Abstract
TRIM5α is a host protein that can bind to incoming retroviral capsid (CA) and inhibit retroviruses in a species-specific manner. The CA protein of HIV-1 also interacts with high affinity to the host protein cyclophilin A (CypA). This binding has been shown to positively affect some early stage of the viral life-cycle in human cells. However, the CypA/CA interaction also renders HIV-1 more susceptible to rhesus TRIM5α (rhTRIM5α) restriction. We find that the ability of old world monkey TRIM5α genes to restrict HIV-1 in a CypA-dependent manner is widespread. On the other hand, we find that simian immunodeficiency viruses from tantalus monkeys (SIVagmTAN), is unlike HIV-1 in that CypA does not enhance the rhTRIM5α restriction against the virus even though the CA of this virus, like HIV-1, does bind CypA. Mapping of the determinants for this phenotype by swapping regions on CA between SIVagmTAN and HIV-1 showed that when SIVagmTAN contains loops between helices 4/5 (4–5 loop) and 6/7 (6–7 loop) from HIV-1 CA, it becomes susceptible to the CypA-enhanced rhTRIM5α restriction. Surprisingly, when SIVagmTAN contains either loop from HIV-1 CA, it gains sensitivity to TRIM5α from species which originally have no effect on the wild-type virus. Moreover, we find that CypA/CA interaction occurs early after viral entry but the CypA-enhanced restriction mostly acts on the stage after reverse transcription.
Introduction
Hosts apply many strategies to counteract viral infections. In addition to the well known responses of the innate and adaptive immune systems, other host proteins that are constitutively expressed and are cell autonomous have also been shown to protect cells from retroviral infections (Bieniasz, 2004). Among these factors, the tripartite motif 5 isoform alpha (TRIM5α) protein can inhibit retroviral infection in a species-specific manner (Stremlau et al., 2004). For example, TRIM5α from rhesus macaque (rhTRIM5α) inhibits simian immunodeficiency viruses from tantalus monkeys, a subspecies of African green monkeys (SIVagmTAN) but not SIVs from rhesus macaque (SIVmac). On the other hand, African green monkey TRIM5α (agmTRIM5α) inhibits SIVmac but not SIVagmTAN. Both rh and agmTRIM5α strongly inhibit HIV-1 infection, while human TRIM5α does not restrict SIVmac and SIVagm, and only weakly inhibits HIV-1 (reviewed in (Newman and Johnson, 2007; Towers, 2005)). Binding of TRIM5α to the viral CA is necessary, but is not sufficient for restriction of retroviruses (Javanbakht et al., 2005; Perez-Caballero et al., 2005a).
Cyclophilin A (CypA) is a highly conserved peptidyl prolyl isomerase that also binds to HIV-1 capsid (CA) (Franke, Yuan, and Luban, 1994; Luban et al., 1993; Thali et al., 1994). HIV-1 directly interacts with the CypA active site by virtue of residues in a loop between the fourth and fifth alpha helices of CA (4–5 loop) (Bukovsky et al., 1997; Gamble et al., 1996). This interaction can be blocked by cyclosporine A (CsA), an immunosuppressive drug. CsA binds to the same active site of CypA and disrupts the CypA/CA interaction which then attenuates HIV-1 infectivity by 2–5 fold in some T cell lines (Franke and Luban, 1996; Hatziioannou et al., 2005; Sokolskaja, Sayah, and Luban, 2004). We and others have shown that SIVagmTAN, and feline immunodeficiency viruses encode CA proteins that can interact also with CypA (Diaz-Griffero et al., 2006; Lin and Emerman, 2006; Zhang et al., 2006), although CA of other lentiviruses such as SIVmac and equine infectious anemia viruses do not interact with CypA (Braaten, Franke, and Luban, 1996b; Lin and Emerman, 2006). Therefore, CypA/CA interaction is a common, but not universal phenotype among lentiviruses.
Interestingly, the positive effect of CypA/CA interaction on HIV-1 replication in human cells is reversed in some old world monkey cells. This is because the interaction of CypA with HIV-1 CA renders HIV-1 more susceptible to the restriction from rh and agmTRIM5α proteins (Berthoux et al., 2005; Keckesova, Ylinen, and Towers, 2006; Sokolskaja, Berthoux, and Luban, 2006; Stremlau et al., 2006b). Therefore, the ability of HIV-1 CA to bind CypA enhances virus replication in many human cells, but negatively affects its ability to infect old world primate cells.
In this report, we identified the regions on CA that render HIV-1 more susceptible to the TRIM5α restriction when bound by CypA. First, we show CypA has no effect on enhancing the restriction of another CypA-binding virus, SIVagmTAN. By generating chimeric SIVagmTAN containing fragments from HIV-1 CA, we mapped the determinants on HIV-1 CA that make HIV-1 more sensitive to TRIM5α after CypA-binding to two regions located at the 4–5 loop in CA and another loop between the sixth and seventh helices (6–7 loop). These two regions of HIV-1 CA are both necessary to transfer the CypA-dependent TRIM5α sensitivity to SIVagmTAN. Moreover, we found that these regions determine viral sensitivity to TRIM5αs even in the absence of CypA. Consistent with previous reports (Keckesova, Ylinen, and Towers, 2006; Stremlau et al., 2006b), our data show that TRIM5α restriction of HIV-1 is composed of both CypA-dependent and CypA-independent components. Here we show that the CypA-enhanced TRIM5α restriction effect occurs early after viral entry and persists for 4–8 hours after infection, and appears to occur mostly after reverse transcription. These results identify the viral determinants that render the HIV-1 CA susceptible to a CypA-dependent TRIM5α restriction.
Results
Old world monkey TRIM5α restriction against HIV-1 has both CypA-dependent and CypA-independent components
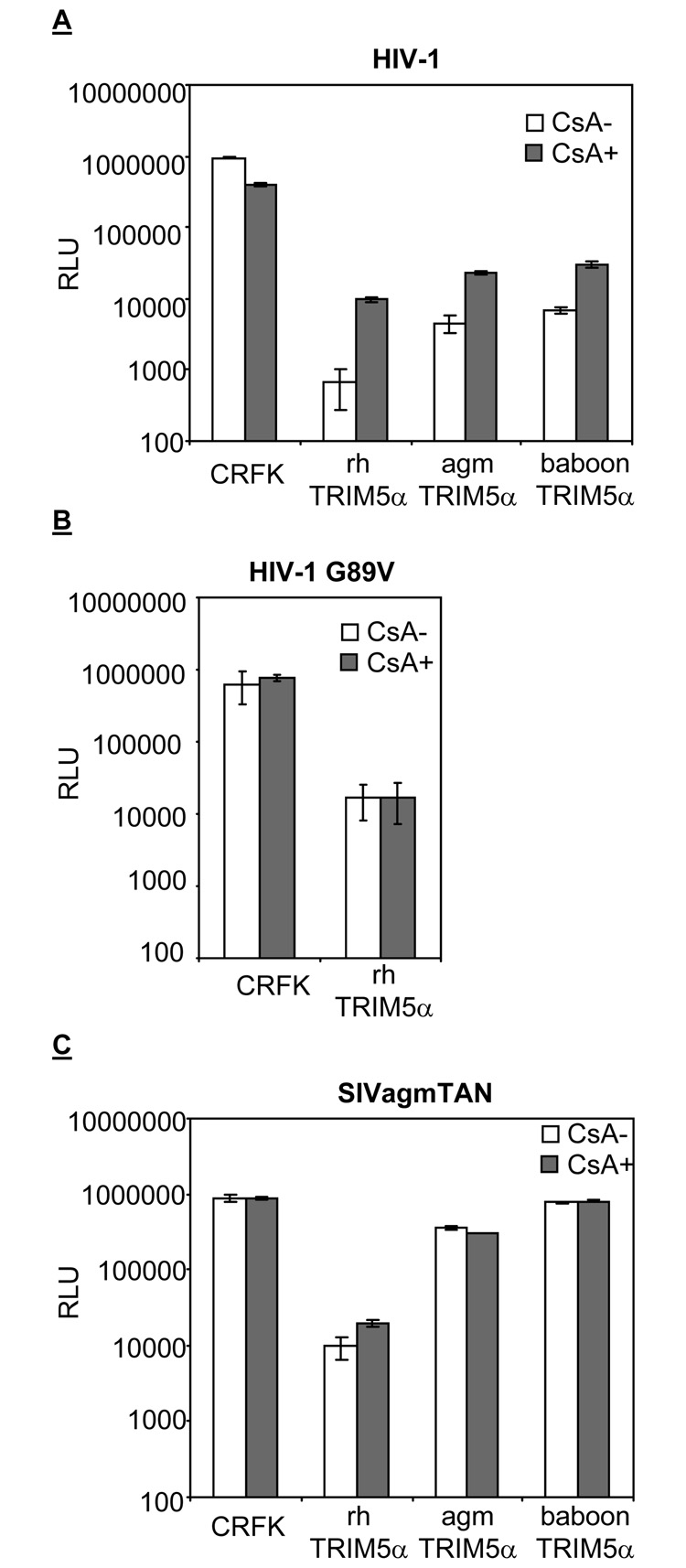
The CypA-dependent TRIM5α restriction against HIV-1 was observed in rhesus macaque and african green monkey cells (Berthoux et al., 2005; Keckesova, Ylinen, and Towers, 2006; Stremlau et al., 2006b). These reports suggest that the CypA/CA interaction renders HIV-1 more susceptible to the restriction by TRIM5αs from old world primates. BaboonTRIM5α also potently inhibits HIV-1 infection (Kaiser, Malik, and Emerman, 2007). Thus, we examined the CypA-dependence of all three TRIM5α restriction factors in parallel in the same cell type. CRFK cells expressing rh, agm, and baboonTRIM5α were generated and infected with VSV-G pseudo-typed wild-type HIV-1 (Figure 1A). CsA was added upon infection to validate the dependence of TRIM5α restriction on CypA/CA interaction. Infections were done with viruses encoding luciferase gene as a reporter, and the relative luminescence unit (RLU) after infection was used as an indicator for infection. The luciferase reporter gives us a longer dynamic assay range than the GFP reporter system, therefore the low level of infectivity under TRIM5α restriction can still be within the linear range of the assay.
Figure 1. Cyclophilin A/capsid interaction renders HIV-1, but not SIVagmTAN, more susceptible to rh, agm, and baboonTRIM5α restriction.
CRFK and CRFK cells expressing rh, agm, and baboonTRIM5α were infected with wild-type HIV-1 (A), HIV-1 G89V (B), and SIVagmTAN (C) in the presence (CsA+, filled boxes) or absence (CsA-, open boxes) of CsA. The infected cells were analyzed by luminometer and the infectivity is presented as relative luciferase unit (RLU, on a log scale).
Consistent with previous reports, CypA/CA interaction renders HIV-1 more susceptible to rhTRIM5α restriction (Berthoux et al., 2005). That is, addition of CsA to block CypA/CA interaction rescues HIV-1 infection from rhTRIM5α restriction by at least 10 fold (t-test, p value< 0.05). As a control we also used HIV-1 G89V, a mutant that does not bind CypA (Franke, Yuan, and Luban, 1994). Consistent with previous reports that used other readouts for infection (Berthoux et al., 2005; Keckesova, Ylinen, and Towers, 2006; Sokolskaja, Berthoux, and Luban, 2006; Stremlau et al., 2006b), HIV-1 G89V is susceptible to the rhTRIM5α inhibition, but the level of infectivity is not affected by the addition of CsA (Figure 1B). On the other hand, adding CsA to block CypA/CA interaction can rescue HIV-1 infection from agm and baboonTRIM5α by about 4 fold (t-test, p <0.05). This suggests that CypA-dependent restriction of TRIM5α is wide-spread among the old world primate TRIM5α genes.
The data in Figure 1 also shows that disruption of CypA/CA interaction with CsA is not able to fully rescue HIV-1 infectivity in CRFK cells. Although the disruption of CypA/CA interaction by CsA rescuse HIV-1 infection by 10 fold, it is still 100-fold lower than that in CRFK cells that express no TRIM5α (Figure 1A). This finding is consistent with previous reports (Keckesova, Ylinen, and Towers, 2006; Stremlau et al., 2006b). The same phenotype is also observed in agm and baboonTRIM5α restriction against HIV-1 infection (Figure 1A). Therefore, these data suggest that rh, agm, and baboonTRIM5α are potent inhibitors against HIV-1 infection, but that the restriction consists of both CypA-independent and CypA-dependent stages.
TRIM5α restriction against SIVagmTAN is not CypA-dependent
SIVagmTAN is susceptible to rhTRIM5α, slightly inhibited by agmTRIM5α, and resistant to baboonTRIM5α restriction (Figure 1C). Because SIVagmTAN, like HIV-1, is able to bind CypA (Lin and Emerman, 2006; Zhang et al., 2006) and is susceptible to rhTRIM5α restriction, we examined whether or not the CypA/CA interaction in SIVagmTAN also renders the virus more susceptible to TRIM5α restriction. We found that, unlike HIV-1, the restriction of SIVagmTAN by rhTRIM5α is not affected by CsA (Figure 1C). In rhTRIM5α-expressing cells, the infectivity of SIVagmTAN only increases by 2-fold in the presence of CsA (p value>0.05, Figure 1C). We verified that CsA does block CypA/CA interaction in SIVagmTAN by examining the infection of the virus in TRIMCyp-expressing CRFK cells with or without CypA ((Lin and Emerman, 2006) and data not shown)). This indicates that old world monkey TRIM5α restriction is not universally dependent on CypA, but rather is virus-specific. The ability of viral CA to interact with CypA does not necessarily result in the CypA-dependent restriction.
The CypA-dependent TRIM5α restriction is determined by loops between the 4th/5th and the 6th/7th helices on viral capsid
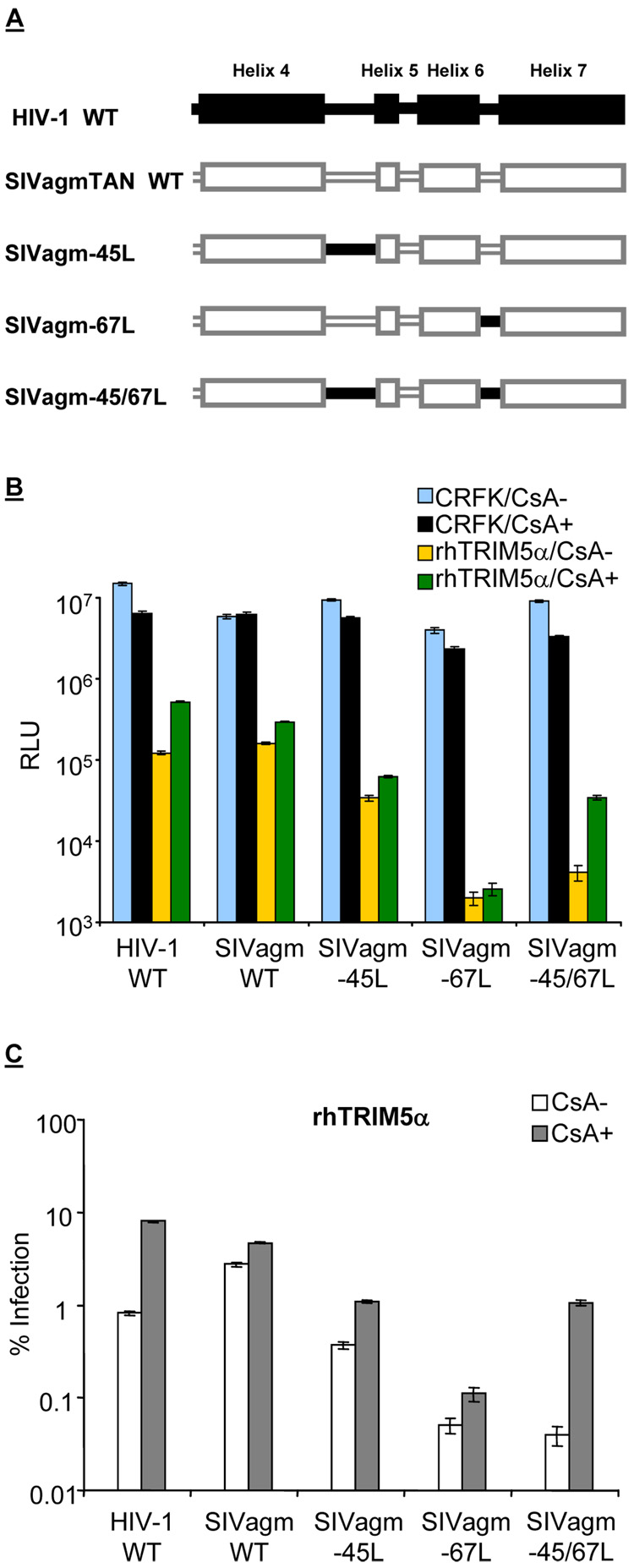
Since HIV-1 and SIVagmTAN both bind CypA but their susceptibilities to CypA-dependent rhTRIM5α restriction are different, we speculated that viral capsid determines this phenotypic variation. Hatziioannou et al. reported that the 4–5 loop and another loop between the 6th and 7th helices (6–7 loop) on HIV-1 CA modulate HIV-1 tropism in different primate cells (Hatziioannou et al., 2004). Therefore, we investigated if these loops are determinants for the CypA-dependent TRIM5α restriction with chimeric SIVagmTAN where the 4–5 loop, 6–7 loop, and both 4–5 and 6–7 loops were replaced by those parallel ones from HIV-1 CA (Figure 2A). Accordingly, these chimeric SIVagmTAN were named SIVagm-45L, SIVagm-67L, and SIVagm-45/67L (Figure 2A). These recombinant SIVagm were pseudotyped with VSV-G and used to infect cells expressing rhTRIM5α in the presence or absence of CsA (Figure 2B). The amount of virus that gives about 107 RLU was used to infect CRFK cells (blue boxes, Figure 2B). Similar to the CypA effect for HIV-1 replication in human cells, the infections of wild-type HIV-1, SIVagm-45L, SIVagm-67L, and SIVagm-45/67L were slightly affected in CRFK cells in the presence of CsA (compare blue boxes with black boxes, Figure 2B).
Figure 2. HIV-1 susceptibility to the CypA-enhanced TRIM5α restriction is determined by the loop between the 4th and 5th helices and the loop between 6th and 7th helices on viral capsid.
(A) Diagram of chimeric SIVagm-45L, SIVagm-67L, and SIVagm-45/67L in which the loop between the 4th and 5th helices or the loop between 6th and 7th helices or both loops on SIVagmTAN capsid were replaced by those from HIV-1. (B) CRFK and CRFK cells expressing rhTRIM5α were infected with wild-type HIV-1, SIVagmTAN, and recombinant SIVagm in the presence or absence of CsA. Infected cells were harvested and analyzed by luminometer, and the infectivity is presented as relative luciferase unit (RLU, on a log scale). (C) The normalization of infection from (B). The open boxes were obtained by dividing the RLU from the yellow boxes of each infection in Panel B by the blue boxes in Panel B times 100 ,and the filled boxes were by dividing the RLU from the green boxes of each infection in Panel B by the black boxes in Panel B times 100.
We find that each of the chimeric SIVagm viruses (Figure 2A) is more susceptible to rhTRIM5α inhibition than wild-type SIVagmTAN (Figure 2B, yellow boxes compared to black boxes). Because of this observation and because there are different viruses in the experiment, we set the viral infectivity in CRFK cells (without a primate TRIM5α) as 100% and normalized the infectivity in TRIM5α-expressing cells based on it (Figure 2C). That is, the infectivity in rhTRIM5α/CsA- (yellow boxes, Figure 2B) was normalized to the infectivity in CRFK/CsA- (blue boxes, Figure 2B) to obtain the open boxes in Figure 2C, and the infectivity in rhTRIM5α/CsA+ (green boxes, Figure 2B) was normalized to the infectivity in CRFK/CsA+ (black boxes, Figure 2B) to obtain the filled boxes in Figure 2C so that the relative effects of CsA on the TRIM5α inhibition can be directly compared.
We find that the level of the CypA-dependent rhTRIM5α restriction against chimeric SIVagm-45L is similar to that of SIVagmTAN, not HIV-1 (filled boxes, Figure 2C). This suggests that the 4–5 loop, which is important for the CypA/CA interaction, is not sufficient for the viral susceptibility to the CypA-dependent TRIM5α restriction. Likewise, the level of the CypA-dependent rhTRIM5α restriction against chimeric SIVagm-67L remains similar to that of SIVagmTAN, not HIV-1 (filled boxes, Figure 2C). Remarkably, however, the chimeric SIVagm-45/67L (with both the 4–5 and 6–7 loops exchanged from HIV-1, Figure 2A) not only became more sensitive to TRIM5α but also gained the susceptibility to the CypA-dependent restriction (Figure 2C). These data suggest that the 4–5 and 6–7 loops form a domain that confers cyclophilin-dependent viral susceptibility to TRIM5α restriction.
Loops between the 4th/5th and the 6th/7th helices on viral capsid affect viral susceptibility to TRIM5α restriction
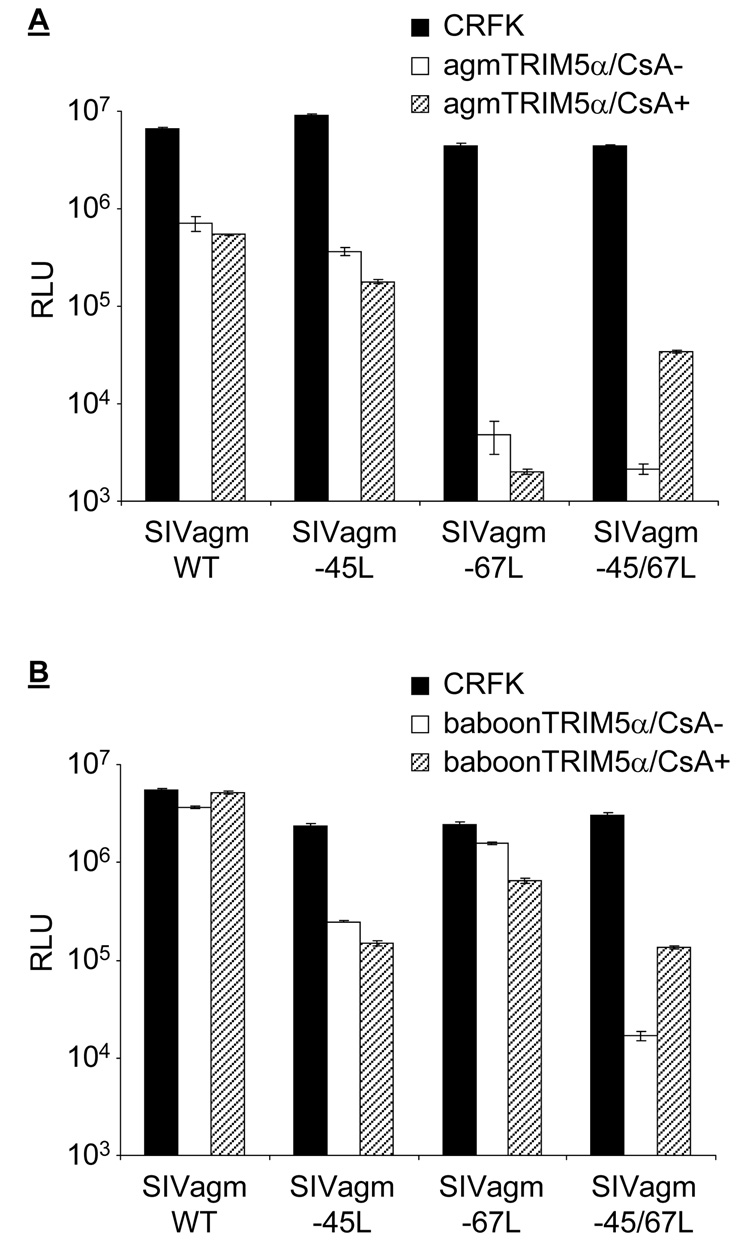
We noticed that chimeric SIVagm-45L, SIVagm-67L, and SIVagm-45/67L are more sensitive than wild-type SIVagmTAN to rhTRIM5α restriction (Figure 2C; notice the open boxes of each SIVagm chimeric virus compared to SIVagm WT). To examine whether these recombinant viruses are generally more sensitive to TRIM5αs, SIVagm-45L, SIVagm-67L and SIVagm-45/67L were used to infect CRFK and CRFK cells which express agm or baboonTRIM5α (Figure 3). When the 4–5 loop on SIVagm CA was replaced by that from HIV-1, the susceptibility of SIVagm-45L to agmTRIM5α is similar to that of wild-type SIVagm (Figure 3A, compare the open boxes). However, when 6–7 or both 4–5 and 6–7 loops on SIVagm CA were replaced, the chimeric SIVagm viruses become more susceptible than the wild-type virus to agmTRIM5α restriction (open boxes, Figure 3A). On the other hand, the wild-type SIVagmTAN and the chimeric SIVagm-67L are resistant to the inhibition from baboonTRIM5α, but recombinant SIVagm with the 4–5 or both 4–5 and 6–7 loops from HIV-1 CA gain susceptibility to baboonTRIM5α restriction (open boxes, Figure 3B). Thus, both CA loops from HIV-1 independently confer different sensitivities to TRIM5α from different old world monkeys.
Figure 3. Viral susceptibility to TRIM5α restriction is affected by loops between the 4th/5th and 6th/7th helices on capsid.
Infection of wild-type SIVagmTAN, chimeric SIVagm-45L, SIVagm-67L, and SIVagm-45/67L in agmTRIM5α (A) and baboonTRIM5α (B)-expressing cells. Infected cells were harvested and analyzed by luminometer, and the infectivity is presented as relative luciferase unit (RLU, on a log scale).
We also examined the dependence of CypA on these Trim5α restrictions. The CypA/CA interaction does not enhance agm and baboonTRIM5α restriction against wild-type SIVagmTAN, SIVagm-45L, and SIVagm-67L (Figure 3, hatched boxes). However, when both the 4–5 and 6–7 loops on SIVagmTAN (SIVagm-45/67L) were replaced by those from HIV-1 CA, the CypA/CA interaction sensitizes this mutant virus to the CypA-dependent TRIM5α restriction. These results indicate that while the 4–5 loop and the 6–7 loop can each alter the species-specific susceptibility of viruses to TRIM5αs from different species, both loops are necessary for the viral susceptibility to the CypA-dependent TRIM5α restriction for all three of the TRIM5α genes tested (Figure 2 and Figure 3). Therefore, the phenotype of HIV-1 that encodes a capsid which becomes more susceptible to TRIM5α proteins after binding to CypA can be transferred to another retrovirus by transferring parts of the CA protein in addition to transferring susceptibility in general to TRIM5α from old world monkeys.
The CypA-dependent TRIM5α restriction occurs early after viral entry, but is mostly post-reverse transcription
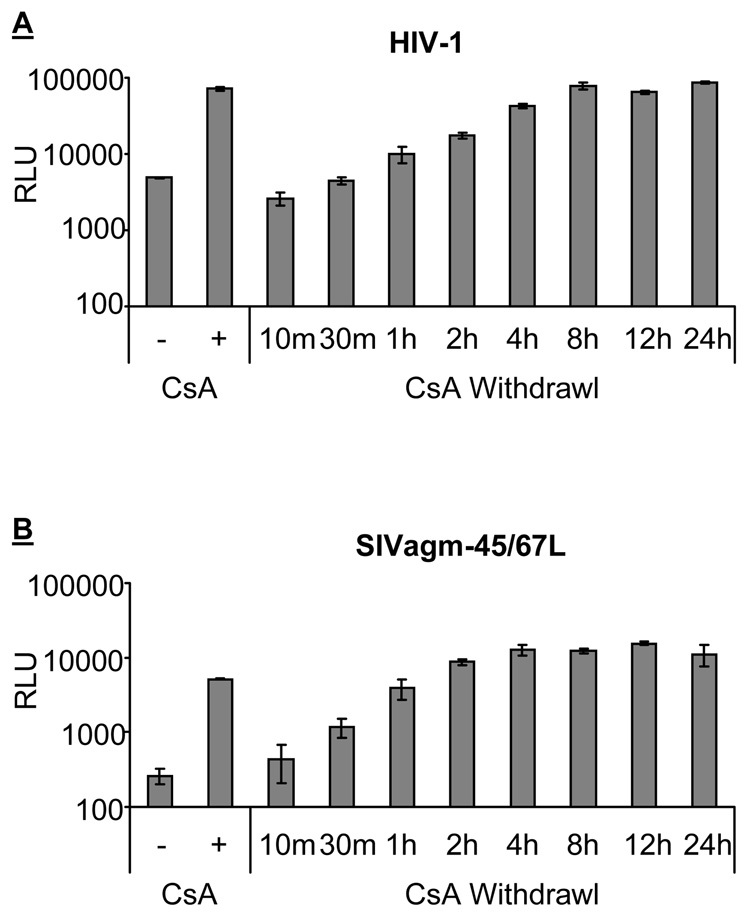
In human cells, CypA is important for the early step of HIV-1 replication (Braaten, Franke, and Luban, 1996a). To determine how early the CypA-dependent TRIM5α restriction affects HIV-1 replication, we conducted a time course experiment by withdrawing CsA from infected rhesus cells. CsA was present when adding HIV-1 to cells and was removed at different time points (Figure 4A). When CsA was withdrawn 10 minutes or 30 minutes after infection, the infectivity of HIV-1 is similar to the infectivity in cells without the presence of CsA. This indicates that 30 minutes of CsA treatment is not enough to overcome the CypA-dependent rhTRIM5α restriction. When CsA was removed 1 hour after infection, the infectivity of HIV-1 increases by 2-fold when compared to the infectivity in cells without the addition of CsA (p value<0.05, Figure 4A). The level of HIV-1 infectivity gradually increases with the time of CsA treatment. When CsA was withdrawn at 8 hours after infection, the infectivity of HIV-1 reached the level where CsA was present throughout the experiment. This data suggests that the CypA-dependent TRIM5α restriction against HIV-1 infection occurs early and persists at least the first 8 hours of replication cycle.
Figure 4. Cyclophilin A-enhanced TRIM5α restriction occurs in early viral life cycle.
Time course experiment of withdrawing CsA from wild-type HIV-1 (A) and SIVagm-45/67L (B) infected rhesus macaque (SMAGI) cells. Viruses were spinoculated at 8°C for 20 minutes and transferred to a 37°C incubator to synchronize viral entry. The presence (CsA+) and absence (CsA−) of cyclosporin A (CsA) throughout the experiment was used as controls. For the withdrawal groups, CsA was added to cells when transferring plates to a 37°C incubator, and the time was set as time 0. CsA was then withdrawn at 10 minutes, 30 minutes, 1 hour, 2 hours, 4 hours, 8 hours, 12 hours, or 24 hours after time 0. Infected cells were harvested after 48 hours and analyzed by luminameter. The infectivity is presented as RLU (on a log scale).
We also examined the time course of rhTRIM5α restriction against SIVagm-45/67L by withdrawing CsA at different time points after infection (Figure 4B). The time course of the CypA-dependent rhTRIM5α restriction against SIVagm-45/67L shows similar pattern as that of HIV-1, although the kinetics of CsA-withdrawn effect in SIVagm-45/67L is faster than that in HIV-1. The presence of CsA in the first 30 minutes already starts to rescue SIVagm-45/67L from the CypA-dependent rhTRIM5α restriction. The level of SIVagm-45/67L infectivity also gradually increases with the time when CsA was removed. Taken together, the data suggest that CypA-dependent TRIM5α restriction against chimeric SIVagm-45/67L occurs early after viral entry, and CypA/CA interaction no longer enhances rhTRIM5α two hours after viral entry.
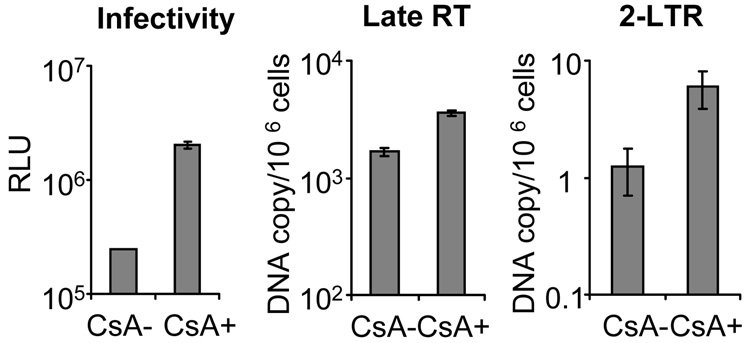
It has been proposed that TRIM5α restriction against retroviral infection can be divided to the inhibition before reverse transcription and the inhibition after reverse transcription (Anderson et al., 2006; Wu et al., 2006). To examine which phase of the HIV-1 life cycle is affected by the CypA-dependent TRIM5α restriction, we used a real-time PCR assay to study the late reverse transcription products (late RT) and 2-LTR circles under rhTRIM5α restriction by infection of rhesus cells (endogenous rhTRIM5α) with or without CsA (Figure 5). The infectivity of HIV-1 is inhibited by rhTRIM5α, and the addition of CsA upon infection can rescue HIV-1 from the restriction by 10-fold. However, when we examined the late RT harvested from HIV-1 infected rhTRIM5α-expressing cells, we found that the CsA addition brings up the RT product level by only about 2-fold. On the other hand, the addition of CsA increases about 5-fold more nuclear 2-LTR circles when compared to the group without CsA treatment. Similar results were observed in CRFK cells that exogenously express rhTRIM5α (data not shown). These results suggest that the CypA-dependent rhTRIM5α restriction can impact HIV-1 replication at reverse transcription but mostly affects the stages after reverse transcription.
Figure 5. Cyclophilin A-enhanced TRIM5α restriction impacts viral replication in the stage after reverse transcription.
SMAGI cells were infected with wild-type HIV-1 in the presence (CsA+) or absence (CsA−) cyclosporine A. Infected cells were harvested 24 hours after infection and the viral DNA were collected for the detection of late reverse transcription products (late RT) and 2-LTR circles (2-LTR) by real-time PCR. The overall infectivity was analyzed by luminometer 48 hours after infection and presented as RLU (on a log scale).
Discussion
The capsid protein from HIV-1 and SIVagmTAN can both interact with CypA (Lin and Emerman, 2006; Zhang et al., 2006). However, in this report we show that SIVagmTAN, unlike HIV-1, is restricted by rhTRIM5α independent of CypA/CA interaction. We used this difference to map the viral determinant for this phenotype, and showed that two loops on HIV-1 capsid, one between the 4th and 5th helices and the other between the 6th and 7th helices, are responsible for the HIV-1 susceptibility to the CypA-dependent TRIM5α restriction. Interestingly, just by replacing one of the two loops from HIV-1, SIVagmTAN gains susceptibility to the TRIM5αs that originally had little effect on the wild-type virus. Moreover, we report here that the CypA-dependent TRIM5α restriction against lentiviral infection occurs early and affects replication mostly after reverse transcription.
It has been reported that in the absence of the vif gene, CypA is incorporated into SIVagm from human cells and can inhibit virus infection in the next round of infection (Takeuchi et al., 2007). Although CypA does not enhance TRIM5α restriction against wild-type SIVagmTAN in target cells, it is possible that the CypA or TRIM5α from producer cells could confound our findings. However, since our virus encodes a functional vif gene, it is not likely that our results can be explained by CypA or TRIM5α proteins from the producer cells. Rather, CypA and TRIM5α in the target cells are more likely to have an effect on the SIVagmTAN mutants in which portions of the HIV-1 CA have been substituted. These results reflect the findings that CypA is more important in target cells than in producer cells for HIV-1 replication (Hatziioannou et al., 2005; Sokolskaja, Sayah, and Luban, 2004).
In some human cells, the CypA/CA interaction has been shown to assist HIV-1 replication (Franke and Luban, 1996; Hatziioannou et al., 2005; Sokolskaja, Sayah, and Luban, 2004; Thali et al., 1994). It has been hypothesized that humans encode an unknown factor that can negatively affect HIV-1 replication, and CypA-binding to HIV-1 CA can protect HIV-1 from this unknown factor (Sokolskaja, Berthoux, and Luban, 2006; Towers et al., 2003). On the other hand, CypA functions negatively for HIV-1 replication in old world monkey cells because CypA/CA interaction sensitizes HIV-1 to the restriction from old world monkey TRIM5α proteins (Berthoux et al., 2005; Keckesova, Ylinen, and Towers, 2006; Stremlau et al., 2006b). These findings complicate the complex puzzle of the CypA role in HIV-1 life cycle. A mechanism has been proposed that CypA binding to HIV-1 CA induces the conformational change of viral core and renders HIV-1 CA more readily recognizable by the C-terminal B30.2 domain of TRIM5α (Berthoux et al., 2005; Keckesova, Ylinen, and Towers, 2006). Our data suggests that if the conformation theory is true, then the domain consisting of both the 4–5 and 6–7 loops on HIV-1 CA is the key region that determines whether or not a TRIM5α restriction can be enhanced by CypA/CA interaction. Because the 4–5 and 6–7 loops are the regions on viral CA that are exposed to the intracellular environment, they encounter many intracellular factors, and the conformational change of this area can affect the affinity of viral CA to these factors. After the conformational change induced by CypA-binding, the domain formed by the 4–5 and 6–7 loops could become more accessible to TRIM5α.
Nonetheless, the CypA-induced conformational change of viral core only explains part of the TRIM5α restriction. If a virus does not bind CypA (e.g. G89V HIV-1 or SIVmac), this virus is still susceptible to the TRIM5α restriction in a CypA-independent manner (Figure 1B and (Stremlau et al., 2006b)). This suggests that if the TRIM5α restriction contains both CypA-independent and CypA-dependent components, then the CypA-dependent stage might inhibit viruses after the capsid conformation has been changed by CypA-binding, while the CypA-independent stage might inhibit susceptible viruses irregardless of CypA-binding status.
Moreover, our results of the recombinant SIVagm containing the 4–5 or 6–7 loop from HIV-1 CA showed that the degree of viral sensitivity to TRIM5α does not necessarily correlate with the CypA-dependent TRIM5α restriction (Figure 2 and Figure 3). The CypA-enhancement of TRIM5α restriction is only detected in viruses that contain both the 4–5 and 6–7 loops from HIV-1 CA. These data suggest that the domain formed by the 4–5 and 6–7 loops is the region directly recognized by TRIM5α. Replacing only one loop can already affect the CypA-independent TRIM5α restriction, but it requires two loops to change the CypA-dependency of TRIM5α restriction.
Interestingly, transferring this domain from HIV-1 CA to another retrovirus transfers not only the susceptibility to the CypA-dependent TRIM5α restriction but also the sensitivity in general to TRIM5α from old world monkeys (Figure 2 and Figure 3). These further confirm that HIV-1 encodes a CA that is susceptible to both CypA-independent and CypA-dependent TRIM5α restriction. Hatziioannou et al. also reported similar observations in that when they replaced the 4–5, 6–7 loops and the β-turn on HIV-1 capsid with those from SIVmac, these exposed variable regions on capsid modulate HIV-1 tropism which is mediated by host intracellular restriction factors such as TRIM5α (Hatziioannou et al., 2004). Furthermore, it has been shown that swapping only the 4–5 loop between two closely related HIV-2 and SIVmac can transfer the viral susceptibility and insensitivity to TRIM5α (Ylinen et al., 2005). Our results presented here show that, in more distantly related viruses (HIV-1 and SIVagm), the 6–7 loop is also independently involved in the transfer of TRIM5α susceptibility.
In addition to these regions on CA, the ridge formed by helices 3 and 6 on CA has also been reported to determine viral susceptible to the TRIM5α restriction (Owens et al., 2004). It has been shown that helix 6 interacts with the CypA-binding loop through hydrogen-bonding (Gitti et al., 1996; Tang, Ndassa, and Summers, 2002). It is possible that CypA interacts with the 4–5 loop, inducing the conformational change, and subsequently affecting areas surrounding the 4–5 loop, such as the 6–7 loop. Therefore, retroviral CA determines the species-specific tropism after entry, and the 4–5 and 6–7 loops are probably the target domain recognized by host intracellular factors, such as CypA and TRIM5α, that are able to affect the infection.
It has been shown that TRIM5α can only target the incoming viral core, not monomeric capsid protein, for restriction (Dodding et al., 2005; Sebastian and Luban, 2005; Stremlau et al., 2006a). Because viral core quickly begins uncoating once in a cell, TRIM5α rapidly loses the target ligand of the viral core (multimeric capsids). Therefore, in order to enhance the TRIM5α restriction, CypA has to interact with the incoming core before it undergoes uncoating.. However, the persistence of 8 hours of CypA-enhancement for TRIM5α function suggests that the CypA-dependent TRIM5α inhibition acts on a later event after reverse transcription. This hypothesis is supported by the real time PCR data that the CypA-dependent restriction can inhibit reverse transcription, but it mostly restricts the step after the completion of reverse transcription (Figure 5).
In conclusion, we report here that HIV-1 encodes a CA that renders the virus more susceptible to old world monkey TRIM5α restriction when bound by CypA, and further determined the regions on CA that are responsible for the phenotype. The fact that HIV-1 is unusual in its susceptibility to the CypA-dependent TRIM5α restriction suggests that this may be an important component of its host-species restriction, or its adaptation to hominoids.
Materials and Methods
Cells
293T cells were used for generation of vesicular stomatitis virus G protein (VSV-G) pseudotyped HIV-1, SIVagmTAN, and SIVagm-45L, SIVagm-67L, and SIVagm-45/67L. CRFK (Crandall Feline Kidney) cells expressing rhTRIM5α, agmTRIM5α, and baboon TRIM5α proteins were generated as described previously (Malik, and Emerman, 2007). SMAGI (rhesus) cells were a kind gift of Julie Overbaugh (Chackerian, Haigwood, and Overbaugh, 1995) (Fred Hutchinson Cancer Research Center, Seattle, USA). All cells were cultured in Dulbecco’s modified Eagle medium with 10% fetal bovine serum.
Construction of chimeric viruses
HIV-LaiΔEnv-Luc was generated from wild-type HIV-Lai strain previously in the laboratory (Yamashita and Emerman, 2004). The plasmid of pSIVagmTANΔEnvΔRev-Luc, which was modified from wild-type pSIVagmTan-1 (Cat # 3444, NIH AIDS Research & Reference Reagent Program) (Soares et al., 1997), was a gift of Nathaniel Landau (New York University, New York, USA). For easier manipulation, a pBluescript-SIVagmApaI-2965 containing the fragment between Apa I (850 bp upstream of the 5’ LTR) and the 2965th bp of pSIVagmTANΔEnvΔRev-Luc was constructed. To replace the CypA-binding loop (4–5 loop) on SIVagmTAN with that from HIV-1, three separate PCRs were applied. The first PCR amplified the 5’ end sequence of the 4–5 loop, from 686 bp to 1573 bp of pSIVagmΔEnvΔRev-Luc, with a Kas I restriction site (at the 708th bp) and the HIV-1 4–5 loop sequence flanking at the 3’ end. Primer sets used for the first PCR are 5’-AGGCTGAGAAATCTCCAGCAGTGGCGCCCG-3’ (forward) and 5’-TGGTGCAATAGGCCCTGCATGCACTGGGTGTGTTATATCCCACTGG-3’ (reverse). The second PCR amplified the 3’ end sequence of the 4–5 loop, from 1601 bp to 2965 bp of pSIVagmΔEnvΔRev-Luc, with the HIV-1 4–5 loop sequence flanking at the 5’ end and a SnaB I restriction site (at the 1814th bp). Primer sets used for the second PCR are 5’-CCAGTGCATGCAGGGCCTATTGCACCAGGGCAGCTAAGGGACCC-3’ (forward) and 5’-TCCACTGTGTTTTGTCTTTTTTCCGGATAC-3’ (reverse). The third PCR used the PCR products from the first and second reactions as the template and forward primer from the first and the reverse primer from the second reaction to combine these two fragments. The PCR products were digested with the Kas I and SnaB I restriction enzymes and inserted to the pBluescript-SIVagmApaI-2965, which was subsequently digested with Apa I and Sna BI and transferred to pSIVagmΔEnvΔRev-Luc to generate pSIVagmΔEnvΔRev-Luc-45L.
Same three-step PCR and cloning strategy was applied to replace the loop between the 6th and 7th helices (6–7 loop) on SIVagmTAN with that from HIV-1. Primer sets used for the first PCR are 5’-AGGCTGAGAAATCTCCAGCAGTGGCGCCCG-3’ (forward) and 5’-TGGGATAGGTGGATTATTAAAAGTCCACTCTATTTGCTCAGC-3’ (reverse); primers for the second PCR are 5’-AATAATCCACCTATCCCAGTAGGACGTATCTACAGAGGATGGG-3’ (forward) and 5’-TCCACTGTGTTTTGTCTTTTTTCCGGATAC-3’ (reverse). The first PCR amplified the 5’ end sequence of the 6–7 loop, from 686 bp to 1678 bp of pSIVagmΔEnvΔRev-Luc, with the HIV-1 6–7 loop sequence flanking at the 3’ end. The second PCR amplified the 3’ end sequence of the 6–7 loop, from 1700 bp to 2965 bp of pSIVagmΔEnvΔRev-Luc, with the HIV-1 6–7 loop sequence flanking at the 5’ end. The third PCR used the PCR products from the first and second reactions as the template and forward primer from the first and the reverse primer from the second reaction to combine these two fragments. The PCR products were cloned to pBluescript-SIVagmApaI-2965 first (after Kas I and Sna BI digestion) and transferred to pSIVagmΔEnvΔRev-Luc (after Apa I and Sna BI digestion) to generate pSIVagmΔEnvΔRev-Luc-67L.
To construct the pSIVagmΔEnvΔRev-Luc-45/67L, pSIVagmΔEnvΔRev-Luc-45L was digested with Apa I and Ppu MI and swapped with pSIVagmΔEnvΔRev-Luc-67L. Due to the length of the pSIVagmΔEnvΔRev-Luc plasmid, all three constructed SIVagmTAN plasmids were transformed into Stbl3 competent cells (Invitrogen) and grown under the instruction of the manufacturer’s protocol.
Generation of VSV-G pseudotyped viruses
2.5×105 cells/ml of 293T cells were plated in a 6-well plate (2ml/well) 16 hours prior to transfection. The VSV-G pseudotyped HIV-1 (WT), SIVagmTAN (WT), SIVagm-45L, SIVagm-67L, and SIVagm-45/67L were generated as described previously ( Lin and Emerman, 2006). Culture media were collected on 48 and 72 hours after transfection, and were passed through a 0.2 µm filter (Nalgene) to remove cell debris. All harvested viruses were ultra-centrifuged in an SW28 rotor at 23000 rpm at 4°C for 1.5 hours for concentrating viruses by 100 fold. All viruses were allotted in 1.5 ml micro-tubes and frozen at −80°C until use. The viral titer was tested by infecting CRFK cells with different volumes of the viral stocks, and the amount of virus generating 106 relative luminance units (RLU) was determined.
Infection and TRIM5α restriction assay
CRFK or CRFK cells expressing TRIM5α proteins or SMAGI cells were plated 16 hours before infection at a density of 1×105 cells/ml on a 96-well (0.1 ml/well) plate. In the presence of DEAE/Dextran (20 µg/ml), VSV-G pseudotyped viruses virus that was titered on CRFK cells to give 106 RLU of the were added to target cells in the presence or absence of 3 µM of CsA and spinoculated at 1200 relative centrifugal force (rcf) at room temperature for 20 minutes (O'Doherty, Swiggard, and Malim, 2000). The infected cells were placed at 37°C incubator for 48 hours until the analysis of the infectivity. After aspirating culture media, 100 µl of 1×lysis buffer (Promega) were added to the infected cells and incubated at room temperature for 5 minutes. 100 µl of BrightGlo (Promega) reagent were then added to the lysed cells. The plate was placed at room temperature for 1 minute and the relative luminance units (RLU) were measured with luminometer to determine the infectivity. The CsA-withdrawing experiment was conducted by following a protocol modified from a previously published report (Perez-Caballero et al., 2005b). The spinoculation step was performed at 8°C for 20 minutes. After spinoculation, the inoculants were removed, and cells were washed with cold media for three times. Fresh media, with or without 3 µM of CsA, were added to cells after washing. Cells were then placed at 37°C incubator for 48 hours. Media containing CsA were removed at 10 minutes, 30 minutes, 1 hour, 2 hours, 4 hours, 8 hours, 12 hours, and 24 hours after placing plates at 37°C incubator. Those cells were washed with media for three times, and fresh media were added to wells to maintain the growth of cells. The viral infectivity was analyzed as described above.
Real time PCR
SMAGI cells were plated 16 hours before infection at a density of 2.5×105 cells/ml on a 6-well plate (2 ml/well). The VSV-G pseudotyped HIV-1 (1000 IU) were added to target cells in the presence of 20 µg/ml DEAE/Dextran, and spinoculated at 1200 relative centrifugal force (rcf) at room temperature for 20 minutes. The plates were placed at 37°C incubator until the analysis of the late reverse transcription products, 2-LTR circles, or the infectivity. One day after inoculating viruses, cells were trypsinized and viral DNA was extracted with DNeasy Blood and Tissue Kit (Qiagen). Two days after inoculating viruses, viral infectivity was analyzed with assays described above. Viral late reverse transcription products and 2-LTR circles were measured by using real-time PCR with protocols described previously (Butler, Hansen, and Bushman, 2001; Yamashita and Emerman, 2004).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80(19):9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Sebastian S, Sokolskaja E, Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc Natl Acad Sci U S A. 2005;102(41):14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5(11):1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Braaten D, Franke EK, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996a;70(6):3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten D, Franke EK, Luban J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J Virol. 1996b;70(7):4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukovsky AA, Weimann A, Accola MA, Gottlinger HG. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporine dependence. Proc Natl Acad Sci U S A. 1997;94(20):10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7(5):631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Chackerian B, Haigwood NL, Overbaugh J. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology. 1995;213(2):386–394. doi: 10.1006/viro.1995.0011. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Vandegraaff N, Li Y, McGee-Estrada K, Stremlau M, Welikala S, Si Z, Engelman A, Sodroski J. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006;351(2):404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Dodding MP, Bock M, Yap MW, Stoye JP. Capsid processing requirements for abrogation of Fv1 and Ref1 restriction. J Virol. 2005;79(16):10571–10577. doi: 10.1128/JVI.79.16.10571-10577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke EK, Luban J. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology. 1996;222(1):279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372(6504):359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87(7):1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273(5272):231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- Hatziioannou T, Cowan S, Von Schwedler UK, Sundquist WI, Bieniasz PD. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J Virol. 2004;78(11):6005–6012. doi: 10.1128/JVI.78.11.6005-6012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J Virol. 2005;79(1):176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;280(29):26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316(5832):1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. Cyclophilin A renders human immunodeficiency virus type 1 sensitive to Old World monkey but not human TRIM5 alpha antiviral activity. J Virol. 2006;80(10):4683–4690. doi: 10.1128/JVI.80.10.4683-4690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73(6):1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Newman RM, Johnson WE. A brief history of TRIM5alpha. AIDS Rev. 2007;9(2):114–125. [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CM, Song B, Perron MJ, Yang PC, Stremlau M, Sodroski J. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J Virol. 2004;78(10):5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005a;79(14):8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Zhang F, Cowan S, Bieniasz PD. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol. 2005b;79(24):15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MA, Robertson DL, Hui H, Allan JS, Shaw GM, Hahn BH. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology. 1997;228(2):394–399. doi: 10.1006/viro.1996.8387. [DOI] [PubMed] [Google Scholar]
- Sokolskaja E, Berthoux L, Luban J. Cyclophilin A and TRIM5alpha independently regulate human immunodeficiency virus type 1 infectivity in human cells. J Virol. 2006;80(6):2855–2862. doi: 10.1128/JVI.80.6.2855-2862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Sayah DM, Luban J. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J Virol. 2004;78(23):12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006a;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Song B, Javanbakht H, Perron M, Sodroski J. Cyclophilin A: an auxiliary but not necessary cofactor for TRIM5alpha restriction of HIV-1. Virology. 2006b;351(1):112–120. doi: 10.1016/j.virol.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Buckler-White A, Goila-Gaur R, Miyagi E, Khan MA, Opi S, Kao S, Sokolskaja E, Pertel T, Luban J, Strebel K. Vif counteracts a cyclophilin A-imposed inhibition of simian immunodeficiency viruses in human cells. J Virol. 2007;81(15):8080–8090. doi: 10.1128/JVI.02727-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol. 2002;9(7):537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372(6504):363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Towers GJ. Control of viral infectivity by tripartite motif proteins. Hum Gene Ther. 2005;16(10):1125–1132. doi: 10.1089/hum.2005.16.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9(9):1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A. 2006;103(19):7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78(11):5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylinen LM, Keckesova Z, Wilson SJ, Ranasinghe S, Towers GJ. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J Virol. 2005;79(18):11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353(2):396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]