Abstract
A key set of reactions for the initiation of new DNA strands during herpes simplex virus-1 replication consists of the primase-catalyzed synthesis of short RNA primers followed by polymerase-catalyzed DNA synthesis (i.e. primase-coupled polymerase activity). Herpes primase (UL5-UL52-UL8) synthesizes products from 2 to ∼13 nucleotides long. However, the herpes polymerase (UL30 or UL30-UL42) only elongates those at least 8 nucleotides long. Surprisingly, coupled activity was remarkably inefficient, even considering only those primers at least 8 nucleotides long, and herpes polymerase typically elongated <2% of the primase-synthesized primers. Of those primers elongated, only 4–26% of the primers were passed directly from the primase to the polymerase (UL30-UL42) without dissociating into solution. Comparing RNA primer-templates and DNA primer-templates of identical sequence showed that herpes polymerase greatly preferred to elongate the DNA primer by 650–26,000-fold, thus accounting for the extremely low efficiency with which herpes polymerase elongated primase-synthesized primers. Curiously, one of the DNA polymerases of the host cell, polymerase α (p70-p180 or p49-p58-p70-p180 complex), extended herpes primase-synthesized RNA primers much more efficiently than the viral polymerase, raising the possibility that the viral polymerase may not be the only one involved in herpes DNA replication.
Herpes simplex virus 1 (HSV-1)2 encodes seven proteins essential for replicating its double-stranded DNA genome; five of these encode the heterotrimeric helicase-primase (UL5-UL52-UL8 gene products) and the heterodimeric polymerase (UL30-UL42 gene products) (1, 2). The helicase-primase unwinds the DNA at the replication fork and generates single-stranded DNA for both leading and lagging strand synthesis. Primase synthesizes short RNA primers on the lagging strand that the polymerase presumably elongates using dNTPs (i.e. primase-coupled polymerase activity). These two protein complexes are thought to replicate the viral genome on both the leading and lagging strands (1, 2).
Previous studies have focused on the helicase-primase and polymerase separately. The helicase-primase contains three subunits, UL5, UL52, and UL8 in a 1:1:1 ratio (3–5). The UL5 subunit has helicase-like motifs and the UL52 subunit has primase-like motifs, yet the minimal active complex that demonstrates either helicase or primase activities contains both UL5 and UL52 (6, 7). Although the UL8 subunit has no known catalytic activity, several functions have been proposed, including enhancing helicase and primase activities, enhancing primer synthesis on ICP8 (the HSV-1 single-stranded binding protein)-coated DNA strands, and facilitating formation of the replisome (8–12). Although primase will synthesize short (2–3 nucleotides long) primers on a variety of template sequences, synthesis of longer primers up to 13 nucleotides long requires the template sequence, 3′-deoxyguanidine-pyrimidine-pyrimidine-5′ (13). Primase initiates synthesis at the first pyrimidine via the polymerization of two purine NTPs (13). Even after initiation at this sequence, however, the vast majority of products are only 2–3 nucleotides long (13, 14).
The herpes polymerase consists of the UL30 subunit, which has polymerase and 3′ → 5′ exonuclease activities (1, 2), and the UL42 subunit, which serves as a processivity factor (15–17). Unlike most processivity factors that encircle the DNA, the UL42 protein binds double-stranded DNA and thus directly tethers the polymerase to the DNA (18). Using pre-existing DNA primer-templates as the substrate, the heterodimeric polymerase (UL30-UL42) incorporates dNTPs at a rate of 150 s–1, a rate much faster than primer synthesis (for primers >7 nucleotides long, 0.0002–0.01 s–1) (19, 20).
We examined primase-coupled polymerase activity by the herpes primase and polymerase complexes. Although herpes primase synthesizes RNA primers 2–13 nucleotides long, the polymerase only effectively elongates those at least 8 nucleotides long. Surprisingly, the polymerase elongated only a small fraction of the primase-synthesized primers (<1–2%), likely because of the polymerase elongating RNA primer-templates much less efficiently than DNA primer-templates. In contrast, human DNA polymerase α (pol α) elongated the herpes primase-synthesized primers very efficiently. The biological significance of these data is discussed.
EXPERIMENTAL PROCEDURES
Reagents and Protein Purification—HSV-1 helicase-primase (UL8-UL5-UL52) and His-UL8 were expressed in baculovirus-infected Sf9 cells grown at the Tissue Culture Core Facility at the University of Colorado Health Sciences Center and purified as described previously (13). Baculoviruses expressing the UL5 and UL52 subunits were generously provided by Dr. Robert Lehman (Stanford University), and a baculovirus expression vector for a His9-tagged UL8 subunit was generously provided by Dr. Heidi Giordano (Tularik). The baculovirus expressing the helicase-deficient UL5 mutant was generously provided by Dr. Sandra Weller (University of Connecticut Health Sciences Center). The subcomplex, UL5-UL52, was purified according to the procedure described previously (21). Binase was a generous gift from Dr. Eric Zuiderweg (University of Michigan).
Exonuclease-deficient (exo–) UL30 was generously provided by Dr. Deborah Parris (Ohio State University) (22). Dr. Parris also generously provided us with the gene for UL42, to which we added a His6 tag and tobacco etch virus-protease site to the C terminus (cloning was performed by GenScript) (23, 24). The resulting plasmid was cloned into bacmid DNA, and expression of the new baculovirus was performed at the Tissue Culture Core Facility. Purification of the polymerase complex, UL30(exo–)-UL42, by nickel nitrilotriacetic acid column chromatography followed the same protocol as described previously (13).
We added a His6 tag and tobacco etch virus-protease site to the N terminus (cloning was performed by GenScript) of the p70 subunit of human polymerase α (25). The resulting plasmid was cloned into bacmid DNA and expression of the new baculovirus was performed at the Tissue Culture Core Facility. Purification of pol α (p70-p180 complex) and polymerase α-primase (p49-p58-p70-p180) by nickel nitrilotriacetic acid column chromatography followed the same protocol as described previously (13).
Unlabeled NTPs and dNTPs were from Sigma, and radiolabeled NTPs and dNTPs were from PerkinElmer Life Sciences. Synthetic DNA oligonucleotides of defined sequence (Table 1) were obtained from Oligos, Etc., or BioSearch Technologies, Inc.; RNA oligonucleotides (Table 1) were obtained from Dharmacon. Oligonucleotide concentrations were determined spectrally and are reported in terms of 5′ termini. All other reagents were of the highest purity available.
TABLE 1.
DNA template and primer-template sequences
Primase Assay—Primase assays were performed as described previously (13) and typically contained 15 μm ssDNA template, 1 mm [α-32P]NTPs, 50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 5% glycerol, 1 mm dithiothreitol, and 0.1 mg/ml bovine serum albumin. Reactions were initiated by adding 100 nm helicase-primase and quenched after 30 min at 37 °C. Products were separated by denaturing gel electrophoresis (20% acrylamide, 7.5 m urea) and analyzed by phosphorimagery (GE Healthcare).
Primase-coupled Polymerase Activity Assay—Primase-coupled polymerase activity was measured in assays as described above, except that they also contained 10 μm [α-32P]dNTPs, and the NTPs were unlabeled. Reactions were initiated by the addition of 100 nm helicase-primase and 100 nm polymerase (herpes polymerase or pol α) and incubated for 30 min at 37 °C. After quenching the assays with an equal volume of gel loading buffer (90% formamide), products were separated by denaturing gel electrophoresis (20% acrylamide, 7.5 m urea) and analyzed by phosphorimagery (GE Healthcare). In some cases, we also included an RNase, binase, in an amount (33 μm) sufficient to hydrolyze any RNA primers released free in solution by primase before the polymerase could elongate them.
Primer Length Assay—To determine the length of RNA primers elongated by the polymerase, coupled products were visualized by phosphorimagery, followed by gel extraction using double-distilled H2O and filtration through micro-spin cellulose acetate membranes (PerkinElmer Life Sciences) (26). Purified products were treated with 33 μm binase for 30 min at 37 °C and reanalyzed by gel electrophoresis followed by phosphorimagery (GE Healthcare).
DNA Polymerase Activity Assay—DNA polymerase activity was measured as described previously (27). Unless noted otherwise, assays typically contained 1–10 μm (DNA or RNA) primer-template, 50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 1 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, 5% glycerol, and [α-32P]dNTPs. Reactions were initiated with 1–2 nm polymerase for the reactions containing DNA primer-template and 20 nm polymerase for reactions containing RNA primer-templates. After 10 min at 37 °C, reactions were quenched by adding an equal volume of gel loading buffer. Products were separated by denaturing gel electrophoresis (20% acrylamide, 7.5 m urea) and analyzed by phosphorimagery (GE Healthcare). dNTP polymerization rates were normalized to 1 nm enzyme concentration. Kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using the program Prism (GraphPad), and errors were determined from the goodness-of-fit.
Fraction of Primers Elongated by the DNA Polymerase in Coupled Activity—Primer utilization values were determined by comparing the total moles of product synthesized in the coupled assays (measured using [α-32P]dNTPs) to the total moles of RNA primers >7 nucleotides long synthesized in the primase assays (measured using [α-32P]NTPs) under otherwise identical conditions.
Fraction of Primers Transferred Intramolecularly—The fraction of primers transferred intramolecularly was determined by measuring the amount of coupled products formed in assays containing binase divided by the amount of coupled products formed in assays lacking binase.
RESULTS
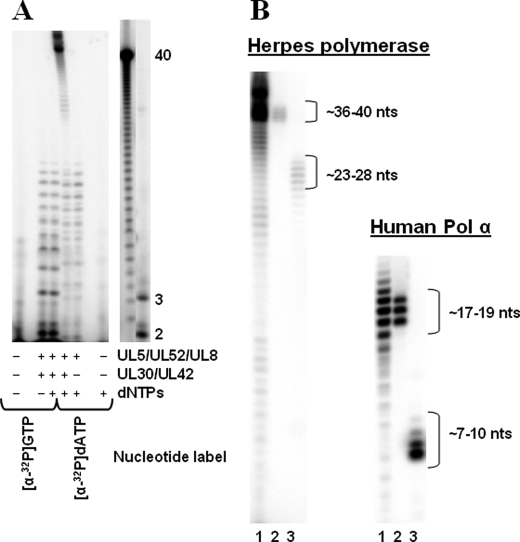
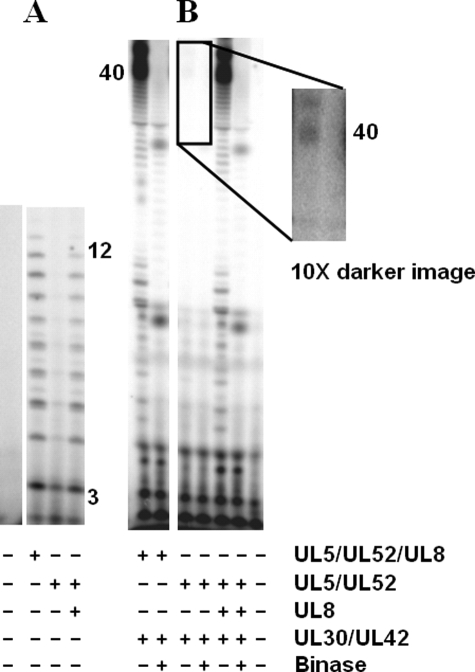
Initiation of new DNA strands typically requires synthesis of an RNA primer by primase and subsequent elongation of the primer by a DNA polymerase (i.e. primase-coupled polymerase activity). To better understand the mechanism by which the herpes replication machinery accomplishes this task, we analyzed primase-coupled polymerase activity in assays containing the primase-helicase complex, DNA polymerase, NTPs to allow primer synthesis, and dNTPs for primer elongation. In the presence of [α-32P]NTPs and the herpes polymerase, primase synthesizes primers 2–13 nucleotides long (Fig. 1A), similar to the results with assays containing just the primase-helicase complex. When assays contained NTPs and [α-32P]dNTPs, radiolabeled products ∼35–40 nucleotides long were produced on the ssDNA template, 3′-d(T20GTCCT36)-5′ (Table 1). This length corresponds to the distance from the primase initiation site (3′-GTC) to the 5′-end of the template. Omitting either the NTPs, primase-helicase complex, or the polymerase eliminated these products, indicating that the formation of these long products requires both primase and polymerase activity (Fig. 1A).3
FIGURE 1.
Herpes primase and primase-coupled polymerase activities on 3′-d(T20GTCCT36)-5′. A, products of primase-coupled polymerase assays were measured in assays containing enzymes as noted, and either [α-32P]GTP or [α-32P]dATP as noted, and were performed as described under “Experimental Procedures.” Product lengths (shown) were determined using 5′-[32P]dA40 as a marker, as well as pppApG and pppApGpA synthesized using human primase, ATP, and [α-32P]GTP. B, length of the RNA primers elongated by the polymerase during primase-coupled polymerase activity was determined in assays containing primase-helicase, 3′-d(T20GTCCT36)-5′, NTPs, [α-32P]dNTPs, and either herpes polymerase (UL30-UL42) or human pol α. Lane 1 shows the coupled products. A small fraction of the coupled products were gel-extracted (lane 2) and treated with binase (lane 3) as described under “Experimental Procedures.” Product lengths (shown) for herpes polymerase-herpes primase-coupled activity were determined using 5′-[32P]dA40 as a marker. Product lengths (shown) for pol α-herpes primase coupled activity were estimated using 5′-[32P]dC30. In this, and subsequent gel images, sections taken from different parts of a larger gel are separated. nts, nucleotides.
Herpes Polymerase Only Elongates Primase-synthesized Primers at Least 8 Nucleotides Long—To determine the length of RNA primers necessary for elongation by the herpes polymerase, we added an RNase, binase,4 to the full-length products of the coupled assay to hydrolyze both unelongated RNA primers and the RNA portion of the polymerase-elongated RNA primers. Fig. 1B shows that binase treatment shortens the full-length products of coupled activity by at least 8 nucleotides, indicating that the herpes polymerase only efficiently elongates primase-synthesized RNA primers >7 nucleotides long to full-length product.
Herpes Polymerase Very Inefficiently Elongates Primase-synthesized Primers—The rate of primase-coupled polymerase activity on the template 3′-d(T20GTCCT36)-5′ was remarkably slow. Assuming that the full-length products are on average 40 nucleotides long and the average primer length associated with these products is 10 nucleotides long, the rate of full-length product synthesis was <1% of the rate at which primase generated primers >7 nucleotides long (see Fig. 1A). These data suggest that the herpes polymerase elongated less than 1% of the longer primers. Similar low efficiencies of primer utilization were observed on three other templates (Table 2).
TABLE 2.
Primase-coupled polymerase activity with the herpes polymerase (UL30 or UL30-UL42)
Primase-coupled polymerase activity (% primers utilized and % primers transferred) was measured for each primase/polymerase pair at equivalent molar concentrations of each enzyme with the exception of studies using UL5-UL52 that contained 400 nm UL5-UL52 and 100 nm polymerase.
| Primase | Polymerase | DNA template | Primers utilized | Primers transferred intramolecularlya |
|---|---|---|---|---|
| % | % | |||
| UL5-UL52-UL8 | UL30-UL42 | T20GTCCT36 | 0.6 ± 1.1 | 0.039 ± 0.026 |
| UL5-UL52-UL8 | UL30 | T20GTCCT36 | 0.3 ± 0.1 | NDb |
| UL5-UL52-UL8 (Hel-) | UL30-UL42 | T20GTCCT36 | 1.6 | 0.26 |
| UL5-UL52-UL8 (Hel-) | UL30 | T20GTCCT36 | 0.3 | ND |
| UL5-UL52 | UL30-UL42 | T20GTCCT36 | <0.2 | ND |
| UL5-UL52 + UL8 | UL30-UL42 | T20GTCCT36 | 0.9 ± 0.5 | 0.036 |
| UL5-UL52c | UL30-UL42 | T20GTCCT36 | 0.0015 ± 0.0005 | ND |
| UL5-UL52 + UL8c | UL30-UL42 | T20GTCCT36 | 0.34 ± 0.07 | 0.025 ± 0.018 |
| UL5-UL52/UL8 | UL30-UL42 | T20GCCCCAT17 | 3.6 ± 0.4 | 0.94 |
| UL5-UL52-UL8 (Hel-) | UL30-UL42 | T20GCCCCAT17 | 2.6 | 0.23 |
| UL5-UL52c | UL30-UL42 | T20GCCCCAT17 | 0.008 ± 0.002 | 0.004 ± 0.0005 |
| UL5-UL52 + UL8c | UL30-UL42 | T20GCCCCAT17 | 0.34 ± 0.02 | 0.05 ± 0.01 |
| UL5-UL52-UL8 | UL30-UL42 | C20GTCCA19 | <0.2 | ND |
| UL5-UL52-UL8 | UL30-UL42 | (TCTG)15 | 0.4 | ND |
The percentage of primers transferred was calculated as a percentage of the primers utilized by the polymerase in each case, i.e. of the 0.6% of the primers synthesized by UL5-UL52-UL8 and further elongated by UL30/UL42 on 3′-d(T20GTCCT36)-5′, only 0.039% were transferred intramolecularly to UL30-UL42.
ND indicates no detectable products in the presence of binase.
These assays contained 1 μm dNTPs instead of 10 μm dNTPs to increase the specific activity of the [α-32P]dNTPs in order to allow quantitation of primase-coupled polymerase activity.
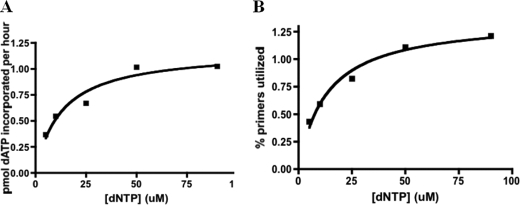
We considered the possibility that inefficient coupled activity resulted from the relatively low dNTP concentration used (10 μm). Varying the dNTP concentration from 10 to 90 μm slightly increased the rate of coupled activity and the fraction of primers elongated (around 2-fold in each case, Fig. 2). However, even at 90 μm dNTPs, the herpes polymerase elongated only <2% of the primase-synthesized primers.
FIGURE 2.
Effects of increasing the dNTP concentration on primase-coupled polymerase activity. Assays contained primase-helicase, polymerase (UL30-UL42), 3′-d(T20GTCCT36)-5′, and either [α-32P]NTPs or NTPs and [α-32P]dNTPs to measure primase activity and primase-coupled polymerase activity, respectively. Coupled activity was measured in terms of pmol of dATP incorporated. The fraction of primers elongated was determined as described under “Experimental Procedures.”
To further test this conclusion, we performed primase and primase-coupled polymerase assays using [α-32P]NTPs ± unlabeled dNTPs to directly observe primer synthesis. Including dNTPs to allow the polymerase to elongate primers resulted in no detectable change in the amount of long primers, and the amount of radiolabel in products ∼40 nucleotides long was below the limits of detection (control experiments that included [α-32P]dNTPs showed that the polymerase-elongated products were formed, Fig. 1A).5 Moreover, titrating in increasing amounts of polymerase (to a 4-fold molar excess over primase) did not significantly increase the amount of long RNA primers utilized (≤1% in all cases, data not shown). Thus, the herpes polymerase only elongates a surprisingly small fraction of the primase-synthesized RNA primers.
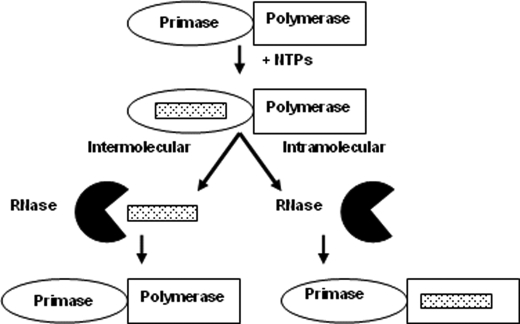
Primer Transfer from Herpes Helicase-Primase to the Polymerase (UL30-UL42) Is Partially Intramolecular—To provide insights into both the mechanism and low efficiency of primase-coupled polymerase activity, we examined the mechanism of RNA primer transfer from the primase to the polymerase active site. To determine whether primer transfer occurred intramolecularly or intermolecularly, we included an RNase, binase, in the coupled reaction. Binase serves to hydrolyze any RNA primers released into solution prior to elongation by the polymerase (Scheme 1) (28). If the RNA primer is transferred intramolecularly such that the two complexes prevent binase from hydrolyzing the primer (i.e.“channeled” from primase directly to the polymerase), the polymerase will still elongate the primer even in the presence of binase. Alternatively, if either the RNA primer is transferred intermolecularly (i.e. primase releases the primer into solution and the polymerase then binds and elongates it) or the primer becomes accessible to binase during intramolecular transfer, then binase will hydrolyze the primer before the polymerase can elongate it.
SCHEME 1.
Differentiating intramolecular and intermolecular primer transfer.
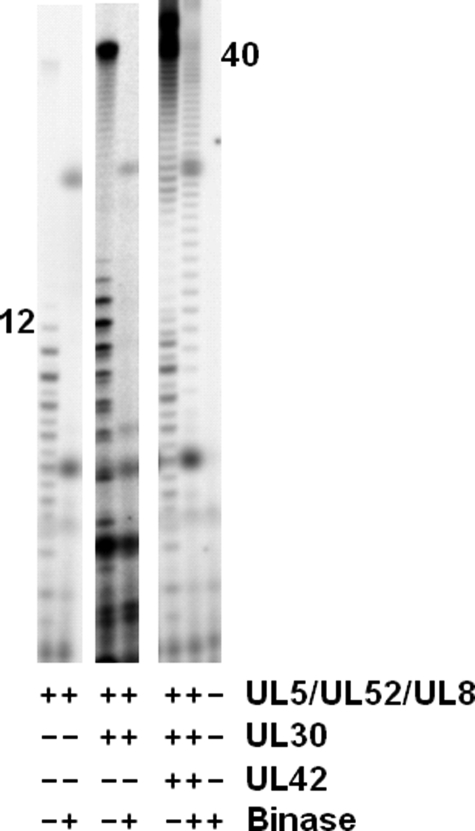
Including binase in either primase assays containing just UL5-UL52-UL8 or coupled activity assays containing UL5-UL52-UL8 and just the UL30 polymerase subunit resulted in no detectable products (Fig. 3). Thus, a 330-fold molar excess of binase will completely hydrolyze the primase-synthesized primers and ensure hydrolysis of all the RNA primers before the polymerase can elongate them. Additionally, these data indicate that primer transfer between primase and UL30 solely occurred either via an intermolecular process or the primers became accessible to binase during an intramolecular transfer. On the other hand, when the coupled activity assays contained the heterodimeric polymerase (UL30-UL42), 7% of those primers elongated by the polymerase were protected from binase during coupled assays on the template 3′-d(T20GTCCT36)-5′ (Fig. 3 and Table 2).6 On the template 3′-d(T20GCCCCAT17)-5′, up to 26% of the RNA primers used by the polymerase were transferred intramolecularly. Thus, although primer transfer between the primase and the polymerase proceeded solely via an intermolecular process when the isolated UL30 polymerase was used, the transfer occurred at least partially via an intramolecular process when the dimeric polymerase (UL30-UL42) was used. Furthermore, preincubating the primase and polymerase complexes with each other and/or the ssDNA template did not affect the mechanism or efficiency of transfer, indicating the two enzyme complexes do not need to interact before primer synthesis for intramolecular primer transfer to occur (data not shown and Fig. 3).
FIGURE 3.
Mechanism of primer transfer during primase-coupled polymerase activity. Assays were performed as described under “Experimental Procedures” and contained the template 3′-d(T20GTCCT36)-5′, NTPs, [α-32P]dNTPs, and the noted enzymes. Product lengths (shown) were determined as described in Fig. 1A. The spots present in the lanes, including binase, are most likely a result from binase modifying a product generated by the interaction of primase with either [α-32P]dNTPs or with a radiolabeled contaminant in the [α-32P]dNTPs (2nd lane), because they do not appear in the binase-only lane (1st lane).
Effects of Helicase Activity on Primase-coupled Polymerase Activity—The herpes helicase and primase activities are not independent of one another (6, 7, 29). Helicase and primase both require the UL5 and UL52 subunits, and mutating UL5 to block helicase activity can stimulate primase activity of the resulting UL5-UL52 complexes (30). Additionally, the helicase could potentially dissociate the RNA primer-template generated by the primase, thereby interfering with the ability of the polymerase to elongate the primer. To determine whether helicase activity affects primase-polymerase coupled activity, we utilized a heterotrimeric primase (UL5-UL52-UL8) lacking helicase activity because of a point mutation (G102V) in the helicase-like motif I in the UL5 subunit (30).
In coupled assays on the template 3′-d(T20GTCCT36)-5′ containing the helicase-deficient UL5-UL52-UL8 complex, the polymerase (UL30-UL42) still elongated very few of the >7 nucleotide-long RNA primers (1.6%), slightly higher than during coupled activity with the wild-type helicase-primase (0.6%, see Table 2). Of those primers that the polymerase had elongated, 16% of them were transferred intramolecularly to the dimeric polymerase (UL30-UL42). In contrast, only 6.5% of the primers elongated by the polymerase were transferred intramolecularly from wild-type primase-helicase to the polymerase. However, on another DNA template, 3′-d(T20GCCCCAT17)-5′, only 9% of the RNA primers elongated by the polymerase had been passed intramolecularly in assays containing the mutant helicase-primase, whereas 26% were transferred intramolecularly in assays containing the wild-type helicase-primase. These data suggest that depending upon the template, an active helicase may interfere with direct transfer of primers between the helicase-primase and polymerase.
Effect of UL8 on Coupled Activity—To determine whether coupled activity requires UL8, we examined primase-coupled polymerase activity using the minimal helicase-primase complex lacking UL8 (i.e. UL5-UL52). Because of the greatly decreased primase activity of the UL5-UL52 complex as compared with the UL5-UL52-UL8 complex, the amount of polymerase-elongated primers could not be accurately quantified in assays containing 10 μm [α-32P]dNTPs. Thus, to quantitatively compare coupled activity using either UL5-UL52 or UL5-UL52-UL8, assays contained 1 μm [α-32P]dNTPs of a much greater specific activity than if they contained 10 μm dNTPs, although the lower dNTP concentration decreases the efficiency of primer utilization (Fig. 4 and Table 2). Consistent with previous studies (9, 31), adding UL8 to primase assays (Fig. 4A) greatly stimulated primase activity. Additionally, including an equimolar concentration of UL8 in coupled assays containing UL5-UL52 increased the fraction of primers elongated by UL30-UL42 (∼40–230-fold (Table 2)), although this fraction remained pathetically low (<1% primers used). Thus, UL8 both stimulates primer synthesis by herpes primase along with primer utilization by the herpes polymerase.
FIGURE 4.
UL8 stimulates primase-coupled polymerase activity with the core primase-helicase (UL5-UL52). Primase-coupled polymerase assays contained 3′-d(T20GTCCT36)-5′, either UL5-UL52-UL8 or UL5-UL52, and also contained other proteins (UL8, UL30-UL42, and binase) as noted. A, relative primase activity using [α-32P]NTPs. B, primase-coupled polymerase activity using [α-32P]dNTPs. Product lengths (shown) were determined using 5′-[32P]dA40 and 5′-[32P]dC30 as markers.
UL30 and UL30-UL42 Prefer to Elongate DNA Primers Rather than RNA Primers—Previous studies on herpes polymerase have used DNA primer-templates, whereas primase produces RNA primer-templates. Because an inability to efficiently elongate RNA primers could explain the surprisingly low efficiency of coupled activity, we determined whether the type of primer used by the polymerase, RNA or DNA, affected polymerase activity. Polymerase activity was measured on chemically synthesized DNA and RNA primers having the same sequence, 5′-d(GGGGTAAA)-3′ and 5′-GGGGUAAA-3′, hybridized to the same DNA template, 3′-d(T20GCCCCATTTT31)-5′. This primer and template were chosen because upon annealing to the template, the resulting primer-template would mimic the sequence of a herpes primase-generated primer-template (Table 1). Additionally, this template also gave the most efficient primer usage by the herpes polymerase during primase-coupled polymerase activity. Assays contained either the monomeric UL30 or dimeric UL30-UL42 polymerase. Importantly, both UL30 and UL30-UL42 polymerized dNTPs much more efficiently onto DNA primers than onto RNA primers (by 650- and 26,000-fold, respectively, Table 3). The huge preference for DNA primer-templates did not result from the concentration of primer-template in the reaction. In assays containing 1, 5, or 10 μm primer-template, herpes polymerase showed a similar preference for the DNA primer-template (Table 3).
TABLE 3.
Polymerase activity measured on DNA primer-templates and RNA primer-templates (see Table 1 for sequence information)
Polymerase assays contained 1 nm of the noted enzyme.
| Enzyme | Primer-template | Vmax | KM (dNTP) | Vmax/KM | Vmax/KM (DNA)/Vmax/KM (RNA) |
|---|---|---|---|---|---|
| pmol dNTP/nm enzyme/h | μm | ||||
| UL30 | DNA1 (5 μm) | 39.1 (2.0) | 9.40 (1.65) | 4.2 | 650 |
| UL30 | RNA1 (5 μm) | 73.5 (11.5) | 11,400 (2900) | 0.0065 | |
| UL30-UL42 | DNA1 (10 μm) | 1.92 (0.10) | 0.17 (0.04) | 11.3 | 26,000 |
| UL30-UL42 | RNA1 (10 μm) | 3.61 (0.64) | 8200 (2600) | 0.00044 | |
| UL30-UL42 | DNA1 (1 μm) | 0.57 (0.02) | 0.21 (0.03) | 2.7 | 10,000 |
| UL30-UL42 | RNA1 (1 μm) | 1.93 (0.53) | 7300 (3700) | 0.00026 | |
| pol α | DNA1 (5 μm) | 7.22 (0.17) | 8.18 (0.68) | 0.88 | 0.85 |
| pol α | RNA1 (5 μm) | 7.50 (0.20) | 7.20 (0.69) | 1.04 | |
| UL30 | DNA2 (5 μm) | 11.5 (0.4) | 3.84 (0.59) | 3.0 | 2200 |
| UL30 | RNA2 (5 μm) | NMa | NM | 0.00138 | |
| UL30-UL42 | DNA2 (5 μm) | 1.15 (0.04) | 0.39 (0.04) | 2.9 | 13,000 |
| UL30-UL42 | RNA2 (5 μm) | 0.41 (0.04) | 1800 (550) | 0.00023 | |
| UL30-UL42 | DNA2 (1 μm) | 0.35 (0.04) | 0.52 (0.25) | 0.67 | 4500 |
| UL30-UL42 | RNA2 (1 μm) | 0.60 (0.04) | 4000 (610) | 0.00015 | |
| pol α | DNA2 (5 μm) | 2.86 (0.10) | 3.17 (0.47) | 0.90 | 1.0 |
| pol α | RNA2 (5 μm) | 2.34 (0.06) | 2.62 (0.30) | 0.89 |
NM indicates not meaningful because the rate increased linearly with increasing dNTP concentrations.
To control for the possibility that these data resulted from some unusual property of the specific RNA primer-template tested, we also examined the ability of human pol α, an enzyme previously shown to efficiently elongate both RNA and DNA primers, to use these primer-templates (32). Consistent with previous work, pol α showed only a slight preference for RNA primers (Table 3). Thus, the differences between herpes polymerase-catalyzed elongation of RNA and DNA primers likely does not result from the RNA primer adopting an unusual structure.
We also tested the possibility that the sequence of the DNA template being copied resulted in the preference of herpes polymerase for the DNA primer. However, herpes polymerase also greatly preferred the DNA primer on the template 3′-d(T20GCCCCATTT(TCTA)6)-5′, indicating that the template sequence being copied does not affect the preference for DNA primers (Table 3).
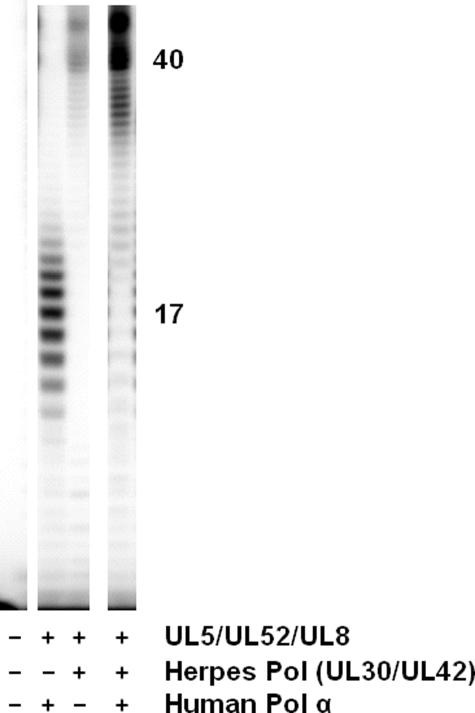
Human pol α Utilizes Herpes Primase-synthesized RNA Primers More Efficiently than the Herpes Polymerase—Because human pol α (as the p70-p180 complex) uses RNA primer-templates much more efficiently than the herpes polymerase, we examined whether pol α could also elongate herpes primase-synthesized RNA primers. Replacing the herpes polymerase with pol α in the coupled reactions resulted in a profound increase in the amount of primase-coupled polymerase activity, consistent with the ability of pol α to use RNA primer-templates more efficiently than UL30-UL42 (Fig. 5 and Table 4). In comparison, an equimolar concentration of another DNA polymerase, Klenow fragment (exo–) of Escherichia coli DNA polymerase I, only elongated 1/6 as many herpes primase-generated primers as did pol α on 3′-d(T20GTCCT36)-5′. Furthermore, if one corrects for the much higher activity of Klenow fragment than of pol α, Klenow fragment elongated 50-fold fewer primers than did pol α in assays containing equal amounts of dNTP polymerizing activity.
FIGURE 5.
Primase-coupled polymerase activity between herpes primase and human pol α. Assays were performed as described under “Experimental Procedures” and contained the template 3′-d(T20GTCCT36)-5′, NTPs, [α-32P]dNTPs, and the noted enzymes. Product lengths (shown) were estimated using 5′-[32P]dA40 and 5′-[32P]dC30 as markers.
TABLE 4.
Primase-coupled polymerase activity with human pol α
| Primase | Polymerase | DNA template | Primers utilized | Primers Transferred Intramolecularly |
|---|---|---|---|---|
| % | % | |||
| UL5-UL52-UL8 (Hel-) | pol α | T20GTCCT36 | 19 | <0.001a |
| UL5-UL52-UL8 | pol α | T20GTCCT36 | 29 ± 11 | <0.001 |
| UL5-UL52-UL8 | pol α-primase | T20GTCCT36 | 18 ± 5 | -b |
| UL5-UL52-UL8 | UL30-UL42 + pol α | T20GTCCT36 | 10 ± 5 | 0.12 ± 0.06 |
| UL5-UL52-UL8 | UL30-UL42 + pol α-primase | T20GTCCT36 | 8 ± 3 | -b |
| UL5-UL52 | pol α | T20GTCCT36 | 7.9 ± 0.7 | <0.001 |
| UL5-UL52 + UL8 | pol α | T20GTCCT36 | 31 ± 9 | <0.001 |
| UL5-UL52-UL8 | pol α | (TCTG)15 | 5.5 ± 1.7 | NDc |
| UL5-UL52 | pol α | (TCTG)15 | 1.6 ± 0.9 | ND |
| UL5-UL52 + UL8 | pol α | (TCTG)15 | 3.3 ± 0.7 | ND |
Data are below the limit of detection.
In assays containing pol α-primase, the human primase dose generate small amounts of primers that pol α elongates, and primer transfer between human primase and pol α occurs intramolecularly (28). This small amount of coupled activity between the human primase and pol α would therefore confound attempts to identify direct primer transfer from herpes primase to pol α.
ND indicates no detectable products in the presence of binase.
Coupled activity between the herpes primase and pol α, however, resulted in shorter products than coupled activity between herpes primase and herpes polymerase, presumably because of the relatively low processivity of pol α (Fig. 5) (33). Although the primers were transferred intermolecularly from herpes primase to pol α (>99%), pol α utilized 20–30% of the primase-synthesized primers compared with the 1% used by the herpes polymerase (Fig. 5 and Table 4). Eliminating the helicase activity of the primase-helicase complex did not affect coupled activity between herpes primase and pol α. However, comparing primase-coupled polymerase assays containing the UL5-UL52 primase complex versus the UL5-UL52-UL8 complex revealed that the presence of UL8 increased primer utilization by pol α 2–4-fold on two different templates (Table 4). These data suggest that UL8 may enhance utilization of UL5-UL52-synthesized primers by pol α. Under all conditions tested, pol α utilized the primase-synthesized RNA primers much more efficiently than the herpes polymerase.
In vivo, pol α typically forms a complex with the cellular primase (p49-p58). To determine whether the presence of the human primase subunits affected the capacity of pol α to elongate herpes-primase-synthesized primers, we tested pol α-primase (p49-p58-p70-p180) in the coupled assays. Because assays would now contain two potential sources of primers (herpes and human primase), we needed to control for the possibility that products would result from the combined activity of human primase and pol α. Importantly, control experiments containing only pol α-primase showed that the human primase did not synthesize detectable amounts of primers in assays containing [α-32P]NTPs. In primase-coupled polymerase assays containing only pol α-primase, NTPs, and [α-32P]dNTPs, only small amounts of coupled products were detected (<1% of those observed in assays containing herpes primase and pol α). The ability to detect products from primase-coupled polymerase activity but not primase activity alone likely results from the much higher specific activity of the [α-32P]dNTPs than [α-32P]NTPs, and that each coupled product contains many more radiolabeled dNMPs than NMPs. Pol α-primase showed the same enhanced coupled activity with herpes primase and utilized 15–25% of the herpes primase-synthesized RNA primers as did pol α by itself (Table 4).
In light of the highly efficient utilization of herpes primase-synthesized primers by pol α, albeit with low processivity, and the ability of the much higher processivity herpes polymerase to completely copy the template, we examined the possibility that a two-polymerase system containing both pol α and UL30-UL42 would give highly efficient herpes primase-dependent replication of these templates. In assays containing herpes primase-helicase, UL30-UL42, and either pol α (p70-p180) or pol α-primase (p49-p58-p70-p180), two primary changes occurred relative to assays lacking one of the polymerases. First, compared with assays containing only pol α, the relatively short products due to coupled activity between herpes primase and pol α greatly decreased. Second, the amount of full-length products (i.e. those due to complete copying of the template from the primer initiation site to the 5′-end of the template) increased dramatically relative to assays that contained only herpes polymerase (Fig. 5). Thus, the combined activities of herpes primase, pol α, and herpes polymerase provide a relatively efficient system for replicating templates containing an initiation site for herpes primase.
Including binase in the two polymerase-coupled assays showed that of the primers elongated by the polymerases, only 1.2–1.9% of them had been transferred intramolecularly to one of the polymerases, a much lower percentage of intramolecular transfer than when assays contained just herpes polymerase. This value reflects both the small amount of primase-synthesized primers directly elongated by herpes polymerase as well as the much larger amount elongated by pol α. Thus, the low percentage presumably indicates that in the two polymerase-coupled assays, most of the elongated primers were initially transferred intermolecularly to pol α and relatively few were transferred intramolecularly to herpes polymerase.
DISCUSSION
When HSV-1 infects eukaryotic cells, it shuts down replication of the host genome and turns on replication of its own genome during its lytic cycle (1). To achieve viral DNA replication, herpes encodes seven essential proteins, including its own helicase-primase (UL5-UL52-UL8) and DNA polymerase (UL30-UL42) (1, 2). Here we explored the elongation of herpes primase-synthesized RNA primers by both herpes polymerase and the nuclear DNA pol α.
Surprisingly, the herpes polymerase was remarkably inefficient at using primers synthesized de novo by herpes helicase-primase. Even though primase predominantly synthesizes dinucleotides and trinucleotides, the polymerase only elongates those primers at least 8 nucleotides long. More surprisingly, at equimolar concentrations, the polymerase typically utilizes <1% of these longer RNA primers (>7 nucleotides). This occurs even though the polymerase polymerizes dNTPs onto a DNA primer much faster than primase synthesizes longer RNA primers (13, 20). Addition of the polymerase processivity factor, UL42, does not significantly change either the rate of dNTP polymerization or the fraction of RNA primers elongated in primase-coupled polymerase assays (Table 2). Therefore, the vast majority of primase-synthesized primers is simply released into solution.
A curious feature of herpes polymerase, both in the presence and absence of UL42, is its relative inability to elongate RNA primers. Because initiation of the leading strand and lagging strand synthesis both presumably require elongation of an RNA primer, and herpes provides its own primase activity, one would have expected herpes polymerase to effectively elongate both RNA and DNA primers. Potentially, this inefficient elongation of RNA primers could result from either the 2′-hydroxyl of the ribose directly interfering with the polymerase, or the very different structures that DNA:DNA and RNA:DNA duplexes adopt (34, 35). Importantly, however, this low efficiency of RNA primer utilization can account for the low efficiency of coupled activity between herpes primase and herpes polymerase.
The slow rate of elongation may partially result from inefficient transfer of the RNA primer from the primase complex to the polymerase. Including an RNase in the assays to hydrolyze any primers released into solution by the primase completely blocked primer elongation by the monomeric UL30 polymerase, indicating that primase does not directly transfer primers to UL30. However, when the polymerase is complexed with UL42, a portion of RNA primer transfer occurs intramolecularly, without the primers dissociating from primase before transfer to the polymerase. Thus, UL42 alters the mechanism of primer transfer. However, even though some of the primers are transferred intramolecularly, the process still remains remarkably inefficient (<1% of the long RNA primers elongated).
The direct, intramolecular transfer from primase to UL30-UL42 indicates that the polymerase and primase complexes must directly interact. Consistent with this requirement, previous studies have shown direct interactions between the UL30 of the polymerase complex and UL8 of the helicase-primase complex (36). Indeed, this interaction could account for the increased utilization of primase-generated primer-templates by the UL5-UL8-UL52 complex (or UL5-UL52 + UL8) as compared with the UL5-UL52 complex.
It remains unclear why intramolecular primer transfer to the polymerase only occurred in the presence of UL42. Because both UL30 and UL30-UL42 utilize RNA primer-templates rather poorly, intramolecular transfer is not a consequence of more efficient primer utilization by the UL30-UL42 complex compared with UL30 alone. In light of the ability of UL42 to directly bind duplex DNA, one possibility is that UL42 also interacts with the helicase-primase complex to form an extended channel that serves to direct the primer-template between the primase and UL30 active sites, thereby protecting it from hydrolysis (37). UL42 might also be located very near the active site of primase such that when primase releases a newly generated primer-template, proximity allows UL42 to bind it. Alternatively, binding of UL42 to UL30 may alter how UL30 interacts with the helicase-primase complex.
The utilization of relatively long primers during primase-coupled polymerase activity is a feature of nuclear DNA replication. Pol α only efficiently elongates RNA primers at least 7 nucleotides long even though eukaryotic primase synthesizes large amounts of primers as short as 2 nucleotides long (28, 38). Additionally, primers are transferred intramolecularly between pol α and primase (28), analogous to the results with the herpes primase and polymerase (UL30-UL42) complexes. In the T7 bacteriophage, studies have similarly suggested direct “handoff” of the RNA primer from gp4 helicase-primase to the lagging strand polymerase (39, 40).
Analogous to the role UL42 plays to assist in intramolecular primer transfer, it has been suggested that the processivity factor of pol III, β, may also facilitate RNA primer transfer during DNA replication in E. coli (40). DnaG primase synthesizes primers 8–14 nucleotides long during lagging strand synthesis (41, 42). However, when pol III is added, shorter primers (<10 nucleotides) are more predominant (40). As a result of this change in primer length, it has been proposed that primer lengths may be controlled in Okazaki fragment synthesis, and the processivity factor, β, facilitates the access of pol III to the 3′-end of the primer and facilitates the transfer of the RNA primer (40).
Perhaps the most intriguing result was the efficient elongation of herpes primase-synthesized primers by human pol α. In eukaryotic cells, pol α exists in a tightly bound pol α-primase complex (p49-p58-p70-p180) and is thought to initiate synthesis of all new DNA strands via primase-coupled pol α activity (43, 44). Consistent with this role, pol α elongates RNA primer-templates and DNA primer-templates with similar efficiency (see below and Ref. 32). Pol α also elongates herpes primase-synthesized RNA primers much more efficiently than herpes polymerase. Whereas either UL30 or UL30-UL42 typically elongated <1% of the herpes primase-synthesized primers, an equal amount of pol α (measured in terms of dNTP polymerizing activity) elongated a much higher percentage (20–30%) of these primers. Unlike when UL30-UL42 elongates the primers, however, binase very effectively prevented pol α from elongating the primer-templates. Thus, either the RNA primers are transferred intermolecularly to pol α (i.e. being released by primase and pol α re-binding the primer) or primer transfer occurs intramolecularly, but the primer becomes solvent-exposed such that binase can hydrolyze it. The presence of human primase neither enhanced nor inhibited utilization of herpes primase generated primer-templates, consistent with previous studies demonstrating the independence of individual pol α and primase activities (28).
A priori, it remains unclear why herpes polymerase elongates primase-synthesized primers so inefficiently, particularly because herpes primase is a relatively slow enzyme (14). Possibly, herpes replication requires minimal levels of coupled activity, especially if initiation of new DNA strands largely occurs via a recombination mechanism (45). Alternatively, this low efficiency may indicate that these minimal assays containing just the helicase-primase and polymerase complexes lack critical component(s). For example, herpes encodes other replication proteins (UL9, UL29, etc.) that could modulate the ability of the polymerase to elongate primase-synthesized primers, or the herpes replication machinery might recruit a cellular factor to enable it to efficiently elongate an RNA primer.
Finally, and perhaps the most intriguing hypothesis, is that herpes replication requires pol α activity. This mechanism would be analogous to eukaryotic nuclear replication where (at least) two different polymerases are involved in synthesizing Okazaki fragments on the lagging strand and, presumably, leading strand initiation. After primase synthesizes an RNA primer, pol α polymerizes ∼15 dNTPs to generate a “DNA primer.” Pol α now dissociates, and pol δ/ε continues DNA synthesis (46). In the case of herpes replication, herpes primase would synthesize a primer that pol α elongates. After pol α dissociates, herpes polymerase, a high processivity polymerase analogous to the cellular pol δ/ε, performs the bulk of the DNA synthesis. Interestingly, when primase-coupled polymerase assays contained both pol α and herpes polymerase, templates containing a herpes primase initiation site were efficiently and completely replicated, a result very different from when assays lacked either DNA polymerase. Finally, any proposal to involve pol α in herpes replication would likely require the formation of a complex involving herpes polymerase, pol α, and herpes primase-helicase. This would be analogous to cellular replication that appears to involve large, multienzyme complexes containing three DNA polymerases (i.e. pol α-primase, pol δ, and pol ε in the human DNA synthesome (47, 48)).
We are not aware of any in vivo data that directly demonstrates a requirement for pol α activity during herpes DNA replication, and studies using inhibitors are equivocal on this issue. For example, treating herpes-infected cells with either aphidicolin or araA, two compounds that inhibit both herpes polymerase and pol α (in the case of araA, araATP is the inhibitor), results in the selection of resistant virus due to mutations in the polymerase. Compared with wild-type viruses, the respective resistant viruses are around 15-fold less sensitive to aphidicolin and 4-fold less sensitive to araA (49, 50). Unfortunately, the sensitivity of the polymerases from the resistant viruses to these inhibitors has not been characterized, and hence it remains unclear if changes in polymerase sensitivity correlate with changes in viral drug resistance. Thus, the partial resistance to these compounds could be due to either the herpes polymerase having a relatively low level of resistance or that herpes replication involves another drug-sensitive polymerase. Two other observations, however, provide support for the possible involvement of pol α. First, Lehman and co-workers (51) demonstrated that UL9, the HSV-1 origin-binding protein, interacts with pol α, thus providing a potential mechanism to recruit pol α to initiation sites. Second, immunofluorescence studies indicate that herpes replication compartments contain pol α (52). Although it is conceivable that pol α accidentally becomes entrained in replication compartments, it seems more likely that herpes specifically recruits the enzyme. We are presently exploring the possibility that pol α plays an essential catalytic function in herpes replication.
Acknowledgments
We thank Drs. Kathryn Ramirez-Aguilar, Deborah S. Parris, and Sandra Weller for helpful discussions during the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI59764 and GM073832. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HSV-1, herpes simplex virus 1; araA, 9-β-d-arabinofuranosyl adenosine; pol α, human DNA polymerase α; exo–, exonuclease-deficient; Sf9, Spodoptera frugiperda; ssDNA, single-stranded DNA.
Herpes primase can polymerize dNTPs as well as NTPs, although incorporation of dNTPs results in strong chain termination (Fig. 1A) (53). Incorporation of a dNTP into a short product also changes its electrophoretic mobility. Thus, the new radiolabeled products observed in assays containing [α-32P]dNTPs result from incorporation of a dNTP by primase.
Binase is a single-stranded RNase very closely related to barnase (54) and will hydrolyze RNA after both G and A. The ability of binase to hydrolyze double strand RNA primer-templates presumably arises from the transient fraying of the ends of the primer along with the high concentrations used in these assays.
Products due to primase-coupled polymerase activity can be detected much more readily when using [α-32P]dNTPs due to their having an ∼100-fold higher specific activity than the [α-32P]NTPs and that each product will contain ∼30 [32P]dNMPs versus just 2 [32P]NMPs.
Two additional control reactions indicate that the amount of binase used was sufficient to prevent the polymerase from using any primase-synthesized primers released into solution. First, varying the binase concentration from 3.3 to 33 μm did not alter the fraction of primase-synthesized primers elongated by UL30-UL42. Second, and as will be described in greater detail later, binase prevented pol α, an enzyme that uses RNA primer-templates orders of magnitude more efficiently than herpes polymerase, from elongating the herpes primase-synthesized primers.
References
- 1.Boehmer, P. E., and Lehman, I. R. (1997) Annu. Rev. Biochem. 66 347–384 [DOI] [PubMed] [Google Scholar]
- 2.Lehman, I. R., and Boehmer, P. E. (1999) J. Biol. Chem. 274 28059–28062 [DOI] [PubMed] [Google Scholar]
- 3.Crute, J. J., Tsurumi, T., Zhu, L. A., Weller, S. K., Olivo, P. D., Challberg, M. D., Mocarski, E. S., and Lehman, I. R. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 2186–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crute, J. J., and Lehman, I. R. (1991) J. Biol. Chem. 266 4484–4488 [PubMed] [Google Scholar]
- 5.Dodson, M. S., Crute, J. J., Bruckner, R. C., and Lehman, I. R. (1989) J. Biol. Chem. 264 20835–20838 [PubMed] [Google Scholar]
- 6.Dodson, M. S., and Lehman, I. R. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calder, J. M., and Stow, N. D. (1990) Nucleic Acids Res. 18 3573–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanguy Le Gac, N., Villani, G., Hoffmann, J. S., and Boehmer, P. E. (1996) J. Biol. Chem. 271 21645–21651 [DOI] [PubMed] [Google Scholar]
- 9.Tenney, D. J., Hurlburt, W. W., Micheletti, P. A., Bifano, M., and Hamatake, R. K. (1994) J. Biol. Chem. 269 5030–5035 [PubMed] [Google Scholar]
- 10.Marsden, H. S., McLean, G. W., Barnard, E. C., Francis, G. J., MacEachran, K., Murphy, M., McVey, G., Cross, A., Abbotts, A. P., and Stow, N. D. (1997) J. Virol. 71 6390–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkenberg, M., Bushnell, D. A., Elias, P., and Lehman, I. R. (1997) J. Biol. Chem. 272 22766–22770 [DOI] [PubMed] [Google Scholar]
- 12.Carmichael, E. P., and Weller, S. K. (1989) J. Virol. 63 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez-Aguilar, K. A., Low-Nam, N. A., and Kuchta, R. D. (2002) Biochemistry 41 14569–14579 [DOI] [PubMed] [Google Scholar]
- 14.Ramirez-Aguilar, K. A., and Kuchta, R. D. (2004) Biochemistry 43 1754–1762 [DOI] [PubMed] [Google Scholar]
- 15.Hernandez, T. R., and Lehman, I. R. (1990) J. Biol. Chem. 265 11227–11232 [PubMed] [Google Scholar]
- 16.Gottlieb, J., Marcy, A. I., Coen, D. M., and Challberg, M. D. (1990) J. Virol. 64 5976–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallo, M. L., Dorsky, D. I., Crumpacker, C. S., and Parris, D. S. (1989) J. Virol. 63 5023–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow, C. S., and Coen, D. M. (1995) J. Virol. 69 6965–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisshart, K., Chow, C. S., and Coen, D. M. (1999) J. Virol. 73 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhuri, M., Song, L., and Parris, D. S. (2003) J. Biol. Chem. 278 8996–9004 [DOI] [PubMed] [Google Scholar]
- 21.Biswas, N., and Weller, S. K. (1999) J. Biol. Chem. 274 8068–8076 [DOI] [PubMed] [Google Scholar]
- 22.Arana, M. E., Song, L., Tanguy Le Gac, N., Parris, D. S., Villani, G., and Boehmer, P. E. (2004) DNA Repair 3 659–669 [DOI] [PubMed] [Google Scholar]
- 23.Tenney, D. J., Hurlburt, W. W., Bifano, M., Stevens, J. T., Micheletti, P. A., Hamatake, R. K., and Cordingley, M. G. (1993) J. Virol. 67 1959–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallo, M. L., Jackwood, D. H., Murphy, M., Marsden, H. S., and Parris, D. S. (1988) J. Virol. 62 2874–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, D. K., Stigger, E., and Lee, S. H. (1996) J. Biol. Chem. 271 15124–15129 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. 5.49–5.53, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 27.Kincaid, K., Beckman, J., Zivkovic, A., Halcomb, R. L., Engels, J. W., and Kuchta, R. D. (2005) Nucleic Acids Res. 33 2620–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheaff, R. J., Kuchta, R. D., and Ilsley, D. (1994) Biochemistry 33 2247–2254 [DOI] [PubMed] [Google Scholar]
- 29.Chen, Y., Carrington-Lawrence, S. D., Bai, P., and Weller, S. K. (2005) J. Virol. 79 9088–9096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graves-Woodward, K. L., Gottlieb, J., Challberg, M. D., and Weller, S. K. (1997) J. Biol. Chem. 272 4623–4630 [DOI] [PubMed] [Google Scholar]
- 31.Sherman, G., Gottlieb, J., and Challberg, M. D. (1992) J. Virol. 66 4884–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, H. C., Sheaff, R. J., and Kuchta, R. D. (1995) Nucleic Acids Res. 23 4109–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel, T. A. (1985) J. Biol. Chem. 260 12866–12874 [PubMed] [Google Scholar]
- 34.Lesnik, E. A., and Freier, S. M. (1995) Biochemistry 34 10807–10815 [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto, N., Nakano, S., Katoh, M., Matsumura, A., Nakamuta, H., Ohmichi, T., Yoneyama, M., and Sasaki, M. (1995) Biochemistry 34 11211–11216 [DOI] [PubMed] [Google Scholar]
- 36.Marsden, H. S., Cross, A. M., Francis, G. J., Patel, A. H., MacEachran, K., Murphy, M., McVey, G., Haydon, D., Abbotts, A., and Stow, N. D. (1996) J. Gen. Virol. 77 2241–2249 [DOI] [PubMed] [Google Scholar]
- 37.Gottlieb, J., and Challberg, M. D. (1994) J. Virol. 68 4937–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vishwanatha, J. K., Yamaguchi, M., DePamphilis, M. L., and Baril, E. F. (1986) Nucleic Acids Res. 14 7305–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusakabe, T., Baradaran, K., Lee, J., and Richardson, C. C. (1998) EMBO J. 17 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benkovic, S. J., Valentine, A. M., and Salinas, F. (2001) Annu. Rev. Biochem. 70 181–208 [DOI] [PubMed] [Google Scholar]
- 41.Kitani, T., Yoda, K., Ogawa, T., and Okazaki, T. (1985) J. Mol. Biol. 184 45–52 [DOI] [PubMed] [Google Scholar]
- 42.Yoda, K., Yasuda, H., Jiang, X. W., and Okazaki, T. (1988) Nucleic Acids Res. 16 6531–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaguni, L. S., Rossignol, J. M., Conaway, R. C., and Lehman, I. R. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 2221–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu, S. Z., Wang, T. S., and Korn, D. (1984) J. Biol. Chem. 259 2602–2609 [PubMed] [Google Scholar]
- 45.Nimonkar, A. V., and Boehmer, P. E. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Podust, V. N., and Hubscher, U. (1993) Nucleic Acids Res. 21 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coll, J. M., Hickey, R. J., Cronkey, E. A., Jiang, H. Y., Schnaper, L., Lee, M. Y., Uitto, L., Syvaoja, J. E., and Malkas, L. H. (1997) Oncol. Res. 9 629–639 [PubMed] [Google Scholar]
- 48.Jiang, H. Y., Hickey, R. J., Abdel-Aziz, W., Tom, T. D., Wills, P. W., Liu, J., and Malkas, L. H. (2002) J. Cell. Biochem. 85 762–774 [DOI] [PubMed] [Google Scholar]
- 49.Tsurumi, T., Maeno, K., and Nishiyama, Y. (1987) J. Virol. 61 388–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall, J. D., Wang, Y. S., Pierpont, J., Berlin, M. S., Rundlett, S. E., and Woodward, S. (1989) Nucleic Acids Res. 17 9231–9244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, S. S., Dong, Q., Wang, T. S., and Lehman, I. R. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7882–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilcock, D., and Lane, D. P. (1991) Nature 349 429–431 [DOI] [PubMed] [Google Scholar]
- 53.Keller, K. E., Cavanaugh, N., and Kuchta, R. D. (2008) Biochemistry 47 8977–8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okorokov, A. L., Panov, K. I., Offen, W. A., Mukhortov, V. G., Antson, A. A., Karpeisky, M., Wilkinson, A. J., and Dodson, G. G. (1997) Protein Eng. 10 273–278 [DOI] [PubMed] [Google Scholar]