Abstract
Prostate cancer (PCa), like most human cancers, features dysregulated CD44 expression. It loses expression of CD44 standard (CD44s), present in benign epithelium, and overexpresses a less abundant splice isoform, CD44v7-10. MicroRNAs 373 and 520c putatively regulate CD44. The levels of these two microRNAs were measured in matched benign and malignant patient tissues and in prostate cell lines. The effects of their transfection on CD44 mRNA and protein were documented. Whether these miRNAs act on CD44 promoter, or its 3′ untranslated region (UTR), was studied with luciferase reporter constructs and their influences on migration and invasion were determined in PC-3M cells. miR-373 and miR-520c expression were decreased in PCa cell lines and tissues, in proportion to their decreases in total CD44 mRNA. Exogenous miR-373 caused a dose-dependent increase in total CD44 RNA, but a decrease in CD44v7-10 RNA, with an optimal dose at 6 nM. At the protein level, however, both microRNAs suppressed CD44. Both migration and invasion were stimulated by miR-373 and miR-520c. The microRNAs had no effect on the CD44 promoter, but did exhibit 3′UTR binding. In conclusion, miR-373 and miR-520c exert their effect in PCa by preventing the translation of CD44 RNA, rather than by degrading the RNA. Despite this observation, they exert pro-invasive functional effects, as previously described in breast cancer cells. Their effects are mediated by binding CD44 3′UTR.
Keywords: MicroRNA, miR-373, miR-520c, prostatic neoplasms, CD44, invasion
Introduction
About 30% of cases of prostate cancer (PCa) undergo a transition from quiescent to aggressive. This transition requires changes in adhesion glycoproteins such as CD44 that allow tumor cells to detach, interact with proteins that digest stromal matrix, and migrate through matrix and intravasate into lymphovascular channels. CD44 is a transmembrane, cell adhesion glycoprotein that mediates cell-cell and cell-stromal interactions, binds hyaluronan and several other matrix substrates, and controls cell shape through the cytoskeleton. CD44 is expressed as a ubiquitous standard (CD44s) isoform, but in epithelial cells, gene products of variant (CD44v) exons get included that lengthen the extracellular portion of CD44. In either isoform, oligomerization at the extracellular domain is required for CD44 function. Global dysregulation of alternate splicing is common in cancer, and in PCa CD44 expression is lost while splicing is altered in favor of a variant isoform which causes invasion [1]. The probable mechanism is that inclusion of abnormal variant sequences alters CD44's ability to oligomerize and its ligand binding [2], potentiating tumor growth, invasion, and metastasis.
CD44, along with 30% of human genes [3], is regulated by at least 851 human microRNAs (miRNAs). Dysregulation of this recently discovered class of noncoding RNAs is also common in PCa [4]. miRNAs that may interact with CD44 have been studied only in breast cancer, not PCa. Recently, Huang et al described miR-373 and miR-520c, members of the same miRNA family sharing similar seed sequences [5], as functional oncomiRs in breast cancer [6]. They bound specifically to the CD44 3′ untranslated region (3′UTR) and suppressed CD44; both CD44 knockdown and miR-373 stimulated tumor invasion and migration. This action can be explained because CD44, particularly CD44s, is a tumor suppressor in breast cancer as it is in PCa. However, CD44 functional implications differ by tumor type: a CD44v confers invasive ability in PCa [1], but CD44s has pro-invasive properties in some tumors such as colon cancer [7]. Thus, we tested whether similar mechanisms and effects were operative in PCa. Because miR-373 targets the E-cadherin promoter [8] and might also target the CD44 promoter according to our sequence analysis, we explored whether its mode of action was by the promoter or 3′UTR.
Materials and Methods
Cell Lines and Tissues
Benign PrEC and BPH-1, and LNCaP and PC-3 prostate cancer lines and MCF-7 breast cancer cells were from American Type Culture Collection (Manassas, VA). The culture medium for all these cell lines was RPMI 1640 (Invitrogen, Carlsbad, CA) with 10% fetal calf serum. PC-3M cells, a metastasis-derived variant of PC-3, were obtained from Dr. Girish Shah, U. of Louisiana—Monroe. They were grown in RPMI 1640 (Invitrogen) with 12% horse serum, 5% fetal calf serum and antibiotics. Cells were grown in 5% CO2 incubator at 37°C. For each experiment, cells in a flask were trypsinized, washed with sterile PBS to remove trypsin, resuspended in basal medium, and counted after dilution with Trypan blue dye using the grid method [2].
Laser Capture Microdissection
We isolated pure benign glandular cells and benign stromal cells. RNA from cryostat sections was prepared using the PicoPure RNA Isolation Kit for frozen tissues (Arcturus, Mountain View, CA) with columns designed to capture short RNA.
Quantitative RT-PCR (qRT-PCR)
Total RNA was prepared from cell pellets using Trizol (Invitrogen) as described by the manufacturer. RNA was further purified by isopropanol precipitation, resuspended in RNase-free water, and its concentration measured. Complementary DNA (cDNA) was synthesized from 4 µg total RNA in 20 μl reaction mixture as described previously [1]. qRT-PCR reactions used 4 μg cDNA plus the manufacturer’s master mix and primer/probe sets (Applied Biosystems, Foster City, CA) in a volume of 20 μl.
For miRNA, the qRT-PCR was performed in two different ways for selected samples. In the first (primer + probe) approach, cDNA was prepared with MultiScribe (Applied Biosystems), with a special recombinant Moloney murine leukemia virus (rMoMuLV) reverse transcriptase in an optimal buffer. Unlike mRNAs, miRNAs are not polyadenylated, so hairpin-loop forming primer + probe sets for miR-373 and miR-520c (Applied Biosystems) were used. The cycling conditions for both mRNA and miRNA were: hold 50°C 2 min, 95°C 10 min, and then 40 cycles of 95°C for 15 sec and 60°C for 1 min, as previously described [9], using the ABI Prism 7500 cycler (Perkin-Elmer, Waltham, MA).
A second, confirmatory qRT-PCR approach used the NCode kit (Invitrogen). After RT, RNA was tailed with poly (A) polymerase, not normally present on miRNA, then a forward primer specific to each miRNA was used together with universal poly-T reverse primer. Detection was done by SYBR green without a probe. Cycling settings were: 50°C for 2 min, and then 40 cycles of 95°C for 25 sec and 60°C for 1 min.
For CD44, samples were run in triplicates with a primer/probe set for all CD44v that brackets the entire variant region [1], one for CD44 total whose probe binds a standard exon, and 18S ribosomal RNA. Primer/probe sets for CD44v were: forward, 5′-AACGCTTCAGCCTACTGCAAA-3′; reverse, 5′-TCTTCCAAGCCTTCATGTGATG-3′; probe, 5′-GATTTGGACAGGACAGGACCTCTTTCAATG-3′. For CD44 total, we used forward, 5′-CAACTCCATCTGTGCAGCAAA-3′; reverse, 5′-GTAACCTCCTGAAGTGCTGCTC-3′; probe, 5′-CATATTGCTTCAATGCTTCAGCTCCACCTG-3′. Primer and probe sets for 18S were proprietary to the manufacturer.
MicroRNA Transfection
A flask of PC-3M cells at 80% confluence was lipofected with miR-373 or miR-520c-3p (IDT, Coralville, IA) or the irrelevant miR Negative Control #2 (Ambion, Austin, TX) for 6 hours using Trans-IT (Mirus, Madison, WI). Cells were allowed at least 24 hours to recover from the transfection before experiments.
Western Blot Analysis
Cultured cells were directly lysed in their wells using RIPA buffer (Upstate Biologicals, Lake Placid, NY) plus the protease inhibitor mini tablets (Applied Science, Indianapolis, IN). Protein concentration of the cell lysate was estimated by Bradford method. SDS-PAGE was performed on 25 μg sample/lane according to the Laemmli method using the NuPAGE system (Invitrogen, Carlsbad, CA). 5 μl of Kaleidoscope Precision Plus Protein Standards (Bio-Rad) was run in at least one lane. After electrophoresis for 2 hr, the protein was transferred to PVDF. Three primary antibodies were used. To assess CD44v9 (the largest component of the overexpressed CD44v7-10), the membrane was reacted with neat supernatant from the hybridoma cell line HB-258 (ATCC). CD44 total (standard + variant) was assessed using anti-HCAM (DF1485, Santa Cruz Biologicals, Santa Cruz, CA). Anti-β-actin-HRP antibody (Sigma, St. Louis) was used at a dilution of 1:10,000. Membranes were washed 3 × 15 min in TBS with 0.1% Tween 20% and 1:1000 dilution of goat anti-mouse IgG antibody labeled with HRP (Bio-Rad, Hercules, CA) was added in 5% skim milk + TBST for 1 hr. Reactivity was detected using the SuperSignal West Pico Substrate chemiluminescent system (Pierce Biotechnology, Rockford, IL). Each experimental run was conducted at least twice.
CD44 Promoter and 3′UTR Luciferase Constructs
Using the PXP2 plasmid, 1150 bases of CD44 sequence including the start site, and beginning 964 bases upstream to the start site, was cloned in between Xho I and Hind III sites. Because one of the most effective positive controls to test the promoter responsiveness is HOXC6 expression [10], we obtained a plasmid to overexpress HOXC6 in prostate cells (Gift of Dr. Jim Lambert). The overexpression was documented by Western blot analysis using an anti-HoxC6 rabbit polyclonal antibody (Aviva Systems Biology, San Diego, CA) at 1:500 dilution, with goat anti-rabbit secondary antibody at 1:10,000 (Bio-Rad, Hercules, CA), as above. The CD44 3′UTR luciferase reporter plasmid was a gift of Dr. Qihong Huang from Wistar Institute, Philadelphia [6]. Luciferase activity was measured 48 hours after transfection using the firefly luciferase assay (Gold Biotechnologies, St. Louis, MO). The cells were harvested in 20 mM K2HPO4 pH 7.8 with 5 mM MgCl2 and 0.5% Triton X-100 buffer for 15 min on ice, and the mixture was centrifuged for 10 min at 4°C. 50 μl of lysate plus 350 μl of luciferase assay buffer as per the manufacturer. The relative luciferase units (RLU) were normalized to protein concentration as determined by Bradford assay.
Invasion and Migration Assays
Invasion was assessed with triplicate 24-well Matrigel two-tier invasion chambers, and migration was assessed with triplicate control inserts, both with 8.0 μm pores (Collaborative Biomedical Products, Bedford, MA) [1, 9]. Cells were seeded at 30,000 per well, untreated or after microRNA transfection. Cells in the upper insert were in serum-free basal medium (RPMI 1640 with 4 mM L-glutamine, 100 μg/mL each of penicillin G and streptomycin). The lower chamber contained chemoattractant medium consisting of 10% fetal bovine serum, 20% conditioned medium from subconfluent culture, and 70% complete medium. The incubation was carried out 24 h in 5% CO2 incubator at 37°C. The medium from the upper inserts, together with any residual cells were removed off the upper Matrigel surface. The membrane was fixed in methanol and stained with May-Grunwald stain (Sigma, St. Louis, MO) according to manufacturer's protocol. Experiments were repeated twice.
Results
Laser Capture Microdissection
The prior study of miR-373 and miR-520c in breast tissue samples did not distinguish its origin from breast glandular cells or stromal cells [6]. To make this determination in prostate, benign glandular cells and benign stromal cells were microdissected. Taking the log2 of -(CT value normalized to β-actin), miR-373 glandular expression was 0.003377; stromal expression was 0.005048. miR-520c was below detection limits in glands and stroma.
Quantitative RT-PCR (qRT-PCR)
To correlate CD44 standard and v7-10 expression with miRNA expression in cells and tissues, qRT-PCR was performed. Total CD44 mRNA was lower in all cancer cell lines than benign ones, and in most cancer tissue samples compared with benign tissue from the same patient (Figure 1).
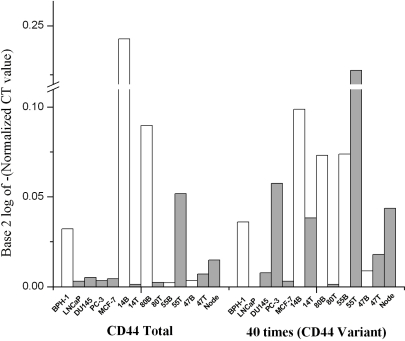
Figure 1.
qRT-PCR of CD44 total (left) and variant (right) in 5 cell lines and 5 clinical prostatectomy specimens normalized to 18S RNA. Left: CD44 total, comprising the more abundant CD44s isoform, is higher in benign BPH-1 cell line than in androgen-sensitive LNCaP cancer or androgen-independent DU-145 or PC-3, consistent with previous findings of ours [13] and others [15, 16]. In MCF-7 breast cancer cells, it is low as expected [6]. In matched, frozen section-confirmed, benign (B) and tumor (T) tissues from 4 patients, CD44 is downregulated in the tumor component of 2. In microdissected metastatic prostate cancer from a lymph node (Node), CD44 is low. Right: The less abundant CD44 v7-10 is expressed in androgen-independent prostate cell lines but very low in MCF-7. In 5 tissues, CD44v7-10 is highly expressed in most tumor (T) components and in lymph node with metastasis (Node), as expected from our prior work [1, 12].
By probe + primer method, miR-373 was highest in the BPH-1 cell line (Figure 2a), intermediate in androgen-sensitive, slow-growing LNCaP cells, and very low in androgen-independent PCa as well as in MCF-7, confirming the finding of Huang et al who used MCF-7 as a negative control [6]. In matched tissues, miR-373 was again generally higher in benign tissue than in the tissue sample with cancer (Figure 2b).
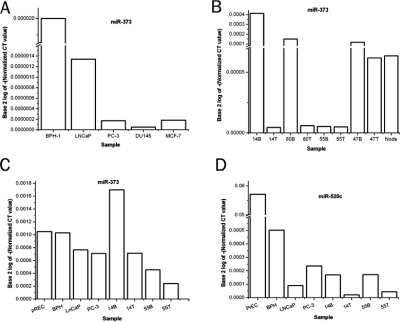
Figure 2.
qRT-PCR for miR-373 by primer and probe method normalized to 18S rRNA (A and B). A. Among cell lines, benign BPH-1 cells have the most miR-373, while slow-growing, androgen-dependent LNCaP cancer cells have decreased but detectable miR-373. Androgen-independent cell lines PC-3 and DU145 have nearly absent miR-373. Level is low in breast cancer cell line MCF-7, consistent with others’ findings [5]. B. Matched (same patient) tissue specimens. miR-373 is higher in the benign (B) component than in tumors (T) for 3 of 4 cases. C. qRT-PCR by SYBRgreen method for confirmation of miR-373 expression. Values are higher because they are normalized to β-actin which has lower copy number. miR-373 is downregulated in cancer cell lines compared to benign PrEC and BPH cells. In 2 patients’ tissues, there is downregulation of miR-373 in the tumor (T) component. D. qRT-PCR for miR-520c by SYBRgreen method normalized to β-actin. Average expression levels of miR-520c were at ⅕ or less the level of miR-373. 2 malignant cell lines have less miR-520c than 2 benign ones. In 2 patients’ tissues, there is downregulation of miR-520c in the tumor (T) component.
The differences were greater than could be accounted for merely by miR-373 being 1.5 times as prevalent in stroma as in glands, and the presence of proportionally less stroma in tumor. These trends were confirmed in separate SYBRgreen qRT-PCR experiments (Figure 2c). A similar trend applied to miR-520c in cell lines and tissues. miR-520c RNA was decreased in primary tumors and metastasis using both a SYBRgreen method (Figure 2d) and by using primers with a specific probe (not shown).
Effects on CD44 Promoter and 3′UTR
PC-3M cells were transfected with our CD44 luciferase construct that tests for binding to the promoter region. HOXC6 is documented to regulate the CD44 promoter in PCa cells [10]. We achieved its overexpression by transfection (Figure 3a). Cells that were co-transfected with CD44 promoter and HOXC6 demonstrated a strong inhibition of promoter activity by HOXC6 (Figure 3b). Overexpression of miR-373 or miR-520c caused ∼85% decreases in luminescence readings generated by the 3′UTR construct (Figure 3c), signifying that both microRNAs target the CD44 3′UTR.
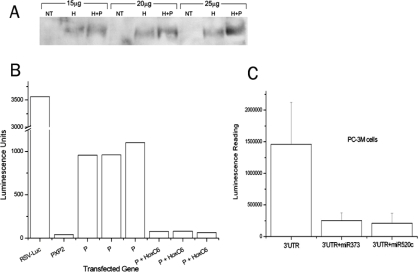
Figure 3.
A. Successful overexpression of HOXC6 (primary Ab) by Western blot analysis using 15-25 μg protein. PC-3M cells were transfected with either HOXC6 (H) alone or H + CD44 promoter (P), compared to no treatment (NT). B. Cells from A were subjected to luciferase activity assay for CD44 promoter (P) activity. RSV is the positive control and PXP2 is the negative control. Promoter activity is inhibited by HOXC6. Experiments were performed in triplicates. C. Optimized doses of miR-373 or miR-520c were transfected into PC-3M cells subsequent to transfection of a luciferase construct containing the 3′UTR of CD44. Both suppressed gene expression, indicating that CD44 is a direct target of miR-373 and miR-520c.
Effects on CD44 Expression
Transfection of 6 nM synthetic miR-373 was optimal for detecting its expression (Figure 4a). Doses up to 6 nM caused dose-dependent overexpression of CD44 total RNA normalized to untreated cells (using a primer set and probe that detects CD44 total = standard + variants). Thus 6 nM was considered an optimal dose. At the RNA level, divergent effects were noted on total CD44 (stimulation) and on CD44 variant (inhibition, Figure 4a). Because miRNA can either degrade mRNA or reversibly inhibit its translation [11], we used western blot analyses to determine whether the directionality of the protein effect was the same. miR-373 and miR-520c both suppressed total CD44 protein compared with untreated positive control or with transfection of an irrelevant, scrambled miRNA (Figure 4b).
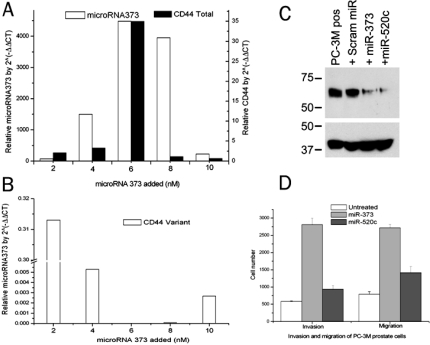
Figure 4.
A. Left y-axis: Transfection of 6 nM dose of synthetic miR-373 maximized its expression by qRT-PCR. Right y-axis: Doses up to 6 nM caused dose-dependent 35-fold overexpression of CD44 total RNA, normalized to untreated cells (using a primer and probe set that detects CD44 total = standard + variants). B. The same dose suppressed CD44v7-10 variant expression. C. Top: Western blot analysis using primary antibody for total CD44. Lane 1: PC-3M cells as positive (pos) control, with strong CD44. Lane 2, PC-3M cells transfected with scrambled (+Scram), irrelevant microRNA. Lanes 3 and 4: PC-3M cells transfected with miR-373 or miR-520c respectively, showing CD44 suppression. Bottom: β-actin loading control. D. 6 nM dose of exogenous miR-373 also has functional effects: more than 5-fold increase in migration and more than 3-fold increase in invasion of 30,000 PC-3M cells compared to untreated cells. Migration and invasion almost doubled with miR-520c.
Effects on Invasion and Migration
Overexpression of miR-373 increased migrating PC-3M cells more than 5 folds and invading cells more than 3 folds. Moreover, the invaders as a percent of migrators increased from 85% to >100% after miR-373 transfection. Overexpression of miR-520c nearly doubled invasion and migration (Figure 4c).
Discussion
Both PCa cells and tumor from prostatectomy show downregulation of miR-373 and miR-520c compared with their benign counterparts. Exogenous doses cause an increase in total CD44 RNA. At the protein level, however, both miR-373 and miR-520c suppress total CD44. This means that miR-373 and miR-520c, like most miRNAs, inhibit, not stimulate, their target; and since CD44 is a tumor suppressor, miR-373 and miR-520c behave as oncomiRs. The protein finding in our study is concordant with that of Huang et al [6], which demonstrated that miRNAs 373 and 520c exerted marked CD44 suppression in MCF-7 breast cancer cells by western blot analysis. Thus, miR-373 and miR-520c function by fundamentally different mechanisms in PC-3M and MCF-7 cells but cause the same phenotypic effect on CD44 protein. The findings of Huang et al suggest RNA degradation, whereas our findings suggest the opposite mechanism: RNA accumulation but paradoxically translational repression. It has been noted that mRNA whose repression is mediated by miRNA gets stored in P bodies and conditions of stress can reverse the repression [11]. In hepatoma cells, cationic amino acid transporter 1 (CAT-1) mRNA and reporters bearing the CAT-1 3′UTR or its fragments were able to be relieved from the miRNA miR-122-induced inhibition by subjection to different stress conditions [11]. Whether placing prostate cancer cells under stress might affect miR-373 and miR-520c effects on CD44 would be of interest.
Most of CD44 is normally CD44s, a tumor suppressor lost in PCa [2, 13-16]. A curious finding was that miR-373 stimulated total CD44 RNA but also suppressed the pro-invasive CD44v7-10 variant. This suggests an effect occurring prior to splicing, or a direct effect on splicing. This effect also bespeaks a role of these miRNAs as oncomiRs, like their role in breast cancer, as studied with non-invasive MCF-7 cells and in tissues. In those studies, miR-373 and miR-520c suppressed RNA and protein of the anti-invasive CD44 (total versus variant was not examined), increased migration and invasion, and were upregulated in metastases; expression levels in benign breast were not examined [6]. We have not examined miR-373 and miR-520c in metastases, except for just one case.
Functionally, miR-373 and miR-520c also behave as oncomiRs in PCa despite their downregulation. They stimulated cells to migrate and invade. miRNA downregulation is in fact the trend in PCa, applying to 76 of 85 detectable miRNAs in one study [4]. There are only a few reportedly upregulated miRNAs in PCa. The pro-invasive effect of these miRNAs suggests that, although downregulated in PCa, they target other invasion-promoting genes [6]. We determined by microdissection of whole, benign prostate that at least miR-373 is present in both glandular and stromal cells of prostate tissue. This distinction was not addressed in breast tissue [6], but suggests that the stromal contribution of miRNA in glandular organs is not negligible. Of course in tumor, the glandular component is much more prevalent, so a lesser stromal contribution might be expected.
Most miRNAs bind a response element in the 3′ untranslated region (3′UTR) of targeted mRNA. This either reversibly represses its translation or directs sequence specific degradation [11]. Because some influences on miR-373 and miR-520c and CD44 were fundamentally different from those described before, we examined the possibility that their actions were mediated by the CD44 promoter, using a promoter luciferase construct. We demonstrated that this interaction is through the CD44 3′UTR, again suggesting a unique role in PCa pathophysiology. Certain miRNAs can target complementary DNA promoter sequences to induce gene expression [8]. A recent paper, using a prostate cancer model, identified a miR-373 binding site in the promoter of E-cadherin [8]. Our sequence analysis also suggested a theoretical miR-373 binding site in the CD44 promoter; but, we ruled out a promoter site of action of miR-373 and miR-520c in PCa cells. Their effect seems to be exerted only by binding to the 3′UTR.
Our prior work demonstrated a unique role for CD44v7-10 in PCa. By isolating RNA from clinical PCa specimens, we discovered that expression of CD44v7-10 constitutes a unique PCa signature, consistently expressed in both primary PCa and PCa metastatic to other tissues [1, 12, 13]. Interference against CD44v caused a 69% reduction in invasion index compared to untreated control cells [13]. Moreover, PCa loses the splicing ability to produce the standard isoform expressed in benign prostate [1, 14-16]. The discovery of downregulation of miR-373 and miR-520c in PCa is consistent with the dysregulated expression of miRNAs in PCa [3, 17] and the upregulation of the enzyme Dicer [18].
In conclusion, in benign prostate, higher miR-373 and miR-520c correlate with the predominance of CD44s isoform. In prostate cancer, the downregulation of total CD44 and upregulation of its less abundant isoform CD44v correlated with loss of miR-373 and miR-520c. With miR-373, an exogenous dose increased CD44 RNA. However, at the protein level miR-373 and miR-520c suppressed total CD44, indicating translational repression. These actions are most likely mediated through the CD44 3′UTR. The loss of CD44 total protein, most of which is CD44s, is probably functionally important in mediating pro-invasive effects of miR-373 and miR-520c. Further work is needed to determine whether these miRNAs prevent or target CD44 splicing factors or splicing.
Acknowledgments
This work was supported by Department of Defense Prostate Cancer Research Program, Grant PC060671 to K.A.I. We thank Dr. Dawn R. Cochrane for advice and technical assistance.
References
- 1.Omara-Opyene AL, Qiu J, Shah GV, Iczkowski KA. Prostate cancer invasion is influenced more by expression of a CD44 isoform including variant 9 than by Muc18. Lab Invest. 2004;84:894–907. doi: 10.1038/labinvest.3700112. [DOI] [PubMed] [Google Scholar]
- 2.Iczkowski KA, Omara-Opyene AL, Shah GV. The predominant CD44 splice variant in prostate cancer binds fibronectin, and calcitonin stimulates its expression. Anticancer Res. 2006;26:2863–2872. [PubMed] [Google Scholar]
- 3.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–44. [PubMed] [Google Scholar]
- 4.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 5.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 6.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 7.Harada N, Mizoi T, Kinouchi M, Hoshi K, Ishii S, Shiiba K, Sasaki I, Matsuno S. Introduction of antisense CD44s cDNA down-regulates expression of overall CD44 isoforms and inhibits tumor growth and metastasis in highly metastatic colon carcinoma cells. Int J Cancer. 2001;91:67–75. doi: 10.1002/1097-0215(20010101)91:1<67::aid-ijc1011>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 8.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iczkowski KA, Omara-Opyene AL, Kulkarni TR, Pansara M, Shah GV. Paracrine calcitonin in prostate cancer is linked to CD44 variant expression and invasion. Anticancer Res. 2005;25:2075–2083. [PubMed] [Google Scholar]
- 10.McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 12.Iczkowski KA, Bai S, Pantazis CG. Prostate cancer overexpresses CD44 variants 7-9 at the messenger RNA and protein level. Anticancer Res. 2003;23:3129–3140. [PubMed] [Google Scholar]
- 13.Iczkowski KA, Pantazis CG, Collins J. The loss of expression of CD44 standard and variant isoforms is related to prostatic carcinoma development and tumor progression. J Urol Pathol. 1997;6:119–129. [Google Scholar]
- 14.Vis AN, van Rhijn BW, Noordzij MA, Schroder FH, van der Kwast TH. Value of tissue markers p27(kip1), MIB-1, and CD44s for the pre-operative prediction of tumour features in screen-detected prostate cancer. J Pathol. 2002;197:148–154. doi: 10.1002/path.1084. [DOI] [PubMed] [Google Scholar]
- 15.Vis AN, Noordzij MA, Fitoz K, Wildhagen MF, Schroder FH, van der Kwast TH. Prognostic value of cell cycle proteins p27(kip1) and MIB-1, and the cell adhesion protein CD44s in surgically treated patients with prostate cancer. J Urol. 2000;164:2156–2161. [PubMed] [Google Scholar]
- 16.Kallakury BV, Sheehan CE, Ross JS. Co-downregulation of cell adhesion proteins alpha- and beta-catenins, p120CTN, E-cadherin, and CD44 in prostatic adenocarcinomas. Hum Pathol. 2001;32:849–855. doi: 10.1053/hupa.2001.26463. [DOI] [PubMed] [Google Scholar]
- 17.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria Ruggero The mir-15a-mir-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 18.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]