Abstract
Previous sodium ion transport results obtained by NMR methods are compared with those obtained by a new ISE technique and are found to correlate well with each other and with the biological activity against E. coli and B. subtilis.
Natural proteins that form ion-conducting channels1,2 or pores have been known for a century and have been actively studied for the last five decades.3 Ion-conducting synthetic peptides4,5 and non-peptide ion channels were developed relatively recently but the number of examples is increasing rapidly.6-9 Our effort to develop cation-conducting channels that we call hydraphiles10 has involved extensive efforts to characterize their position within the bilayer11-13 and their function. These model channel systems have proved successful in several important senses. They insert into the bilayer and they transport ions at a high rate. Moreover, they show the classical open–shut behavior typical of protein channels that conduct ions. A particular success of our channels was the identification of what we have called a “central relay unit” in the KcsA channel of Streptomyces lividans.14 Such a structural subunit was dictated in our channel design by chemical intuition but its presence in natural channels was unknown prior to this important solid state structure.
The hydraphile channels have four essential features. The two distal (outer) macrocycles serve as headgroups in the amphiphilic sense.15 They also serve as entry portals, although whether the transported cations pass through them16 or by them12 is unclear and may be channel-dependent. Covalent spacers determine the channel's overall span. The central relay links the two covalent spacer chains and provides a polar site within the bilayer's midplane. In this case, a third diaza-18-crown-6 macrocycle serves as the central relay.10 The efficacy of central relay units has been confirmed by structure–activity studies.17 Additional structure–activity studies have made it clear that when the central relay unit is a macrocycle, it is parallel to the membrane's hydrocarbon regime rather than parallel to the membrane surfaces.18 The fourth structural feature of this design comprises sidearms that are linked to the distal macrocycles. These are expected to insert into the bilayer and, depending on their shape and properties, align with the bilayer's hydrocarbon chains.
The hydraphile channels can be defined schematically by using the following structural shorthand:18 PhCH2<N18N>(CH2)n<N18N>(CH2)n<N18N>CH2Ph. We refer to this particular compound as “benzyl channel” 3. Wehave previously reported compounds 1–5;19 6 and 720 were prepared analogously and found to give satisfactory analytical data.
Assessing ion transport in synthetic channels has proved to be a challenge for all laboratories engaged in this effort.8 Early measurements of sodium transport were done for the hydraphile system in liposomes using 23Na NMR to detect the ion transport.21,22 The NMR method was remarkably useful in evaluating transport but it is experimentally complex. An important issue is that the results one obtains by this method depend on a very well-tuned NMR instrument, on buffer identity, and on the concentrations chosen for study. Notwithstanding the complexity of the method, we used it to demonstrate a clear, length dependence for transport activity in vesicles. As an alternative to this complex and instrument intensive method, we sought an assay that would be less operator-dependent and still be a reliable, convenient, and realistic assay for ion transport. We now report that ion-selective electrodes serve as the basis for this methodology and that the results obtained show an excellent correspondence with biological studies that were conducted independently.
Measurement of hydraphile-mediated Na+ transport using ion selective electrodes
The present methodology builds in part on recent work by Silberstein et al. who examined membrane destabilization, as mediated by small peptides, using a K+ selective electrode.23 In the studies reported here, large unilamellar vesicles (LUVs) were prepared using the reverse evaporation method of Skoza and Papahadjopoulos.24 A dry lipid film of 1,2-dioleoyl-sn-glycero-3-phosphocholine was dissolved in ether and a sodium-containing internal buffer (750 mM NaCl/15 mM N-2-hydroxyethylpiperazine (HEPES), pH 7.0) was added. Following ∼20 s sonication, the organic solvent was removed and the liposomes sized through a 200 μM polycarbonate filter. The internal buffer solution was exchanged for a sodium-free external buffer (750 mM cholineCl/15 mM HEPES, pH 7.0). Lipid concentration was assayed chemically25 and vesicle size was measured on a Coulter N4MD dynamic light scattering instrument.
Na+ transport measurements
Sodium transport was measured using a Micro-Combination pH/sodium electrode (Thermo Orion). After equilibration in an aqueous sodium-free external buffer, the electrode was placed in a 5 mL disposable beaker containing external buffer and vesicle solution to achieve a lipid concentration of 0.4 mM in a total volume of 2 mL. The voltage was recorded for five minutes prior to the addition of channel (12 μM). The hydraphile was added as a solution in 2-propanol and the conductivity was measured for 25 minutes. Addition of 200 μL of 10% aqueous n-octylglucoside induced complete lysis of the vesicles to achieve total sodium release. All data were normalized to total Na+ release and converted from mV to a concentration based on the calibration curve for the electrode.
The previously reported length dependence of Na+ transport for 1–5 is illustrated in the top panel of Fig. 1 (open squares, dotted line). These values were determined by the 23Na NMR method. They are included to calibrate the new data, obtained by the ISE method, for the expanded set of compounds 1–7. The ISE data were acquired as described above and are shown as filled circles (solid line). The connecting lines in both cases are to guide the eye and do not represent a mathematical fit.
Fig. 1.
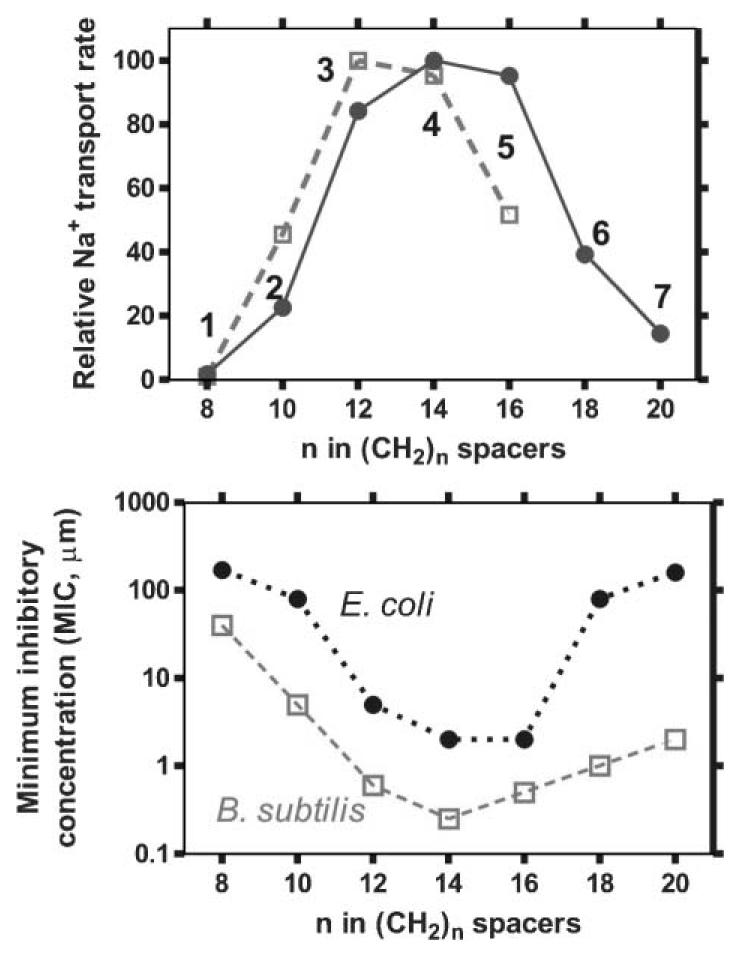
Top. Plot (open squares) of Na+ transport from liposomes mediated by 1–5 as determined by the 23Na NMR method. Plot (filled circles) of fractional Na+ release at 1500 seconds from phospholipid vesicles vs. the number of methylenes in the spacer chains as measured for 1–7 by ion selective electrode methods. Bottom. Activity (expressed as minimum inhibitory concentration, or MIC, in μM) of 1–7 against the Gram-negative bacterium E. coli (filled circles) and the Gram-positive bacterium Bacillus subtilis (open squares).
The ISE data were acquired as described herein in liposomes prepared from 1,2-dioleoyl-sn-glycero-3-phosphocholine. The NMR experiments were done using liposomes prepared from phosphatidyl glycerol and phosphatidyl choline (1 : 2 w/w). Despite the differences in vesicles and methodology, the chain length dependence of channel function determined by the two methods agrees well. Further, the two additional, longer-chained channels (6 and 7) show the declining activity trend only suggested by the previously reported NMR results. The consistency of these results confirms the value of the ISE method for the assay of synthetic ion channel function.
The function of the benzyl channel was of particular interest because we recently found that it is toxic to E. coli.26 It seemed possible that channel function in liposomal bilayers might parallel biological activity for an array of channel compounds. We therefore determined the minimum inhibitory concentration (MIC, in μM) for compounds 1–7 as the lowest two-fold dilution of hydraphile that prevented bacterial growth, as outlined by the NCCLS.27 The results are plotted in the lower panel of Fig. 1. Note that the ordinate is logarithmic and that 4 (C14) and 5 (C16) are more than 100-fold more toxic to all three organisms than 1 (C8) and 2 (C10). The activity of the hydraphiles diminishes as the compounds are further lengthened to C18 (6) and C20 (7) spacer chains. Compound 4 is the most toxic to both organisms, with extremely low MIC values of 0.5 μM for B. subtilis and 2 μM for E. coli. For comparison, the most active synthetic channel compounds reported previously are the cyclic peptides of Ghadiri et al., which kill B. subtilis at concentrations as low as 2.5 μM and E. coli as low as 5.5 μM.28
To our knowledge, this is the first example of a synthetic channel series that shows parallel, length-dependent behavior both in synthetic liposomes and living organisms. Indeed, the sum of these results indicates that, as far as synthetic ion channels are concerned, length truly does matter.
Acknowledgments
We warmly thank the laboratory of Shin-Ichiro Imai M.D., PhD for their generosity with biological equipment and supplies. We thank the NIH for a grant (GM-36262) that supported this work and for Training Grant Fellowships under grants T32-GM08785 and T32-GM04892 to MEW and WML, respectively.
Notes and references
- 1.Aidley DJ, Stanfield PR. Ion Channels: Molecules in Action. Cambridge University Press; Cambridge: 1996. [Google Scholar]
- 2.Hille B. Ionic Channels of Excitable Membranes 3/e. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- 3.Katz B. Arch. Sci. Physiol. 1949;3:285–299. [Google Scholar]
- 4.Åkerfeldt KS, Lear JD, Wasserman ZR, Chung LA, DeGrado WF. Acc. Chem. Res. 1993;26:191–197. [Google Scholar]
- 5.Montal M. Annu. Rev. Biophys. Biomol. Struct. 1995;24:31–57. doi: 10.1146/annurev.bb.24.060195.000335. [DOI] [PubMed] [Google Scholar]
- 6.Cross GG, Fyles TM, Montoya-Pelaez PJ, Van Straaten-Nijenhuis WF, Zhou X. Interfacial Design and Chemical Sensing. ACS Symp. Ser. 1994;561:38–48. Chem. Abstr.: 122:39529. [Google Scholar]
- 7.Gokel GW, Murillo O. Acc. Chem. Res. 1996;29:425–432. [Google Scholar]
- 8.Gokel GW, Mukhopadhyay A. Chem. Soc. Rev. 2001;30:274–286. [Google Scholar]
- 9.Fyles TM, Van Straaten-Nijenhuis WF. In: Comprehensive Supramolecular Chemistry. Reinhoudt DN, editor. Vol. 10. Oxford; Pergamon: 1996. pp. 53–77. [Google Scholar]
- 10.Gokel GW. Chem. Commun. 2000:1–9. [Google Scholar]
- 11.Abel E, Maguire GEM, Meadows ES, Murillo O, Jin T, Gokel GW. J. Am. Chem. Soc. 1997;119:9061–9062. [Google Scholar]
- 12.Murillo O, Suzuki I, Abel E, Gokel GW. J. Am. Chem. Soc. 1996;118:7628–7629. [Google Scholar]
- 13.Gokel GW, Ferdani R, Liu J, Pajewski R, Shabany H, Uetrecht P. Chem.–Eur. J. 2001;7:33–38. doi: 10.1002/1521-3765(20010105)7:1<33::aid-chem33>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 15.Muñoz S, Mallén J, Nakano A, Chen Z, Gay I, Echegoyen L, Gokel GW. J. Am. Chem. Soc. 1993;115:1705–1711. [Google Scholar]
- 16.Murillo O, Abel E, Maguire GEM, Gokel GW. Chem. Commun. 1996:2147–2148. [Google Scholar]
- 17.Murray CL, Shabany H, Gokel GW. Chem. Commun. 2000:2371–2372. [Google Scholar]
- 18.Murillo O, Watanabe S, Nakano A, Gokel GW. J. Am. Chem. Soc. 1995;117:7665–7679. [Google Scholar]
- 19.Murray CL, Gokel GW. J. Supramol. Chem. 2001;1:23–30. [Google Scholar]
- 20.Weber ME, Schlesinger PH, Gokel GW. J. Am. Chem. Soc. doi: 10.1021/ja044936+. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riddell FG, Arumugam S, Cox BG. J. Chem. Soc., Chem. Commun. 1987:1890–1891. [Google Scholar]
- 22.Riddell FG, Arumugam S. Biochim. Biophys. Acta. 1989;984:6–10. [Google Scholar]
- 23.Silberstein A, Mirzabekov T, Anderson WF, Rozenberg Y. Biochim. Biophys. Acta. 1999;1461:103–112. doi: 10.1016/s0005-2736(99)00152-2. [DOI] [PubMed] [Google Scholar]
- 24.Szoka FJ, Papahadjopoulos D. Proc. Natl. Acad. Sci. USA. 1978;75:4194–4198. doi: 10.1073/pnas.75.9.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart JCM. Anal. Biochem. 1980;104:10–14. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- 26.Leevy WM, Donato GM, Ferdani R, Goldman WE, Schlesinger PH, Gokel GW. J. Am. Chem. Soc. 2002;124:9022–3. doi: 10.1021/ja017052o. [DOI] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards (NCCLS) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that grow Aerobically. 5th edn. M7-A5. Wayne; Pennsylvania: 2000. [Google Scholar]
- 28.This value is based on a MW of 1196 for KQRWLWLW @ 3 μg mL−1 for B. subtilis, and a MW of 910 for RRLWLW for E. coli as reported in: Fernandez-Lopez S, Kim HS, Choi EC, Delgado M, Granja JR, Khasanov A, Kraehenbuehl K, Long G, Weinberger DA, Wilcoxen KM, Ghadiri MR. Nature. 2001;412:452–455. doi: 10.1038/35086601.


