following PRNA and pIpC transfection, we prepared nuclear extracts from PKR+ and PKRkd cells at 4 h post-transfection and carried out electrophoretic mobility shift analysis using a NF-κB oligonucleotide probe. Either lipid alone or transfected chemically synthesized siRNA gave a low level of NF-κB band shift with extract from either cell type (Fig. 6B, lanes 1, 2, 6, and 7), whereas PsiLUC50 and pIpC transfection induced significant complex formation with extract from the parental PKR+ cells (lanes 3 and 4). However, the intensity of the NF-κB band shift complex seen with extracts prepared from PKRkd cells (lanes 8 and 9) compared with untransfected cells was lower than PKR+ cells (lanes 3 and 4) following PsiLUC50 and pIpC transfection, suggesting that PKR is required for maximal NF-κB activation following transfection with these RNAs. As an independent measure of NF-κB activation (45), the level of the IκB-α inhibitor of NF-κB was measured. IκB-α protein levels decreased in PKR+ cells to nearly undetectable levels by 4 h after transfection with PsiLUC50 and pIpC, whereas in PKRkd cells substantial levels of IκB-α remained even at 6 h after transfection (Fig. 6C). By contrast, TNF-α treatment gave a similar NF-κB gel shift response in both PKR+ and PKRkd cells (Fig. 6B, lanes 5 and 10), and IκB-α levels were comparable in PKR+ and PKRkd cells following TNF-α treatment (Fig. 6C, bottom panel), suggesting PKR is not an essential mediator of NF-κB activation in the TNF-α pathway as previously shown (30).
DISCUSSION
PKR is firmly established to play a central role in the antiviral actions of IFN through phosphorylation of eIF-2α, a modification that alters the translational pattern in cells and leads to an inhibition of protein synthesis in virus-infected, IFN-treated cells (3, 27). The results presented herein also implicate PKR as a key recognition receptor of foreign RNA that enhances a TLR-independent signal transduction response through the cytosolic RNA sensor pathway leading to increased IFN-β synthesis. Our initial observations of nonspecific gene silencing using siRNAs that possess a 5′-triphosphate synthesized with T7 phage polymerase but not chemically synthesized siRNAs that lack a 5′-triphosphate led us to examine the activation of PKR, the production of IFN-β and the induction of apoptosis by these RNAs compared with pIpC. In HeLa cells we found that 50-nt-long RNAs with sufficient structure to activate PKR induced IFN-β and apoptosis when the RNAs were synthesized enzymatically. We also found that optimal induction of both IFN-β and apoptosis were dependent upon the presence of 5′-terminal phosphate moieties, because CIP phosphatase treatment uniformly reduced IFN-β induction, effector caspase activation, and PARP cleavage, but not PKR activation. Shorter PRNAs and chemically synthesized 20-nt dsRNA did not significantly increase either IFN-β message levels or apoptosis in transfected cells.
What is the relationship of PKR as an RNA sensor relative to the well established roles of the RLR and TLR3, 7, and 8 sensor proteins? Importantly, our studies with cells deficient in PKR, including both transient knockdown and stable knockdown of the kinase in HeLa cells, established that PKR is a necessary component required to achieve both optimal RNA-mediated induction of IFN-β and maximal activation of apoptosis in cells transfected with structured RNAs containing a 5′-phosphate. Knockdown of IPS-1 and IRF3 likewise demonstrated the importance of these proteins for the induction of IFN-β in RNA-transfected cells, which is consistent with current understanding of the RLR signaling pathway activated by dsRNA (11) but inconsistent with IPS-1-independent signaling as would occur via TLR7 and 8 (12). Furthermore, knockdown of TRIF, the adaptor protein required for TLR3-induced IFN production, had little effect on IFN-β induction by transfected RNAs. Several studies implicate RIG-I as the RLR cytoplasmic sensor that detects 5′-triphosphate containing RNAs and MDA5 as the sensor for long pIpC dsRNA, and both RIG-I and MDA5 are known to signal through IPS-1, leading to the production of type I IFNs (7–9, 21–24). The fact that a similar reduction in induction of IFN-β and effector caspases was seen in our studies following IRF3 knockdown as IPS-1 knockdown is consistent with the known activation of IRF3 by RNA-mediated signaling via RLR and the IPS-1 adaptor (11).
Among our most striking results are those obtained with the human HeLa cells stably knocked down for PKR (30). Not only was there less activation of apoptosis in the PKR-deficient cells, there also was a significant reduction in IFN-β induction. This phenotype was not due to a clonal variation of the stable PKRkd cells, because a similar PKR dependence of IFN-β production was observed in cell populations transiently knocked down for PKR by CsiRNA. Furthermore, we found that the level of residual PKR protein in knockdown experiments correlated well with the level of effector caspase induction seen following RNA transfection. PKR and IPS-1 conceivably could exert their effects through either the same or different signaling pathways. When the stable PKR knockdown cells were transiently knocked down for IPS-1, we found a further reduction in both IFN-β and apoptosis induction, suggesting that these proteins may act in part through different signal transduction pathways. The possibility of distinct pathways for PKR and IPS-1 is further supported by our observations that IRF3 nuclear localization following RNA transfection was similar in parental PKR and PKR knockdown cells. Furthermore, we found that PKR was necessary for maximal NF-κB activation following transfection of activating RNAs, consistent with the notion that PKR may act as a signaling sensor in addition to its role as an eIF-2α kinase inhibiting translation.
Our results are consistent with and extend the findings of Kim et al. (46), who originally identified the 5′-triphosphate of phage polymerase-synthesized RNAs as an inducer of IFN, suggesting that activation of the IFN system contributes to the nonspecific gene silencing in RNAi studies. We found RNA size dependence of the 5′-terminal phosphate a requirement, as neither single-stranded nor double-stranded RNA of 20-nt induced IFN-β, whereas a 50-bp PRNA did. Likewise, pIpC, a long dsRNA without a 5′-triphosphate, also induced IFN-β as expected. Recent reports show that both the 5′-triphosphate moiety and the A/U composition of ssRNA are determinants of their ability to function as RIG-I ligands (18, 19, 21). Interestingly, one of the 50-nt single-stranded PRNAs was a potent inducer of IFN-β, whereas the other was not, and CIP treatment of this PRNA, while reducing induction of both IFN-β and apoptosis, did not affect PKR or eIF-2α phosphorylation as has been shown for other RNAs (47). However, it has been known for nearly 20 years from studies with the reovirus single-stranded and dsRNAs and pIpC that a 5′-triphosphate is neither necessary nor sufficient for PKR activation (48). Although the single-stranded PRNAs used herein were gel-purified, we cannot exclude the possibility of small amounts of contaminating double-stranded RNA from phage polymerase-synthesized RNA preparations, even following gel purification, which may be able to activate PKR (49) and possibly other cytoplasmic RNA sensors. In addition to inducing IFN-β, activating apoptosis, and inhibiting total cellular translation, the structured 50-nt and 50-bp PRNAs activated PKR and resulted in eIF-2α phosphorylation, whereas the 20-nt and 20-bp PRNAs did not. These findings are consistent with the notion that PKR activation and its subsequent phosphorylation of eIF-2α are important contributors to the nonspecific silencing effects mediated by PRNAs in HeLa cells.
Our data suggest that transfected PRNAs, with sufficient length and structure to activate PKR, induce apoptosis in a manner dependent upon PKR, IPS-1, and IRF3 proteins and independent of TLR3 signaling via TRIF. PRNA and pIpC induced IFN-β induction also was dependent upon PKR, IPS-1, and IRF3, but TRIF-independent. IPS-1 and IRF3 are known mediators of IFN-β production in response to RIG-I and MDA5 activation (6, 11). Recently they have also been implicated as important mediators of apoptosis, for example IPS-1 when activated by reovirus infection (50) and IRF3 when activated by Sendai virus (51, 52) or vaccinia virus ΔE3L mutant (53) infection. The significant role of PKR as an RNA sensor leading to IFN-β induction, although initially surprising, is consistent with the implication of PKR involvement in IFN production in bone marrow dendritic cells following transfection of pIpC (54). PKR also is implicated in the activation of NF-κB-mediated signal transduction (29, 55) and as an interacting partner with TRAF (56, 57). Because PKR was required for maximal NF-κB activation following PRNA and pIpC transfection as measured by both electrophoretic mobility shift assay with a NF-κB probe and by Western blotting for IκB-α and because the IFN-β promoter contains NF-κB as well as IRF-binding sites (43), it is reasonable to assume that NF-κB activation by PKR in response to the PRNAs accounts at least in part for the enhanced IFN-β production seen.
We observed that phosphatase treatment of PRNAs reduced both IFN-β induction as expected (46) and also caspase 3/7 activity but importantly did not affect either PKR autophosphorylation or eIF-2α phosphorylation. These differences, together with the IPS-1 dependence of the PKR-dependent signaling responses, suggest that although PKR is needed for maximal induction of IFN-β and apoptosis, PKR catalytic activity alone is not sufficient to fully activate these responses without a concomitant activation of IPS-1. PKR may act in an alternative manner in response to transfected RNA, perhaps as an adaptor or integral structural component of an IPS-1 activated complex that selectively affects downstream effects, which would be consistent with our observation that PKR knockdown did not affect the nuclear localization of IRF3 but did affect NF-κB signaling following PRNA transfection. Finally, the requirement of PKR, IPS-1, and IRF3 for maximal apoptosis induction both by polymerase-synthesized RNAs and by pIpC suggests that the apoptotic pathway, like IFN-β induction, is a fundamental antiviral response of mammalian cells to cytoplasmic RNAs with 5′-terminal phosphates and significant RNA secondary or double-stranded character.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-12520 and AI-20611. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RIG-I, retinoic acid-inducible protein; RLR, RIG-I-like receptor; PKR, RNA-activated protein kinase; IFN, interferon; ds, double-stranded; ss, single-stranded; IRF, IFN regulatory factor; PRNA, T7 phage polymerase-synthesized RNA; pIpC, polyinosinic-polycytidylic acid; TRIF, Toll/IL-1 receptor domain-containing adaptor-inducing IFN-β; TLR, Toll-like receptor; RNAi, RNA interference; MDA, melanoma differentiation-associated gene; eIF, eukaryotic translation initiation factor; MEF, mouse embryonic fibroblast; nt, nucleotide; CHO, Chinese hamster ovary; PBS, phosphate-buffered saline; PARP, poly(ADP-ribose) polymerase; CIP, calf intestinal phosphatase.
References
- 1.Lampson, G. P., Tytell, A. A., Field, A. K., Nemes, M. M., and Hilleman, M. R. (1967) Proc. Natl. Acad. Sci. U. S. A. 58 782-789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499-511 [DOI] [PubMed] [Google Scholar]
- 3.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14 778-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sioud, M. (2006) Trends Mol. Med. 12 167-176 [DOI] [PubMed] [Google Scholar]
- 5.Honda, K., Takaoka, A., and Taniguchi, T. (2006) Immunity 25 349-360 [DOI] [PubMed] [Google Scholar]
- 6.Meylan, E., and Tschopp, J. (2006) Mol. Cell 22 561-569 [DOI] [PubMed] [Google Scholar]
- 7.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K.-K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994-997 [DOI] [PubMed] [Google Scholar]
- 8.Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., and Reis e Sousa, C. (2006) Science 314 997-1001 [DOI] [PubMed] [Google Scholar]
- 9.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981-988 [DOI] [PubMed] [Google Scholar]
- 10.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C.-S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101-105 [DOI] [PubMed] [Google Scholar]
- 11.Yoneyama, M., and Fujita, T. (2007) J. Biol. Chem. 282 15315-15318 [DOI] [PubMed] [Google Scholar]
- 12.Uematsu, S., and Akira, S. (2007) J. Biol. Chem. 282 15319-15323 [DOI] [PubMed] [Google Scholar]
- 13.Samuel, C. E. (2004) Nat. Biotech. 22 280-282 [DOI] [PubMed] [Google Scholar]
- 14.Robbins, M. A., and Rossi, J. J. (2005) Nat. Med. 11 250-251 [DOI] [PubMed] [Google Scholar]
- 15.Thompson, A. J. V., and Locarnini, S. A. (2007) Immunol. Cell Biol. 85 435-445 [DOI] [PubMed] [Google Scholar]
- 16.Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001) Nature 413 732-738 [DOI] [PubMed] [Google Scholar]
- 17.Marshall-Clarke, S., Downes, J. E., Haga, I. R., Bowie, A. G., Borrow, P., Pennock, J. L., Grencis, R. K., and Rothwell, P. (2007) J. Biol. Chem. 282 24759-24766 [DOI] [PubMed] [Google Scholar]
- 18.Saito, T., Owen, D. M., Jiang, F., Marcotrigiano, J., and Gale, M., Jr. (2008) Nature 454 523-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui, S., Eisenächer, K., Kirchhofer, A., Brzózka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K.-K., Krug, A., and Hopfner, K.-P. (2008) Mol. Cell 29 169-179 [DOI] [PubMed] [Google Scholar]
- 20.Kang, D.-C., Gopalkrishnan, R. V., Aaron Randolph, L. L., Valerie, K., Pestka, S., and Fisher, P. B. (2004) Oncogene 23 1789-1800 [DOI] [PubMed] [Google Scholar]
- 21.Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., Hiiragi, A., Dermody, T. S., Fujita, T., and Akira, S. (2008) J. Exp. Med. 205 1601-1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seth, R. B., Sun, L., Ea, C.-K., and Chen, Z. J. (2005) Cell 122 669-682 [DOI] [PubMed] [Google Scholar]
- 23.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, J. (2005) Nature 437 1167-1172 [DOI] [PubMed] [Google Scholar]
- 24.Xu, L.-G., Wang, Y.-Y., Han, K.-J., Li, L.-Y., Zhai, Z., and Shu, H.-B. (2005) Mol. Cell 19 727-740 [DOI] [PubMed] [Google Scholar]
- 25.McCormack, S. J., Thomis, D. C., and Samuel, C. E. (1992) Virology 188 47-56 [DOI] [PubMed] [Google Scholar]
- 26.Bevilacqua, P. C., George, C. X., Samuel, C. E., and Cech, T. R. (1998) Biochemistry 37 6303-6316 [DOI] [PubMed] [Google Scholar]
- 27.Samuel, C. E. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 600-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil, J., and Esteban, M. (2000) Apoptosis 5 107-114 [DOI] [PubMed] [Google Scholar]
- 29.Garcia, M. A., Gil, J., Ventoso, I., Guerra, S., Domingo, E., Rivas, C., and Esteban, M. (2006) Microbiol. Mol. Biol. Rev. 70 1032-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, P., and Samuel, C. E. (2007) J. Virol. 81 8192-8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Q., and Carmichael, G. G. (2004) Microbiol. Mol. Biol. Rev. 68 432-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fire, A. (1998) Nature 391 806-811 [DOI] [PubMed] [Google Scholar]
- 33.Caplen, N. J., Parrish, S., Imani, F., Fire, A., and Morgan, R. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9742-9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meister, G., and Tuschl, T. (2004) Nature 431 343-349 [DOI] [PubMed] [Google Scholar]
- 35.Elbashir, S. M. (2001) Nature 411 494-498 [DOI] [PubMed] [Google Scholar]
- 36.Marques, J. T., Devosse, T., Wang, D., Zamanian-Daryoush, M., Serbinowski, P., Hartmann, R., Fujita, T., Behlke, M. A., and Williams, B. R. (2006) Nat. Biotechnol. 24 559-565 [DOI] [PubMed] [Google Scholar]
- 37.Das, S., Ward, S. V., Tacke, R. S., Suske, G., and Samuel, C. E. (2006) J. Biol. Chem. 281 3244-3253 [DOI] [PubMed] [Google Scholar]
- 38.Bouxsein, N. F., McAllister, C. S., Ewert, K. K., Samuel, C. E., and Safinya, C. R. (2007) Biochemistry 46 4785-4792 [DOI] [PubMed] [Google Scholar]
- 39.Matsukura, S., Stellato, C., Plitt, J. R., Bickel, C., Miura, K., Georas, S. N., Casolaro, V., and Schleimer, R. P. (1999) J. Immunol. 163 6876-6883 [PubMed] [Google Scholar]
- 40.Ui-Tei, K., Zenno, S., Miyata, Y., and Saigo, K. (2000) FEBS Lett. 479 79-82 [DOI] [PubMed] [Google Scholar]
- 41.Samuel, C. E. (1993) J. Biol. Chem. 268 7603-7606 [PubMed] [Google Scholar]
- 42.Panne, D., Maniatis, T., and Harrison, S. C. (2007) Cell 129 1111-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garoufalis, E., Kwan, I., Lin, R., Mustafa, A., Pepin, N., Roulston, A., Lacoste, J., and Hiscott, J. (1994) J. Virol. 68 4707-4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visvanathan, K. V. G., S. (1989) EMBO J. 8 1129-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alkalay, I., Yaron, A., Hatzubai, A., Orian, A., Ciechanover, A., and Ben-Neriah, Y. (1995) Proc. Natl. Acad. Sci. U. S. A. 7 10599-10603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, D.-H., Longo, M., Han, Y., Lundberg, P., Cantin, E., and Rossi, J. J. (2004) Nat. Biotechnol. 22 321-325 [DOI] [PubMed] [Google Scholar]
- 47.Nallagatla, S. R., Hwang, J., Toroney, R., Zheng, X., Cameron, C. E., and Bevilacqua, P. C. (2007) Science 318 1455-1458 [DOI] [PubMed] [Google Scholar]
- 48.Bischoff, J. R., and Samuel, C. E. (1989) Virology 172 106-115 [DOI] [PubMed] [Google Scholar]
- 49.Mellits, K. H., Pe'ery, T., Manche, L., Robertson, H. D., and Mathews, M. B. (1990) Nucleic Acids Res. 18 5401-5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm, G. H., Zurney, J., Tumilasci, V., Leveille, S., Danthi, P., Hiscott, J., Sherry, B., and Dermody, T. S. (2007) J. Biol. Chem. 282 21953-21961 [DOI] [PubMed] [Google Scholar]
- 51.Heylbroeck, C., Balachandran, S., Servant, M. J., DeLuca, C., Barber, G. N., Lin, R., and Hiscott, J. (2000) J. Virol. 74 3781-3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters, K., Chattopadhyay, S., and Sen, G. C. (2008) J. Virol. 82 3500-3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, P., and Samuel, C. E. (2008) J. Biol. Chem. 283 34580-34587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P., and Reis e Sousa, C. (2003) Nature 424 324-328 [DOI] [PubMed] [Google Scholar]
- 55.Jiang, Z., Zamanian-Daryoush, M., Nie, H., Silva, A. M., Williams, B. R. G., and Li, X. (2003) J. Biol. Chem. 278 16713-16719 [DOI] [PubMed] [Google Scholar]
- 56.Gil, J., Garcia, M. A., Gomez-Puertas, P., Guerra, S., Rullas, J., Nakano, H., Alcami, J., and Esteban, M. (2004) Mol. Cell. Biol. 24 4502-4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2006) Nature 439 208-211 [DOI] [PubMed] [Google Scholar]